Colistin-Resistant mcr-1-Positive Escherichia coli on Public Beaches, an Infectious Threat Emerging in Recreational Waters (original) (raw)
ABSTRACT
The emergence and rapid spread of colistin-resistant Escherichia coli carrying the mcr-1 gene have generated an urgent need to strengthen surveillance. We performed a meticulous investigation of strains of this sort, which resulted in the identification of international clones of E. coli carrying IncX4-plasmid-mediated mcr-1 and _bla_CTX-M genes in recreational waters of public urban beaches in cities with high tourist turnover, highlighting a new environmental reservoir.
KEYWORDS: MCR-1, ESBL, CTX-M, IncX4, polymyxins, Brazil
TEXT
The emergence and rapid spread of colistin-resistant Enterobacteriaceae carrying the mcr-1 gene have generated a profound sense of public alarm (1). Escherichia coli, one of the bacterial species that is most widely distributed and exchanged between the environment, animals, and humans, has been the main host of mcr-1 (2, 3). In South America, the occurrence of E. coli carrying mcr-1 and _bla_CTX-M genes in human (4–6) and wild animal (7) infections and food-producing animals (8) has created an urgent need to strengthen epidemiological surveillance. Using a whole-genome sequencing (WGS) approach, we performed a meticulous investigation of strains of this sort, which resulted in the identification of international clones of E. coli carrying mcr-1 and _bla_CTX-M-type genes in recreational waters of public urban beaches and highlighted a new source of transmission of this infectious threat.
In September 2016, coastal water samples were collected from 11 different public beaches (in the southeastern Brazilian continental margin of São Paulo State) surrounding urban counties with a population of about 800,000 inhabitants, which can double during the summer. Following standard methods for the examination of water and wastewater (http://www.standardmethods.org), 500-ml surface water samples were collected, on the same day, in sterile bottles, transported to the laboratory in cooled containers (at about 4°C to 10°C), and processed within 6 h. From each water sample, 100 ml was concentrated by filtration through sterile membrane filters with a pore size of 0.45 μm. The filters were placed on MacConkey agar plates and incubated for 24 h at 37°C. Next, the membrane filters were aseptically removed and placed separately in sterile tubes that had been filled previously with 10 ml of sterile Mueller-Hinton broth. After vortex mixing, an aliquot (100 μl) of each culture was streaked on MacConkey agar plates supplemented with colistin (2 μg/ml).
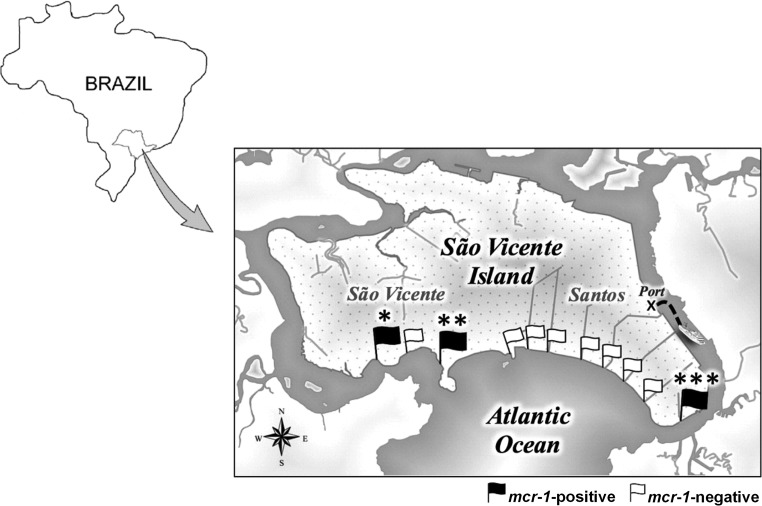
Three colistin-resistant E. coli strains were recovered from different beaches located in the cities of São Vicente and Santos (Fig. 1); the latter is the major beachfront city of the region, with the largest shipping terminal in Latin America. The isolates were identified by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) analysis, and antimicrobial susceptibility profiles and polymyxin MICs were determined by using the disc diffusion and broth microdilution methods, respectively (9, 10). Additionally, imipenem and meropenem MICs were determined by using the Etest method, and all isolates displayed susceptibility to imipenem (MICs of ≤0.19 μg/ml) and meropenem (MICs of ≤0.032 μg/ml).
FIG 1.
Map showing sampling locations (represented by flags) on public beaches surrounding the area of Santos and São Vicente cities, in the southeastern Brazilian continental margin of São Paulo State. MCR-1-positive E. coli strains (black flags) were isolated from seawater at recreational beaches in São Vicente (*, ICBEC2AM [location, −23.974697S, −46.395060W]; **, ICBEC3AM [location, −23.974995S, −46.371613W]) and Santos (***, ICBEC13AM [location, −23.986450S, −46.309086W]).
DNA libraries from ICBEC2AM and ICBEC3AM E. coli isolates were sequenced using the NextSeq platform with paired-end reads (Illumina), whereas the DNA library from ICBEC13AM E. coli was sequenced using the MiSeq platform with paired-end reads (Illumina). Serotypes (STs), multilocus sequence typing (MLST), plasmid replicons, antimicrobial resistance genes, and E. coli virulence genes were identified or performed using multiple databases, i.e., SerotypeFinder 1.1, MLST 1.8, PlasmidFinder 1.3, ResFinder 2.1, and VirulenceFinder 1.5, respectively, available from the Center for Genomic Epidemiology.
The presence of mcr-1 and other clinically important resistance genes, including the extended-spectrum β-lactamase (ESBL) genes _bla_CTX-8 and _bla_CTX-M-1, conferred a multidrug resistance (MDR) phenotype to E. coli strains belonging to the globally reported sequence types ST10, ST46, and ST1638 (Table 1). ST10 and ST46 encompass pathogenic strains responsible for human and animal infections, as reported for E. coli (7, 11, 12). Interestingly, the isolation of an E. coli ST10 strain carrying the mcr-1 gene from a water sample collected from a public beach on the coast of Santos city and the isolation of an E. coli ST10 strain from an infected migratory Magellanic penguin suffering from pododermatitis, in the same area, in an earlier study by our group (7) suggest that the ubiquitous ST10 survives easily and also spreads in the marine environment. Indeed, all E. coli stains identified in this study showed tolerance to NaCl concentrations up to 10% (Table 1). Recent studies have reported observation of the coexistence of mcr-1 and _bla_CTX-M in MDR E. coli strains belonging to the ST10 complex in well water in rural China (13), identification of environmental _mcr-1_-positive E. coli isolates surrounding German swine farm areas (14), and isolation of _mcr-1_-positive E. coli strains from diseased food-producing animals in China (15) and in France and Italy (16), supporting the rapid adaptation of these lineages to different hosts and ecosystems.
TABLE 1.
Characteristics of colistin-resistant Escherichia coli strains carrying the mcr-1 gene from Brazil
| Characteristica | ICBEC2AM | ICBEC3AM | ICBEC13AM | ICBEC7P | ICBEC72H |
|---|---|---|---|---|---|
| Source | Seawater | Seawater | Seawater | Infected migratory penguin | Human infection |
| Location | −23.974697S, −46.395060W | −23.974995S, −46.371613W | −23.986450S, −46.309086W | −23.986306S, −46.308361W | −5.779257S, −35.200916W |
| Isolation date | September 2016 | September 2016 | September 2016 | June 2013 | March 2016 |
| NaCl tolerance (%) | 10 | 10 | 10 | 10 | 10 |
| Serotype | ONT:H55 | O9:H4 | O54:H32 | ONT:H32 | ONT:H9 |
| ST/CC | 1638 | 46/46 | 10/10 | 10/10 | 101/43 |
| Virulence genes | Not detected | iss, gad, mchF | gad | gad | iroN, mcmA, mchB, mchC, mchF, lpfA, iss |
| Phylogroup | B1 | B1 | B1 | A | B1 |
| Resistance | AMO, AMP, CAZ, CEF, CRO, CTF, CTX, DOX, NAL, SUL, TET | AMO, CEF, CLO, NAL, SUL, SXT | AMO, AMP, ATM, CAZ, CEF, CRO, CTX, DOX, NAL, SUL, SXT, TET | AMK, AMO, AMP, ATM, CAZ, CEF, CIP, CTF, CTX, ENR, FEP, GEN, NAL, SXT, TET | AMO, AMP, ATM, CEF, CTX, FEP |
| Colistin/polymyxin MIC (μg/ml) | 4/4 | 4/4 | 4/4 | 8/8 | 4/4 |
| Resistance genotype | mcr-1, _bla_CTX-M-8, qnrB19, aadA2, strA, strB, sul2 | mcr-1, _bla_TEM-1B, qnrB19, catA1, aadA1, strA, strB, sul1, sul2, tetA, dfrA1, dfrA8 | mcr-1, _bla_CTX-M-1, aadA1, sul2, tetA, tetB | mcr-1, _bla_CTX-M-1, aadA1, sul2, tetA, tetB | mcr-1, _bla_CTX-M-8 |
| Plasmids (Inc)b | I1, ColRNAI, X4 | FIB, Q1, X4 | HI2, I1, N, X4 | FIN, HI2, HI2A, I1, N, X4 | I1, X4 |
IncX4 plasmids (∼33 kb) were identified by WGS analysis in all strains carrying the mcr-1 gene. After de novo assembly, plasmid sequences were manually annotated using Geneious R9 software, and then PlasmidFinder 1.3 was used to identify incompatibility groups. For comparative analysis, plasmid sequences were aligned against the nonredundant database using the MegaBLAST algorithm (NCBI BLAST), with default settings for the parameters. The plasmids pICBEC2AM and pICBEC3AM displayed 91% and 100% nucleotide identity, respectively, to the plasmid pICBEC72Hmcr (GenBank accession number CP015977), which originated from a human patient (5), and pICBEC13AM displayed 100% identity to the plasmid pICBEC7Pmcr (GenBank accession number CP017246), which was identified in the E. coli ST10 isolate from the infected penguin (7), confirming an epidemiological link (Table 1); IncX4 plasmids are key vectors responsible for dissemination of the mcr-1 gene (5, 7, 17).
The coexistence of mcr-1 and/or plasmid-mediated quinolone resistance (PMQR) and ESBL-encoding genes, such as qnrB19 and _bla_CTX-M-type variants, is of great concern, because the occurrence of mcr-1 and other clinically significant resistance genes in E. coli would seriously compromise treatment options (18). These results suggest that MCR-1-positive E. coli isolates are able to recruit other resistance genes, becoming MDR.
In summary, we report the occurrence of colistin-resistant, MCR-1-producing, E. coli lineages in recreational coastal waters of anthropogenically affected public beaches (19). In this situation, it is possible that residents, tourists, and wildlife could be exposed to this infectious threat directly from water exposure, from contact with sand, or through food consumption on the beach. Therefore, epidemiological studies addressing the consequences for human health of environmental dissemination of E. coli strains carrying the mcr-1 gene are necessary.
Accession number(s).
Complete plasmid sequences were deposited in GenBank under accession numbers KY770023 (pICBEC2AM), KY770024 (pICBEC3AM), and KY770025 (pICBEC13AM).
ACKNOWLEDGMENTS
This work was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (grant 2016/08593-9) and by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (grant 462042/2014-6). M.R.F. and N.L. are research grant fellows of the Fundação de Amparo à Pesquisa do Estado de São Paulo and the Conselho Nacional de Desenvolvimento Científico e Tecnológico, respectively.
We thank Cefar Diagnóstica (Brazil) for kindly supplying antibiotic discs for susceptibility testing.
We have no conflicts of interest to declare.
REFERENCES
- 1.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 2.Nordmann P, Poirel L. 2016. Plasmid-mediated colistin resistance: an additional antibiotic resistance menace. Clin Microbiol Infect 22:398–400. doi: 10.1016/j.cmi.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 3.Ovejero CM, Delgado-Blas JF, Calero-Caceres W, Muniesa M, Gonzalez-Zorn B. 2017. Spread of _mcr-1_-carrying Enterobacteriaceae in sewage water from Spain. J Antimicrob Chemother 72:1050–1053. doi: 10.1093/jac/dkw533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rapoport M, Faccone D, Pasteran F, Ceriana P, Albornoz E, Petroni A, MCR Group, Corso A. 2016. First description of _mcr-1_-mediated colistin resistance in human infections caused by Escherichia coli in Latin America. Antimicrob Agents Chemother 60:4412–4413. doi: 10.1128/AAC.00573-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandes MR, McCulloch JA, Vianello MA, Moura Q, Pérez-Chaparro PJ, Esposito F, Sartori L, Dropa M, Matté MH, Lira DP, Mamizuka EM, Lincopan N. 2016. First report of the globally disseminated IncX4 plasmid carrying the mcr-1 gene in a colistin-resistant Escherichia coli sequence type 101 isolate from a human infection in Brazil. Antimicrob Agents Chemother 60:6415–6417. doi: 10.1128/AAC.01325-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ortega-Paredes D, Barba P, Zurita J. 2016. Colistin-resistant Escherichia coli clinical isolate harbouring the mcr-1 gene in Ecuador. Epidemiol Infect 144:2967–2970. doi: 10.1017/S0950268816001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sellera FP, Fernandes MR, Sartori L, Carvalho MP, Esposito F, Nascimento CL, Dutra GH, Mamizuka EM, Pérez-Chaparro PJ, McCulloch JA, Lincopan N. 2017. Escherichia coli carrying IncX4 plasmid-mediated mcr-1 and _bla_CTX-M genes in infected migratory Magellanic penguins (Spheniscus magellanicus). J Antimicrob Chemother 72:1255–1256. doi: 10.1093/jac/dkw543. [DOI] [PubMed] [Google Scholar]
- 8.Fernandes MR, Moura Q, Sartori L, Silva KC, Cunha MP, Esposito F, Lopes R, Otutumi LK, Gonçalves DD, Dropa M, Matté MH, Monte DF, Landgraf M, Francisco GR, Bueno MF, de Oliveira Garcia D, Knöbl T, Moreno AM, Lincopan N. 2016. Silent dissemination of colistin-resistant Escherichia coli in South America could contribute to the global spread of the mcr-1 gene. Euro Surveill 21(17):pii=30214. doi: 10.2807/1560-7917.ES.2016.21.17.30214. [DOI] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. CLSI document M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 10.European Committee on Antimicrobial Susceptibility Testing. 2016. Breakpoint tables for interpretation of MICs and zone diameters, version 6. www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_6.0_Breakpoint_table.pdf.
- 11.Maluta RP, Logue CM, Casas MR, Meng T, Guastalli EA, Rojas TC, Montelli AC, Sadatsune T, de Carvalho Ramos M, Nolan LK, da Silveira WD. 2014. Overlapped sequence types (STs) and serogroups of avian pathogenic (APEC) and human extra-intestinal pathogenic (ExPEC) Escherichia coli isolated in Brazil. PLoS One 9:e105016. doi: 10.1371/journal.pone.0105016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mshana SE, Imirzalioglu C, Hain T, Domann E, Lyamuya EF, Chakraborty T. 2011. Multiple ST clonal complexes, with a predominance of ST131, of Escherichia coli harbouring _bla_CTX-M-15 in a tertiary hospital in Tanzania. Clin Microbiol Infect 17:1279–1282. doi: 10.1111/j.1469-0691.2011.03518.x. [DOI] [PubMed] [Google Scholar]
- 13.Sun P, Bi Z, Nilsson M, Zheng B, Berglund B, Stålsby Lundborg C, Börjesson S, Li X, Chen B, Yin H, Nilsson LE. 2017. Occurrence of _bla_KPC-2, _bla_CTX-M and mcr-1 in Enterobacteriaceae from well water in rural China. Antimicrob Agents Chemother 61:e02569-16. doi: 10.1128/AAC.02569-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guenther S, Falgenhauer L, Semmler T, Imirzalioglu C, Chakraborty T, Roesler U, Roschanski N. 2017. Environmental emission of multiresistant Escherichia coli carrying the colistin resistance gene mcr-1 from German swine farms. J Antimicrob Chemother doi: 10.1093/jac/dkw585. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Zhang R, Li J, Wu Z, Yin W, Schwarz S, Tyrrell JM, Zheng Y, Wang S, Shen Z, Liu Z, Liu J, Lei L, Li M, Zhang Q, Wu C, Zhang Q, Wu Y, Walsh TR, Shen J. 2017. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbiol 2:16260. doi: 10.1038/nmicrobiol.2016.260. [DOI] [PubMed] [Google Scholar]
- 16.El Garch F, Sauget M, Hocquet D, LeChaudee D, Woehrle F, Bertrand X. 2017. mcr-1 is borne by highly diverse Escherichia coli isolates since 2004 in food-producing animals in Europe. Clin Microbiol Infect 23:51.e1–51.e4. doi: 10.1016/j.cmi.2016.08.033. [DOI] [PubMed] [Google Scholar]
- 17.Li R, Xie M, Zhang J, Yang Z, Liu L, Liu X, Zheng Z, Chan EW, Chen S. 2017. Genetic characterization of _mcr-1_-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J Antimicrob Chemother 72:393–401. doi: 10.1093/jac/dkw411. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Tian GB, Zhang R, Shen Y, Tyrrell JM, Huang X, Zhou H, Lei L, Li HY, Doi Y, Fang Y, Ren H, Zhong LL, Shen Z, Zeng KJ, Wang S, Liu JH, Wu C, Walsh TR, Shen J. 2017. Prevalence, risk factors, outcomes, and molecular epidemiology of _mcr-1_-positive Enterobacteriaceae in patients and healthy adults from China: an epidemiological and clinical study. Lancet Infect Dis 17:390–399. doi: 10.1016/S1473-3099(16)30527-8. [DOI] [PubMed] [Google Scholar]
- 19.Lamparelli CC, Pogreba-Brown K, Verhougstraete M, Sato MI, Bruni Ade C, Wade TJ, Eisenberg JN. 2015. Are fecal indicator bacteria appropriate measures of recreational water risks in the tropics: a cohort study of beach goers in Brazil? Water Res 87:59–68. doi: 10.1016/j.watres.2015.09.001. [DOI] [PubMed] [Google Scholar]