The identification of Hoxc8 target genes (original) (raw)
Abstract
Hox genes encode transcription factors that control spatial patterning during embryogenesis. To date, downstream targets of Hox genes have proven difficult to identify. Here, we describe studies designed to identify target genes under the control of the murine transcription factor Hoxc8. We used a mouse 16,463 gene oligonucleotide microarray to identify mRNAs whose expression was altered by the overexpression of Hoxc8 in C57BL/6J mouse embryo fibroblasts (MEF) in cell culture (in vitro). We identified a total of 34 genes whose expression was changed by 2-fold or greater: 16 genes were up-regulated, and 18 genes were down-regulated. The majority of genes encoded proteins involved in critical biological processes, such as cell adhesion, migration, metabolism, apoptosis, and tumorigenesis. Two genes showed high levels of regulation: (i) secreted phosphoprotein 1 (Spp1), also known as osteopontin (OPN), was down-regulated 4.8-fold, and (ii) frizzled homolog 2 (Drosophila) (Fzd2) was up-regulated 4.4-fold. Chromatin immunoprecipitation (ChIP) analysis confirmed the direct interaction between the OPN promoter and Hoxc8 protein in vivo, supporting the view that OPN is a direct transcriptional target of Hoxc8.
Keywords: chromatin immunoprecipitation, microarray, osteopontin
Hox genes regulate anterior/posterior developmental patterning in an extensive domain, extending from the midbrain/hindbrain junction to the tail. The individual genes are expressed in an overlapping array, each regulating differentiation and morphogenesis in their individual expression domains along the anterior/posterior axis. The Hoxc8 gene has been studied in considerable detail by us and others (1–4). Expression analysis shows that the gene is expressed initially at 8 days postconception (dpc) in the tail bud and then extends to an anterior position at the level of the forelimbs. A posterior limit of expression is defined later at the junction between the thoracic and lumbar regions. The gene is expressed in both the neural tube and the somites in the prospective thorax (5–7). Null mutants of Hoxc8 show neuromuscular defects in the forelimb and skeletal defects in the ribs and vertebrae of the thorax (8). We have shown recently that a retardation of Hoxc8 expression results in the phenocopy of _Hoxc8_-null mutations, demonstrating the criticality of expression timing for the Hox transcription factors (9).
The tissue-specific overexpression of Hoxc8 has been shown to inhibit chondrocyte maturation and stimulate chondrocyte proliferation (10). Bone morphogenetic protein (BMP) is a potent osteotropic protein that induces osteoblast differentiation and bone formation. Hox-binding elements (ATTA) are common in promoters of osteoblast differentiation marker genes, especially those that rapidly respond to BMP stimulation, such as osteoprotegerin, BMP-4, and osteonectin (11–14). Gel retardation studies have shown that Hoxc8 binds to ATTA-rich sites in the osteopontin (OPN) promoter domain (15), but evidence for functional interaction in vivo is lacking. BMP stimulation activates gene transcription by depressing Hoxc8 protein through the interaction of Smad1 and Hoxc8 proteins. These results suggest that direct interaction between Smad1 and Hoxc8 proteins represents a major mechanism of osteoblast differentiation in BMP-induced skeletal development (15).
Hox gene deregulation is implicated in human cancers including leukemia and colorectal, breast, renal, and lung cancers (16–21). Expression of Hoxc8 correlates with higher Gleason scores in prostate cancers (22). Hoxc8 is selectively activated in cervical cancer cells (23). Hematopoietic progenitor cells show abnormalities in _Hoxc8_-null mutant mice (24). However, it is not clear how Hox gene deregulation specifically effects neoplastic inception and progression. Few studies have established direct functional roles for Hox genes in carcinogenesis.
In the present study, we overexpressed mouse Hoxc8 gene in the C57BL/6J mouse embryo fibroblasts (MEF) cells and then applied microarray assay to identify possible Hoxc8 target genes. Expression of candidate genes was also examined by semiquantitative RT-PCR, and these data correlated well with the array data. Chromatin immunoprecipitation (ChIP) assay confirmed that OPN is a direct transcriptional target of Hoxc8 in vivo. Most of the 34 identified candidate target genes are involved in proliferation, adhesion, migration, metabolism, and related cellular processes, and can be viewed as global regulators of growth and differentiation (25). In general, our results suggest that Hox genes may play important roles in cancer progression by serving as modulators in neoplastic pathways.
Materials and Methods
Cell Culture, Plasmid Construction, and Transfection. Cells were cultured in DMEM (GIBCO/BRL) supplemented with 10% FCS (HyClone) and 100 μg/ml penicillin–streptomycin–glutamine (GIBCO/BRL). MEF cell lines were cultured from 15.5-dpc C57BL/6J embryos and were immortalized by transfecting an SV40 T-antigen plasmid (pPVU0neo) (26). Cells were plated in six-well plates and transfected with 1 μg of pPVU0neo plasmid DNA by using FuGENE 6 Transfection Reagent (Roche Molecular Biochemicals). Stable transfectants were selected in media containing 500 μg/ml G-418 (GIBCO/BRL). _Hoxc8_-overexpressing cells were produced by transfecting immortalized MEFs with the pcDNA4/Hoxc8 plasmid. The plasmid pcDNA4/Hoxc8 was constructed by RT-PCR amplification of Hoxc8 cDNA and cloning the amplicon into the _Kpn_I/_Eco_RI site of the zeomycin-resistant expression vector pcDNA4 (Invitrogen). The insert was sequenced to affirm that no enzymatic errors had been introduced. Immortalized MEF cells were transfected with the pcDNA4/Hoxc8 plasmid to produce _Hoxc8_-positive cell lines or blank pcDNA4 vector to produce _Hoxc8_-negative control cell lines by using FuGENE 6 transfection reagent (Roche Molecular Biochemicals). Zeomycin (GIBCO/BRL) at 500 μg/ml was added to the cell culture medium to select for stable _Hoxc8_-overexpressing cell lines and control cell lines. Three _Hoxc8_-positive and three _Hoxc8_-negative colonies were selected at random and paired into three independent positive and negative sets. Each set was subjected to microarray analysis.
RNA Preparation, Real-Time PCR Quantification, and Semiquantitative RT-PCR. Total RNA was isolated with RNeasy miniprep columns and an RNase-free DNaseI kit (Qiagen, Valencia, CA). The cDNA for real-time PCR and semiquantitative RT-PCR was made by using the OmniScript kit (Qiagen). Real-time PCR was performed in triplicate by using TaqMan probes and the ABI Prism 7900 sequence detection system (Applied Biosystems). The DNA Taq polymerase (Qiagen) was used to carry out PCR amplification. Primers and the expected product sizes for the semiquantitative PCR are detailed in Table 1. All primers were synthesized by Invitrogen. The PCR products were resolved on a 1.5% agarose gel containing ethidium bromide.
Table 1. Primers for RT-PCR and ChIP analysis.
| Gene | Forward primer | Reverse primer | Size, bp |
|---|---|---|---|
| GAPDH | CATCACCATCTTCCAGGAGC | ATGCCAGTGAGCTTCCCGTC | 500 |
| Cdh11 | CGGGAACCATTTTTGTGATT | TCCACCGAGAAATAGGGTTG | 343 |
| OPN | TCTGATGAGACCGTCACTGC | TCTCCTGGCTCTCTTTGGAA | 349 |
| PRDC | CTGTCTCCAGCTCCTTCCAC | GATCTGGTGATGCCACCTCT | 394 |
| Pmp22 | GCCAGCTCTTCACTCTCACC | AACAGCAATCCCCACTCAAC | 394 |
| Fzd2 | ATCTTTCTGTCCGGCTGCTA | GCCAAGATGGTGATGGTTTT | 326 |
| Gas1 | GCGAATCGGTCAAAGAGAAC | TACAAGTGTGACCCGAGCAG | 349 |
| Zac1 | TGAAGAACCACCTCCAGACC | CTCTGGGCACAGAACTGACA | 346 |
| Hoxc8 | CTACCAGCAGAACCCATGCT | GGCGCTTTCTGGTCAAATAA | 323 |
| OPN promoter for ChIP | GAAAGTTCTGCCGAGACAGC | TGAGGTTTTTGCCACTACCC | 371 |
Gene Expression Analysis. We used the RNeasy miniprep columns (Qiagen) to isolate total RNA from three sets of _Hoxc8_-positive and -negative cells as above. Each set was subjected to microarray analysis. The mouse 16K 70-mer oligo-operon microarray (OMM16K) was obtained from the Keck facility at Yale University. The oligonucleotide array gene set is described at http://keck.med.yale.edu/dnaarrays/genelists. The biotinylated cRNA preparation, hybridization, and scanning of the microarrays were performed according to the manufacturer's protocols. In brief, a total of 50 μg of RNA was reverse-transcribed by using Superscript II reagents (Invitrogen) at 42°C for 2 h. After RNA removal and cDNA probe purification, Cy5 and Cy3 fluorescent dyes were coupled to aa-dUTP-labeled samples and controls, respectively, and cohybridized to the microarray slides. Microarray protocols, including probe-labeling, cleaning, hybridization, and posthybridization washing procedures, can be found at http://keck.med.yale.edu/dnaarrays/protocols.htm. Hybridized slides were scanned with a GenePix 4000A scanner (Axon Instruments, Union City, CA), and the acquired images were analyzed with genepix pro 3.0. Only those genes that showed a minimum 2-fold increase or decrease in all three sets were included in the results presented in Table 2. The final values of increase or decrease were calculated as the average of the values for the three independent sets.
Table 2. Table of genes deregulated by overexpression of Hoxc8 in C57BL/6J MEF cell lines.
| GenBank accession no. | Entrez definition | Fold change | Chromosome no. |
|---|---|---|---|
| mRNA that decreases in the overexpression of Hoxc8 | |||
| NM_009263 | _Mus musculus_-secreted phosphoprotein 1 (Spp1, OPN) | 4.8 | 5 |
| NM_013468 | ankyrin repeat domain 1 (cardiac muscle) (CARP) | 2.1 | 19 |
| NM_008471 | Mus musculus keratin complex 1, acidic, gene 19 (Krt1-19) | 2.2 | 11 |
| NM_080288 | engulfment and cell motility 1, ced-12 homolog (Ced-12h) | 2.7 | 13 |
| NM_021274 | Mus musculus chemokine (C-X-C motif) ligand 10 (Cxcl10) | 3.3 | 5 |
| AF147785 | Mus musculus zinc finger protein regulator of apoptosis (Zacl) | 2.1 | 10 |
| NM_009922 | Mus musculus calponin 1 (Cnn1) | 2.4 | 9 |
| X15052 | Mus musculus neural cell adhesion molecule 1 (NCAM) | 2.4 | 9 |
| D50410 | Mus musculus mRNA for meltrin beta (ADAM19) | 2.2 | 11 |
| NM_008086 | Mus musculus growth-arrest-specific 1 (Gas1) | 2.2 | 13 |
| NM_011825 | Mus musculus protein related to DAC and cerberus (PRDC) | 2.4 | 1 |
| NM_011526 | Mus musculus transgelin (Tagln) | 2.3 | 8 |
| NM_011340 | Serine (or cysteine) proteinase inhibitor (Serpinf1) | 3.0 | 11 |
| BC022623 | Mus musculus cDNA sequence BC022623 | 2.4 | 16 |
| AK002886 | Mus musculus adult male kidney cDNA, clone:0610041G09 | 2.6 | 4 |
| BC019134 | Mus musculus, clone IMAGE:5037334, mRNA, partial cds | 2.3 | 9 |
| NM_008706 | Mus musculus NAD(P)H dehydrogenase, quinone 1 (Nqo1) | 2.1 | 8 |
| NM_007494 | Mus musculus argininosuccinate synthetase 1 (Ass1) | 2.3 | 13 |
| mRNA that increases in the overexpression of Hoxc8 | |||
| NM_020510 | Mus musculus frizzled homolog 2 (Drosophila) (Fzd2) | 4.4 | 11 |
| NM_030696 | Mus musculus monocarboxylate transporter 4 (MCTs) | 2.5 | 11 |
| NM_010330 | Mus musculus embigin (Emb), mRNA | 3.0 | 13 |
| NM_010555 | Mus musculus interleukin 1 receptor, type II (l11r2), mRNA | 2.3 | 1 |
| NM_009866 | Mus musculus cadherin 11 (Cdh11), mRNA | 3.0 | 8 |
| NM_013654 | Mus musculus chemokine (C-C motif) ligand 7 (Cc17), mRNA | 2.4 | 11 |
| NM_011333 | Mus musculus chemokine (C-C motif) ligand 2 (Ccl2), mRNA | 3.0 | 11 |
| NM_018857 | Mus musculus mesothelin (Msln), mRNA | 2.5 | 17 |
| NM_010738 | Mus musculus lymphocyte antigen 6 complex, locus A (Ly6a) | 2.9 | 15 |
| NM_008530 | Mus musculus lymphocyte antigen 6 complex, locus F (Ly6f) | 3.2 | 15 |
| NM_010741 | Mus musculus lymphocyte antigen 6 complex, locus C (Ly6c) | 2.8 | 15 |
| NM_008236 | Mus musculus hairy and enhancer of split 2 (Hes2) | 2.2 | 4 |
| NM_012011 | eukaryotic translation initiation factor 2, subunit 3 (Eif2s3y) | 3.8 | Y |
| NM_008885 | Mus musculus peripheral myelin protein (Pmp22), mRNA | 2.1 | 11 |
| NM_053188 | Mus musculus steroid 5 alpha-reductase 2 (Srd5a2), mRNA | 2.3 | 17 |
| AK014614 | Mus musculus day 0 neonate skin cDNA, clone:4633401122 | 3.3 | 2 |
ChIP Assay. The ChIP protocol was modified from the ChIP Assay kit (Upstate Biotechnology, Lake Placid, NY). To improve specificity, we performed two sequential ChIPs using the same antibody for both the first and second steps. Hoxc8 monoclonal anibody (MMS-266R) was purchased from Covance Research Products (Princeton). Anti-mouse IgG antibody (A-4312) was purchased from Sigma.
Results
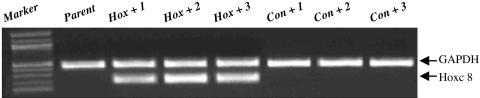
Overexpression of Hoxc8 in C57BL/6J MEF Cell Lines. To identify the Hoxc8 candidate target genes, we first generated primary C57BL/6J MEF cell lines. The primary MEF cells prepared from 15.5-dpc mouse embryo bodies were immortalized by transfecting an SV-40 T antigen-containing plasmid. Stable transfectants were selected in media containing 500 μg/ml G-418 (GIBCO/BRL). Thirty colonies were typically obtained from transfection. Real-time PCR was used to quantitate the Hoxc8 expression level in each cell line. The lowest-expressing Hoxc8 cell line was selected for further study (real-time PCR data not shown). A Hoxc8 expression vector, pcDNA4/Hoxc8, was transfected into the above selected MEF cell line to generate the stable _Hoxc8_-overexpressing cell line. As a control, the same cell line was transfected with empty pcDNA4 vectors. Total RNA was obtained from the parental cells, three sets of Hoxc8 transfectants, and control transfectants, and the expression levels of Hoxc8 were compared by RT-PCR. In the Hoxc8 transfectants, higher levels of Hoxc8 expression were observed in the Hox+1, Hox+2, and Hox+3 cells than in the parent cells and the control transfectants, Con+1, Con+2, and Con+3 cells (Fig. 1). The specificity of the RT-PCR products was confirmed by sequencing the amplified PCR bands. Real-time PCR results showed that the Hoxc8 expression level was increased >10,000 times in the three Hoxc8 transfectants compared with the three control transfectants.
Fig. 1.
Semiquantitative RT-PCR analysis of the Hoxc8 gene expression in parent C57BL/6J MEF cells and zeomycin-selected transfectants. Total cellular RNA was isolated from parent cells and three sets of pcDNA4/Hoxc8 (Hox) or pcDNA4 (Con) transfectants, and, after reverse transcription, PCR was performed with _Hoxc8_- and _GAPDH_-specific primers. The up-regulation of the 323-bp _Hoxc8_-specific band was detected in cells transfected with pcDNA4/Hoxc8 plasmid. A 100-bp DNA ladder (Biolabs) was used for size markers.
Expression Array Analysis. We used a mouse 16K oligonucleotide microarray to analyze gene expression profiles between the overexpressing Hoxc8 cell lines and the control cell lines. Three groups of total RNA samples were prepared from MEF cells transfected with plasmid pcDNA4/Hoxc8 or control empty pcDNA4 vector (see Materials and Methods). Probe labeling, hybridization, and scanning were done by the Keck facility (Yale University). We identified 34 genes that showed a minimum 2-fold change in expression (Table 2). Sixteen genes were up-regulated 2-fold or more, and 18 genes were down-regulated 2-fold or more. The majority of the identified genes are known to play roles in cell proliferation, differentiation, integration, apoptosis, metabolism, and carcinogenesis, with the exception of four genes whose function is unknown.
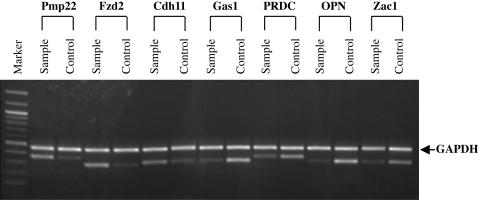
Semiquantitative RT-PCR of Selected Differentially Expressed Genes Identified by cDNA Microarray. To corroborate the cDNA microarray data, semiquantitative RT-PCR analysis was performed on seven selected genes identified as being differentially expressed (Fig. 2). These included osteopontin (OPN), growth-arrest-specific 1 (Gas1), peripheral myelin protein 22 (Pmp22), zinc finger protein regulator of apoptosis (Zac1), protein related to DAN and Cerberus (PRDC), cadherin 11 (Cdh11), and frizzled homolog 2 (Fzd2). All PCR products were sequenced to prove specificity. In general, the RT-PCR data were consistent with the cDNA microarray data, showing that expression of Fzd2 and Pmp22 was significantly increased and expression of OPN and Gas1 was decreased in _Hoxc8_-overexpression cells when compared with control cells.
Fig. 2.
Semiquantitative RT-PCR analysis of gene expressions of Pmp22, Fzd2, Cdh11, Gas1, PRDC, OPN, and Zac1 in pcDNA4/Hoxc8 transfectant cells (Sample) and pcDNA4 empty-vector transfectant cells (Control). Total RNA was extracted from the cultured cells, and, after reverse transcription, PCR was performed with _Pmp22_-, _Fzd2_-, _Cdh11_-, _Gas1_-, _PRDC_-, _OPN_-, _Zac1_-, and _GAPDH_-specific primers. A 100-bp DNA ladder (Biolabs) was used for size markers.
ChIP Assay Confirms OPN as a Direct Target of Hoxc8 in Vivo. Mouse-secreted phosphoprotein 1, also known as ostepontin (OPN), was found to be down-regulated ≈5-fold in the microarray analysis. Semiquantitative RT-PCR further confirmed that OPN is down-regulated in the _Hoxc8_-overexpressing cell lines (Fig. 2). Shi et al. (15) reported that, in Cos-1 cells, Hoxc8 interacts with Smad1 and that this interaction specifically activates OPN gene transcription in response to BMP stimulation. Five putative Hox-binding sites were identified within the first 382 bp of the 5′-flanking region in the OPN gene. A gel-shift assay was performed to demonstrate that Hoxc8 binds to the OPN promoter in vitro (15).
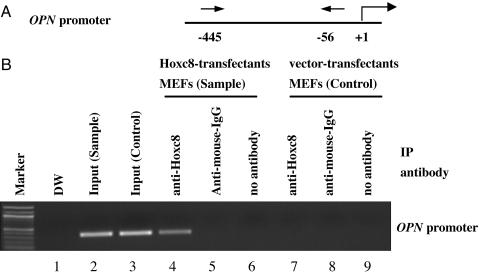
To determine whether the Hoxc8 protein can bind directly to the OPN promoter in vivo, we performed a ChIP assay of the OPN promoter (Fig. 3). The ChIP assay relies on the ability of specific antibodies to immunoprecipitate DNA-binding proteins along with the associated genomic DNA. Immunoprecipitation of DNA–protein complex by using an antibody against Hoxc8 was performed on formaldehyde-crosslinked extract from overexpressed Hoxc8 MEF cell lines or control MEF cell lines. We then measured the abundance of genomic DNA containing the OPN promoter sequence within the immunoprecipitate complex by PCR amplification. PCR products after ChIP assay were sequenced to verify the identity of the amplified DNA. ChIP assay showed that the OPN promoter sequence was present in a complex immunoprecipitated by an antibody against Hoxc8 but not by an anti-mouse IgG antibody (Fig. 3_B_, lanes 4–6). OPN promoter was detected in the input-positive control for PCR but not in the immunoprecipitated sample from the control MEF cells with an antibody against Hoxc8 (Fig. 3_B_, lanes 3 and 7). Our results show that the Hoxc8 protein can interact with the OPN promoter in vivo.
Fig. 3.
The mouse OPN gene has a Hoxc8-responsive element in the promoter region. (A) Shown is a schematic of the approximate location of the primers (arrows) used in the ChIP experiments. The numbers are relative to +1 being the 5′ end of the mRNA (indicated by a bent arrow). (B) A ChIP experiment was performed from Hoxc8-transfectant MEF cells (Sample) or vector-transfectant MEF cells (Control). Crosslinked Hoxc8 protein–DNA complexes were immunoprecipitated by a Hoxc8 monoclonal antibody (lanes 4 and 7). PCR amplification of the immunopriecipitated samples was performed by using the primers that flank the OPN promoter (see A). Immnoprecipitates with an anti-mouse IgG (lanes 5 and 8) or in the absence of antibody (lanes 6 and 9) were used for controls. Input chromatin represents a portion of the sonicated chromatin before immunoprecipitation (lanes 2 and 3). DW indicates a no-template control (lane 1). A 100-bp DNA ladder (Biolabs) was used for size markers.
Discussion
Hox genes regulate pattern formation during early development. Pattern formation itself is mediated by the coordinated control of cell proliferation, migration, adhesion, and differentiation. Thus, Hox genes must directly and/or indirectly exert control over such mediators. For this reason, it is necessary to identify the gene targets of Hox genes to properly understand pattern formation. We reason that a plethora of target genes serve as targets, but, to date, only a few have been properly identified. In this report, we identify 34 candidate gene targets for Hoxc8. In the specific case of OPN, we present evidence based on ChIP that supports its direct in vivo regulation by Hoxc8.
Whereas cell proliferation, migration, adhesion, and differentiation are mediators of normal development, they are also involved in abnormal growth when aberrantly regulated, as in neoplasia. It should come as no surprise that master control genes such as the Hox genes should also be involved in atypical growth, given the hierarchical relationship between Hox genes and their mediators. In fact, Hoxc8 has been shown to play a significant role in the progression of leukemias and lung and prostate cancers, among others, and possibly in precancerous-like conditions such as polycystic kidney disease.
In the present study, the use of a gene-expression microarray permitted us to identify those genes that are positively or negatively changed when Hoxc8 was overexpressed in MEF cells. Overall, we identified 34 genes that were at least 2-fold up- or down-regulated in comparisons between overexpressed Hoxc8 cells and control cells. Semiquantitative RT-PCR of selected genes performed on the same samples also confirmed the changed levels of expression detected on the microarray analysis. Some genes involved in the induction of apoptosis, such as growth arrest specific (Gas1), zinc finger protein regulator of apoptosis (Zac1), cardiac responsive adriamycin protein (CARP), serine (or cysteine) proteinase inhibitor (Serpinf1), NAD(P)H, and quinone oxidoreductase1 (Nqo1), were found in the present study to be down-regulated by Hoxc8 overexpression (Table 2). The proliferation rate of a cell population reflects a balance between cell division, cell-cycle arrest, differentiation, and apoptosis. The neural cell adhesion molecule (NCAM), cadherin 11 (Cdh11), and embigin (Emb) play important roles in cell–cell adhesion. NCAM was down-regulated, whereas Cdh11 and Emb were up-regulated (Table 2). Calponin h1 (Cnn1), a protein related to DAN and Cerberus (PRDC), engulfment and cell motility 1 (Ced-12h), and transgelin (Tagln) have been categorized as genes related to cell migration, motility, and proliferation. These genes were down-regulated by Hoxc8 (Table 2).
Significantly, a majority of the 34 Hoxc8 target genes identified here are tumor-related genes. In general, our results support the role of Hox genes as modulators of neoplastic progression. Hox genes are global regulators of growth and differentiation. They can interact with different classes of genes at different developmental stages and in specific tissue types (25). It is particularly interesting to find that OPN, the major noncollagenous bone matrix protein associated with osteoblastic cell adhesion and abundantly expressed during the early stages of osteoblast differentiation, is down-regulated in our microarray analysis. Semiquantitative RT-PCR proved that OPN is dramatically down-regulated by Hoxc8 overexpression (Fig. 2). OPN expression is rapidly induced by both BMP and transforming growth factor β (TGF-β). BMPs, members of the TGF-β superfamily, play a pivotal role in signaling networks and are involved in nearly all processes associated with limb development (27). It is likely that Hoxc8 is involved in both osteo- and chondrogenic processes in development, because Hoxc8 knockout mice display skeletal abnormalities in ribs, sternum, and vertebra (8). Moreover, expression of Hoxc8 in skeletal tissue results in an accumulation of blastogenic cells in hypertrophic areas (10).
Shi et al. (15) have demonstrated that Hoxc8 functions as an OPN transcription repressor and that the interaction of Smad1 and Smad4 with Hoxc8 can stimulate OPN transcription in response to BMP stimulation. They also performed gel-shift assays that support the binding of Hoxc8 to the OPN promoter in vitro. In this report, we confirm and extend their results by showing, by means of ChIP analysis, that Hoxc8 interacts with the OPN promoter in vivo. This result strengthens our argument that Hox genes may serve as modulators of neoplastic progression. Studies in vitro and in animal models of cancer have clearly indicated that OPN can function to regulate tumor growth and progression. Numerous reports of elevated OPN expression in human cancers support the idea that OPN should be considered as a potential prognostic marker for a variety of human cancers (28).
OPN is strongly regulated by Hoxc8 expression, showing a 4.8-fold reduction in the experiments reported here. We also demonstrate a direct interaction between Hoxc8 and the OPN promoter in vivo. All of the other candidate genes reported here, with one exception, show significantly lower levels of change. Possibly, Hoxc8 indirectly influences those genes showing lower levels of modulation through intermediary genes. The exceptional gene is Fzd2, which is a cell-surface receptor in the WNT–β-catenin–TCF-signaling pathway. This pathway is important in dorsal patterning during early development and in the progression of intestinal cancers, among others (29, 30). Fzd2 is up-regulated 4.4-fold, and we might speculate that it is under direct control by Hoxc8. Our future aim is to determine the intimate regulatory interactions involving Hoxc8, the candidate genes described here, and additional candidates genes as they are identified.
Acknowledgments
We thank Prof. Mary J. Tevethia (Pennsylvania State University, Hershey, PA) for providing pPVU0neo plasmid. This work was supported by National Institutes of Health Grant GM09966.
Abbreviations: dpc, days postconception; BMP, bone morphogenetic protein; OPN, osteopontin; MEF, mouse embryo fibroblast; ChIP, chromatin immunoprecipitation; Fzd2, frizzled homolog 2.
References
- 1.McGinnis, W. & Krumlauf, R. (1992) Cell 68**,** 283-302. [DOI] [PubMed] [Google Scholar]
- 2.Krumlauf, R. (1994) Cell 78**,** 191-201. [DOI] [PubMed] [Google Scholar]
- 3.Belting, H. G., Shashikant, C. S. & Ruddle, F. H. (1998) Proc. Natl. Acad. Sci. USA 95**,** 2355-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shashikant, C. S., Bieberich, C. J., Belting, H. G., Wang, J. C. H., Borbély, M. A. & Ruddle, F. H. (1995) Development (Cambridge, U.K.) 121**,** 4339-4347. [DOI] [PubMed] [Google Scholar]
- 5.Bradshaw, M. S., Shashikant, C. S., Belting, H. G., Bollekens, J. A. & Ruddle, F. H. (1996) Proc. Natl. Acad. Sci. USA 93**,** 2426-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belting, H. G., Shashikant, C. S. & Ruddle, F. H. (1998) J. Exp. Zool. 282**,** 196-222. [PubMed] [Google Scholar]
- 7.Shashikant, C. S. & Ruddle, F. H. (1996) Proc. Natl. Acad. Sci. USA 93**,** 12364-12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mouellic, H. L., Lallemand, Y. & Brulet, P. (1992) Cell 69**,** 251-264. [DOI] [PubMed] [Google Scholar]
- 9.Juan, A. H. & Ruddle, F. H. (2003) Development (Cambridge, U.K.) 130**,** 4823-4834. [DOI] [PubMed] [Google Scholar]
- 10.Yueh, Y. G., Gardner, D. P. & Kappen, C. (1998) Proc. Natl. Acad. Sci. USA 95**,** 9956-9961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofbauer, L. C., Dunstan, C. R., Spelsberg, T. C., Riggs, B. L. & Khosla, S. (1998) Biochem. Biophys. Res. Commun. 250**,** 776-781. [DOI] [PubMed] [Google Scholar]
- 12.Feng. J., Chen, D., Cooney, A. J., Tsai, M., Harris, M. A., Tsai, S. Y., Feng, M., Mundy, G. R. & Harris, S. E. (1995) J. Biol. Chem. 270**,** 28364-28373. [DOI] [PubMed] [Google Scholar]
- 13.Zhou, H., Hammonds, R. G., Jr., Findlay, D. M., Martin, T. J. & Ng, K. W. (1993) J. Cell. Physiol. 155**,** 112-119. [DOI] [PubMed] [Google Scholar]
- 14.Wan, M., Shi, X., Feng, X. & Cao, X. (2001) J. Biol. Chem. 276**,** 10119-10125. [DOI] [PubMed] [Google Scholar]
- 15.Shi, X., Yang, X., Chen, D., Chang, Z. & Cao, X. (1999) J. Biol. Chem. 274**,** 13711-13717. [DOI] [PubMed] [Google Scholar]
- 16.Celetti, A., Barba, P., Cillo, C., Rotoli, B., Boncinelli, E. & Magli, M. C. (1993) Int. J. Cancer 53**,** 237-244. [DOI] [PubMed] [Google Scholar]
- 17.DeVita, G., Barba, P., Odartchenki, N., Givel, J.-C., Freschi, G., Bucciarelli, G., Magli, M., Boncinelli, E. & Cillo, C. (1993) Eur. J. Cancer 29A**,** 887-893. [DOI] [PubMed] [Google Scholar]
- 18.Chariot, A. & Castronovo, V. (1996) Biochem. Biophys. Res. Commun. 222**,** 292-297. [DOI] [PubMed] [Google Scholar]
- 19.Cillo, C., Barba, P., Freschi, G., Bucciarelli, G., Magli, M. C. & Boncinelli, E. (1992) Int. J. Cancer 51**,** 892-897. [DOI] [PubMed] [Google Scholar]
- 20.Hamada, J.-I., Omatsu, T., Okada, F., Furuuchi, K., Okubo, Y., Takahashi, Y., Tada, M., Miyazaki, Y. J., Taniguchi, Y., Shirato, H., et al. (2001) Int. J. Cancer 93**,** 516-525. [DOI] [PubMed] [Google Scholar]
- 21.Miller, G. J., Miller, H. L., Bokhoven, A. V., Lambert, J. R., Werahera, P. N., Schirripa, O., Lucia, M. S. & Nordeen, S. K. (2003) Cancer Res. 63**,** 5879-5888. [PubMed] [Google Scholar]
- 22.Waltregny, D., Alami, Y., Clausse, N., de Leval, J. & Castronovo, V. (2002) Prostate 50**,** 162-169. [DOI] [PubMed] [Google Scholar]
- 23.Alami, Y., Castronovo, V., Belotti, D., Flagiello, D. & Clausse, N. (1999) Biochem. Biophys. Res. Commun. 257**,** 738-745. [DOI] [PubMed] [Google Scholar]
- 24.Shimamoto, T., Tang, Y., Naot, Y., Nardi, M., Brulet, P., Bieberich, C. J. & Takeshita, K. (1999) J. Exp. Zool. 283**,** 186-193. [PubMed] [Google Scholar]
- 25.Shen, C. A. (2002) Nat. Rev. Cancer 2**,** 777-785. [DOI] [PubMed] [Google Scholar]
- 26.Kierstead, T. D. & Tevethia, M. J. (1993) J. Virol. 67**,** 1817-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheifetz, S., Li, I. W., McCulloch, C. A., Sampath, K. & Sodek, J. (1996) Connect. Tissue Res. 35**,** 71-78. [DOI] [PubMed] [Google Scholar]
- 28.Rittiing, S. R. & Chambers, A. F. (2004) Br. J. Cancer 90**,** 1877-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brennan, K. R. & Brown, A. M. (2004) J. Mammary Gland Biol. Neoplasia 9**,** 119-131. [DOI] [PubMed] [Google Scholar]
- 30.Sancho, E., Batle, E. & Clevers, H. (2004) Annu. Rev. Cell Dev. Biol. 20**,** 695-723. [DOI] [PubMed] [Google Scholar]