Inducible DNA-loop formation blocks transcriptional activation by an SV40 enhancer (original) (raw)
Abstract
It is well established that gene expression in eukaryotes is controlled by sequence-dependent binding of _trans_-acting proteins to regulatory elements like promoters, enhancers or silencers. A less well understood level of gene regulation is governed by the various structural and functional states of chromatin, which have been ascribed to changes in covalent modification of core histone proteins. And, much on how topological domains in the genome take part in establishing and maintaining distinct gene expression patterns is still unknown. Here we present a set of regulatory proteins that allow to reversibly alter the DNA structure in vivo and in vitro by adding low molecular weight effectors that control their oligomerization and DNA binding. Using this approach, we completely regulate the activity of an SV40 enhancer in HeLa cells by reversible loop formation to topologically separate it from the promoter. This result establishes a new mechanism for DNA-structure-dependent gene regulation in vivo and provides evidence supporting the structural model of insulator function.
Keywords: conditional gene regulation, dimerizer, DNA-loop formation, SV40 enhancer, tet
Introduction
Growth and differentiation of multicellular organisms is accompanied by changes in the transcriptional pattern of their genome. Cell-type-specific transcription of genes is often regulated by enhancer sequences that act over large distances or even in trans upon promoters on a different chromosome (Blackwood and Kadonaga, 1998). This long-range regulatory potential of enhancers poses the question of how their sphere of influence is restricted to the cognate promoters and whether or how their range of activity can be regulated.
Insulator elements are one example for eukaryotic DNA sequences that represent boundaries between transcriptionally independent genetic units in the genome. They are functionally defined by their ability to block the activity of an enhancer in a position-dependent manner and/or by their capacity to protect a transgene against position effects (Gerasimova and Corces, 2001). The modes of action of insulators remain unresolved and different models have been proposed (Kuhn and Geyer, 2003). One of them, the so-called structural model, is based on the observation that the eukaryotic genome is physically separated into topological domains (Fransz et al, 2002; Mahy et al, 2002). Insulators might initially participate in structuring the DNA, which—as a secondary effect—might result in the establishment of functionally independent domains (Gerasimova and Corces, 1998), for example, by the formation of DNA loops (Udvardy et al, 1985; Vazquez and Schedl, 1994).
Since it is not known whether DNA loops have a regulatory impact, we analyze here, in transiently transfected HeLa cells, the effect of conditionally looping out an enhancer on gene expression from a distal promoter as a potential mechanism for DNA structure-dependent gene regulation.
Results and discussion
The homodimerization system can completely repress SV40 enhancer activity
To analyze the regulatory effect of looping out an enhancer, we constructed an inducible homodimerization system based on doxycycline (dox)-dependent components (Berens and Hillen, 2003): The reporter plasmid pWHE206 is based on plasmids previously used to analyze enhancer-blocking elements (Recillas-Targa et al, 1999). It encodes an SV40 enhancer which drives expression of a luciferase reporter gene from a distal SV40 promoter (Figure 1A). In order to topologically separate the enhancer from the promoter, it was flanked by repeats of seven tet operators (Gossen and Bujard, 1992). The center-to-center distance of the innermost tetO palindromes flanking the enhancer is 344 base pairs (bp). They are 596 bp for the middle pairs and 848 bp for the outermost pairs (Figure 1A). The two tetO arrays are thus centered around the ‘optimal separation distance for interaction by looping', as calculated by Rippe for supercoiled and linearized nonchromatinized naked DNA (Rippe, 2001). Since this distance is also more than twice the persistence length of DNA (Lu et al, 2001), it should be possible to form a DNA loop without torsionally straining the DNA. A regulator plasmid encodes a dox-controlled dimerizer (tD) which consists of either a GCN4- (tDG) (Morii et al, 2002) or a LexA-dimerization domain (tDL) (Schnarr et al, 1988) fused to the C-terminal end of a monomerized single-chain Tet repressor (sc TetR) (Krueger et al, 2003) (Figure 1B). Binding of tD to the tet operators of the reporter plasmid should lead to structural looping-out of the enhancer due to interaction between the dimerization domains, resulting in reduced luciferase expression. Addition of dox causes tD to dissociate from the DNA, thereby dismantling the loop to yield full luciferase expression. HeLa cells were co-transfected with one reporter and one regulator plasmid each (Figure 1C). Unenhanced basal luciferase activity was determined by solely transfecting pGL3-promoter, containing neither enhancer nor tet operators. Both tDG and tDL repress with factors of 6.8 for tDG and 5.4 for tDL, reducing luciferase activity in the absence of dox to a level just 1.6- or 2.6-fold above that of pGL3-promoter. In contrast, the repression factor of sc TetR*, the transregulator control lacking a dimerization domain, is 2.6 and the residual luciferase activity remains six-fold higher than that of the pGL3-promoter control. In the presence of dox, repression is relieved in all cases to the level of the unrepressed control. Quantification of the Western blot analysis showed that tDG is expressed to a 2.2-fold higher level than tDL (Figure 1D). Therefore, slight differences in the regulatory activity of tDG and tDL might simply be explained by differences in their steady-state protein levels. However, a higher association constant of the dimerization domain derived from GCN4 (_K_a=2.2−3.1 × 107 M−1) (Okahata et al, 1998) compared to LexA (_K_a=2.1 × 104 M−1) (Schnarr et al, 1988) might also add to this difference.
Figure 1.
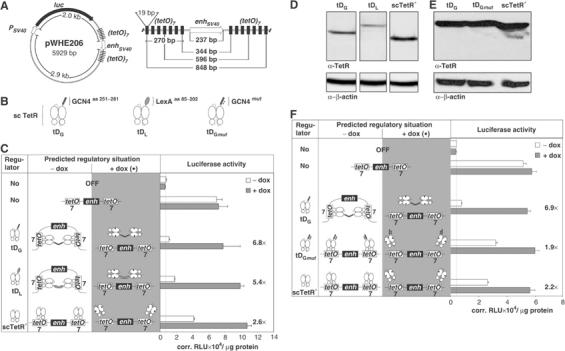
Dimerization activity of tDs is necessary and sufficient for regulation of enhancer activity. (A) Outline of the reporter plasmid pWHE206 used for the analysis of tD-dependent looping out of an enhancer. An SV40 enhancer located 2.0 kb downstream, respectively 2.9 kb upstream of an SV40 promoter, activates the transcription of a luciferase reporter gene. The enhancer is flanked by a repeat of seven TetR-binding sides (tetO; 19 bp). The two (tetO)7 boxes are separated by 327 bp including the 237 bp SV40 enhancer. (B) Tet-dimerizers (tD) consist of a GCN4- (tDG), LexA- (tDL) or a dimerization-deficient GCN4- (tDG_mut_) dimerization domain fused to the C-terminal end of sc TetR. Presence (C) and functionality (F) of the dimerization domain are necessary to regulate the activity of an enhancer. Transregulators are indicated schematically in column one. The predicted regulatory situations are shown in column two. Cells were cultured in the absence (white bars) or presence (gray bars) of dox. Repression factors and basal luciferase activity (thin line) are indicated. Values represent the means of triplicate samples with standard deviations given in corrected relative light units (corr. RLU) per μg of total cell protein. (D, E) Western blot analysis to detect transregulators in HeLa cell extracts.
The regulatory effect of sc TetR* is unspecific and dependent on its expression level
The sc TetR* control without dimerization domain exhibits slight but measurable regulation of enhancer activity, as shown in Figures 1C and F. Its steady-state protein level as determined by quantification of Western blot data is 1.6- and 3.5-fold higher than tDG and tDL, respectively (Figure 1D). To compare the regulatory properties of tDG and sc TetR*, we titrated the amounts of transiently transfected plasmids expressing transregulators (1–100 ng) (Figure 2). In the absence of dox, the results indicate for the sc TetR*-expressing plasmid that increasing the amount of transfected transregulator-expressing DNA leads to a steady decrease in luciferase activity (Figure 2A). Accordingly, increasing amounts of transfected transregulator-expressing DNA lead to slightly, but steadily higher repression factors (Figure 2B). In contrast, enhancer repression by tDG increases strongly and reaches a plateau at 10–30 ng transfected transregulator-expressing DNA. Western blot analysis reveals for both sc TetR* and tDG that rising amounts of transfected transregulators correlate with increasing steady-state protein levels (Figure 2C).
Figure 2.
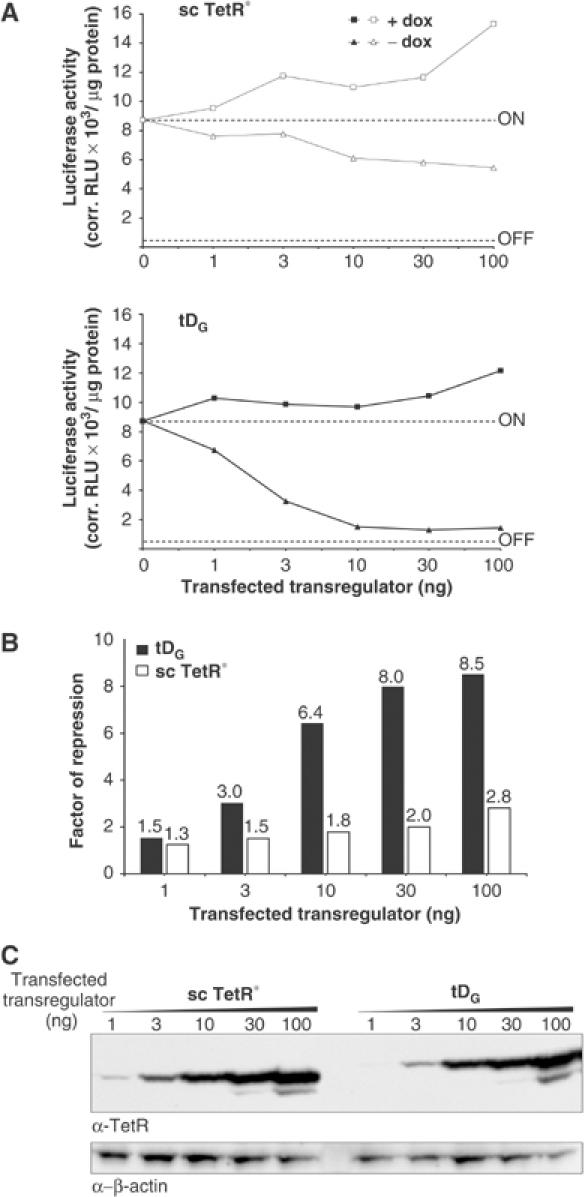
Regulatory effect of sc TetR* is nonspecific and depends on the amount of transregulator expressing DNA transfected. (A) tDG and sc TetR* exhibit different regulatory behaviors. As controls, pGL3-promoter (OFF) lacking the SV40 enhancer and tet operators or pWHE206 (ON) were transfected without transregulator (indicated by dotted lines). Cells were cultured in the absence (filled/open triangles) or in the presence (filled/open squares) of dox. Values represent the means of triplicate samples given in corrected relative ray light units (corr. RLU) per μg of total cell protein. (B) Factor of repression was determined for both tDG and sc TetR* by comparison of luciferase activity in the presence and the absence of dox and plotted against different amounts of transfected transregulator-expressing DNA. (C) Western blot analysis to detect the respective transregulators in HeLa cell extracts. Higher amounts of DNA transfected lead to higher amounts of transregulator protein.
These differences in the repression profiles of tDG and sc TetR* suggest that the more pronounced repression mediated by tDG is primarily due to the additional functional domain and not solely due to protein binding to the tet operators, whereas sc TetR* acts in an expression level-dependent, nonspecific manner. As a consequence, for the homodimerization system all experiments shown in the manuscript were performed using 20–30 ng transregulator-expressing plasmids and the respective repression factors are indicated. The DNA-kinking activity of TetR (Tovar and Hillen, 1989; Orth et al, 2000) could additionally contribute to the sc TetR* effect. Assuming 10.5 bp per helical turn of DNA (Hayes et al, 1990), all tet operators are located on the same side of the DNA helix. Such additive kinking and distortion of DNA after binding of TetR to tetO might influence enhancer-mediated gene activation in the topologically constrained context of a supercoiled plasmid.
A functional GCN4-dimerization domain is necessary for the activity of tDG
To analyze if the regulatory activity of tDG requires a functional GCN4-dimerization domain, tDG_mut_ was constructed by introducing the exchanges LP253 and LP260 (Figure 1B). Each of these single amino-acid replacements alone abolishes dimerization of a GCN4-fusion protein (Hu et al, 1990). We transiently transfected HeLa cells with pWHE206 and plasmids expressing either tDG_mut_ or tDG (Figure 1F). The latter again displays strong, 6.9-fold repression of luciferase activity. In contrast, tDG_mut_ exhibits only 1.9-fold repression, similar to the 2.2-fold repression obtained with sc TetR*. Repression is, once again, fully relieved in the presence of dox. Their expression levels are similar (see Figure 1E), demonstrating that the regulatory properties of tDG depend on its functional dimerization domain rather than on the added molecular mass.
tDL interacts with tet operators on both sides of the SV40 enhancer at the same time in vitro
In order to further analyze whether the positioning of tet operators in the reporter plasmid pWHE206 allows the formation of a DNA loop in the presence of tDs, we made use of an assay that has previously been used to determine DNA-loop formation in vitro (Bondarenko et al, 2003). Supercoiled pWHE206 bearing seven tet operators flanking the SV40 enhancer (Figure 3A) was incubated in the presence of tDL, with or without dox, and then digested with restriction enzymes producing 718 and 337 bp fragments, each containing seven tet operators (Figure 3A). If tDL dimers interact with (tetO)7 elements on both sides of the SV40 enhancer at the same time, both DNA fragments are expected to migrate as one complex in a native gel. After preincubation of supercoiled pWHE206 with tDL and subsequent digestion, no other DNA bands but a complex with slower mobility was detected (compare Figure 3B, lanes 1, 2 and 3). The retarded complex forms a diffuse band in the gel, which is most likely due to the multiple tet operators present in each array. Individual complexes will most likely differ in the total number of tDL molecules bound and the interaction of the tDL molecules with the two arrays will not always be within the same register. These differences probably emerge as variations in the migration distance of individual complexes and lead to the observed diffuse band. Analysis of DNA extracted from this complex indicates that it contains both 718 and 337 bp fragments at an even molar ratio (Figure 3C). Additionally, formation of this tDL-based complex is entirely abolished in the presence of dox (Figure 3B, lane 5). Similar results were obtained with a construct bearing single tet operators flanking the enhancer, except that here only one distinct band containing the two DNA fragments was detected (Supplementary Figure S1). This clearly shows that tDL dimers are able to bind two independent tetO elements at the same time in vitro. As, furthermore, an active GCN4-dimerization domain (Figure 1F) was also shown to be necessary for enhancer blocking in vivo, tDs assemble stable DNA loops enclosing the SV40 enhancer on supercoiled DNA.
Figure 3.
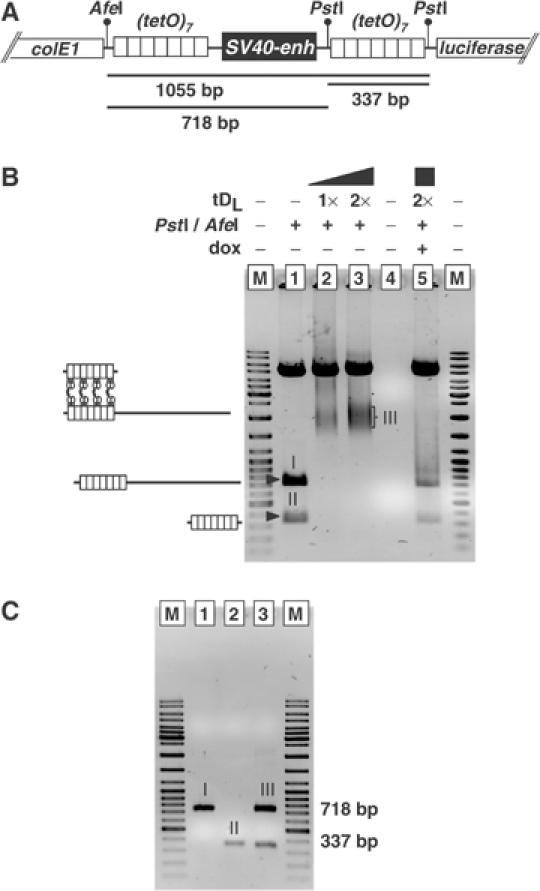
tDG interacts with tet operators on both sides of the SV40 enhancer in vitro. (A) Partial restriction map of the pWHE206 plasmid. If tDL interacts with tet operators on both sides of the SV40 enhancer on supercoiled DNA, two DNA fragments (718 and 337 bp) generated after digestion with restriction enzymes _Afe_I and _Pst_I are expected to be bound to tDL and migrate as a single DNA–protein complex in a native gel. (B) Analysis of tDL-induced DNA–protein complex formation on supercoiled DNA: supercoiled pWHE206 was pre-incubated with or without tDL, then digested and analyzed in a native agarose gel. The expected products of digestion are shown on the left. Different bands in the gel are arbitrarily numbered. M, peqGOLD DNA ladder (PEQLAB, Germany). (C) Analysis of DNA composition of the tDL-dependent DNA–protein complex. DNA was purified from different bands (I–III in (B)) and analyzed on a 1.2% agarose gel. M, peqGOLD DNA ladder (PEQLAB, Germany).
Regulation of the enhancer is not restricted to the supercoiled state of reporter plasmids
Next, we asked if superhelicity is required for enhancer blocking in vivo. We therefore linearized the reporter plasmids (Figure 4A) for the enhancer-blocking assay (Figure 4B). The maximum repression factor spanned by the controls is 7.9. Sc TetR* shows only marginal 1.4-fold regulation. In contrast, tDG exhibits complete repression of the enhancer in the absence of dox and full luciferase expression in its presence. The observation that sc TetR* shows no repression with the linearized reporter plasmids reinforces the assumption that the regulatory activity of sc TetR* in the supercoiled reporter plasmid (Figures 1 and 2) is due to topological effects as a result of regulator binding.
Figure 4.
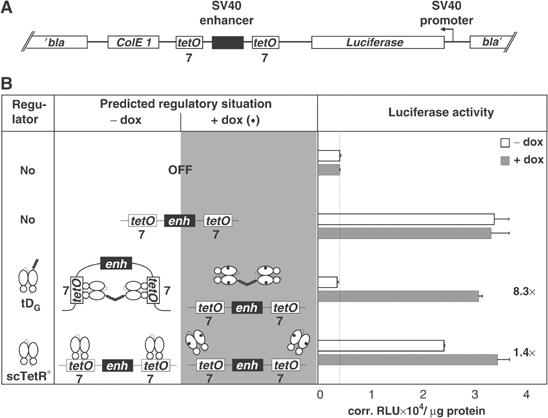
Regulation of enhancer activity is not restricted to the supercoiled state of plasmids. (A) Reporter plasmid pWHE206, encoding an SV40 enhancer flanked with seven tet operators and a luciferase gene under the control of an SV40-promoter, was linearized. (B) Transregulators are indicated schematically in column one. The predicted regulatory situations are shown in column two. Cells were cultured in the absence (white bars) or presence (gray bars) of dox. Repression factors and basal luciferase activity (dotted line) are indicated. Values represent the means of triplicate samples with standard deviations given in corrected relative light units (corr. RLU) per μg of total cell protein.
In summary, an SV40 enhancer can be regulated by dox-dependent dimerizers if it is flanked with tet operators, irrespective of the superhelicity of the reporter plasmid DNA, whereas the simple formation of nucleoprotein complexes, like DNA-bound sc TetR*, between enhancer and the corresponding promoter is not sufficient to effectively reduce enhancer activity.
Suitable models for the mode of action of the homodimerization system
Multiple tet operators are present and necessary for efficient regulation (Supplementary Figure S2) and the sc TetR* control also leads to a slight reduction of enhancer activity. Even though the in vitro data on linking of different DNA fragments through tDL support loop formation, we cannot presuppose that the in vitro situation will be identical in vivo in cultured cells. Thus, the three models shown in Figure 5 could formally explain enhancer silencing via tDs. (A) The internal contact model assumes that tet operator-bound tDs dimerize on one side of the enhancer, leading to the formation of snarls on both sides of the enhancer which might affect enhancer activity. (B) The bead string model could be a consequence of high intracellular amounts of tD and the ensuing saturation of tet operators. Consequentially, one tD dimer would be bound to only one tet operator and not to two, thereby preventing loop formation. These nucleoprotein complexes could, nonetheless, interfere with the signaling properties of the enhancer. (C) The loop model features interaction of tDs bound to tet operators on different sides of the enhancer, thereby enclosing the enhancer within a loop. This would silence the enhancer, probably by generating a structurally and functionally independent DNA segment.
Figure 5.
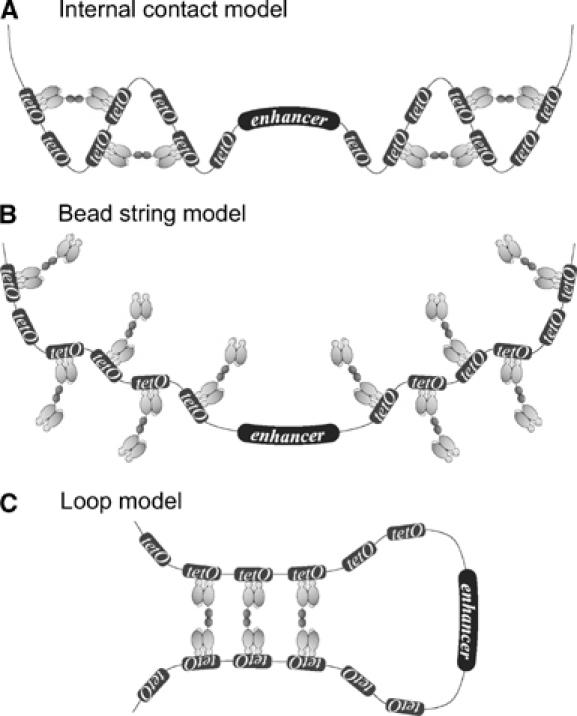
Different models can explain the operating mode of the homodimerization system. (A) Internal contact model; (B) bead string model; (C) loop model.
The formation of ‘internal contacts' does not contribute to the regulation of enhancer activity
To distinguish between these three models, we developed a heterodimerization system: The reporter plasmid (pWHE367) differs from pWHE206 only in the sequence of the tet operators flanking the enhancer, as it contains seven copies of _tetO_-4C on one side and seven copies of _tetO_-6C on the other side (Baron et al, 1999). In addition, transregulators with different DNA-binding specificities and heterodimerization domains, termed dox-controlled heterodimerizers (Tet heterodimerizer (tHD)) (Figure 6A), were used. The different DNA-binding specificities were realized by using 4C and 6C variants of sc TetR, which carry mutations in the DNA-binding heads, leading to specific and exclusive recognition of the respective tetO variants (Baron et al, 1999), while maintaining inducibility by dox. The respective sc TetR variants were fused to the heterodimerization domains FRB(TL2098) and FKBP (Chong et al, 2002). These domains interact with each other only in the presence of AP21967, a chemically modified form of rapamycin.
Figure 6.

Exclusion of internal contact and bead string models for enhancer regulation. (A) tHD with different DNA-binding and heterodimerization domains. (B, D) Transregulators are indicated schematically in column one. The predicted regulatory situations are shown in column two. Cells were cultured in the absence (white bars), in the presence (dark gray bars) of dox, in the presence of AP21967 (light gray bars) or in the presence of dox and AP21967 (black bars). Repression factors and basal luciferase activity are indicated. Values represent the means of triplicate samples with standard deviations given in corrected relative light units (corr. RLU) per μg of total cell protein. (C, E) Western blot analysis to detect the respective transregulators in HeLa cell extracts.
Combining tHD4C/FRB and tHD6C/FKBP or tHD4C/FKBP and tHD6C/FRB leads to the binding of the transregulators to the respective tet operators in the absence of dox and AP21967. Under such conditions, they do not interact with each other via their heterodimerization domains and thereby should not lead to decreased luciferase expression. When AP21967 is added, it induces dimerization of the tetO bound tHDs with different DNA-binding specificities, resulting in loop formation, thereby reducing luciferase expression. Addition of dox dissociates tHDs from the DNA and luciferase is fully expressed. As the heterodimerization system does not permit interaction between tHDs bound to identical tet operator variants, it allows evaluation of the internal contact model.
Combinations of plasmids expressing tHD4C/FRB and tHD6C/FKBP, or tHD4C/FKBP and tHD6C/FRB, were transfected transiently with the corresponding reporter plasmid (pWHE367) in HeLa cells (Figure 6B). In the absence of AP21967, a combination of tHD4C/FRB and tHD6C/FKBP exhibits no reduction of luciferase expression, even though both transregulators are expressed (Figure 6C). After addition of AP21967, the same combination of tHDs shows 3.1-fold repression, which is relieved upon addition of dox. The inverse combination of tHD4C/FKBP and tHD6C/FRB is equally active, with a repression factor of 3.3. Repressed luciferase activities are only 1.4- or 1.8-fold higher than the unenhanced control, demonstrating almost complete repression by both transregulator combinations. This clearly shows that internal contacts do not contribute to the regulation of enhancer activity.
Productive interactions between tet operators on both sides of the enhancer are necessary for regulation
If productive interactions between operators on both sides of the enhancer are necessary to regulate enhancer activity, while interactions within an operator array interfere with regulation of enhancer activity, then a combination of all four tHDs should reduce the repression factor due to the competition between productive and nonproductive interactions. To analyze this, HeLa cells were transfected with combinations of appropriate transregulators together with the reporter plasmid pWHE367. pGL3-promoter was transfected to determine unenhanced basal luciferase expression. The results are shown in Table I.
Table 1.
Productive interactions between tet operators on both sides of the enhancer are necessary for regulation
| Reporter | Combination of dox-controlled dimerizers | Luciferase activity (corr. RLU/μg protein) | Regulatory factor | |
|---|---|---|---|---|
| +dox | −dox | |||
| pGL3-promoter | None | 1590 (±118) | 1620 (±31) | — |
| pWHE367 | None | 8091 (±957) | 8002 (±152) | — |
| pWHE367 | tHD4C/FKBP+tHD6C/FRB | 7319 (±504) | 6487 (±270) | — |
| +dox/+AP21967 | −dox/+Ap21967 | |||
| pWHE367 | tHD4C/FKBP+tHD6C/FRB | 7041 (±405) | 2236 (±98) | 3.1 |
| pWHE367 | tHD4C/FRB+tHD6C/FKBP | 9725 (±172) | 2920 (±37) | 3.3 |
| pWHE367 | tHD4C/FKBP+tHD6C/FRB+tHD4C/FRB+tHD6C/FKBP | 6974 (±116) | 5069 (±459) | 1.4 |
| pWHE367 | tD4C/GCN4+tD6C/GCN4 | 6720 (±53) | 4246 (±168) | 1.6 |
A combination of tHDs that allow only productive interactions between tet operators on different sides of the enhancer (tHD4C/FRB and tHD6C/FKBP, or tHD4C/FKBP and tHD6C/FRB) leads to nearly complete regulation of the enhancer (regulatory factor: 3.1 or 3.3). In contrast, a combination containing all four tHDs results in the expected decrease of the regulatory factor to 1.4. Western blot analysis revealed that the decrease in regulatory activity of the transregulators is not due to lower expression levels of the respective transregulators (data not shown). The same effect is observed when the GCN4-derived dimerization domain is fused to 4C and 6C variants of sc TetR. When plasmids expressing both transregulators are transfected into HeLa cells, three types of dimer can be formed: a 4C/6C heterodimer that can only form interactions between the operators flanking the enhancer, and 4C/4C or 6C/6C homodimers that can only form interactions within one of the tetO arrays. Repression of the heterodimer-specific reporter gene plasmid pWHE367 is 1.6-fold, close to the 1.4-fold determined for the combination of all four tHDs. Clearly, interactions between tet operators on both sides of the enhancer are productive (Figures 1C and 6B), while interactions within an array of tet operators on only one side of the enhancer are nonproductive (see Table I).
Taken together, not only are productive interactions between tet operators on both sides of the enhancer necessary for regulation of enhancer activity via the dimerization systems, but also nonproductive interactions within a tet operator array on one side of the enhancer interfere with this regulation.
The bead string model does not explain regulation of enhancer activity
Although the data of the heterodimerization system strongly support the loop model, one cannot exclude that high protein levels of tHDs prevent interactions between tet operators, leading to a situation predicted by the bead string model. To address this possibility, we modified the heterodimerization system by combining the two transregulators tHD4C/FKBP and tHD6C/FKBP with an sc TetR mutant deficient in DNA binding (Berens et al, 1995) fused to an FRB domain (tHDWTΔ/FRB) (Figure 6A). In the absence of dox and presence of AP21967, this transregulator combination should lead to the binding of tHD dimers to only one tet operator, as predicted by the bead string model. Transfection of HeLa cells with a combination of tHD4C/FKBP and tHD6C/FRB together with the reporter plasmid pWHE367 (Figure 6D) leads to 5.3-fold repression. In contrast, a combination of tHD4C/FKBP, tHD6C/FKBP and tHDWTΔ/FRB results only in 1.4-fold repression. Their overall, rather high dox-independent, reduction in luciferase activity is most likely caused by the large amounts of transfected DNA since, even in the presence of dox, the luciferase activity is clearly reduced compared to luciferase activities determined in the absence of a transregulator. Quantification of Western blot analysis shows that tHDWTΔ/FRB (lower band) is present in an amount that is 1.6-fold of a combination of tHD4C/FKBP and tHD6C/FKBP (upper band) (Figure 6E), ensuring that each FKBP-containing tHD will be bound by a tHDWTΔ/FRB molecule, which is a necessity required for evaluating the bead string model.
Taken together, the data clearly show that the internal contact model does not contribute to enhancer silencing, while the bead string model is not sufficient to explain this effect.
Connecting looping-dependent enhancer regulation with insulator properties
Here, we report that enclosure of an enhancer in a DNA loop can block activation of gene expression. Not only does the in vitro data show that tDL is able to link different _tetO_-bearing DNA fragments, but also in vivo repression of luciferase activity is only observed when tDs form productive interactions between two tetO elements flanking an SV40 enhancer. This repression is strictly dependent on an active dimerization domain. This approach not only represents a new strategy for gene regulation in eukaryotes based on reversible alteration of DNA structure, but also allows the direct analysis of the regulatory effect of DNA-loop formation. Up to now, the data on genomic loops, which are intensely discussed in the literature, is generally derived from indirect assays (Chambeyron and Bickmore, 2004).
Our results are fully consistent with a recently described chromatin-loop model for the imprinted expression of Igf2 (Murrell et al, 2004). Here, parent-specific interactions between differentially methylated regions lead to the formation of alternative chromatin loops. When the Igf2 promoters are in an active compartment with the H19 enhancer, they are active. Transcription does not occur, if they are located in an inactive domain, separated from the enhancer. Furthermore, our results are consistent with recent studies on in vitro blocking of prokaryotic enhancer activity (Bondarenko et al, 2003), as well as analyses of insulator elements: (I) The proteins Zw5 and BEAF, which bind to the insulator elements scs and _scs_′, interact with each other in vitro and their binding sites are located in close proximity to each other in vivo (Blanton et al, 2003). (II) Recillas-Targa et al (1999) reported enhancer-blocking action, one of the defining characteristics of insulators (Chung et al, 1993, 1997), in a transient reporter assay similar to our reporter plasmids, using cHS4 insulator sequences instead of tet operators to flank the enhancer. These cHS4 elements are bound by the ubiquitous vertebrate transcription factor CTCF (Bell et al, 1999), which has been shown to interact with itself (Pant et al, 2004; Yusufzai et al, 2004). (III) Several intensely studied insulator elements, like cHS4 from the chicken β-globin locus and su(Hw) or scs/_scs_′ from Drosophila, do not require a chromosomal context for enhancer blocking, but are also active when present on plasmids (Holdridge and Dorsett, 1991; Dunaway et al, 1997; Recillas-Targa et al, 1999). As we demonstrate, loop formation would be able to explain these observations.
Materials and methods
Plasmid constructions
(a) Tet dimerizers. A DNA fragment encoding the GCN4-dimerization domain (aa 251–281) flanked by a 5′-_Ngo_MIV- and a 3′-_Sma_I site was generated by standard PCR with the primers ‘GCN4-1' (CATTGGGCCGGCCCTGCAGAGGGTGAAGCAGCTG GAGGACAAGGTG), ‘GCN4-2' (GTGGTAGTTCTTGCTGAGCAGTTCCTCCACCTTG TCCTCCAGCTG), ‘GCN4-3' (CTCAGCAAGAACTACCACCTGGAGAACGAGGTGG CCCGGCTGAAGAAAC) and ‘GCN4-4' (TGCATTCCCGGGTCACCGCTCGCCCACCAGTTTC TTCAGCCGGGCCACC). The LexA-dimerization domain (aa 85–202) was amplified by colony-PCR, using the Escherichia coli strain DH5α (Invitrogen, 1986) as template source for lexA and the primers ‘LexA-5′' (CATTGGGCCGGCCGGTGAACCACTTCTGGCGCAA CAGC) and ‘LexA-3′' (TGCATTCCCGGGTTACAGCCAGTCGCCGTTGCGA ATAACCCC) to introduce a 5′-_Ngo_MIV- and a 3′-_Sma_I site. The fragments were digested with _Ngo_MIV/_Sma_I and ligated into equally restricted pWHE120(sB+B) (Krueger et al, 2003), resulting in pWHE351 encoding tDG and pWHE352 encoding tDL. In addition, the GCN4-dimerization domain was likewise inserted into pWHE120(sB+B)4C and pWHE120(sB+B)6C (to be described elsewhere), resulting in pWHE368 encoding tD4C/GCN4 and pWHE369 encoding tD6C/GCN4.
The sc TetR*-encoding plasmid pWHE355 was constructed via digestion of pWHE120(sB+B) with _Ngo_MIV/_Sma_I, a fill-in of the 5′-overhangs with T4-DNA-polymerase and subsequent religation.
The double mutation LP253/LP260 was introduced into the GCN4-dimerization domain by two-step PCR with the primers ‘GCN4-mut' (GAGGGTGAAGCAGCCCGAGGACAAGGTGGAGGAA CCCCTCAGCAAGAAC), ‘GCN4-4' and ‘P2' (AAAACAGTATGAAACTCTCG) using pWHE351 as template. The _Ngo_MIV/Sma_I restricted fragment was then ligated into equally cut pWHE352, resulting in pWHE360 which encodes tDG_mut.
(b) Tet heterodimerizers. The heterodimerization domains FRB(T2098L) and FKBP12 were PCR-amplified with the primers ‘5′-FRB-Dom' (CATGGCGCCGGCAATCCTCTGGCATGAGATGTGG ) and ‘FRB-Dom-3′' (GTACGGCCCGGGTCACTTTGAGATTCGTCGGAAC ACATG) or ‘5′-FKBP' (AAAGGTGCCGGCAGGAGTGCAGGTGGAAACCATC ) and ‘FKBP-3′' (TTCTCACCCGGGTTAATAACTAGTTTCCAGTTTT AG), respectively, from the templates pC4EN-F1 and pC4-RHE of the _ARGENT_™ Regulated Heterodimerization Kit of ARIAD Pharmaceuticals (www.ariad.com/regulationkits). The amplification reactions introduced 5′-_Ngo_MIV and 3′-_Sma_I sites. The fragments were ligated after digestion with _Ngo_MIV/_Sma_I into equally restricted pWHE120(sB+B)4C, resulting in pWHE361 encoding tHD4C/FRB or pWHE362 encoding tHD4C/FKBP. The _Ngo_MIV/_Sma_I-restricted FRB(T2098L) fragment was also ligated with equally restricted pWHE120(sB+B)6C, resulting in pWHE363 encoding tHD6C/FRB. The tHD6C/FKBP encoding pWHE364 was constructed by cutting the FKBP12 domain out of pWHE362 by digestion with _Ngo_MIV/_Hpa_I and ligating this fragment into equally restricted pWHE120(sB+B)6C.
The DNA fragment encoding TetR(B)Δ26–53 was amplified by PCR using ‘P7' (CGCCGTACTGCCCGCTTGG) and ‘TP2' (CTCTGCACCTTGGTGATCAA) as primers and pWH1919Δ26–53 (Berens et al, 1995) as template. Restriction of the fragment with _Xba_I and _Mlu_I, followed by ligation into equally digested pWHE120(B+sB) (Krueger et al, 2003), resulted in pWHE120(B+sB)Δ26–53a. Ligation of _Ngo_MIV- and _Sma_I-digested FRB(T2098L) domain into equally restricted pWHE120(B+sB)Δ26–53a resulted in pWHE291.
(c) Reporter plasmids. pGL3-promoter and pGL3-control were purchased from Promega. A _Pst_I site was introduced into pGL3-control by overlap extension PCR using the primers ‘luc+3′-forward' (GCAAGAAAAATCAGAGAGATCC), ‘pGL3-PstI-back' (CGTTCAGATCCTTCTGCAGTTTACCACATTTGTA GAGG), ‘pGL3-_Pst_I-forward' (CCTCTACAAATGTGGTAAACTGCAGAAGGATCTG AACG) and ‘RVprimer4' (GACGATAGTCATGCCCCGCG). Restriction of the resulting fragment with _Xba_I/_Bam_HI and ligation into equally digested pGL3-control resulted in pWHE200, which is similar to an unnamed construct published in Recillas-Targa et al (1999).
A fragment encoding seven tet operators and flanking _Sal_I sites was amplified by PCR with the primers ‘5′-_Sal_I-tetO' (ATATTGTCGACCTTTCGTCTTCAAGAATTCCTCG ) and ‘tetO-_Sal_I-3′' (TTATATGTCGACCCGGGTACCGAGCTCG) from the template pUHC13-3 (Gossen and Bujard, 1992). Restriction of the fragment with _Sal_I and ligation into equally digested pWHE200 resulted in pWHE202, encoding the SV40 enhancer bordered by a 3′-(tetO)7 box. A fragment encoding seven tet operators and flanking _Pst_I sites was amplified by PCR with the primers ‘5′-_Pst_I-tetO' (ATATTCTGCAGCTTTCGTCTTCAAGAATTCCTCG ) and ‘tetO-_Pst_I-3′' (TTATATCTGCAGCCGGGTACCGAGCTCG) from the template pUHC13-3 (Gossen and Bujard, 1992). Restriction of the fragment with _Pst_I and ligation into equally digested pWHE200 resulted in pWHE203, encoding the SV40 enhancer bordered by a 5′-(tetO)7 box. The reporter plasmid with (tetO)7 boxes flanking the SV40 enhancer was constructed by cutting the SV40 enhancer together with the seven upstream tet operators out of pWHE203 via digestion with _Bam_HI and _Xba_I. This fragment was ligated into equally digested pWHE202 resulting in pWHE206, encoding a SV40 enhancer flanked with seven tet operators.
The (_tetO_-6C)7 box was amplified by PCR with the primers ‘5′-_Sal_I-tetO' and ‘tetO4C-_Sal_I-3′' (GGCCTCGTCGACTACACGCCTACCTCGAC) from the template pUHC13-9 (Baron et al, 1999). The fragment was digested with _Sal_I and ligated into equally restricted pWHE200, leading to pWHE366. The (_tetO_-4C)7-element was amplified by PCR with the primers ‘5′-_Pst_I-tetO' and ‘tetO4C-PstI-3′' (GGCCTCCTGCAGTACACGCCTACCTCGAC) from the template pUHC13-8 (Baron et al, 1999). The fragment was digested with _Pst_I and ligated into equally restricted pWHE366, resulting in the plasmid pWHE367.
The reporter plasmid encoding the SV40 enhancer flanked with a single tet operator (pWHE228) was generated as follows: A fragment encoding a single tet operator was generated by PCR using ‘1 × tetO2-_Pst_I' (ATCCTTCTGCAGTCTCTATCACTGATAGGGATTT ACCACATTTGTAGAGGTTTTACTTGC) and ‘luc+3′-forward' as primers as well as pWHE200 as template. This fragment was digested with _Xba_I and _Pst_I and ligated into equally cut pWHE200 resulting in pWHE208. Hybridization of the primers ‘tetO2-_Sal_Iup' (TCGACTCCCTATCAGTGATAGAGAAAAGTGAAAG TCGAGTTC) and ‘tetO2-_Sal_Idown' (TCGAGAACTCGACTTTCACTTTTCTCTATCACTG ATAGGGAG) led to a fragment encoding a single tet operator. This fragment was ligated into _Sal_I-digested pWHE200, resulting in pWHE223. The tetO box in pWHE208 was cut out with _Xba_I and _Pst_I and ligated into equally cut pWHE223, which resulted in pWHE228.
The reporter plasmids encoding the SV40 enhancer flanked with either two or three tetO boxes (pWHE229/pWHE230) were generated as follows: fragments encoding single tet operators were generated by hybridization of the primers ‘tetO2-_Pst_Iup' (CAAAGTCGAGTTTACCACTCCCTATCAGTGATAG AGACTGCA)/‘tetO2-_Pst_Idown' (GTCTCTATCACTGATAGGGAGTGGTAAACTCGAC TTTGTGCA) and ‘tetO2-_Sal_Iup'/‘tetO2-_Sal_Idown'. Ligation of these fragments into _Pst_I cut pWHE208 or _Sal_I cut pWHE223 resulted in pWHE221 and pWHE224, respectively. Two tetO boxes were cut out of pWHE221 with _Xba_I and _Pst_I and ligated into equally digested pWHE224, which resulted in pWHE229. A third tet operator was introduced to each tetO box by ligation of the hybridized primers ‘tetO2-_Pst_Iup'/‘tetO2-_Pst_Idown' and ‘tetO2-_Sal_Iup'/‘tetO2-_Sal_Idown' into _Pst_I cut pWHE221 or _Sal_I cut pWHE224, which resulted in pWHE222 and pWHE225, respectively. Three tetO boxes were cut out of pWHE222 with _Xba_I and _Pst_I and ligated into equally digested pWHE225, resulting in pWHE230.
All reporter plasmids were isolated from the _recABC_-deficient E. coli strain JC5547 (Willetts and Clark, 1969) to avoid recombination between tet operator elements.
Purification of tDL
The coding region of tDL was cut out of pWHE352 with _Xba_I/_Hpa_I and ligated into equally restricted pWH610(B+sB) (Ettner et al, 1996; P Schubert and W Hillen, unpublished data), resulting in pWH610tDL. E. coli RB791 (Brent and Ptashne, 1981) was transformed with pWH610tDL. Cells were grown at room temperature in LB supplemented with 100 μg/ml ampicillin. Gene expression was induced at an optical density of 0.4 at 600 nm by adjusting the broth to 1 mM isopropyl β-D-thiogalactopyranoside and incubation was continued for 3 h. Cells were harvested by centrifugation and ruptured in 50 mM NaCl; 20 mM phosphate buffer, pH 6.8; 2 mM dithiothreitol, by using a French press. The soluble proteins obtained after centrifugation at 4°C for 60 min at 40 000 r.p.m. (Beckmann L7–55; TI60) were loaded on a POROS HS/M 20 cation exchange column (BioCad, Vision). Proteins were eluted with a linear gradient of 50–1000 mM NaCl. Fractions were collected and analyzed by SDS–PAGE. tDL-containing fractions were pooled and further purified via gel filtration (HiLoad™ G200, Äkta Prime) as described (Ettner et al, 1996). The protein concentration was determined via Bio-Rad Protein Assay and saturation titration with tetracycline as described (Henssler et al, 2004).
Gel-shift assay for DNA-loop detection
In all, 500 fmol of supercoiled pWHE206 was incubated without or with tDL (7 and 14 pmol) in buffer NEB4 (50 mM potassium acetate, 20 mM Tris-acetate, 10 mM magnesium acetate, 1 mM dithiothreitol (pH 7.9)) in 20 μl aliquots. Doxycycline (0.1 mg/ml) was added to a final concentration of 5 μg/ml as control. Probes were incubated at 37°C for 30 min to allow DNA–protein complex formation. Then, 20 U _Afe_I together with 10 U _Pst_I were added and restriction was performed at 37°C for 60 min. The reaction was terminated by adding 5 μl of 5 × loading buffer containing 50 mM EDTA, 5 × TAE buffer (0.2 M Tris-phosphate, 0.1 M sodium acetate, 8.25 mM EDTA, pH 8.3) and 80% glycerol. The samples were immediately analyzed in a 1.2% agarose gel in TAE buffer. The gel was stained with ethidium bromide and DNA fragments of interest were extracted from the gel by using NucleoSpin Extract 2 in 1 (Macherey-Nagel, Germany). The purified DNA fragments were separated in a 1.2% agarose gel in TAE buffer and detected by ethidium bromide staining.
Cell culture and transfections
Transfection of HeLa cells were performed at 30–60% confluency with 1–1.3 μg of total DNA and a respective volume of Lipofectamine (Invitrogen) or Perfectin (PEQLAB) in 35-mm dishes according to the instructions of the producer.
Transfection of the homodimerization system was performed as follows. The DNA mixtures for the homodimerization system with supercoiled reporter plasmid (Figure 1C and F) consisted of 100 ng reporter plasmid, 30 ng transregulator-expressing plasmid, 400 ng lacZ expression vector pUHD16-1 (Gossen and Bujard, 1992) and pWHE121 as unspecific DNA (Krueger et al, 2003) to a total of 1 μg DNA. The DNA mixtures for the homodimerization system with linearized reporter plasmid (Figure 4) differ by containing 300 ng reporter plasmid and 20 ng transregulator-expressing plasmid. In both cases, doxycycline (Sigma) was added at 5 μg/ml for induction.
Transfection of heterodimerization systems were performed as follows: The DNA mixtures for the analysis of the internal loop model (Figure 5B) or of the necessity of productive interactions between tet operators on both sides of the enhancer (Table I) consisted of 100 ng reporter plasmid (pWHE367), 150 ng transregulator-expressing plasmid, 400 ng pUHD16-1 and pWHE121 to a total of 1.3 μg DNA. The DNA mixtures for the analysis of the bead string model (Figure 6D) consisted of either 50 ng of each tHD4C/FKBP and tHD6C/FRB or 50 ng of each tHD4C/FKBP and tHD6C/FKBP plus 600 ng tHDWTΔ/FRB as transregulator-expressing DNA, 100 ng reporter plasmid (pWHE367), 400 ng pUHD16-1 and pWHE121 to a total of 1.3 μg DNA. Doxycycline was added in both cases at 5 μg/ml and AP21967 (ARIAD) at 300 nM for induction.
Determination of luciferase activity and Western blot analysis of Tet-based transregulators were performed as described previously (Krueger et al, 2003).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Acknowledgments
We acknowledge Christel Krueger, Christina Danke, Christiane Lang and Peter Schubert for providing plasmids. The work was supported by the DFG (SFB 473; Graduiertenkolleg 805) and the FCI Deutschland. Competing commercial interests statement The authors declare that they have no competing commercial interests.
References
- Baron U, Schnappinger D, Helbl V, Gossen M, Hillen W, Bujard H (1999) Generation of conditional mutants in higher eukaryotes by switching between the expression of two genes. Proc Natl Acad Sci USA 96: 1013–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G (1999) The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98: 387–396 [DOI] [PubMed] [Google Scholar]
- Berens C, Hillen W (2003) Gene regulation by tetracyclines. Constraints of resistance regulation in bacteria shape TetR for application in eukaryotes. Eur J Biochem 270: 3109–3121 [DOI] [PubMed] [Google Scholar]
- Berens C, Pfleiderer K, Helbl V, Hillen W (1995) Deletion mutagenesis of Tn_10_ Tet repressor localization of regions important for dimerization and inducibility in vivo. Mol Microbiol 18: 437–448 [DOI] [PubMed] [Google Scholar]
- Blackwood EM, Kadonaga JT (1998) Going the distance: a current view of enhancer action. Science 281: 60–63 [DOI] [PubMed] [Google Scholar]
- Blanton J, Gaszner M, Schedl P (2003) Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev 17: 664–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondarenko VA, Jiang YI, Studitsky VM (2003) Rationally designed insulator-like elements can block enhancer action in vitro. EMBO J 22: 4728–4737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent R, Ptashne M (1981) Mechanism of action of the lexA gene product. Proc Natl Acad Sci USA 78: 4204–4208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA (2004) Does looping and clustering in the nucleus regulate gene expression? Curr Opin Cell Biol 16: 256–262 [DOI] [PubMed] [Google Scholar]
- Chong H, Ruchatz A, Clackson T, Rivera VM, Vile RG (2002) A system for small-molecule control of conditionally replication-competent adenoviral vectors. Mol Ther 5: 195–203 [DOI] [PubMed] [Google Scholar]
- Chung JH, Bell AC, Felsenfeld G (1997) Characterization of the chicken β-globin insulator. Proc Natl Acad Sci USA 94: 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JH, Whiteley M, Felsenfeld G (1993) A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74: 505–514 [DOI] [PubMed] [Google Scholar]
- Dunaway M, Hwang JY, Xiong M, Yuen HL (1997) The activity of the scs and _scs_′ insulator elements is not dependent on chromosomal context. Mol Cell Biol 17: 182–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettner N, Müller G, Berens C, Backes H, Schnappinger D, Schreppel T, Pfleiderer K, Hillen W (1996) Fast large-scale purification of tetracycline repressor variants from overproducing Escherichia coli strains. J Chromatogr A 742: 95–105 [DOI] [PubMed] [Google Scholar]
- Fransz P, De Jong JH, Lysak M, Castiglione MR, Schubert I (2002) Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc Natl Acad Sci USA 99: 14584–14589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova TI, Corces VG (1998) Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell 92: 511–521 [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Corces VG (2001) Chromatin insulators and boundaries: effects on transcription and nuclear organization. Annu Rev Genet 35: 193–208 [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA 89: 5547–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JJ, Tullius TD, Wolffe AP (1990) The structure of DNA in a nucleosome. Proc Natl Acad Sci USA 87: 7405–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henssler EM, Scholz O, Lochner S, Gmeiner P, Hillen W (2004) Structure-based design of Tet repressor to optimize a new inducer specificity. Biochemistry 43: 9512–9518 [DOI] [PubMed] [Google Scholar]
- Holdridge C, Dorsett D (1991) Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol Cell Biol 11: 1894–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JC, O'Shea EK, Kim PS, Sauer RT (1990) Sequence requirements for coiled-coils: analysis with λ repressor-GCN4 leucine zipper fusions. Science 250: 1400–1403 [DOI] [PubMed] [Google Scholar]
- Invitrogen (1986) BRL pUC host: E. coli DH5α competent cells. Focus 8: 9 [Google Scholar]
- Krueger C, Berens C, Schmidt A, Schnappinger D, Hillen W (2003) Single-chain Tet transregulators. Nucleic Acids Res 31: 3050–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn EJ, Geyer PK (2003) Genomic insulators: connecting properties to mechanism. Curr Opin Cell Biol 15: 259–265 [DOI] [PubMed] [Google Scholar]
- Lu Y, Weers B, Stellwagen NC (2001) DNA persistence length revisited. Biopolymers 61: 261–275 [DOI] [PubMed] [Google Scholar]
- Mahy NL, Perry PE, Bickmore WA (2002) Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J Cell Biol 159: 753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morii T, Sato S, Hagihara M, Mori Y, Imoto K, Makino K (2002) Structure-based design of a leucine zipper protein with new DNA contacting region. Biochemistry 41: 2177–2183 [DOI] [PubMed] [Google Scholar]
- Murrell A, Heeson S, Reik W (2004) Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet 36: 889–893 [DOI] [PubMed] [Google Scholar]
- Okahata Y, Niikura K, Sugiura Y, Sawada M, Morii T (1998) Kinetic studies of sequence-specific binding of GCN4-bZIP peptides to DNA strands immobilized on a 27-MHz quartz-crystal microbalance. Biochemistry 37: 5666–5672 [DOI] [PubMed] [Google Scholar]
- Orth P, Schnappinger D, Hillen W, Saenger W, Hinrichs W (2000) Structural basis of gene regulation by the tetracycline inducible Tet repressor–operator system. Nat Struct Biol 7: 215–219 [DOI] [PubMed] [Google Scholar]
- Pant V, Kurukuti S, Pugacheva E, Shamsuddin S, Mariano P, Renkawitz R, Klenova E, Lobanenkov V, Ohlsson R (2004) Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol Cell Biol 24: 3497–3504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recillas-Targa F, Bell AC, Felsenfeld G (1999) Positional enhancer-blocking activity of the chicken β-globin insulator in transiently transfected cells. Proc Natl Acad Sci USA 96: 14354–14359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe K (2001) Making contacts on a nucleic acid polymer. Trends Biochem Sci 26: 733–740 [DOI] [PubMed] [Google Scholar]
- Schnarr M, Granger-Schnarr M, Hurstel S, Pouyet J (1988) The carboxy-terminal domain of the LexA repressor oligomerises essentially as the entire protein. FEBS Lett 234: 56–60 [DOI] [PubMed] [Google Scholar]
- Tovar K, Hillen W (1989) Tet repressor binding induced curvature of tet operator DNA. Nucleic Acids Res 17: 6515–6522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardy A, Maine E, Schedl P (1985) The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J Mol Biol 185: 341–358 [DOI] [PubMed] [Google Scholar]
- Vazquez J, Schedl P (1994) Sequences required for enhancer blocking activity of scs are located within two nuclease-hypersensitive regions. EMBO J 13: 5984–5993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts NS, Clark AJ (1969) Characteristics of some multiply recombination-deficient strains of Escherichia coli. J Bacteriol 100: 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G (2004) CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell 13: 291–298 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2