The association of epsin with ubiquitinated cargo along the endocytic pathway is negatively regulated by its interaction with clathrin (original) (raw)
Abstract
Monoubiquitination of plasma membrane proteins is a mechanism to control their endocytic trafficking by promoting their interaction with cytosolic adaptor proteins that contain ubiquitin (Ub)-binding domains. Epsin, which contains Ub interaction motifs (UIMs), as well as binding sites for the clathrin coat and clathrin accessory factors, is thought to function as one of such adaptors. The importance of clathrin in the internalization of ubiquitinated cargo, however, has been questioned. Here, we show that a GFP-Ub chimera directly targeted to the plasma membrane via a lipid-based interaction is efficiently taken up by endocytosis and delivered to the same endosomes that accumulate internalized EGF. Internalization of the chimera requires integrity of the UIM binding interface of Ub, but does not require clathrin. Surprisingly, WT epsin showed little colocalization with this chimera, whereas UIM-containing epsin constructs that lack the clathrin and AP2 binding region, strikingly colocalized with this chimera on endocytic vacuoles. In addition, extensive colocalization of WT epsin with the chimera on endocytic structures could be observed in cells where clathrin levels were drastically reduced by RNA interference. Our results reveal an important regulatory mechanism in epsin function. The mutually exclusive colocalization of epsin with membrane-bound Ub or clathrin may play a role in controlling the endocytic route taken by ubiquitinated cargo.
Keywords: ubiquitin, endocytosis, Rab5, endosome, FYVE domain
Protein ubiquitination plays pleotropic roles in cell physiology. Polyubiquitination of cytosolic proteins directs them for destruction by the proteasome. Reversible monoubiquitination regulates protein location, function, and interactions (1–3). In the case of membrane proteins, monoubiquitination triggers their interaction with adaptor proteins that control their intracellular traffic. More specifically, monoubiquitination of plasma membrane (PM) proteins tags them for internalization and subsequent sorting to late endosomes/lysosomes for degradation (2, 4). PM-associated ubiquitin (Ub) is recognized by endocytic adaptors, among which epsin has been extensively characterized (5–11). Epsin has a modular structure comprising an epsin N-terminal homology (ENTH) domain, which binds phosphoinositides, Ub interaction motifs (UIMs), and a long flexible region that includes binding sites for clathrin heavy chain, the clathrin adaptor AP-2, and EH domain-containing proteins such as Eps15, another UIM containing endocytic adaptor (8, 12–14). The UIM region of epsin, in addition to mediating binding to Ub, is required for the ubiquitination of epsin itself (6). This reaction, which occurs constitutively and is further stimulated by growth factors (6), inhibits the property of epsin to interact with the clathrin coat and the membrane (15). The importance of Ub metabolism in epsin function is further supported by genetic studies in Drosophila (16–19). The presence in epsin of binding sites for both Ub and core components of the clathrin coat, together with the localization of epsin at clathrin-coated pits, suggested that a function of this protein is to recruit ubiquitinated cargo to clathrin-coated pits, thus triggering its internalization (13). Surprisingly, however, the endocytosis of a chimeric protein composed of the extracellular and transmembrane domains of the EGF receptor fused to a cytoplasmically exposed Ub does not require clathrin, questioning the role of clathrin-mediated budding in the internalization of ubiquitinated cargo (20).
Here, we used a GFP-fusion protein of Ub that is directly targeted to the PM via lipid-based interactions, to further elucidate mechanisms in epsin-mediated sorting of PM-associated Ub.
Methods
Antibodies, Constructs, and Reagents. Antibody directed against epsin 1 was generated in our laboratory as described (5). Mouse mAbs against clathrin heavy chain, GFP, the hemagglutinin (HA) epitope, the Xpress epitope, and the Flag epitope were from Affinity BioReagents (Golden, CO), Clontech, Roche Applied Science, Invitrogen, and Sigma, respectively. Human antibody directed against EEA1 was a kind gift of Harald Stenmark (Norwegian Radium Hospital, Oslo). Rabbit antibody directed against Rabankyrin-5 was a kind gift of Marino Zerial (Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany). Xpress-tagged full-length epsin 1 and HA-tagged amphiphysin A1 fragment have been described (5, 21). PM-GFP-Ub was made by PCR using PM-GFP in a pcDNA3 vector (a kind gift of Tobias Meyer, Stanford University, Stanford, CA) as a template, followed by subcloning in a pcDNA3-HA-Ub vector as described (15) except that the HA tag was removed in the final construct. PM-GFP-UbΔGG was generated by replacement of Ub in PM-GFP-Ub with a mutant Ub fragment obtained by PCR using a reverse primer that lacks the last two glycine residues in Ub cDNA sequence. GFP-UbΔGG was generated by subcloning the mutant Ub lacking the last two glycine residues in pEGFP-C2 vector (Clontech). PM-GFP-UbΔGG-K48R and PM-GFP-UbΔGG-I44A double mutants were obtained from PM-GFP-UbΔGG by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's instruction with primers harboring the corresponding mutation sequences. GFP-FYVE(Hrs)-UbΔGG was constructed by subcloning the mutant Ub lacking the last two glycine residues in a pEGFP-FYVE vector (a kind gift from Harald Stenmark). GFP-PH(FAPP1)-Ub was generated by subcloning Ub in a pEGFP-PH vector (a gift from Dario Alessi, University of Dundee, Dundee, Scotland). A mutant epsin 1 lacking the clathrin and AP-2 binding region was generated by ligating together two pieces of the rat epsin 1 cDNA sequence (5) corresponding to amino acids 1–254 and 401–576. Blunt-end ligation was followed by subcloning in a pcDNA3-Flag vector. The ENTH-UIM construct, corresponding to amino acids 1–254 of epsin 1, was generated by PCR followed by subcloning in pcDNA3-Flag vector. The UIM construct, encoding amino acids 181–254 of epsin 1, was obtained by PCR followed by subcloning in pcDNA3-HA vector. All constructs were confirmed by DNA sequencing. Rhodamine-conjugated EGF and Alexa Fluor 594-conjugated dextran were purchased from Molecular Probes.
Cell Culture and cDNA Transfection. HeLa or CHO cells were grown at 37°C and 5% CO2 in DMEM with 10% (vol/vol) FBS, 10 mM Hepes, 1 mM sodium pyruvate, 100 units/ml penicillin, and 100 μg/ml streptomycin. cDNA transfection was performed with Lipofectamine (Invitrogen). Twenty-four hours later, cells were fixed in 4% formaldehyde and processed for immunofluorescence. In some experiments, cells were incubated with rhodamine-conjugated EGF or Alexa Fluor 594-conjugated dextrin for 20 min followed by rapid wash in PBS three times before fixation.
Clathrin Heavy Chain RNA Interference (RNAi). Small interfering RNA (siRNA) pairs corresponding to the sequences GAAAGA ATCTGTAGAGA A A and GCA ATGAGCTGT T TGAAGA of clathrin heavy chain (22, 23) were synthesized and purchased from Integrated DNA Technologies (Coralville, IA). HeLa cells were transfected with 50 nM each of the two siRNA pairs or with 50 nM of a control siRNA pair corresponding to nucleotides 695–715 of the firefly luciferase (U31240) (AAGAATATTGTTGCACGATTT), using Oligofectamine (Invitrogen). Three days later, cells were transfected with PM-GFP-UbΔGG or Xpress-tagged epsin 1 cDNA in a pcDNA3 vector with Lipofectamine and processed for immunofluorescence studies.
Miscellaneous Procedures. SDS/PAGE, Western blotting, and immunoprecipitation were performed according to standard procedures. Immunofluorescence microscopy was performed with an Axioplan 2FS microscope (Zeiss) using ×63 oil-immersion objectives. Images were acquired with a charge-coupled device camera (ORAC II, Hamamatsu, Middlesex, NJ) and analyzed with metamorph software (Universal Imaging, Downingtown, PA). Confocal images were obtained with a Zeiss 510 laser scanning confocal microscope.
Results
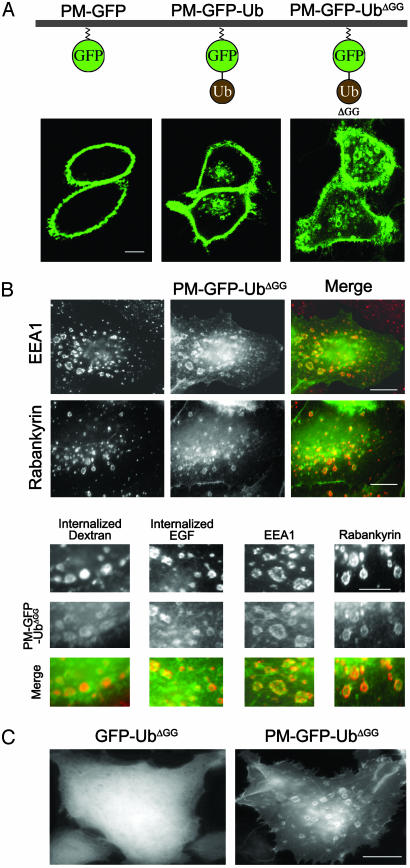
Monoubiquitin Targeted to the PM by a Lipid Anchor Is Efficiently Internalized. To determine mechanisms in the internalization of PM-bound Ub we generated a reporter construct (PM-GFP-Ub) represented by Ub fused in-frame to the COOH terminus of PM-targeted GFP (PM-GFP). PM-GFP is a modified form of GFP resulting from the addition to its NH2 terminus of the short amino acid sequence (10 aa) that directs the myristoylation and palmitoylation, and therefore PM targeting of Lyn kinase (24). We used a lipid-bound Ub chimera rather than Ub fused to a transmembrane protein to avoid potential effects caused by traffic of the chimera through biosynthetic compartments. As expected, when expressed in HeLa or CHO cells, PM-GFP accumulated selectively at the PM, consistent with a direct targeting to this membrane independent of the secretory pathway (Fig. 1_A_ and data not shown). In contrast, PM-GFP-Ub also accumulated on internal vacuolar structures (Fig. 1 A).
Fig. 1.
PM-GFP is selectively localized at the PM, whereas PM-GFP-Ub and PM-GFP-UbΔGG are internalized and also accumulate on intracellular endocytic compartments. (A) Confocal microscopy images of HeLa cells. (B) HeLa cells transfected with PM-GFP-UbΔGG were incubated with or without Alexa Fluor 594-conjugated dextran or rhodamine-conjugated EGF, then fixed and either processed by immunofluorescence for the endosomal proteins EEA1 and rabankyrin or examined directly for the internalized dye-conjugated endocytic probes. (C) Epifluorescence microscopy of HeLa cells transfected with the constructs indicated. Note the diffuse cytosolic localization of GFP-UbΔGG that lacks the short amino acid sequence directing acylation and PM localization. (Bars: 12 μm.)
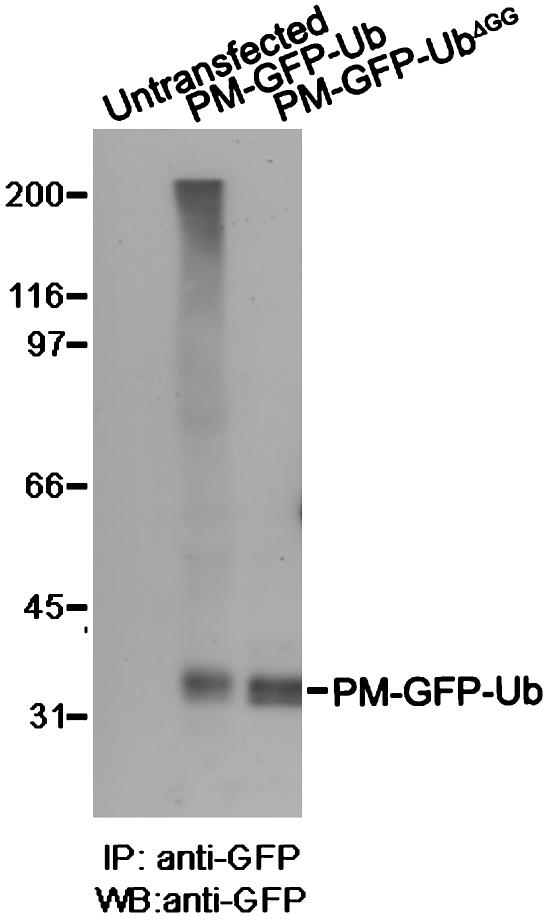
Anti-GFP Western blots of anti-GFP immunoprecipitates from cells transfected with PM-GFP-Ub revealed a smear characteristic of polyubiquitin-containing conjugates, in addition to the PM-GFP-Ub band, suggesting that this construct participates in the formation of polyubiquitin chains (Fig. 2). This smear was absent when WT Ub in PM-GFP-Ub was replaced by a mutant Ub that lacks the last two glycine residues (UbΔGG). UbΔGG cannot be conjugated to lysine residues of ubiquitinated substrates because this conjugation requires a COOH-terminal glycine (25). Similar results were obtained with an Ub mutant that contained an additional lysine-to-alanine substitution at position 48, a major acceptor site for the COOH-terminal glycine of another Ub (25). Like PM-GFP-Ub, both PM-GFP-UbΔGG and PM-GFP-UbΔGG-K48R were targeted to internal vacuoles (Fig. 1 A and data not shown). PM-GFP-UbΔGG was used for most subsequent experiments.
Fig. 2.
PM-GFP-Ub, but not PM-GFP-UbΔGG, is incorporated into polyubiquitin chains. Anti-GFP immunoprecipitates from detergent extracts of untransfected cells and cells transfected with GFP constructs were processed by Western blotting for GFP immunoreactivity. IP, immunoprecipitation; WB, Western blot.
Extracellular applications of Alexa Fluor 594-conjugated dextran (100 μg/ml for 20 min), a hydrophilic polysaccharide that does not cross the bilayer, and rhodamine-conjugated EGF (40 ng/ml for 20 min) resulted in the accumulation of these probes in the PM-GFP-UbΔGG-positive vacuoles, proving their endocytic nature. Furthermore, partial overlap (with some variability from cells to cells) was observed between these vacuoles and the endosomal proteins EEA1 and Rabankyrin-5, two Rab5 effectors (26, 27) (Fig. 1_B_). The large size of these endocytic structures may be related to the accumulation of the exogenous protein. To confirm that PM-GFP-UbΔGG found on endocytic vacuolar structures is not recruited directly to these membranes via endosomal-associated Ub-binding proteins, a GFP-UbΔGG chimera was used as a control. This chimera, which lacks the sequence directing its acylation, and therefore PM targeting, had a diffuse localization in the cytosol (Fig. 1_C_).
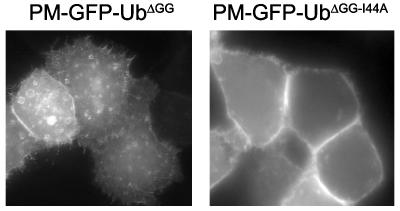
Interactions with Endocytic Adaptors, but Not Clathrin, Are Required for the Endocytosis of Membrane-Targeted Ub. Studies in yeast and cells of higher eukaryotes have demonstrated that isoleucine 44 of Ub plays a critical role in endocytosis (28, 29), and biochemical and structural studies have confirmed the important role of this residue in the binding of Ub to the UIM motifs of endocytic adaptors (28–30). Accordingly, a PM-GFP-Ub chimera harboring an isoleucine-to-alanine substitution at position 44 (PM-GFP-UbΔGG-I44A) accumulated selectively at the PM and failed to be internalized (Fig. 3), thus implicating adaptors comprising UIMs or other Ub-binding domains in its endocytosis.
Fig. 3.
The internalization of PM-GFP-UbΔGG is abolished by the substitution of isoleucine 44 with alanine as examined by epifluorescence microscopy of HeLa cells. (Magnification: ×500.)
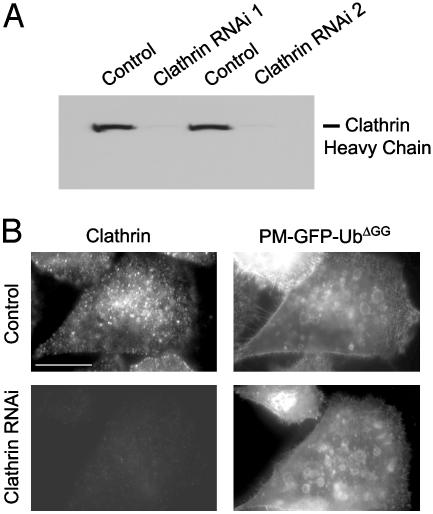
An EGF receptor-Ub chimera can be internalized in a clathrin-independent way (20). In agreement with this finding, disruption of clathrin function by siRNA or dominant negative interference did not block the internalization of PM-GFP-UbΔGG. Thus, addition to HeLa cells of two different clathrin heavy chain-specific siRNA pairs, which alone were sufficient to nearly deplete clathrin, as shown by Western blotting (Fig. 4_A_) and immunofluorescence (Fig. 4_B_), blocked the internalization of transferrin as expected (data not shown), but did not prevent the accumulation of PM-GFP-UbΔGG on endosomal vacuoles (Fig. 4_B_). In contrast, PM-GFP-UbΔGG still accumulated on endosomes in cells treated with a control siRNA pair (Fig. 4_B_). Furthermore, transfection of the fragment of amphiphysin 1 that comprises the binding sites for clathrin and the endocytic clathrin adaptor AP-2 (fragment A1) did not prevent accumulation of PM-GFP-UbΔGG on endosomes (data not shown). This construct has a powerful inhibitory effect on clathrin-mediated endocytosis because it titrates out available clathrin and AP-2 (21). At variance with results obtained with the EGF receptor-Ub chimera (20), however, dynamin, a GTPase that mediates the fission steps in a variety of endocytic reactions (31), was also not required for the endocytosis of PM-GFP-UbΔGG. This reporter protein was found on endosomes in the presence of dynaminK44A (32), a mutant dynamin that has a dominant negative effect on the function of endogenous dynamin, or in HeLa cells in which dynamin 2 had been knocked down by RNAi (data not shown).
Fig. 4.
Endocytosis of PM-GFP-Ub does not depend on clathrin. (A) Anticlathrin Western blot of extracts from cells treated with control siRNA and two distinct sets of clathrin siRNA duplexes. (B) HeLa cells treated with control siRNA or a mixture of the two siRNA duplexes used for A were transfected with PM-GFP-UbΔGG, then fixed and analyzed for GFP fluorescence and clathrin immunoreactivity. (Bar: 12 μm.)
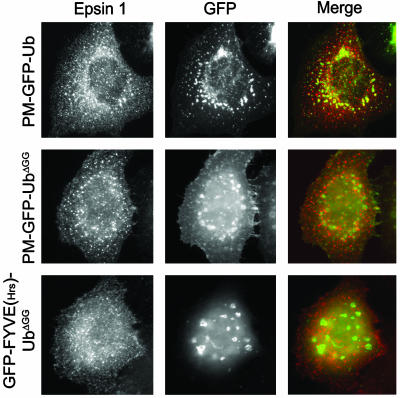
Clathrin Affects the Colocalization of Epsin with Membrane-Targeted Ub. Given the putative role of epsin as an endocytic adaptor for ubiquitinated cargo, we investigated whether epsin colocalizes, at least partially, with PM-GFP-Ub or PM-GFP-UbΔGG. To this aim, we cotransfected cells with Xpress-tagged epsin 1 (5) and either PM-GFP-Ub or PM-GFP-UbΔGG. Epsin immunoreactivity showed little colocalization with either chimera, but displayed primarily the typical, previously reported punctate pattern characteristic of clathrin-coated pits (5, 11) (Fig. 5). This result may reflect the occurrence of regulatory mechanisms in the binding of epsin's UIMs to Ub or the inefficient interaction of epsin with Ub in the context of the cell cytosol.
Fig. 5.
Full-length epsin 1 shows little colocalization with GFP-Ub constructs. CHO cells (Top) or HeLa cells (Middle and Bottom) cotransfected with epitope-tagged full-length epsin 1 and GFP-Ub constructs were fixed and examined for GFP fluorescence and epitope tag immunoreactivity by epifluorescence microscopy. Note the typical punctate pattern of epsin, reflecting clathrin-coated pits localization (5), regardless of the subcellular localization of GFP-Ub constructs. (Magnification: ×750.)
To address these possibilities, fragments of epsin containing the UIMs, including a construct represented by the UIMs alone (Fig. 6), were coexpressed in HeLa cells together with PM-GFP-UbΔGG or other Ub-containing GFP fusion proteins targeted to specific intracellular compartments by lipid-binding domains. Two tandem-arranged FYVE domains of the late endosomal protein Hrs (33), which binds PI(3)P, were used to target Ub to endosomes [GFP-FYVE(Hrs)-UbΔGG], and the PH domain of FAPP1, which binds PI(4)P, was used to target Ub to the Golgi complex [GFP-PH(FAPP1)-Ub] (34, 35). Both of these fusion proteins behaved as expected, although they partially disrupted the morphology of the two organelles, possibly as a result of interactions, or functional interference, with the many intracellular Ub-binding proteins (36–38). GFP-FYVE(Hrs)-UbΔGG accumulated on vacuolar endosome-like structures (often enlarged and/or clustered), whereas GFP-PH(FAPP1)-Ub accumulated on perinuclear particles and, at high levels of expression, disrupted the Golgi complex (Fig. 7_A_).
Fig. 6.
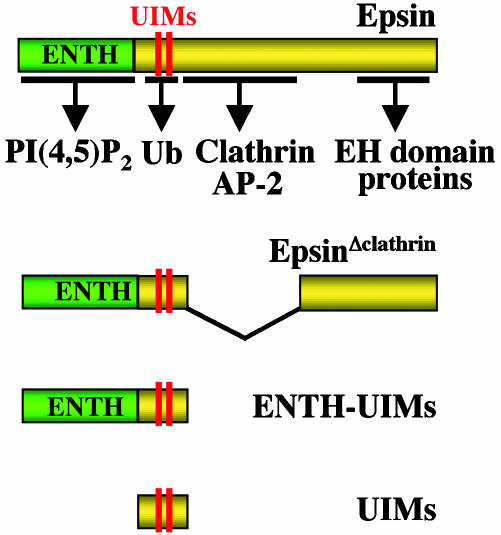
Domain cartoon of epsin and epsin constructs used in this study. Interacting regions for binding partners are indicated by arrows.
Fig. 7.
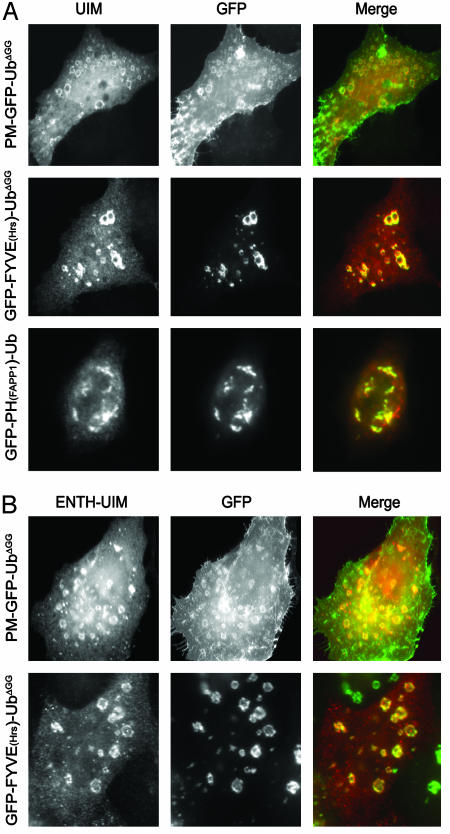
Fragments of epsin containing the UIM (A) or the ENTH-UIM (B) region colocalize extensively with internalized PM-GFP-UbΔGG. HeLa cells cotransfected with GFP-Ub constructs and epitope-tagged UIMs or ENTH-UIMs were fixed and examined for GFP fluorescence and epitope tag immunoreactivity. Note colocalization of UIMs and ENTH-UIMs with GFP-Ub constructs irrespective of their subcellular localization. (Magnification: ×800.)
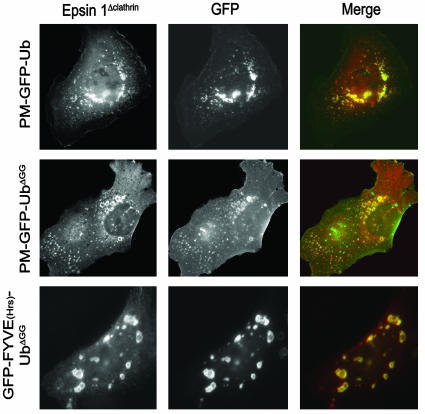
Coexpressed full-length epsin 1 did not colocalize with either FYVE(Hrs)- or PH(FAPP1)-containing constructs, and in all cases had the typical distribution of endocytic clathrin-coated pits (Fig. 5 and data not shown). In contrast, an epitope-tagged epsin 1 fragment composed only of its UIM domains (Fig. 6) strongly colocalized with membrane-targeted Ub fusion proteins irrespective of their localization (Fig. 7_A_). It colocalized with PM-GFP-UbΔGG and GFP-FYVE(Hrs)-UbΔGG on intracellular vacuoles and with GFP-PH(FAPP1)-Ub on the Golgi remnants observed in cells transfected with this domain (Fig. 7_A_). Thus, epsin's UIMs do interact efficiently with Ub and monoubiquitin in the cell cytosol. An epsin 1 construct including both the ENTH domain and the UIMs (Fig. 6) also colocalized with PM-GFP-UbΔGG and GFP-FYVE(Hrs)-UbΔGG, but not with GFP-PH(FAPP1)-Ub (Fig. 7_B_ and data not shown), indicating that the ENTH domain contributes specificity to UIM binding. More important, even full-length epsin 1 with an internal deletion, i.e., deletion of the clathrin and AP-2 binding region (epsinΔclathrin), a predicted unfolded region (Fig. 6), colocalized with PM-GFP-UbΔGG and GFP-FYVE(Hrs)-UbΔGG (Fig. 8). It would therefore appear that the presence of binding sites for core components of the clathrin coat is what prevents the colocalization of epsin with Ub-containing reporter fusions localized on endocytic membranes.
Fig. 8.
An epitope-tagged epsin mutant (EpsinΔclathrin) lacking the clathrin and AP-2 binding region colocalized with PM-GFP-UbΔGG and GFP-FYVE(Hrs)-UbΔGG on endocytic compartments. Cotransfected CHO cells (Top) or HeLa cells (Middle and Bottom) were fixed and examined for GFP fluorescence and epitope tag immunoreactivity. (Magnification: ×750.)
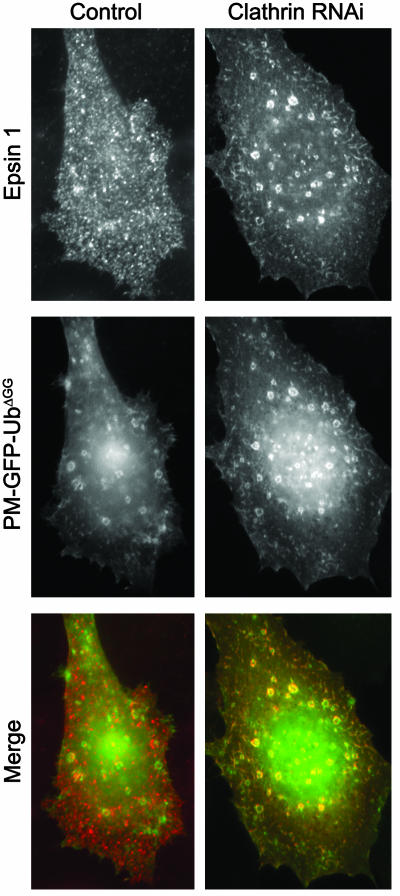
To determine whether this finding resulted from a masking effect of the clathrin-AP-2 binding region of epsin on its UIM domain region or an effect of the binding of clathrin to epsin, the localization of Xpress-tagged epsin was investigated in HeLa cells expressing PM-GFP-UbΔGG after RNAi-mediated clathrin knockdown (see Fig. 4). Strikingly, in these cells, a significant fraction of WT epsin colocalized with PM-GFP-UbΔGG on internal vacuoles (Fig. 9). Thus, binding of epsin to the clathrin coat either prevents its binding to Ub or makes this interaction very transient.
Fig. 9.
A pool of full-length epsin colocalizes with PM-GFP-UbΔGG on intracellular endocytic vacuoles after RNAi-mediated clathrin knockdown. HeLa cells pretreated with control siRNA or clathrin heavy chain-specific siRNA duplexes were transfected with epitope-tagged full-length epsin 1 construct and PM-GFP-UbΔGG. Cells were then fixed and analyzed for GFP fluorescence and epitope tag immunoreactivity. (Magnification: ×1,000.)
Discussion
Our results demonstrate the importance of Ub as an internalization signal at the PM (2) and of epsin in the recognition of this signal (2, 8, 13). In addition, a key conclusion of our study is that the interaction of epsin with Ub is critically regulated by the interaction of epsin with clathrin.
Epsin contains a multiplicity of binding sites, including binding sites for phosphoinositides (39), a transcription factor (40), Ub (6, 14), clathrin (41, 42), the clathrin adaptor AP-2 (5), and other signaling (43) and endocytic proteins (5, 44, 45), which implies the occurrence of regulatory mechanisms to control these interactions. As we have shown previously, some of the interactions of epsin are regulated by its phosphorylation and reversible ubiquitination (15, 46). Here, we report evidence for an additional control mechanism based on its binding to clathrin. Full-length epsin has the typical, previously described (5, 11) clathrin-coated pit localization even in cells expressing a PM-targeted, Ub-containing reporter protein. In contrast, an epsin construct lacking the clathrin and AP-2 binding region (epsinΔclathrin) shows a striking colocalization with this reporter protein throughout the endocytic pathway and also colocalizes with a Ub-reporter protein targeted directly to endosomes. This colocalization is observed for full-length epsin 1 as well, but only in cells where clathrin levels were drastically decreased by RNAi-mediated knockdown. It therefore would appear that the recruitment of epsin to clathrin coats promotes its dissociation from, or prevents its binding to, ubiquitinated cargo. The ubiquitination of epsin itself prevents its interaction with clathrin and AP-2, possibly as a result of an intramolecular interaction (15). It remains to be seen whether this “cis” interaction with an intramolecular Ub occludes clathrin binding in a way mechanistically similar to “trans” binding to another ubiquitinated protein. We note that our present results may not necessarily imply a direct, mutually exclusive interaction of epsin with either ubiquitinated cargo or clathrin, a possibility that, because of technical limitations, we could not test satisfactorily with in vitro biochemical experiments.
Clearly, as shown by Sigismund et al. (20) and our present results, ubiquitinated PM proteins can be internalized in a clathrin-independent way. The simple explanation is that epsin may function as an endocytic adaptor in two completely distinct internalization pathways, one clathrin-dependent but Ub-independent, and the other clathrin-independent but Ub-dependent. However, this explanation seems unlikely, given the striking effect on epsin's localization of its ability to interact with clathrin in the context of the cell cytoplasm. Furthermore, binding sites for both Ub and clathrin are conserved in epsin from yeast to mammals. Thus, an interplay between Ub binding and clathrin binding appears more plausible. A pool of epsin bound to a ubiquitinated cargo may be recruited to clathrin-coated pits, for example, by EH domain endocytic proteins, but then dissociate from Ub, thus allowing other interactions of such cargo with coat components. Interestingly, a recent study demonstrated that epsin is critically needed for the function of the Notch receptor Delta (18, 19), and that this function involves not only the ubiquitination of Delta and its endocytosis, but also the targeting of internalized Delta to a specific endosomal subcompartment (19). Perhaps, epsin functions to recruit to clathrin-coated pits and a recycling endosomal compartment, a pool of ubiquitinated Delta that is otherwise internalized and targeted to lysosomes through a clathrin-independent pathway. Alternatively, epsin may participate in coated-pit assembly (11), and the subsequent ubiquitination of cargo accumulated at coated pits may induce epsin separation form the coat. This possibility is consistent with the lack of enrichment of epsin in purified clathrin-coated vesicle fractions (5). It is also possible that the interplay between clathrin and Ub may occur on the surface of endosomes, at the specialized clathrin coat that has been described on these structure. This coat is thought to play a role in the progression of membrane cargo into luminal vesicles of multivesicular bodies (47, 48). Normally epsin is not detectable at these sites, but a small undetectable pool of epsin on endosomes may be greatly amplified if epsin cannot interact with this clathrin coat.
Acknowledgments
We thank P. P. Di Fiore and S. Polo for communicating their results before publication and for discussion and M. Zerial and H. Stenmark for reagents. This work was supported in part by a Human Frontiers Science Program grant and grants from the National Institutes of Health (to P.D.C.).
Abbreviations: Ub, ubiquitin; UIM, Ub interaction motif; HA, hemagglutinin; PM, plasma membrane; RNAi, RNA interference; siRNA, small interfering RNA; ENTH, epsin N-terminal homology.
See Commentary on page 2679.
References
- 1.Hershko, A. & Ciechanover, A. (1998) Annu. Rev. Biochem. 67**,** 425–479. [DOI] [PubMed] [Google Scholar]
- 2.Hicke, L. & Dunn, R. (2003) Annu. Rev. Cell. Dev. Biol. 19**,** 141–172. [DOI] [PubMed] [Google Scholar]
- 3.Pickart, C. M. (2001) Annu. Rev. Biochem. 70**,** 503–533. [DOI] [PubMed] [Google Scholar]
- 4.Aguilar R. C. & Wendland, B. (2003) Curr. Opin. Cell Biol. 15**,** 184–190. [DOI] [PubMed] [Google Scholar]
- 5.Chen, H., Fre, S., Slepnev, V. I., Capua, M. R., Takei, K., Butler, M. H., Di Fiore, P. P. & De Camilli, P. (1998) Nature 394**,** 793–797. [DOI] [PubMed] [Google Scholar]
- 6.Polo, S., Sigismund, S., Faretta, M., Guidi, M., Capua, M. R., Bossi, G., Chen, H., De Camilli, P. & Di Fiore, P. P. (2002) Nature 416**,** 451–455. [DOI] [PubMed] [Google Scholar]
- 7.Wendland, B., Steece, K. E. & Emr, S. D. (1999) EMBO J. 18**,** 4383–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wendland, B. (2002) Nat. Rev. Mol. Cell Biol. 3**,** 971–977. [DOI] [PubMed] [Google Scholar]
- 9.Aguilar, R. C., Watson, H. A. & Wendland, B. (2003) J. Biol. Chem. 278**,** 10737–10743. [DOI] [PubMed] [Google Scholar]
- 10.Shih, S. C., Katzmann, D. J., Schnell, J. D., Sutanto, M., Emr, S. D. & Hicke, L. (2002) Nat. Cell Biol. 4**,** 389–393. [DOI] [PubMed] [Google Scholar]
- 11.Ford, M. G., Mills, I. G., Peter, B. J., Vallis, Y., Praefcke, G. J., Evans, P. R. & McMahon, H. T. (2002) Nature 419**,** 361–366. [DOI] [PubMed] [Google Scholar]
- 12.De Camilli, P., Chen, H., Hyman, J., Panepucci, E., Bateman, A. & Brunger, A. T. (2002) FEBS Lett. 513**,** 11–18. [DOI] [PubMed] [Google Scholar]
- 13.Polo, S., Confalonieri, S., Salcini, A. E. & Di Fiore, P. P. (2003) Science STKE, re17. [DOI] [PubMed]
- 14.Hofmann, K. & Falquet, L. (2001) Trends Biochem. Sci. 26**,** 347–350. [DOI] [PubMed] [Google Scholar]
- 15.Chen, H., Polo, S., Di Fiore, P. P. & De Camilli, P. V. (2003) Proc. Natl. Acad. Sci. USA 101, 14908–14913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cadavid, A. L., Ginzel, A. & Fischer, J. A. (2000) Development (Cambridge, U.K.) 127**,** 1727–1736. [DOI] [PubMed] [Google Scholar]
- 17.Chen, X., Zhang, B. & Fischer, J. A. (2002) Genes Dev. 16**,** 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Overstreet, E., Fitch, E. & Fischer, J. A. (2004) Development (Cambridge, U.K.) 131**,** 5355–5366. [DOI] [PubMed] [Google Scholar]
- 19.Wang, W. & Struhl, G. (2004) Development (Cambridge, U.K.) 131**,** 5367–5380. [DOI] [PubMed] [Google Scholar]
- 20.Sigismund, S., Woelk, T., Puri, C., Maspero, E., Tacchetti, C., Transidico, P., Di Fiore, P. P. & Polo, S. (2005) Proc. Natl. Acad. Sci. USA 102**,** 2760–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slepnev, V. I., Ochoa, G. C., Butler, M. H. & De Camilli, P. (2000) J. Biol. Chem. 275**,** 17583–17589. [DOI] [PubMed] [Google Scholar]
- 22.Motley, A., Bright, N. A., Seaman, M. N. & Robinson, M. S. (2003) J. Cell Biol. 162**,** 909–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, F., Khvorova, A., Marshall, W. & Sorkin, A. (2004) J. Biol. Chem. 279**,** 16657–16661. [DOI] [PubMed] [Google Scholar]
- 24.Kovarova, M., Tolar, P., Arudchandran, R., Draberova, L., Rivera, J. & Draber, P. (2001) Mol. Cell. Biol. 21**,** 8318–8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haglund, K., Sigismund, S., Polo, S., Szymkiewicz, I., Di Fiore, P. P. & Dikic, I. (2003) Nat. Cell Biol. 5**,** 461–466. [DOI] [PubMed] [Google Scholar]
- 26.Simonsen, A., Lippe, R., Christoforidis, S., Gaullier, J. M., Brech, A., Callaghan, J., Toh, B. H., Murphy, C., Zerial, M. & Stenmark, H. (1998) Nature 394**,** 494–498. [DOI] [PubMed] [Google Scholar]
- 27.Schnatwinkel, C., Christoforidis, S., Lindsay, M. R., Uttenweiler-Joseph, S., Wilm, M., Parton, R. G. & Zerial, M. (2004) PLoS Biol. 2**,** E261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shih, S. C., Sloper-Mould, K. E. & Hicke, L. (2000) EMBO J. 19**,** 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sloper-Mould, K. E., Jemc, J. C., Pickart, C. M. & Hicke, L. (2001) J. Biol. Chem. 276**,** 30483–30489. [DOI] [PubMed] [Google Scholar]
- 30.Kang, R. S., Daniels, C. M., Francis, S. A., Shih, S. C., Salerno, W. J., Hicke, L. & Radhakrishnan, I. (2003) Cell 113**,** 621–630. [DOI] [PubMed] [Google Scholar]
- 31.Hinshaw, J. E. (2000) Annu. Rev. Cell Dev. Biol. 16**,** 483–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Damke, H., Baba, T., Warnock, D. E. & Schmid, S. L. (1994) J. Cell Biol. 127**,** 915–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raiborg, C., Bremnes, B., Mehlum, A., Gillooly, D. J., D'Arrigo, A., Stang, E. & Stenmark, H. (2001) J. Cell Sci. 114**,** 2255–2263. [DOI] [PubMed] [Google Scholar]
- 34.Dowler, S., Currie, R. A., Campbell, D. G., Deak, M., Kular, G., Downes, C. P. & Alessi, D. R. (2000) Biochem. J. 351**,** 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Godi, A., Di Campli, A., Konstantakopoulos, A., Di Tullio, G., Alessi, D. R., Kular, G. S., Daniele, T., Marra, P., Lucocq, J. M. & De Matteis, M. A. (2004) Nat. Cell Biol. 6**,** 393–404. [DOI] [PubMed] [Google Scholar]
- 36.Katzmann, D. J., Babst, M. & Emr, S. D. (2001) Cell 106**,** 145–155. [DOI] [PubMed] [Google Scholar]
- 37.Bonifacino, J. S. (2004) Nat. Rev. Mol. Cell. Biol. 5**,** 23–32. [DOI] [PubMed] [Google Scholar]
- 38.Pelham, H. R. (2004) Curr. Biol. 14**,** R357–R359. [DOI] [PubMed] [Google Scholar]
- 39.Itoh, T., Koshiba, S., Kigawa, T., Kikuchi, A., Yokoyama, S. & Takenawa, T. (2001) Science 291**,** 1047–1051. [DOI] [PubMed] [Google Scholar]
- 40.Hyman, J., Chen, H., Di Fiore, P. P., De Camilli, P. & Brunger, A. T. (2000) J. Cell Biol. 149**,** 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenthal, J. A., Chen, H., Slepnev, V. I., Pellegrini, L., Salcini, A. E., Di Fiore, P. P. & De Camilli, P. (1999) J. Biol. Chem. 274**,** 33959–33965. [DOI] [PubMed] [Google Scholar]
- 42.Drake, M. T., Downs, M. A. & Traub, L. M. (2000) J. Biol. Chem. 275**,** 6479–6489. [DOI] [PubMed] [Google Scholar]
- 43.Rosse, C., L'Hoste, S., Offner, N., Picard, A. & Camonis, J. (2003) J. Biol. Chem. 278**,** 30597–30604. [DOI] [PubMed] [Google Scholar]
- 44.Yamabhai, M., Hoffman, N. G., Hardison, N. L., McPherson, P. S., Castagnoli, L., Cesareni, G. & Kay, B. K. (1998) J. Biol. Chem. 273**,** 31401–31407. [DOI] [PubMed] [Google Scholar]
- 45.Morinaka, K., Koyama, S., Nakashima, S., Hinoi, T., Okawa, K., Iwamatsu, A. & Kikuchi, A. (1999) Oncogene 18**,** 5915–5922. [DOI] [PubMed] [Google Scholar]
- 46.Chen, H., Slepnev, V. I., Di Fiore, P. P. & De Camilli, P. (1999) J. Biol. Chem. 274**,** 3257–3260. [DOI] [PubMed] [Google Scholar]
- 47.Raiborg, C., Bache, K. G., Gillooly, D. J., Madshus, I. H., Stang, E. & Stenmark, H. (2002) Nat. Cell Biol. 4**,** 394–398. [DOI] [PubMed] [Google Scholar]
- 48.Sachse, M., Urbe, S., Oorschot, V., Strous, G. J. & Klumperman, J. (2002) Mol. Biol. Cell 13**,** 1313–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]