The pattern of intron loss (original) (raw)
Abstract
We studied intron loss in 684 groups of orthologous genes from seven fully sequenced eukaryotic genomes. We found that introns closer to the 3′ ends of genes are preferentially lost, as predicted if introns are lost through gene conversion with a reverse transcriptase product of a spliced mRNA. Adjacent introns tend to be lost in concert, as expected if such events span multiple intron positions. Directly contrary to the expectations of some, introns that do not interrupt codons (phase zero) are more, not less, likely to be lost, an intriguing and previously unappreciated result. Adjacent introns with matching phases are not more likely to be retained, as would be expected if they enjoyed a relative selective advantage. The findings of 3′ and phase zero intron loss biases are in direct contradiction to an extremely recent study of fungi intron evolution. All patterns are less pronounced in the lineage leading to Caenorhabditis elegans, suggesting that the process of intron loss may be qualitatively different in nematodes. Our results support a reverse transcriptase-mediated model of intron loss.
Keywords: evolution, genome evolution
Two generalities of the intron-exon structure of eukaryotic genes remain unexplained. First, introns in intron-sparse species or in intron-sparse genes cluster near the 5′ ends of genes (1, 2). Second, intron positions within codons are doubly biased: introns tend to lie between the third base of one codon and the first base of the subsequent codon (phase zero) rather than between the first and second (phase one) or second and third (phase two) bases of a codon; and adjacent introns tend to be of the same phase (3–9).
The 5′ skew could be due to mutation-biased intron loss, selection-biased intron loss, or biased intron gain. If intron loss proceeds by means of gene conversion of the genomic copy of a gene by the reverse transcriptase product of a spliced transcript (RT-mRNAs) (10–15), the 3′ bias of RT products (12) could cause a higher rate of loss for 3′ introns (1, 2). Alternatively, possible preferential retention of 5′ introns could reflect their greater selective importance, possibly due to a greater concentration of regulatory elements (reviewed in ref. 16). Finally, intron gain could favor 5′ ends of genes for some unappreciated reason.
We analyzed intron losses in 684 groups of orthologous genes from seven eukaryotic species, previously analyzed by Rogozin et al. (17). Fig. 1 shows the most likely phylogeny for the species (18). Results calculated assuming the alternative coelomata grouping (19, 20) are similar and provided in Tables 3 and 4 and Figs. 6–9, which are published as supporting information on the PNAS web site. For each lineage, we defined introns known to be ancestral to the lineage (KAL) based on presence in both the sister group of the lineage and an outgroup. For example, introns present in a dipteran (Drosophila melanogaster and Anopheles gambiae) or Caenorhabditis elegans as well as a non-animal are KAL for the lineage leading to Homo sapiens. We found that 3′ KAL introns are preferentially lost for every lineage analyzed except C. elegans, suggesting that the clustering of introns near the 5′ of genes in intron-sparse genomes is due to biased intron loss.
Fig. 1.
The most likely relationship between the analyzed species. The deepest node is not resolved, with Arabidopsis thaliana clustering either with Plasmodium falciparum or with the other species.
To determine whether this 3′ bias is due to RT-mRNA-mediated loss or to differential selection, we analyzed the pattern of loss among adjacent KAL introns. If introns are lost through gene conversion by RT-mRNAs, adjacent introns may sometimes be lost in concert when a gene conversion event spans multiple intron positions (14, 21, 22). For each lineage, we compared the pattern of intron loss with the expectation assuming independent intron loss, and found a signal of concerted loss of adjacent introns for diptera and Saccharomyces pombe, but not C. elegans, evidence that the 5′ bias is due to RT-mRNA-mediated intron loss.
The two phase skews (abundance of phase zero introns and correspondence of adjacent intron phases) could be due to the legacy of gene formation, to insertional bias, to selection, or to alternative splicing. First, the biases could be echoes of gene formation through combination of exons or groups of exons (3–7, 9, 23–26) mediated by ancient phase zero introns (23). Second, intron insertion might be phase-biased (5, 8, 16, 27–34), perhaps due to preferential insertion into sites which themselves happen to be phase-biased (although, see ref. 7). Third, selection could favor phase zero introns due to transcript fidelity, usefulness in exon shuffling, or resilience to intron boundary sliding (17, 35), and the avoidance of downstream frame shifts in cases of errant exon exclusion could favor adjacent introns of the same phase (35). Finally, conditionally spliced exons that must be flanked by same phase introns to avoid downstream frame shifts could cause the bias.
A relative selective advantage for phase zero introns predicts preferential retention of phase zero introns. Instead, we find the opposite-phase zero KAL introns are more likely to be lost in all analyzed lineages except C. elegans. Among introns known to be present at the animal/fungi split, introns in phase zero are retained in fewer animal/fungi taxa than those in phases one and two. Thus, the phase bias persists despite, not because of, intron loss biases. KAL introns with adjacent KAL introns of matching phase are no more likely to be retained, suggesting that adjacent intron phase correspondence is not due to selection.
The pattern of intron loss for 684 eukaryotic groups of orthologs provides evidence that (i) introns are lost through gene conversion by RT-mRNAs; (ii) adjacent introns are often lost in concert; (iii) phase zero introns are lost at higher rates than phase one and two introns, thus selection does not appear to drive phase zero intron abundance; (iv) adjacent introns of the same phase are not preferentially retained, thus selection does not appear to drive adjacent intron phase correspondence; and (v) the lineage leading to C. elegans is exceptional in its intron loss pattern, evidencing none of the biases observed elsewhere.
Methods
Data Set and Programs. We downloaded amino acid level sequence alignments and corresponding intron positions with presence-absence matrices for each intron position in the conserved regions of 684 groups of orthologous genes, compiled by Rogozin et al. (see ref. 17 for details), from the National Center for Biotechnology Information (which can be accessed at ftp://ftp.ncbi.nlm.nih.gov./pub/koonin/intron_evolution). Introns present at the exact same position in two orthologs were assumed to be homologous. S. cerevisiae was excluded because it has extremely few introns and interrupts the lineage running from the animal-fungus split to S. pombe. Analyses were performed by using perl scripts.
KAL Introns. For each lineage, we defined KAL introns as those present in a sister group to the lineage, as well as an outgroup, and which are thus assumed to be present at the base of the lineage [e.g., for the C. elegans lineage, those introns present in a dipteran (sister group) as well as H. sapiens or a non-animal (outgroup)]. Introns present in the studied taxa (e.g., C. elegans) and the sister but no outgroup, or in the studied taxa and an outgroup, but not the sister, are also known to be ancestral to the lineage. However, such introns are only known to be ancestral by virtue of their presence in the studied taxa. If they had been lost in this taxa, they would not be known to be ancestral. Thus, such introns are themselves a retention-biased set, inappropriate for studying the pattern of loss, and were excluded.
Choice of Lineages. The data are summarized in Table 1. We selected lineages with large numbers of KAL introns for further analysis. For analyses of the pattern of adjacent intron loss, only genes with at least one retained KAL intron and at least two lost KAL introns are informative (right column of Table 1). We therefore chose the general diptera lineage, rather than the specific A. gambiae or D. melanogaster lineages, because it has more informative genes. When speaking of intron loss in diptera, “retained” denotes presence in one or both dipteran species, and “lost” denotes absence in both.
Table 1. Summary of the data.
| Total introns | Shared with | KAL introns (retained plus lost) | Genes with P ≥ 2; r ≥ 1 | |||
|---|---|---|---|---|---|---|
| Lineage | Sister taxa | Sister | Non-sister | |||
| D. melanogaster | A. gamblae | 725 | 382 | 489 | 451 (295 + 156) | 11 |
| A. gambiae | D. melanogaster | 675 | 382 | 451 | 489 (295 + 194) | 3 |
| Diptera | C. elegans | 1,016 | 234 | 609 | 634 (198 + 436) | 32 |
| C. elegans | Diptera | 1,468 | 234 | 634 | 609 (198 + 411) | 29 |
| H. sapiens | Diptera, C. elegans | 3,345 | 933 | 907 | 551 (339 + 112) | 4 |
| S. pombe | Animals | 450 | 223 | 158 | 927 (131 + 796) | 30 |
| A. thaliana | Animals, S. pombe | 2,933 | 908 | 97 | 119 (73 + 46) | 0 |
| P. falciparum | Animals, Sp. At | 450 | 143 | — | — | — |
Probability of Sums over a Group of Genes. For many tests, we compare the sum of a function for each gene over a group of genes to the null expectation. For a function 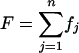 , where fj is some function for the j_th gene of n total, Pr{F = X} = Π_j Pr{fj = xj} summed over all sets of x values for which Σ_j xj_ = X. Expressions for Pr{fj = xj} for a range of functions are given below. P is then the probability that F is as divergent from the expectation as is the real value.
, where fj is some function for the j_th gene of n total, Pr{F = X} = Π_j Pr{fj = xj} summed over all sets of x values for which Σ_j xj_ = X. Expressions for Pr{fj = xj} for a range of functions are given below. P is then the probability that F is as divergent from the expectation as is the real value.
Adjacent Intron Loss. The probability that a gene which randomly loses l and retains r KAL introns through single-intron loss events loses exactly d pairs of lost adjacent introns (where n lost adjacent introns count as n - 1 lost pairs) is
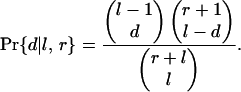 |
[1] |
|---|
The probability that it loses t triples of adjacent KAL introns is:
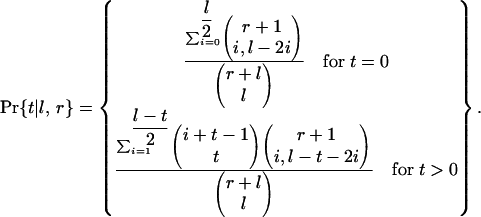 |
[2] |
|---|
The probability that it loses q quartets of adjacent KAL introns is
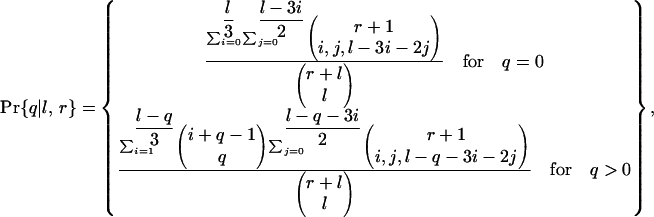 |
[3] |
|---|
where terms including negative factorials are defined to be zero throughout.
If introns are lost independently, the probability that a gene will lose e separate clusters of one or more adjacent introns is
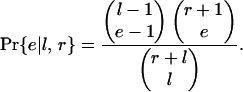 |
[4] |
|---|
If instead adjacent introns tend to be lost in concert, the lost introns will fall in fewer, larger clusters. The chance that all lost KAL introns will be adjacent (forming one cluster) is
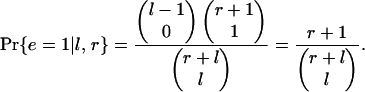 |
[5] |
|---|
Because these calculations require multiple KAL introns to have been lost in a gene, numbers for the H. sapiens lineage were too small for analysis (four genes lost two KAL introns; none of the genes lost more).
Loss by Phase. To test whether introns of different phases in the same gene are equally likely to be lost, we use only genes with KAL introns both in and out of phase zero and both lost and retained. The chance that such a gene loses x out of z total phase zero KAL introns is
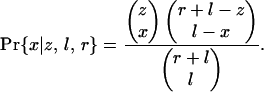 |
[6] |
|---|
3′ Loss Bias. For some groups of orthologs, the termini are unalignable, obscuring the true end of the ancestral protein. Therefore, we discarded genes in which the last conserved region lies 20% or more of the length of the alignment from either end of the alignment.
Results
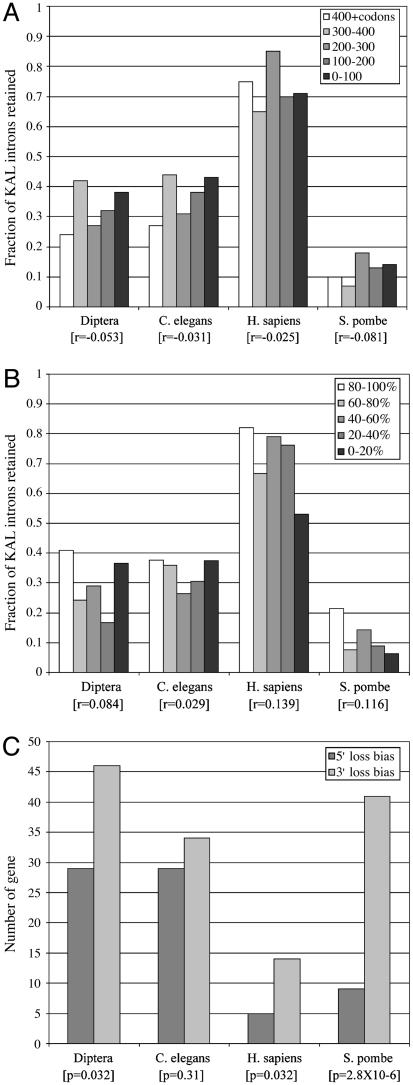
5′-3′ Position. We calculated the correlation between intron position and retention or loss (1, 0 respectively) for the KAL introns for each lineage. We measured intron position either as number of codons from the 3′ end of the gene or as percentage of total coding sequence length from 3′ to 5′ (Fig. 2). There is no significant correlation for the former measure; for the latter, there are significant positive correlations (greater 3′ loss; P < 0.05) for H. sapiens and S. pombe. However, these metrics are problematic. Intergene differences in numbers of transcripts, rates of reverse transcription per transcript, distributions of reverse transcription product lengths, and rates of gene conversion may obscure the signal. To avoid such problems, we then compared introns within the same gene. For each lineage, we counted each gene as an independent test. If a 3′ intron loss bias exists, the number of genes with a positive correlation between retention and distance from the 3′ should be greater than the number with a negative correlation; if not, the numbers should be equal. As Fig. 2_C_ shows, for each lineage, more genes have a positive than a negative correlation. This bias is statistically significant for three of the four lineages (diptera, P = 0.032; H. sapiens, P = 0.032; S. pombe, P = 2.8 × 10-6; C. elegans, P = 0.31 by χ2).
Fig. 2.
3′ intron loss bias. (A) The relationship between intron position measured as the length of coding sequence from the 3′ end of the gene and the fraction of introns retained. (B) The relationship between the intron position and the fraction of introns retained is shown. For each lineage, each bar represents a quintile of gene length: the rightmost bar gives the fraction retained for KAL introns in the C-terminal 20% of the gene, the second rightmost the fraction for introns from 20–40% of the length from 3′ to 5′, etc. (C) Detection of 3′ loss bias per gene. For each lineage, the numbers of genes showing a positive correlation between distance from 3′ and intron retention (3′ loss bias, light bars) and negative correlation (5′ bias, dark bars) are given.
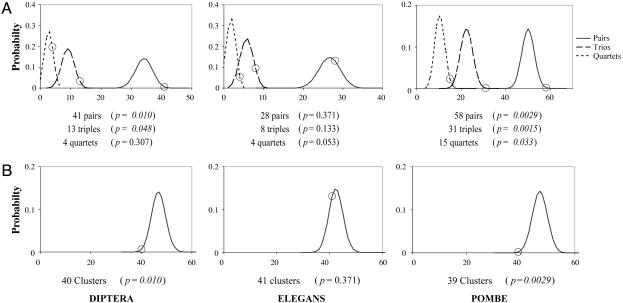
Adjacent Intron Loss. For each lineage, we calculated probability distributions for the total numbers of pairs, trios, and quartets of adjacent KAL introns lost, assuming independent single-intron loss, and compared these distributions to the observed values. For instance, a gene losing two of its three KAL introns will either retain the 5′ intron (100), the middle intron (010), or the 3′ intron (001), thus the chance of losing adjacent introns is 2/3. A gene losing three of five KAL introns may lose no adjacent pairs of introns (01010), one adjacent pair (00101, 00110, 10010, 01001, 01100, and 10100), or two adjacent pairs (00011, 10001, and 11000), with respective probabilities of 1:6:3. This gene also has a 3/10 chance (00011, 10001, and 11000) of losing an adjacent trio. For each lineage, we calculated probability distributions for numbers of lost adjacent pairs, trios, and quartets of KAL introns for each gene and used these values to calculate overall probability distributions for the total numbers for all genes. Fig. 3_A_ gives probability distributions and observations. The S. pombe and diptera lineages show significant excesses of adjacent pairs and trios of lost KAL introns, and S. pombe shows a significant excess of quartets (diptera pairs: P = 0.010, trios, P = 0.048, quartets, P = 0.307; S. pombe: P = 0.0029, P = 0.0015, P = 0.033). C. elegans shows slight but nonsignificant trends.
Fig. 3.
Adjacent introns are lost in concert. (A) The probability distributions for total numbers of pairs, trios, and quartets of adjacent KAL introns lost for all analyzed genes for each lineage, with the observed values given below. (B) The probability distribution for the total numbers of clusters of one or more lost adjacent introns for all analyzed genes, with the observed values given below.
We then asked generally about clustering of KAL intron losses along the gene. For instance, if a gene independently loses three KAL introns of five, the relative probabilities of one, two, and three clusters of one or more adjacent lost KAL introns is 3:6:1 (11000, 10001, and 00011 vs. 01100, 00110, 10100, 10010, 01001, and 00101 vs. 01010). For each lineage, we calculated probability distributions for each gene, and from these distsributions the overall probability distributions for total numbers of clusters for all genes (Fig. 3_B_). All lineages show fewer clusters (greater clustering) than expected, reaching significance for diptera (P = 0.010) and S. pombe (P = 0.0029). There was also a significant excess of genes in which all lost KAL introns were adjacent (one cluster) in diptera (P = 0.013) and S. pombe (P = 0.0013), but not C. elegans (P = 0.425).
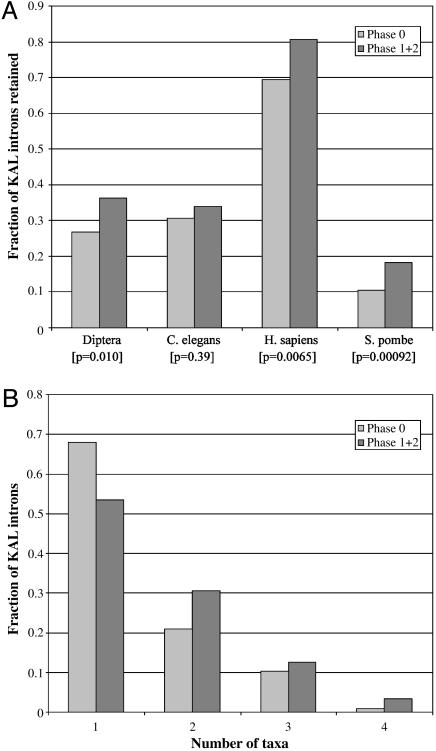
Phase. For each lineage, we calculated the fractions of phase zero and phase one and two KAL introns retained (Fig. 4_A_). In the diptera lineage, 87/327 (26.7%) phase zero and 111/307 (36.2%) phase one and two KAL introns were retained (P = 0.0061). For humans, the numbers are 152/219 (69.4%) and 187/232 (80.6%; P = 0.0041), for S. pombe,52/493 (10.5%) and 79/434 (18.2%; P = 0.00059). Only C. elegans gives a nonsignificant result (P = 0.22).
Fig. 4.
Phase-biased intron loss. (A) The fraction of phase zero, phase one, and two KAL introns retained along each lineage. (B) The distribution of the number of fungi/animal taxa (diptera, C. elegans, H. sapiens, and S. pombe) in which an intron is retained for introns shared between fungi/animals and Arabidopsis/Plasmodium for phase zero and phase one and two introns.
To ensure this result was not due to phase zero introns happening to lie in loss-prone genes, we analyzed the loss pattern by phase for each gene separately. For each informative gene (those with phase zero and non-phase zero KAL introns and that have lost some but not all of their KAL introns) for each lineage, we calculated expectations and probability distributions for losing a given number of phase zero introns given the number of total introns lost. Although the data set is small because of a paucity of informative genes, the general trend holds. For the lineage leading to diptera, 54 phase zero introns were lost vs. 49.0 expected (P = 0.096); for H. sapiens, 11 observed vs. 6.7 expected (P = 0.012); for S. pombe, 55 observed vs. 50.5 expected (P = 0.086).
Another way to look at the data is to analyze introns known to be present at the fungi-animal split (by virtue of presence in fungi and/or animals as well as plants and/or Plasmodium). There are 503 such introns in phase zero and 451 in phase one or two. For each intron we asked how many animal-fungi taxa it was found in (1, 2, 3, or 4) among S. pombe, H. sapiens, C. elegans and diptera. Fig. 4_b_ gives the results. Only 32.0% phase zero introns were found in more than one fungi-animal taxon, compared with 53.8% for phase one and two introns (P = 2.79 × 10-6 by a χ2 test) and only 0.8% compared with 5.8% were found in all four taxa (P = 0.0046). The overall difference between the distributions is significant (P = 1.0 × 10-5 by χ2, 3 df).
Adjacent Intron Phase. For each lineage, we divided KAL introns based on matching or nonmatching adjacent intron phase (for upstream and downstream adjacent KAL introns, separately) and asked whether introns whose adjacent KAL introns are in the same phase were preferentially retained. No pattern of preferential retention of introns which match the adjacent KAL intron in phase is observed (Table 2).
Table 2. The effect of adjacent intron phase on intron loss.
| Percent introns retained | ||||
|---|---|---|---|---|
| Adjacent intron phase | ||||
| Same | Different | P | ||
| Diptera | ||||
| Ph0 | 5′ | 14 (202) | 23 (188) | 0.99 |
| 3′ | 23 (202) | 16 (159) | 0.08 | |
| Ph1 | 5′ | 31 (35) | 13 (112) | 0.01 |
| 3′ | 43 (35) | 27 (128) | 0.06 | |
| Ph2 | 5′ | 25 (32) | 31 (123) | 0.81 |
| 3′ | 23 (32) | 32 (136) | 0.46 | |
| All | 5′ | 17 (269) | 22 (423) | 0.95 |
| 3′ | 27 (269) | 25 (423) | 0.29 | |
| C. elegans | ||||
| Ph0 | 5′ | 17 (139) | 22 (127) | 0.87 |
| 3′ | 22 (139) | 24 (106) | 0.65 | |
| Ph1 | 5′ | 19 (26) | 22 (69) | 0.70 |
| 3′ | 35 (26) | 32 (81) | 0.50 | |
| Ph2 | 5′ | 18 (22) | 17 (83) | 0.55 |
| 3′ | 36 (22) | 16 (92) | 0.04 | |
| All | 5′ | 18 (187) | 20 (279) | 0.92 |
| 3′ | 26 (187) | 24 (279) | 0.35 | |
| H. sapiens | ||||
| Ph0 | 5′ | 67 (24) | 80 (35) | 0.93 |
| 3′ | 75 (24) | 86 (21) | 0.90 | |
| Ph1 | 5′ | 100 (8) | 67 (9) | 0.12 |
| 3′ | 100 (8) | 87 (23) | 0.39 | |
| Ph2 | 5′ | 63 (8) | 85 (20) | 0.96 |
| 3′ | 88 (8) | 80 (20) | 0.55 | |
| All | 5′ | 73 (40) | 80 (64) | 0.86 |
| 3′ | 83 (40) | 84 (64) | 0.72 | |
| S. pombe | ||||
| Ph0 | 5′ | 4 (137) | 9 (122) | 0.96 |
| 3′ | 9 (137) | 10 (104) | 0.60 | |
| Ph1 | 5′ | 9 (23) | 11 (65) | 0.74 |
| 3′ | 4 (23) | 21 (81) | 0.99 | |
| Ph2 | 5′ | 5 (21) | 12 (84) | 0.93 |
| 3′ | 5 (21) | 23 (86) | 0.99 | |
| All | 5′ | 5 (181) | 10 (271) | 0.99 |
| 3′ | 8 (181) | 17 (271) | 1.00 |
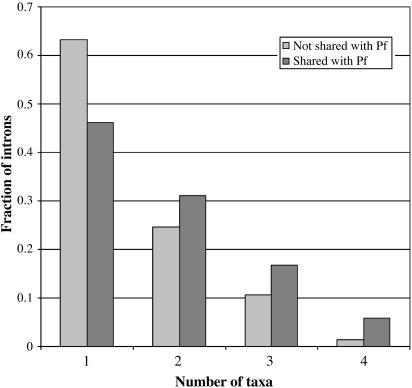
Introns Shared with Plasmodium. If Plasmodium is an outgroup to the plant-animal divergence, introns shared between Plasmodium falciparum and modern plants, animals, or fungi have been maintained for an extremely long time down multiple disparate lineages. Alternatively, if Plasmodium is more closely related to plants, it has lost an impressive 92% percent of its introns since that divergence (S.W.R. and W.G., unpublished data). In either case, introns shared between Plasmodium and animals/fungi/plants are expected to be particularly resilient to intron loss. Among introns present in animals and/or fungi as well as Plasmodium and/or Arabidopsis (and thus known present at the animal/fungi split), 104 are present in Plasmodium, and an additional 835 are present in Arabidopsis. For each such intron, we asked how many animal/fungi taxa it is present in. The data are shown in Fig. 5. Only 36.8% of non-Plasmodium introns, but 53.8% of Plasmodium introns, are present in multiple fungi/animal taxa (P = 0.00031); only 1.4% of non-Plasmodium introns, but 5.9% of Plasmodium introns are present in all four taxa (P = 0.0055). The overall distributions are significantly different (P = 0.00014 by χ2, 3 df).
Fig. 5.
Introns shared with Plasmodium are preferentially retained. The distribution of number of fungi/animal taxa in which an intron is retained for introns shared between fungi/animals and Arabidopsis/Plasmodium for introns shared and not shared with Plasmodium.
Discussion
Our results provide two lines of evidence for a model of intron loss through gene conversion with RT-mRNAs. First, 3′ introns are more likely to be lost in the lineages leading to diptera and S. pombe as predicted either by a model of RT-mRNA-mediated intron loss or by even rates of loss, followed by stronger selection against loss of 5′ introns [possibly due to important intronic regulatory elements, which appear to be enriched in 5′ introns (16)]. Thus, the observation of a greater 3′ bias among introns found in diverse species (36) appears to be due to biases in the pattern of intron loss, not gain. These results are in tension with other recent studies. Patthy and Banyai (37) found instances of intron loss in even the 5′ regions of very long multidomain genes in D. melanogaster and C. elegans. Cho et al. (38) found no 3′ intron bias in the cytoplasmic polyadenylation element-binding genes of nematodes, leading them to suggest than introns might instead be lost by genomic deletion. If in fact introns are lost by genomic deletion, the only other plausible proposed mechanism for intron loss that we are aware of, we should see a fairly high ratio of inexact deletions in which a small number of codons are added or lost from the flanking coding sequence (39) to exact deletions, a prediction that has yet to be tested. However, our finding of a lack of 3′ loss bias in C. elegans suggests that perhaps intron loss in nematodes is subject to different forces, possibly resolving these tensions.
Second, we find that introns that have been lost tend to be adjacent along the gene, suggesting concerted loss of adjacent introns. Alternatively, clustering of intron losses along the gene could reflect differential intron loss rates along the length of the gene. However, in the absence of such evidence, this result also supports the RT-mRNA-mediated loss model.
A second set of results provides evidence against creation of the observed phase biases by selection or biased loss. The tendency of introns to fall between codons (phase zero) has been explained by some as more positive (or less negative) selection on these introns. However, phase zero introns are shown here to be more, not less, likely to be lost. This result is quite general across the eukaryotes studied. These results eliminate selection, as well as a mutational loss bias, as possible explanations for the observed bias. Correspondence of adjacent intron phases also appears non-selection-driven, because we find that adjacent KAL introns whose phases match are no more likely to be retained.
Our findings of phase zero and 3′ biases in the pattern of intron loss contradict the findings of Nielsen et al. (40), who found neither bias in a similar very recent genome-wide study of orthologous groups in four fungal taxa. They analyzed the influence of the 5′-3′ position on intron loss by dividing introns in each gene into five quintiles based on position along the gene and comparing probabilities of loss between groups. However, differences between genes in gene lengths, expression levels, reverse transcription rates, distributions of reverse transcription product lengths, and rates of gene conversion cause problems for such intergene comparisons. These problems may be overcome by comparing introns within the same gene, as we do here. Supporting this interpretation, whereas our intragene analysis shows a strong 3′ loss bias, our intergene analysis, which is very similar to that of Nielsen et al. (40), does not. An intragene analysis of their data could determine whether choice of method can explain this discrepancy.
The discrepancy over phase bias in intron loss is harder to explain. Whereas we find that phase zero introns are preferentially lost, their table 1 in ref. 40 shows that phase zero introns are instead preferentially retained in the lineage leading to Neurospora crassa (P = 0.048 by a Fisher's Exact test on phase zero and non-phase zero, conserved and raw losses) and show no significant bias in the other two lineages. These patterns are surprising and deserve further attention.
A final intriguing result is the deviation in the intron loss pattern in the lineage leading to C. elegans. Of the three observed patterns observed here (3′ loss bias, a signal of adjacent intron loss, and phase zero intron loss bias) none is observed in C. elegans. This result suggests that intron loss may occur through qualitatively different mechanisms in nematodes. Kent and Zahler (41) offer the interesting observation that the 5′ and 3′ boundaries of unique introns between C. elegans and Caenorhabditis briggsae show greater similarity to each other than do the boundaries of control introns, and suggested that introns may be lost by nonhomologous recombination between intron boundaries [although their result could also be explained by preferential intron insertion into sites with a consensus sequence of AG|GT (17, 27–34)]. Comparative analyses of multiple nematode genomes should help to better understand these deviations.
The pattern of intron lost in 684 groups of orthologs diverse eukaryotic taxa suggests that (i) introns are lost through gene conversion by a retrotransposed copy of a spliced transcript of the gene often spanning multiple intron positions, (ii) the phase biases of introns are not due to differential selection or loss biases, and (iii) intron loss in the lineage leading to C. elegans does not exhibit the biases found in other lineages, suggesting that the dynamics of intron evolution may be qualitatively different in nematodes.
Supplementary Material
Supporting Information
Abbreviation: KAL, known to be ancestral to the lineage.
References
- 1.Sakurai, A., Fujimori, S., Kochiwa, H., Kitamura-Abe, S., Washio, T., Saito, R., Carninci, P., Hayashizaki, Y. & Tomita, M. (2002) Gene 300**,** 89-95. [DOI] [PubMed] [Google Scholar]
- 2.Mourier, T. & Jeffares, D. C. (2003) Science 300**,** 1393. [DOI] [PubMed] [Google Scholar]
- 3.Fedorov, A., Suboch, G., Bujakov, M. & Fedorova, L. (1992) Nucleic Acids Res. 20**,** 2553-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long, M., Rosenberg, C. & Gilbert, W. (1995) Proc. Natl. Acad. Sci. USA 92**,** 12495-12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long, M., de Souza, S. J., Rosenberg, C. & Gilbert, W. (1998) Proc. Natl. Acad. Sci. USA 95**,** 219-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fedorov, A., Fedorova, L., Starshenko, V., Filatov, V. & Grigor'ev, E. (1998) J. Mol. Evol. 46**,** 263-271. [DOI] [PubMed] [Google Scholar]
- 7.Long, M. & Rosenberg, C. (2000) Mol. Biol. Evol. 17**,** 1789-1796. [DOI] [PubMed] [Google Scholar]
- 8.Wolf, Y. I., Kondrashov, F. A. & Koonin, E. V. (2001) Trends Genet. 16**,** 333-334. [DOI] [PubMed] [Google Scholar]
- 9.Roy, S. W., Lewis, B. P., Fedorov, A. & Gilbert, W. (2001) Trends Genet. 17**,** 496-501. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein, L. B., Mount, S. M. & Weiner, A. M. (1983) Cell 32**,** 461-472. [DOI] [PubMed] [Google Scholar]
- 11.Lewin, R. (1983) Science 219**,** 1052-1054. [DOI] [PubMed] [Google Scholar]
- 12.Weiner, A. M. (1986) Annu. Rev. Biochem. 55**,** 631-661. [DOI] [PubMed] [Google Scholar]
- 13.Fink, G. R. (1987) Cell 49**,** 5-6. [DOI] [PubMed] [Google Scholar]
- 14.Long, M. & Langley, C. H. (1993) Science 260**,** 91-95. [DOI] [PubMed] [Google Scholar]
- 15.Derr, L. K. (1998) Genetics 148**,** 937-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fedorova, L. & Fedorov, A. (2003) Genetica (The Hague) 118**,** 123-131. [PubMed] [Google Scholar]
- 17.Rogozin, I. B., Wolf, Y. I., Sorokin, A. V., Mirkin, B. G. & Koonin, E. V. (2003) Curr. Biol. 13**,** 1512-1517. [DOI] [PubMed] [Google Scholar]
- 18.Aguinaldo, A. M., Turbeville, J. M., Linford, L. S., Rivera, M. C., Garey, J. R., Raff, R. A. & Lake, J. A. (1997) Nature 387**,** 489-493. [DOI] [PubMed] [Google Scholar]
- 19.Knoll, A. H. & Carroll, S. B. (1999) Science 284**,** 2129-2137. [DOI] [PubMed] [Google Scholar]
- 20.Wolf, Y. I., Rogozin, I. B. & Koonin, E. V. (2004) Genome Res. 14**,** 29-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frugoli, J. A., McPeak, M. A., Thomas, T. L. & McClung, C. R. (1998) Genetics 149**,** 355-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wada, H., Kobayashi, M., Sato, R., Satoh, N., Miyasaka, H. & Shirayama, Y. (2002) J. Mol. Evol. 54**,** 118-128. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert, W. (1987) Cold Spring Harbor Symp. Quant. Biol. 52**,** 901-905. [DOI] [PubMed] [Google Scholar]
- 24.Patthy, L. (1999) Gene 238**,** 103-114. [DOI] [PubMed] [Google Scholar]
- 25.de Souza, S. J., Long, M., Schoenbach, L., Roy, S. W. & Gilbert, W. (1996) Proc. Natl. Acad. Sci. USA 93**,** 14632-14636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy, S. W. (2003) Genetica (The Hague) 118**,** 251-266. [PubMed] [Google Scholar]
- 27.Dibb, N. J. & Newman, A. J. (1989) EMBO J. 8**,** 2015-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarrio, R., Rodriguez-Trelles, F. & Ayala, F. J. (2003) Proc. Natl. Acad. Sci. USA 100**,** 6580-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sverdlov, A. V., Rogozin, I. B., Babenko, V. N. & Koonin, E. V. (2003) Curr. Biol. 13**,** 2170-2174. [DOI] [PubMed] [Google Scholar]
- 30.Sadusky, T., Newman, A. J. & Dibb, N. J. (2004) Curr. Biol. 14**,** 505-509. [DOI] [PubMed] [Google Scholar]
- 31.Qiu, W. G., Schisler, N. & Stoltzfus, A. (2004) Mol. Biol. Evol. 21**,** 1252-1263. [DOI] [PubMed] [Google Scholar]
- 32.Coghlan, A. & Wolfe, K. H. (2004) Proc. Natl. Acad. Sci. USA 101**,** 11362-11367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paquette, S. M., Bak, S. & Feyereisen, R. (2000) DNA Cell Biol. 19**,** 307-317. [DOI] [PubMed] [Google Scholar]
- 34.Logsdon, J. M., Jr. (1998) Curr. Opin. Genet. Dev. 8**,** 637-648. [DOI] [PubMed] [Google Scholar]
- 35.Lynch, M. (2002) Proc. Natl. Acad. Sci. USA 99**,** 6118-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sverdlov, A. V., Babenko, V. N., Rogozin, I. B. & Koonin, E. V. (2004) Gene 338**,** 85-91. [DOI] [PubMed] [Google Scholar]
- 37.Banyai, L. & Patthy, L. (2004) FEBS Lett. 565**,** 127-132. [DOI] [PubMed] [Google Scholar]
- 38.Cho, S., Jin, S. W., Cohen, A. & Ellis, R. E. (2004) Genome Res. 14**,** 1207-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Llopart, A., Comeron, J. M., Brunet, F. G., Lachaise, D. & Long, M. (2002) Proc. Natl. Acad. Sci. USA 99**,** 8121-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen, C. B., Friedman, B., Birren, B., Burge, C. B. & Galagan, J. E. (2004) PloS Biol. 2**,** e422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kent, W. J. & Zahler, A. M. (2000) Genome Res. 10**,** 1115-1125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information