A single-layer platform for Boolean logic and arithmetic through DNA excision in mammalian cells (original) (raw)
. Author manuscript; available in PMC: 2017 Sep 27.
Published in final edited form as: Nat Biotechnol. 2017 Mar 27;35(5):453–462. doi: 10.1038/nbt.3805
Abstract
Genetic circuits engineered for mammalian cells often require extensive fine-tuning to perform their intended functions. To overcome this problem, we present a generalizable biocomputing platform that can engineer genetic circuits which function in human cells with minimal optimization. We used our Boolean Logic and Arithmetic through DNA Excision (BLADE) platform to build more than 100 multi-input-multi-output circuits. We devised a quantitative metric to evaluate the performance of the circuits in human embryonic kidney and Jurkat T cells. Of 113 circuits analysed, 109 functioned (96.5%) with the correct specified behavior without any optimization. We used our platform to build a three-input, two-output Full Adder and six-input, one-output Boolean Logic Look Up Table. We also used BLADE to design circuits with temporal small molecule-mediated inducible control and circuits that incorporate CRISPR/Cas9 to regulate endogenous mammalian genes.
INTRODUCTION
A fundamental goal of synthetic biology is to predictably and efficiently reprogram cells to perform computations and carry out specific biological tasks 1. Cells genetically engineered with biocomputation circuits hold great promise for improving therapeutics 2–5, diagnostics 6, 7, animal models 8, 9 and industrial biotechnological processes 10, 11. Despite rapid advances and promising results in biocomputation design 12–16, implementing simple two-input, single-output Boolean functions in prokaryotic and eukaryotic cells necessitated layering of multiple genetic circuits, requiring extensive construction and tuning of genetic components. Circuits with multiple inputs and multiple outputs remain scarce in scientific literature due to the need of a large library of interoperable genetic parts and the compounding effects of connecting increasingly more transcriptional components together into single cells 17. Curation of genetic circuit components and the development of automated design software are starting to address these problems in bacteria 18–22, but are not currently available for mammalian cells. It is not clear whether either parts or design software can be transferred from bacteria to higher organisms.
To enable complex computation in mammalian cells, we present a biocomputation platform named Boolean Logic and Arithmetic through DNA Excision (BLADE) that can be used to create decision-making circuits using site-specific recombinase technology. To our knowledge, BLADE provides the first generalizable framework for engineering complex logic functions in mammalian cells with minimal optimization by the user. Unlike most circuit designs already published, our platform encodes genetic logic through a single-layer design, which can readily yield circuits with multiple inputs or outputs without increasing the number of transcription units.
We use our platform to design, build, and test more than 100 functionally distinct circuits, including some of the most complex logical operations engineered in any living cell to date. Our platform accommodates multiple outputs without incurring additional design complexity and can be adapted to regulate the expression of messenger RNAs from Pol II promoters or non-coding regulatory RNAs from Pol III promoters, such as CRISPR/Cas9 guide RNAs. Being able to build complex logic circuits that control multiple different guide RNAs could enable exquisite control of endogenous mammalian gene expression.
RESULTS
Characterization of recombinases
Site-specific DNA recombinases, which are a class of DNA-modifying enzymes that recognize a specific DNA sequence and perform cleavage and reunion of DNA 23, show promise for use in building synthetic circuits in eukaryotic cells. This is in part due to their versatile ability to simultaneously function as transcriptional activators and repressors within the same cell on the same transcription unit; a feat that is difficult to achieve with transcription factors. Recombinase-mediated gene expression can be achieved by tyrosine recombinase-mediated excision of a transcription terminator located upstream of a gene of interest (GOI) or serine integrase-mediated inversion of a GOI (analogous to a buffer BUF gate) (Figure 1a). Conversely, termination of gene expression can be achieved by the placement of recombination sites around a GOI and elimination of gene expression using tyrosine recombinase-mediated excision or serine integrase inversion (analogous to a NOT gate).
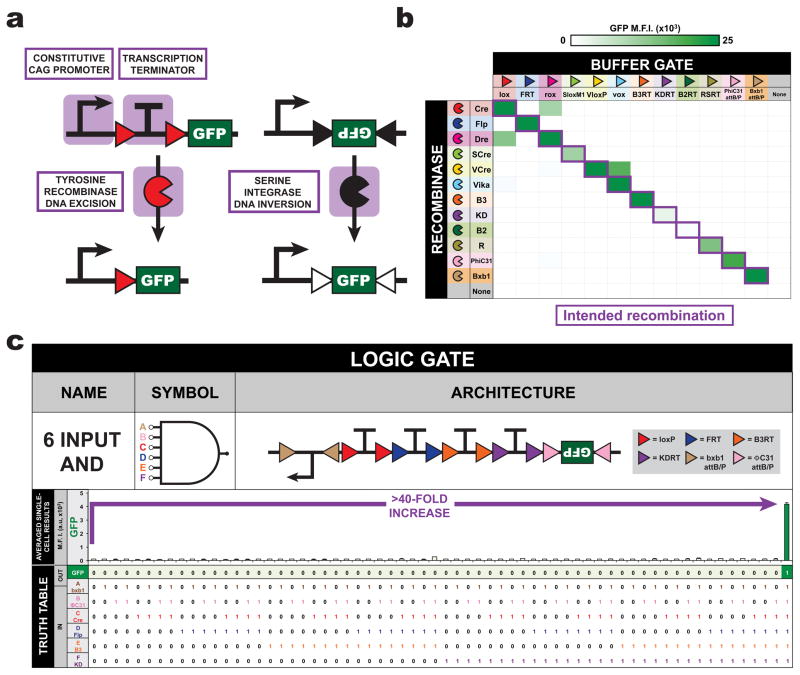
Figure 1. Orthogonal site-specific tyrosine recombinases and serine integrases enable implementation of multi-input AND gates in mammalian cells.
(a) Recombinases can perform simple BUF logic operations, either by tyrosine recombinase-mediated excision (left) or serine integrase-mediated inversion (right). (b) Recombinases are tested for their recombination efficiency and orthogonality on all BUF logic reporters. (c) A 6-input AND-gate that produces GFP when all inputs are present. M.F.I. = mean fluorescence intensity from n = 3 transfected cell cultures; a.u. = arbitrary units. Error bars represent standard error of the mean.
Multi-input recombinase-based biological computational circuits require robust and orthogonal genetic components. Here, twelve recombinases, including both tyrosine recombinase and serine integrase families were tested for activity and orthogonality in a human embryonic kidney cell line (HEK293FT). Through transient transfection of recombinases and their reporters (deletion-based BUF gates for tyrosine recombinases and inversion-based for serine integrases), ten of the enzymes were found to be highly active and sufficiently orthogonal to each other for our circuit design effort (Figure 1b, Supplementary Figure 1a, Supplementary Table 1). It was found that recombinase expressional level could be tuned to minimize cross-talk between non-orthogonal recombinases (Supplementary Figure 2). We focused our efforts on the use of Cre and Flp recombinases due to their extensive prevalence in mammalian cell culture and animal experimentation.
Through an ad-hoc design approach, we created all sixteen possible two-input Boolean logic gates by placing Cre and Flp recombination sites (loxP and FRT, respectively) around termination and coding sequences to yield the intended behaviors (Supplementary Figure 3, Supplementary Table 2). Due to the orthogonality of recombinases, it is possible to generate multi-input AND gates simply by placing more termination sequences in tandem between a promoter and GFP. Indeed, we created a 6-input AND gate that expresses GFP upon the excision of four termination sequences by tyrosine recombinases and the inversion of the EF1α promoter and GFP by two serine integrases (Figure 1c, Supplementary Figure 1b, Supplementary Table 3).
A key element that is required to implement more complex functions on a single transcriptional unit is the use of heterospecific recombination sites. Heterospecific sites, such as loxP and lox2272, differ from one another by only a few base pairs 24, but they retain DNA excision capabilities in the presence of Cre, as long as two of the same sites are present, i.e. loxP with loxP. We validated three sets of lox sites (loxP, lox2272, loxN), three sets of FRT (FRT, F3, F14) and Vlox (VloxP, Vlox2272), which demonstrated heterospecificity using Cre, Flp, and VCre enzymes, respectively (Supplementary Figure 4, Supplementary Table 4). This feature allows the excision of more than one non-connected region of DNA simultaneously, enabling encoding of logic on a single-layer (recombinase inputs recombining with a single DNA strand reporter). This is juxtaposed to transcription-factor based genetic logic, which necessitates cascading connections of multiple transcriptional responses; here, extensive fine-tuning is usually required to engineer functional circuits to balance input-output responses, which grows substantially as the number of inputs and outputs of the circuits increases.
Single-layer multi-input recombinase-based design platform
Although an ad hoc design approach can be successful in creating some recombinase-based circuits as we have demonstrated here and others have in scientific literature,8, 9, 25–27 a generalizable strategy for producing any N-input-M-output logic behavior has not, to our knowledge, been demonstrated before. We exploited the features of site-specific recombinases to establish a platform for N-input-M-output combinatorial computation in mammalian cells named Boolean Logic and Arithmetic through DNA Excision (BLADE), where N and M can be any non-negative integer. Beyond the orthogonality of the recombinases and heterospecific sites, no further characterization of individual components is needed.
Each BLADE circuit is organized as a single transcriptional layer comprising a single promoter upstream of up to 2N regions of DNA sequences (or addresses, Z), which are surrounded by recombination sites. Each BLADE circuit is designed such that when it is presented with recombinase inputs, addresses become transcriptionally active through excision of intervening regions between recombination sites downstream of the promoter. For example, a two-input (N = 2) BLADE circuit, which responds to inputs A and B, can accommodate up to 22 = 4 addresses with possible addresses being Z = ZAB = Z00, Z10, Z01, and Z11, enumerating all combinatorial states of inputs A and B (Figure 2a). The BLADE design is flexible with regards to the outputs that can be generated. Outputs can range from no transcriptional outputs (transcription terminator) at all, an arbitrary combination of outputs separated by ribosomal skip sequences (2A), Boolean functions like BUF that can toggle transcriptional responses ON or OFF through use of additional site-specific recombinases, or even other BLADE topologies. A BLADE design that utilizes 2N addresses permits creation of all possible N-input-M-output combinatorial circuits and thus is necessary for implementing the most complex multi-input-multi-output truth tables 28.
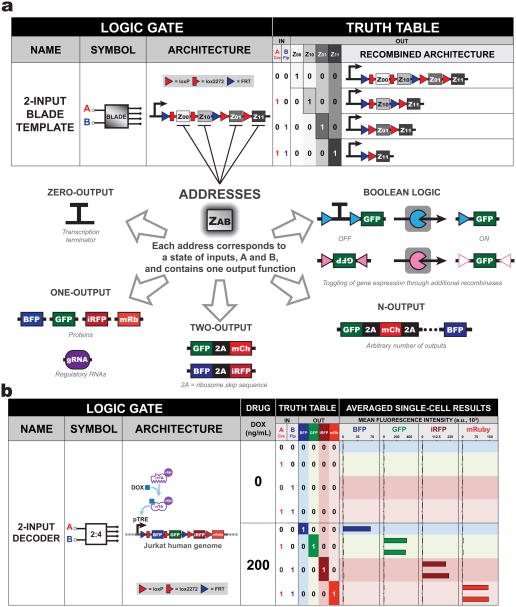
Figure 2. 2-input BLADE platform can produce four distinct output functions based on two inputs.
(a) 2-input BLADE template on one plasmid with a single transcriptional unit. This template contains four distinct regions of DNA (addresses) downstream of a promoter. Each address corresponds to an output function and is accessed or deleted via site-specific DNA recombination. Each address can be programmed from different configurations ranging from zero-inputs to Boolean functions. The first address (Z00), which is the closest to the promoter, corresponds to a state where no recombinase is expressed (A = 0, B = 0). If the Z00 address contains a protein coding sequence, then that gene will be expressed. Gene expression from the other addresses downstream of Z00 will be blocked by the presence of Z00 protein coding region. In the presence of recombinase A, which corresponds to state (A = 1, B = 0), addresses Z00 and Z01 will be removed, thus moving address Z10 directly downstream of the promoter and allowing gene expression of address Z10 only to occur. Similarly, when only recombinase B is present (A = 0, B = 1), addresses Z00 and Z10 are excised, allowing Z01 to be moved directly downstream of the promoter. Finally, when both recombinases are expressed (A = 1, B = 1), addresses Z00, Z01, Z10 are all excised, thus placing Z11 downstream of the promoter unobstructed by the other addresses. (b) Integrated 2-input BLADE decoder with tagBFP, EGFP, iRFP720, and mRuby2 as addresses Z00, Z10, Z01, and Z11 respectively. Plasmids constitutively expressing Cre and/or Flp are then stably integrated in. Three days of doxycycline (DOX) treatment is used to permit the rtTA-VP48 protein to bind to the tetracycline response elements promoter (pTRE) to activate gene expression. Mean fluorescence intensity (MFI) is plotted of either n = 1 or n = 2 independent integrations. a.u. = arbitrary units.
A two-input, four-output circuit
For initial characterization of the 2-input BLADE platform, single transcriptional output functions were tested. We constructed a 2-input, 4-output decoder circuit, which necessitates four addresses, each of which coincides with a distinct output: blue, green, infrared and red fluorescent proteins (tagBFP, EGFP, iRFP720, and mRuby2, respectively) using a modular assembly strategy known as Unique Nucleotide Sequence-Guided Assembly, based on Gibson isothermal assembly 29, 30 (Figure 2b, Supplementary Figures 5–9). One advantage of the BLADE platform is that expression of outputs is driven by a single promoter, thereby permitting additional control through the use of a drug-regulated promoter or encoding of further transcriptional logic. We demonstrated this principle by stable integration of a doxycycline-controlled decoder circuit into Jurkat T lymphocytes using _piggyBac_-mediated transposition. Jurkat T cells were chosen because they are an important suspension cell line for understanding T cell signaling and easy to maintain during long-term passages. Constitutively-expressing recombinases were then integrated into the genome. Following stable integration of the recombinases driven by constitutive promoters, doxycycline was used to regulate final output expression in a dose-dependent manner (Supplementary Figure 10). This strategy provides a facile way of scaling the circuit response while maintaining the logical functionality of the circuit. Furthermore, we showed that BLADE circuit functionality can be maintained for at least two weeks under varying doxycycline conditions (Supplementary Figure 11). Moreover, the decoder circuit performed as predicted through transient transfection of HEK293FT cells with strong fluorescent protein expression for each state of inputs (Supplementary Figure 12, Supplementary Table 5). HEK293FT was chosen here (and for subsequent experiments) because high throughput experimentation is enabled due to the inexpensive and highly efficient polyethylenimine transfection reagent.
Scaling up and quantitation of circuit design
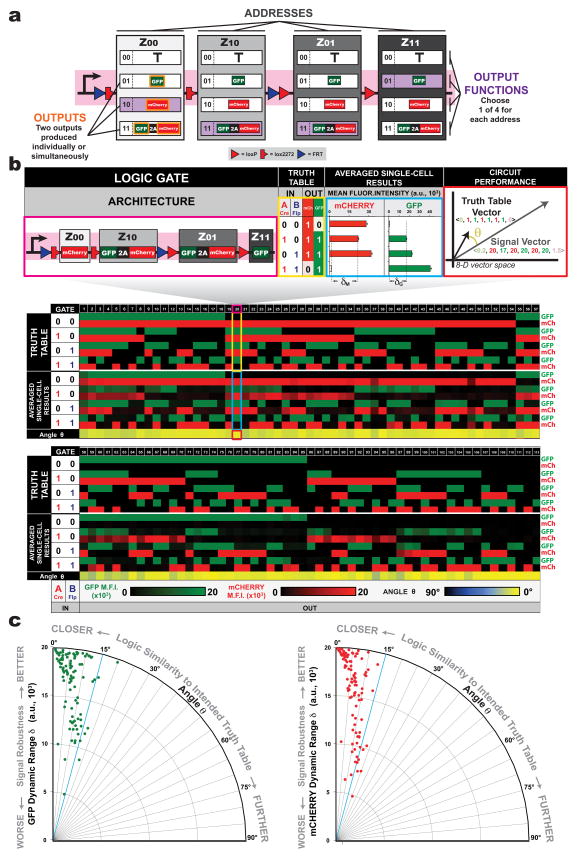
To test the robustness of the BLADE platform on a large scale, we produced the largest collection of functionally unique logic circuits in mammalian cells (a library of 113 circuits with up to two inputs and outputs (Figure 3, Supplementary Figure 13, Supplementary File 1)). This set contains two 2-input circuits: the half adder (Gate 104) and half subtractor (Gate 99), which perform 2-input arithmetic and are widely-used and studied in electronics. Whereas all 113 circuits were qualitatively observed to implement the correct computation, we quantified their functional correctness using a novel Vector Proximity (VP) metric measuring the misalignment between a circuit’s biological implementation and its ideal implementation from its intended truth table (See Experimental Procedures and Supplementary Code) Truth tables and obtained experimental results were represented as vectors, Truth Table and Signal Vectors, respectively, in an 8-dimensional vector space. The angular error between these two vectors (VP angle metric) was calculated with 0° meaning the data represents the intended truth table perfectly and 90° meaning the data demonstrates completely incorrect output (inverted response to the intended truth table). The VP angle metric shows that 93.8% (106/113) of the circuits had an angle no more than 15°, and none had an angle of more than 25° from their ideal implementation.
Figure 3. One hundred and thirteen distinct gene circuits with up to two inputs and two outputs implemented using the 2-input BLADE template.
(a) To generate 2-input, 2-ouput circuits, a 2-input BLADE template can be configured with different combinations of output functions: zero-output (transcription termination sequence), one-output (GFP or mCherry) or two-output (GFP-T2A-mCherry). (b) A diverse library of >100 gene circuits, each shown as an individual column with predicted truth table GFP/mCherry ON/OFF behavior (black = no output, green = GFP ON, red = mCherry ON) and corresponding experimental averaged single-cell results obtained from flow cytometry. (c) Angles between each Signal Vector and corresponding Intended Truth Table Vectors are plotted versus worst-case dynamic range values for GFP (δG) and mCherry (δM) signals. Shown above is an expanded view of one of the logic gates made using this platform. M.F.I. = mean fluorescence intensity from n = 3 transfected cell cultures; a.u. = arbitrary units. Error bars represent standard error of the mean.
We extended the VP measure of correctness to obtain a quantitative and discrete test of whether an implemented circuit is correct or incorrect. For any implemented circuit we measured its VP angle from all possible truth tables, and sorted the results in ascending order. We define the rank of the intended truth table in this sorted list as the circuit’s VP global rank. We call a circuit as functionally valid under this measure, if it has the best (that is, smallest) VP global rank. For our library of 113 circuits, we calculated the VP angle between each Signal Vector and all 255 (up to 2-input, up to 2-output, excluding the 0-input, 0-output FALSE) Truth Table vectors. Global rank values were determined according to how many Truth Table Vectors had lower VP angles than the Intended Truth Table Vector. We found that 96.5% (109/113) of the tested circuits gave the lowest angle between their Signal Vector (global rank = 0/255) and their Intended Truth Table vectors (Supplementary Figures 14 and Supplementary Table 6). This success rate of 96.5% is the highest reported to our knowledge for large scale circuit construction in mammalian cells. Only four circuits had a global rank of 1, meaning there was only one other truth table that yielded a lower VP angle; no circuits had a rank more than 1. To facilitate data sharing and further analysis by other researchers, we have developed an interactive website (datasheets.synbiotools.org) that contains the data all of our 113 circuits summarized in a datasheet format 31.
Re-programmable combinatorial logic
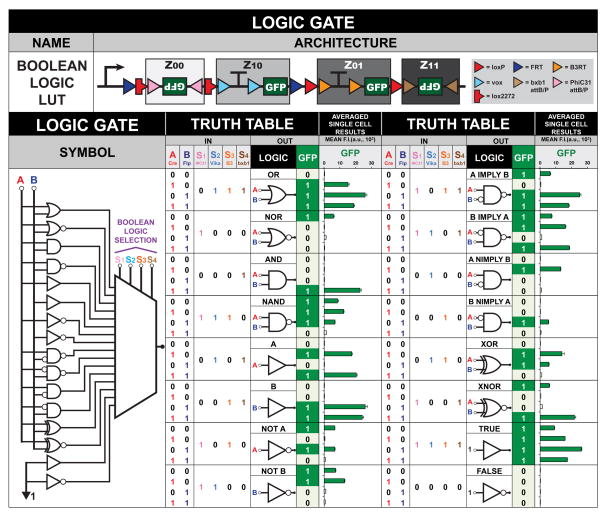
One important class of circuits found in electronics is Field-Programmable Read-Only Memory (FPROM). The input-output behavior of these circuits can be configured in the field post-manufacturing, allowing users to program the function computed by the circuit at a later time. We built a genetic FPROM circuit for living cells termed a Boolean Logic Look-Up Table (LUT) that is based on placing BUF gates into the four addresses of the 2-input BLADE template. (Figure 4, Supplementary Figure 15, Supplementary Table 7). This circuit has two data inputs, A and B, and four select inputs, S1, S2, S3, and S4. Each select input can control which buffer gates are transcriptionally active or not (GFP ON or OFF). Thus, each combination of select inputs configures the circuit to one of sixteen-possible Boolean logic gates with up to two inputs and one output. For instance, an OR function can be achieved using select inputs S2, S3, and S4, keeping address Z00 GFP OFF and setting addresses Z10, Z01, and Z11 GFP ON. Thus, this circuit allows one to reconfigure the computation within living cells without requiring additional DNA assembly. This circuit behaves as expected in HEK293FT cells. To illustrate the flexibility of our platform in terms of recombinase choices, we created an alternative Boolean Logic LUT (Supplementary Figure 16).
Figure 4. Field-programmable storage and retrieval of logic and memory using a Boolean Logic Look-Up Table (LUT).
The Boolean Logic LUT is a six-input-one-output genetic circuit that receives two data inputs, A and B, and is controlled by four select inputs, S1, S2, S3, and S4, producing an output of GFP. The select inputs are used to change data input-output behavior; each combination configures the circuit to any of the sixteen Boolean logic gates. F.I. = fluorescence intensity from n = 3 independent transfections. Error bars represent standard error of the mean.
Arithmetic operations using BLADE
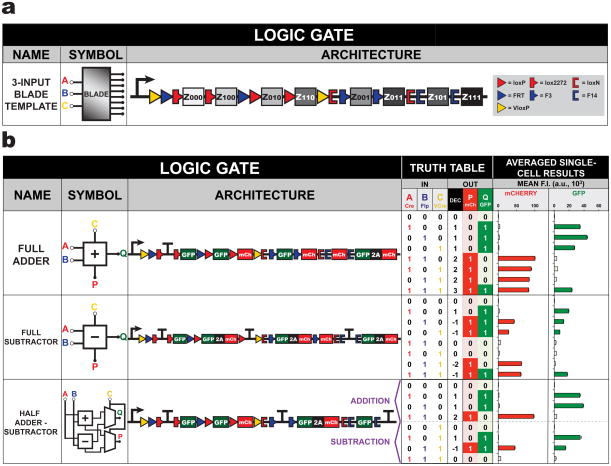
Extending the BLADE framework further, we developed a 3-input BLADE template for constructing sophisticated arithmetic functions in human cells (Figure 5a, Supplementary Figures 17 and 18). This template responds to three inputs (Cre, Flp, and VCre) and contains eight addresses for expression of up to eight distinct transcriptional outputs. This design utilizes three different heterospecific sites for Cre and Flp, but just one site for VCre. Three 3-input-2-ouput arithmetic computational circuits were made and tested in HEK293FT cells from the 3-input BLADE template (Figure 5b, Supplementary Figure 19, Supplementary Table 8). The full adder and full subtractor can perform either binary addition or subtraction of three 1-bit inputs, respectively. Furthermore, the half adder-subtractor is an arithmetic FPROM circuit that can compute addition or subtraction on two data inputs, A and B, depending on the presence of one select input C. BLADE templates with more than three inputs can be generated by using additional recombinases and heterospecific recombination sites that follow a simple recursive design algorithm where recombinase switches are nested within each other yielding designs for implementing any N-input combinatorial logic function (Supplementary Figure 20). This non-intensive computational approach produces designs with the total number of recombination site pairs for N-inputs being 2N-1, for N≥0; this is consistent with the 2N rows needed to specify the truth table of an N-input function.
Figure 5. A 3-input BLADE template can be applied to create 3-input arithmetic computational circuits.
(a) The 3-input BLADE template can receive up to three inputs and produce eight distinct output functions. (b) Three 3-input-2-output binary arithmetic computational circuits made using the 3-input BLADE template. The full adder can add A + B + C while the full subtractor calculates A – B – C. For addition, input C, output P, and output Q represent Carry In, Carry Out and Sum, respectively. For subtraction, input C, output P, and output Q signify Borrow In, Borrow Out and Difference, respectively. The half adder-subtractor performs either binary addition of A + B or binary subtraction of A – B depending on the presence of select input C. F.I. = fluorescence intensity from n = 3 transfected cell cultures; a.u. = arbitrary units. Error bars represent standard error of the mean.
Small molecule inputs for BLADE
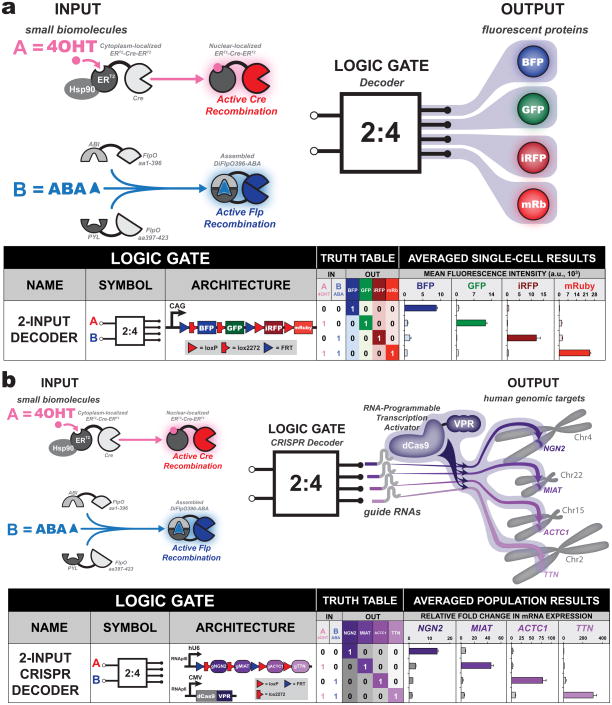
Next, we interfaced the BLADE system with biochemically relevant inputs. To induce Cre recombination, an ERT2-Cre-ERT2 construct was used whereby a mutated estrogen receptor (ERT2) secludes Cre recombinase activity from the nucleus unless a small molecule 4-hydroxytamoxifen (4OHT) is added, which permits translocation of the fusion protein to the nucleus. For Flp induction, a split Flp system was developed that induces Flp recombinase activity upon phytohormone abscisic acid (ABA)-induced heterodimerization (Figure 6a, Supplementary Figure 21, Supplementary Table 9). Logical detection of these two small molecule inputs was successful using the 2-input BLADE decoder with minimal leaky recombinase behavior in HEK293FT cells. Logic induction dynamics were characterized over the course of 48 hours, which revealed early encoding of logic and introduction of leaky recombinase behavior towards the end of the time course (Supplementary Figures 22, 23 and 24). A BLADE decoder affords the ability to combinatorially select an exponential number of DNA regions (e.g. eight regions via three inputs), which is advantageous for the limited set of eukaryotic inducible systems.
Figure 6. Interfacing BLADE with biologically relevant inputs and outputs.
(a) Small molecules, 4-hydroxytamoxifen (4OHT) and abscisic acid (ABA), are used to induce Cre and Flp recombination activities, respectively, on a decoder circuit containing four fluorescent protein outputs. Chemical induction of Cre recombination is achieved through 4OHT-mediated translocation of a Cre protein fused to mutated estrogen nuclear receptors (ERT2) from the cytoplasm to the nucleus. Chemical induction of Flp recombination is achieved through a split Flp recombinase construct fused to ABA-binding domains ABI and PYL. Mean fluorescence intensity is plotted from n = 3 transfected cell cultures and error bars indicate the standard error of the mean. (b) Small molecules, 4OHT and ABA, are used to induce Cre and Flp recombination activities on a decoder circuit interfaced with a dCas9-VPR (VP64, p65, Rta) transcription activator. Four human genomic promoters are targets for activation via association of corresponding guide RNAs (gRNA) with dCas9-VPR. Total RNA was collected and averaged relative fold changes in target mRNA expression were obtained through quantitative real-time PCR of n = 3 transfected cell cultures. Error bars represent the standard error of the mean.
CRISPR/Cas9 combined with BLADE
To test whether the BLADE system can be used to regulate endogenous mammalian gene expression, we interfaced it with the CRISPR/Cas9 system using recombinase-based excision of guide RNA (gRNA) sequences. We first tested whether recombination sites or cloning scar DNA sequences would affect the activity of a Cas9 transcription activator (dCas9-VPR), as these sequences would be directly coded into RNA fused to the gRNA. We found that these sequences (up to 136 bases) added to the 5′ end of the gRNA had no detrimental effect on transcription activation of a mCherry reporter plasmid (Supplementary Figure 25). Next, the two-input decoder was rebuilt to use an RNA polymerase III human U6 promoter and single guide RNAs (gRNAs) as addresses that target promoters of four human endogenous genes (NGN2, MIAT, ACTC1 and TTN). These targets were chosen because of their documented efficient gRNA activity 32. This CRISPR-based two-input decoder system was transfected along with a Cas9 transcription activator (dCas9-VPR) 32 and recombinase activities were induced with 4OHT and ABA (Figure 6b, Supplementary Tables 10 and 11). At the completion of the HEK293FT transient transfection, mRNA fold changes of the target endogenous genes were determined using quantitative real-time PCR and corresponded to the two-input decoder truth table logic. Owing to the easy programmability of the CRISPR/Cas9 system, this system will be very useful for genome-wide transcriptional reprogramming studies, entailing unprecedented control of endogenous human genes and cell states.
DISCUSSION
The BLADE platform that we present here can generate a large number of complex genetic circuits. To demonstrate the application of BLADE in genetic circuit design, we created more than 100 functionally distinct circuits, most of which have never, to our knowledge, been documented before in any living organism. Most of the circuits that we built displayed the intended logic, as quantified by the VP metrics that we devised to quantitate performance. BLADE can be used to regulate messenger and guide RNA expression, thus increasing flexibility in the modulation of cell functions. When combined with drug-inducible control of recombinase activity, we were able to conditionally regulate gene expression and cell states. We created a split Flp protein whose activity can be reconstituted with chemically inducible dimerization systems. This split Flp configuration could be valuable to animal geneticists to complement existing Cre systems. Furthermore, we have shown that the BLADE system is functional in different cell types (e.g. embryonic kidney and Jurkat T cells). Key attributes of the BLADE platform are summarized in Supplementary Table 12.
Composition of simple gates into large hierarchies of modules has been successful in electronic circuit design because components are physically separated from each other, allowing reuse of well-characterized and high-performing parts. This has enabled computing circuits with extremely predictable behaviors to be built. However, applying this electrical engineering strategy to synthetic biology has proven difficult due to the lack of a complete understanding of how different components will behave when connected together in a single cell 17, 33, even when these components were designed to be “plug and play”. As such, aside from some elegant work14, 34, very few synthetic multi-input-multi-output circuits have been reported. Furthermore, this multi-layer circuit design approach also requires many orthogonal biological “wires” which have unpredictable properties. This has driven effort toward parts development 35, 36 at the expense of circuit design. Efforts are underway to provide detailed characterization of components and develop standards and design automation program to allow for better predictability of the interoperability of different parts 18, 31.
The BLADE platform is enabled by the unique chemistry of recombinases (DNA rearrangement) in regulating gene expression. The same recombinase can activate and inhibit gene expression simultaneously in the same transcription unit with equal efficiency, a feature that is very difficult to accomplish with transcription factors. This feature allowed us to design the BLADE platform in a single-layer (single transcription unit) to implement any combinatorial Boolean logic provided sufficient numbers of recombinases and heterospecific sites are available. We define the number of layers in a computing circuit as the maximum number of states along the shortest path in any computation. By “single-layer,” we connote the following property unique to BLADE designs. Unlike other circuit architectures, there is no distinct “molecular wire” needed to communicate the results of a sub-computation in one part of the circuit to any other part of the circuit: the inputs themselves communicate information throughout a computation. Our single-layer BLADE platform can also use fewer protein components to perform a large set of complex logical operations in mammalian cells. Single-layer circuits do not need output signal levels of one gate to be tuned or matched to input signal levels of another. A trade-off of a single-layer design is that the performance of circuits cannot be predicted based on the property of its components.
Some circuits did not pass our stringent VP global rank metric evaluation; however, all circuits yielded results that had close alignment with their intended truth table. A potential source of underperformance could arise from interference of recombinase sites sequences on ribosome activities (e.g. hair-pinning hampering translation initiation or cryptic translation initiation sequences). The RNA Pol III/CRISPR-based BLADE system could be an attractive circuit design to explore since gRNA or other regulatory RNA outputs are not subjected to translation-based failure modes.
Transcription factor-based genetic circuits have played a substantial role in synthetic circuit design 18, 37–39. This is because natural genetic circuits, which commonly serve as the inspiration for synthetic circuit design, are mostly implemented using transcription factors, with only a few documented cases involving recombinases (e.g. fim invertase in Escherichia coli) 40, 41. Unlike transcription-based circuits, recombinase-based circuits are single-use systems, which prevent them from tracking dynamic input signals. However, recombinases are particularly advantageous for engineering logic behaviors. Furthermore, when integrated into the genome, recombinase-based systems can provide stable memory and have proven to be invaluable in numerous experiments, such as in tissue specific gene expression experiments where memory of tissue or condition specific inputs that lead to sustained gene expression (or knockout) is more desirable. In addition, serine integrases have been used to develop temporal logic state machines in bacteria where the circuits can sense and remember the order in which signal inputs appeared 42, 43. It is conceivable that those designs can be refactored to function in mammalian cells. It is also possible to modify the BLADE platform to perform temporal logic computations, thus enabling the use of the more widely adopted tyrosine recombinases in a single-layer circuit topology. The prevalence of recombinases in different systems and organisms illustrates the fact that single-use systems are often sufficient in biomedical research and biotechnological applications 23, 44.
Genome-mining has been performed to uncover and test a large set of serine integrases 45 and many putative recombinases remain to be characterized from structural identification in genome sequence databases. Furthermore, efforts have been made to design and implement recombinases that can be made to recombine different DNA sequences through fusion of recombinase catalytic domains to programmable DNA-binding proteins, such as zinc finger, TAL, and Cas9 sequences 46–48; generation of chimeric recombinases 49; or alteration of specificity through molecular evolution experiments 50. Therefore, we envision that an unlimited set of recombinase parts are likely available for incorporation with BLADE to create biocomputation circuits with unprecedented sophistication.
ONLINE METHODS
DNA assembly
All constructs were transformed and maintained in Top10 Escherichia coli competent cells (Life Technologies) at 37°C or 30°C prior to miniprep (Epoch Life Sciences) or midiprep (Macherey Nagel). All plasmids were created using standard molecular biology practices of ligation, digestion and transformation, in addition to Gibson isothermal assembly and Unique Nucleotide Sequence Guided assembly (a modular extension of Gibson assembly), where DNA fragments that are to be connected to each other are flanked by short homology sequences and are then fused together by a one-pot isothermal digestion, polymerization, and ligation reaction. This latter strategy permits a modular, easy, efficient and fast framework for construction of DNA. In UNS-guided assembly, the homology sequences have been standardized (e.g. U1, U2, U3…UX) and have been computationally optimized for proper assembly (reduction of hairpins, sequence homology, and GC tracts) and ease of use (no start codons or useful restriction sites). Genetic cassettes that are to be connected are cloned into part vectors, which contain the UNSes that surround the insert. Part vectors are then sequenced-verified using Sanger sequencing (Quintara Biosciences). Restriction digests and gel purifications are then performed to isolate the cassettes flanked with the UNSes. Finally, these products are joined to a linearized destination vector via Gibson isothermal assembly. For the 113 circuits displayed in Figure 3, analytical PCRs were performed to verify inserts were assembled correctly through amplifications across the UNS sequences. Construction details are elaborated in Supplementary Figures 5, 7 and 18.
Maintenance and transient DNA transfection of HEK293FT cells
DNA was transfected into the human embryonic kidney cell line (HEK293FT) using a polyethylenimine (PEI) protocol. Cells were plated onto 48- (250μL) or 96-well (100μL) plates the day prior to transfection (200,000 cells/mL), such that the cells were 50–70% confluent the day of transfection. Cells were kept in a humidified incubator at 37°C and 5%CO2 and maintained in DMEM medium (Corning) with 10% Heat Inactivated Fetal Bovine Serum (Life Technologies), 50U.I./mL penicillin/ 50μg/mL streptomycin (Corning), 2mM glutamine (Corning) and 1mM sodium pyruvate (Lonza). PEI stocks were made from linear polyethylenimine (Polysciences 23966-2) and were dissolved at a concentration of 0.323g/L in deionized water with the assistance of concentrated hydrochloric acid and sodium hydroxide and then filtered sterilized (0.22μm). PEI stocks were stored at −80°C until use and warmed to room temperature prior to usage. For 48-well plate transfections, DNA (1000ng, 50ng/μL) was dissolved and brought up to a volume of 50μL using 0.15 M sodium chloride (NaCl, Fisher Scientific). DNA-NaCl solutions were then mixed with 50μL of a PEI-NaCl mixture (8uL PEI: 42uL NaCl). These solutions were then incubated at room temperature for ten minutes and 25μL was carefully dropped into individual wells of HEK293FT cells (250ng DNA/well). Similarly, for 96-well plate transfections, DNA (500ng, 50ng/μL) was dissolved and brought up to a volume of 25μL using 0.15 M sodium chloride (NaCl). DNA-NaCl solutions were then mixed with 25μL of a PEI-NaCl (4uL PEI: 21uL NaCl) mixture. These solutions were then incubated at room temperature for ten minutes and 10μL was carefully dropped into individual wells of HEK293FT cells (100ng DNA/well). Electronic space-adjustable multichannel pipettors (Integra Biosciences) were utilized throughout the process for rapid aspiration and dispensing of molecular and cellular reagents. A Countess II image-based cell counter (Life Technologies) was used for measuring human cell population densities.
Small molecule chemical induction
1000X stock small molecules abscisic acid (100mM, Sigma Aldrich), doxycycline (200μg/mL, Fisher Scientific), and 4-hydroxytamoxifen (1mM, Sigma Aldrich) were dissolved in 100% ethanol and stored at recommended temperatures. For transient transfection induction experiments, two hours post-transfection, small molecules were mixed with mammalian cell medium such the concentrations were 25X; then, proper volumes were dispensed into wells such that the final concentration was 1X.
Maintenance and generation of stable Jurkat T lymphocytes through electroporation, piggyBac-mediated integration, eukaryotic selection, and doxycycline induction
Wild type Jurkat T lymphocyte cells were kept in a humidified incubator at 37°C and 5%CO2 and maintained in RPMI medium (Corning) with 5% Heat Inactivated Fetal Bovine Serum (HI-FBS, Life Technologies), 50U.I./mL penicillin/ 50μg/mL streptomycin (Corning), and 2mM glutamine (Corning). Prior to electroporation, cells were changed with medium containing 10% HI-FBS and without antibiotics. On days of transfection, cells were checked to be within 5–8x105 cells/mL and 2x107 cells were spun down at 300xg and resuspended in 300μL medium per transfection. Cell solutions were mixed with 20μg transposon vector and 4μg of transposase (pCAG-SuperPBase, pBW900). After fifteen minutes of room temperature incubation, DNA/cell mixtures were transferred to a 4mm electroporation cuvette and electroporated in a Harvard Apparatus BTX instrument using a single pulse 300V square wave for ten milliseconds. Cells were then transferred to 10% HI-FBS medium without antibiotics and placed into the incubator. One to two days later, cells were spun down and resuspended in medium with 5% HI-FBS with antibiotic and antieukaryotic chemicals. Puromycin (Thermo Scientific) was used at a final concentration of 2μg/mL and zeocin (Invivogen) at 400μg/mL. Antieukaryotic selections were performed for at least ten days before removing antieukaryotic compounds.
For experiment in Figure 2b, the decoder recombinase reporter (pBW2293) was first integrated and selected with zeocin; then, recombinase expression vectors (hPGK-iCre, hPGK-FlpO, hPGK-FlpO-2A-iCre) were integrated and selected for with puromycin in duplicate (n=2). After final stable line generation, 300,000 Jurkat cells of each line were spun down and resuspended in doxycycline or ethanol-containing medium. Cells were maintained in respective media for given number of time (fourteen days in ethanol and then with or without doxycycline for three days in Figure 2b, square points in Supplementary Figure 11) and then run for cytometric readings.
Flow cytometry
Two days post-transfection and after trypsinization (0.05%Trypsin/0.53mM EDTA, Corning) and resuspension, all HEK293FT cell populations were analysed using a Becton Dickinson (BD) LSRFortessa SORP flow cytometer with HTS, except for data in Figure 5, which were recorded on a BD LSRII. Time-course data was achieved through trypsinization and fixation of cells at particular time points (BD Cytofix) and kept at 4°C until analysed through flow cytometry. The LSRFortessa was equipped for detection of EGFP (488nm laser, 530/30 emission filter, 505 longpass dichroic mirror), tagBFP (405nm laser, 450/50 emission filter), mCherry or mRuby2 (561nm, 610/20 emission filter, 595 longpass dichroic mirror), iRFP-720 (637nm laser, 730/45 emission filter, 685 longpass dichroic mirror) and LSS-mOrange (405nm, 610/20 emission filter, 535 longpass dichroic mirror). The LSRII was similarly equipped for detection of EGFP, tagBFP, mCherry and mRuby2, but with additional channels for iRFP-720 (633nm laser, 720/40 emission filter, 710 longpass dichroic mirror) and LSS-mOrange (405nm, 590/35 emission filter, 505 longpass dichroic mirror). All Jurkat T lymphocyte experiments were run using a Life Technologies Attune Nxt 4-laser acoustic focusing flow cytometer. The Attune Nxt was equipped for detection of EGFP (488nm laser, 510/10 emission filter), tagBFP (405nm laser, 440/50 emission filter), mRuby2 (561nm, 585/16 emission filter), iRFP-720 (638nm laser, 720/30 emission filter) and LSS-mOrange (405nm, 603/48 emission filter).
A gate was applied on FSC-A and SSC-A to remove debris from cell populations in FlowJo (Tree Star) (Supplementary Figure 26). pCAG-tagBFP or pCAG-LSS-mOrange plasmids were used as HEK293FT transient transfection markers and were gated for by applying a gate on the top 0–0.1% wildtype cells in those channels. Figure plots represent at least 5000 events in the transfected cell subset. Compensation was applied for four and five-color experiments (Supplementary Figures 27, 28, and 29). No gates other than a viable cell FSC/SSC were applied for Jurkat T lymphocyte stable line experiments. To demonstrate the digital ON/OFF behavior of the genetic circuits, main text plots have additionally been expressed as percentage of cells in an ON state through application of gates in fluorescent protein channels. Gates were chosen per experiment in an arbitrarily-defined manner, but applied uniformly for all samples in each experiment (Supplementary Figures 1, 6, 13, 15, 19, and 21).
Quantitative real-time polymerase chain reaction analysis
RNA was extracted from HEK293FT cells using the Qiagen RNeasy Plus Mini kit. 500ng of RNA was reverse-transcribed into cDNA using qScript cDNA SuperMix 20uL reaction kit (Quanta BioSciences). cDNA samples were diluted to 100μL using DEPC-treated water. Next, 5μL of diluted cDNA was amplified using primers (Supplementary Table 11) through the use of the LightCycler 480 SYBR Green I Master polymerase kit (Roche) and a LightCycler 480 Instrument II (Roche). Relative fold-changes were determined using the ΔΔCt method.
Vector Proximity computational analysis
Each desired 2-input 2-output Boolean function corresponded to a truth table with four rows and two output columns. The two output columns in each desired Boolean function f were mapped to an 8-dimensional binary vector t, which we call the Truth Table Vector. The fluorescent reporter signals measured from each of the two outputs, for each of the four input conditions from every genetic circuit implementation were also mapped to an 8-dimensional real vector s, which we call the Signal Vector. An Ideal Implementation of t is a circuit whose signal vector satisfies the equation s = c · t, for some positive real number c. We quantified the correctness of a genetic circuit implementing Boolean function f by computing the angle θ between the vectors t and s using the formula θ=cos-1(xy), where x=∑i=18ti·s^i,y=∑i=18ti2·∑i=18s^i2, and t i and s i are the i-th components of the vectors t and s. In computing θ, we capped the signal values s i to a maximum of 2x104 a.u., denoted by ŝi in the formulae above. The angular difference ranges from 0° (best) to 90° (worst). We quantified the strength of the signal vector by computing its Dynamic Range (δ) of a genetic circuit implementation as δ=min{i:ti=1}(si)-max{i:ti=0}(si). We computed the Dynamic Range separately for the two outputs of each circuit and omitted the computation for circuits where the magnitude differences were not defined. Consequently, we had to omit two circuits (always ON GFP/always OFF mCherry, and always ON GFP and mCherry) from the δ computation and 16 GFP or mCherry outputs of circuits (but not both) from the δ computation.
Statistical Methods
All transient gene expression experiments involved transfection of DNA into n = 3 separate cell cultures. Fluorescence intensities for each cell culture population was averaged and the standard error of the mean was taken. Stable integrations were performed once or in duplicate as indicated.
Supplementary Material
1
2
3
Acknowledgments
B.W. acknowledges funding from the NSF Graduate Research Fellowship Program (DGE-1247312) and an NIH/NIGMS fellowship (T32-GM008764). S.B. was supported in part by the National Science Foundation Expeditions in Computing Award No. 1522074, which is part of the “Living Computing Project” (programmingbiology.org). W.W. acknowledges funding from the NIH Director’s New Innovator Award (1DP2CA186574), NSF Expedition in Computing (1522074), NSF CAREER (162457), NSF BBSRC (1614642), and Boston University College of Engineering Dean’s Catalyst Award. We thank C. Bashor, D. Chakravarti, N. Patel, and S. Slomovic for suggestions on the manuscript; A. Belkina and T. Haddock for flow cytometry assistance; J. Torella for help with UNS-guided assembly; and M. Park and J. Eyckmans for RT-qPCR assistance. A. Nagy for the kind gift of the Dre construct.
Footnotes
AUTHOR CONTRIBUTIONS
B.W. made molecular and cellular reagents, performed experiments, analysed data and generated all figures. S.B. conceived the vector proximity analyses for circuit performance and developed the datasheets attribution and website. B.W. and S.B. developed and performed the vector proximity analyses. L.C., N.P., T.L and A.E. made molecular and cellular reagents and performed preliminary experiments. B.W. and W.W. conceived the project. B.W., S.B, and W.W. wrote the paper. All authors commented on and approved the paper.
COMPETING FINANCIAL INTERESTS STATEMENT
Invention disclosures have been filed based on this work.
References
- 1.Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nature reviews. Genetics. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei P, et al. Bacterial virulence proteins as tools to rewire kinase pathways in yeast and immune cells. Nature. 2012;488:384–388. doi: 10.1038/nature11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roybal KT, et al. Precision Tumor Recognition by T Cells With Combinatorial Antigen-Sensing Circuits. Cell. 2016;164:770–779. doi: 10.1016/j.cell.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakravarti D, Wong WW. Synthetic biology in cell-based cancer immunotherapy. Trends in biotechnology. 2015;33:449–461. doi: 10.1016/j.tibtech.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie M, et al. Beta-cell-mimetic designer cells provide closed-loop glycemic control. Science. 2016;354:1296–1301. doi: 10.1126/science.aaf4006. [DOI] [PubMed] [Google Scholar]
- 6.Slomovic S, Collins JJ. DNA sense-and-respond protein modules for mammalian cells. Nature methods. 2015;12:1085–1090. doi: 10.1038/nmeth.3585. [DOI] [PubMed] [Google Scholar]
- 7.Courbet A, Endy D, Renard E, Molina F, Bonnet J. Detection of pathological biomarkers in human clinical samples via amplifying genetic switches and logic gates. Sci Transl Med. 2015;7:289ra283. doi: 10.1126/scitranslmed.aaa3601. [DOI] [PubMed] [Google Scholar]
- 8.Fenno LE, et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nature methods. 2014;11:763–772. doi: 10.1038/nmeth.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madisen L, et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron. 2015;85:942–958. doi: 10.1016/j.neuron.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ro DK, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 11.Bogorad IW, Lin TS, Liao JC. Synthetic non-oxidative glycolysis enables complete carbon conservation. Nature. 2013;502:693–697. doi: 10.1038/nature12575. [DOI] [PubMed] [Google Scholar]
- 12.Gaber R, et al. Designable DNA-binding domains enable construction of logic circuits in mammalian cells. Nature chemical biology. 2014;10:203–208. doi: 10.1038/nchembio.1433. [DOI] [PubMed] [Google Scholar]
- 13.Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science. 2011;333:1307–1311. doi: 10.1126/science.1205527. [DOI] [PubMed] [Google Scholar]
- 14.Guinn M, Bleris L. Biological 2-input decoder circuit in human cells. ACS synthetic biology. 2014;3:627–633. doi: 10.1021/sb4001596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber W, et al. A synthetic time-delay circuit in mammalian cells and mice. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2643–2648. doi: 10.1073/pnas.0606398104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Regot S, et al. Distributed biological computation with multicellular engineered networks. Nature. 2011;469:207–211. doi: 10.1038/nature09679. [DOI] [PubMed] [Google Scholar]
- 17.Brophy JA, Voigt CA. Principles of genetic circuit design. Nature methods. 2014;11:508–520. doi: 10.1038/nmeth.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen AA, et al. Genetic circuit design automation. Science. 2016;352:aac7341. doi: 10.1126/science.aac7341. [DOI] [PubMed] [Google Scholar]
- 19.Appleton E, Tao J, Haddock T, Densmore D. Interactive assembly algorithms for molecular cloning. Nature methods. 2014;11:657–662. doi: 10.1038/nmeth.2939. [DOI] [PubMed] [Google Scholar]
- 20.Rodrigo G, Jaramillo A. AutoBioCAD: full biodesign automation of genetic circuits. ACS synthetic biology. 2013;2:230–236. doi: 10.1021/sb300084h. [DOI] [PubMed] [Google Scholar]
- 21.Huynh L, Kececioglu J, Koppe M, Tagkopoulos I. Automatic design of synthetic gene circuits through mixed integer non-linear programming. PloS one. 2012;7:e35529. doi: 10.1371/journal.pone.0035529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanton BC, et al. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nature chemical biology. 2014;10:99–105. doi: 10.1038/nchembio.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26:99–109. [PubMed] [Google Scholar]
- 24.Lee G, Saito I. Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene. 1998;216:55–65. doi: 10.1016/s0378-1119(98)00325-4. [DOI] [PubMed] [Google Scholar]
- 25.Siuti P, Yazbek J, Lu TK. Synthetic circuits integrating logic and memory in living cells. Nature biotechnology. 2013;31:448–452. doi: 10.1038/nbt.2510. [DOI] [PubMed] [Google Scholar]
- 26.Bonnet J, Yin P, Ortiz ME, Subsoontorn P, Endy D. Amplifying genetic logic gates. Science. 2013;340:599–603. doi: 10.1126/science.1232758. [DOI] [PubMed] [Google Scholar]
- 27.Schonhuber N, et al. A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nature medicine. 2014;20:1340–1347. doi: 10.1038/nm.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shannon CE. The Synthesis of Two-Terminal Switching Circuits. Bell System Technical Journal. 1949;28:59–98. [Google Scholar]
- 29.Torella JP, et al. Rapid construction of insulated genetic circuits via synthetic sequence-guided isothermal assembly. Nucleic acids research. 2014;42:681–689. doi: 10.1093/nar/gkt860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torella JP, et al. Unique nucleotide sequence-guided assembly of repetitive DNA parts for synthetic biology applications. Nature protocols. 2014;9:2075–2089. doi: 10.1038/nprot.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Canton B, Labno A, Endy D. Refinement and standardization of synthetic biological parts and devices. Nature biotechnology. 2008;26:787–793. doi: 10.1038/nbt1413. [DOI] [PubMed] [Google Scholar]
- 32.Chavez A, et al. Highly efficient Cas9-mediated transcriptional programming. Nature methods. 2015;12:326–328. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jayanthi S, Nilgiriwala KS, Del Vecchio D. Retroactivity controls the temporal dynamics of gene transcription. ACS synthetic biology. 2013;2:431–441. doi: 10.1021/sb300098w. [DOI] [PubMed] [Google Scholar]
- 34.Auslander S, Auslander D, Muller M, Wieland M, Fussenegger M. Programmable single-cell mammalian biocomputers. Nature. 2012;487:123–127. doi: 10.1038/nature11149. [DOI] [PubMed] [Google Scholar]
- 35.Khalil AS, et al. A synthetic biology framework for programming eukaryotic transcription functions. Cell. 2012;150:647–658. doi: 10.1016/j.cell.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green AA, Silver PA, Collins JJ, Yin P. Toehold switches: de-novo-designed regulators of gene expression. Cell. 2014;159:925–939. doi: 10.1016/j.cell.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 38.Stricker J, et al. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 40.Johnson RC. Mobile DNA II. American Society of Microbiology; 2002. [Google Scholar]
- 41.Blomfield IC. The regulation of pap and type 1 fimbriation in Escherichia coli. Advances in microbial physiology. 2001;45:1–49. doi: 10.1016/s0065-2911(01)45001-6. [DOI] [PubMed] [Google Scholar]
- 42.Roquet N, Soleimany AP, Ferris AC, Aaronson S, Lu TK. Synthetic recombinase-based state machines in living cells. Science. 2016;353:aad8559. doi: 10.1126/science.aad8559. [DOI] [PubMed] [Google Scholar]
- 43.Hsiao V, Hori Y, Rothemund PW, Murray RM. A population-based temporal logic gate for timing and recording chemical events. Molecular systems biology. 2016;12:869. doi: 10.15252/msb.20156663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Developmental cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 45.Yang L, et al. Permanent genetic memory with >1-byte capacity. Nature methods. 2014 doi: 10.1038/nmeth.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mercer AC, Gaj T, Fuller RP, Barbas CF., 3rd Chimeric TALE recombinases with programmable DNA sequence specificity. Nucleic acids research. 2012;40:11163–11172. doi: 10.1093/nar/gks875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sirk SJ, Gaj T, Jonsson A, Mercer AC, Barbas CF., 3rd Expanding the zinc-finger recombinase repertoire: directed evolution and mutational analysis of serine recombinase specificity determinants. Nucleic acids research. 2014;42:4755–4766. doi: 10.1093/nar/gkt1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chaikind B, Bessen JL, Thompson DB, Hu JH, Liu DR. A programmable Cas9-serine recombinase fusion protein that operates on DNA sequences in mammalian cells. Nucleic acids research. 2016;44:9758–9770. doi: 10.1093/nar/gkw707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shaikh AC, Sadowski PD. Chimeras of the Flp and Cre recombinases: tests of the mode of cleavage by Flp and Cre. Journal of molecular biology. 2000;302:27–48. doi: 10.1006/jmbi.2000.3967. [DOI] [PubMed] [Google Scholar]
- 50.Karpinski J, et al. Directed evolution of a recombinase that excises the provirus of most HIV-1 primary isolates with high specificity. Nature biotechnology. 2016;34:401–409. doi: 10.1038/nbt.3467. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3