Inflammasome complexes: emerging mechanisms and effector functions (original) (raw)
. Author manuscript; available in PMC: 2017 Jul 10.
Abstract
Canonical activation of the inflammasome is critical to promote caspase-1-dependent maturation of the proinflammatory cytokines IL-1β and IL-18, as well as to induce pyroptotic cell death in response to pathogens and endogenous danger signals. Recent discoveries, however, are beginning to unveil new components of the inflammasome machinery, and the full spectrum of inflammasome functions, extending their influence beyond canonical functions, to regulation of eicosanoid storm, autophagy and metabolism. In addition, the receptor components of the inflammasome can also regulate diverse biological processes, such as cellular proliferation, gene transcription and tumorigenesis, all of which are independent of their inflammasome complex-forming capabilities. Here, we review these recent advances that are shaping our understanding of the complex biology of the inflammasome and its constituents.
Inflammasome complex formation and canonical functions
Inflammasomes are large multimolecular complexes best known for their ability to control activation of the proteolytic enzyme caspase-1 (Martinon et al., 2002). Caspase-1 in turn regulates the proteolytic maturation of Interleukin-1β and IL-18, as well as a rapid, noxious, inflammatory form of cell death termed pyroptosis (Rathinam et al., 2012a). Assembly of inflammasome complexes is dependent on cytosolic sensing of pathogen-associated molecular patterns that gain access to the cytosol during microbial infection. In addition, endogenous danger signals (danger-associated molecular patterns) released from damaged or dying cells also activate inflammasomes and drive pathological inflammation in sterile inflammatory diseases including atherosclerosis, Alzheimer’s disease, diabetes and cancer (Latz et al., 2013).
Several distinct inflammasomes have been identified each differentiated by unique activators, NLR/ALR family members and caspase effectors. The classical or canonical inflammasome complex consists of a cytosolic sensor (which can be either a nucleotide binding domain and leucine-rich-repeat- containing (NLR) protein or a member of the AIM2 like receptor (ALR) family), an adaptor protein ASC and an effector caspase pro-caspase-1 (Moltke et al., 2013). ASC is a bipartite molecule that contains both an N-terminal pyrin domain (PYD) and a C-terminal caspase activation and recruitment domain (CARD), enabling it to bridge the sensors (NLRs or ALRs) and the effector pro-caspase-1. Pro-caspase-1 is subsequently activated leading to the cleavage of pro-IL-1β and pro-IL-18 and the generation of the mature biologically active cytokines.
Direct vs indirect activation of the inflammasomes
Depending on the NLR/ALR within the complex, inflammasomes are equipped with the ability to respond to a wide array of signals. Some of these signals act as direct ligands for NLR/ALR proteins and bind them leading to their oligomerization and activation. The NLR apoptosis inhibitory proteins (NAIP)/NLRC4 inflammasome directly recognizes bacterial flagellin and type III secretion system components (Kofoed and Vance, 2011; Zhao et al., 2011). In the case of AIM2, direct binding of double stranded DNA leads to inflammasome complex formation (Fernandes-Alnemri et al., 2009; Hornung et al., 2009; Roberts et al., 2009). Another ALR protein, IFI16, can also bind DNA and engage ASC leading to inflammasome activation (Kerur et al., 2011). IFI16 sensing of DNA and inflammasome dependent pyroptosis is critical during HIV infections as it contributes to the depletion of abortively infected CD4 T cells in lymphoid tissues and consequently, immunosuppression (Monroe et al., 2014). On the other hand, certain inflammasomes respond to cellular perturbations associated with microbial infections and tissue damage. Recently, the Pyrin inflammasome was shown to employ a novel mechanism to recognize bacterial infections. In this system, Pyrin detects bacterial modification of Rho GTPases. Various modifications of Rho GTPases namely glucosylation, adenylylation, ADP-ribosylation and deamidation at different amino acid residues by bacterial toxins such as Clostridium difficile TcdB and Clostridium botulinum C3 activate the pyrin inflammasome (Xu et al., 2014). The NLRP3 inflammasome responds to a multitude of signals of diverse physicochemical nature (e.g. ATP, potassium ionophores, particulate matters etc.) (Latz et al., 2013). However, the exact mechanism(s) by which NLRP3 is activated by these diverse ligands is surprisingly still unclear. There is no evidence that NLRP3 binds directly to any of its diverse activators. Although it has been shown that known NLRP3 stimuli converge on potassium ion efflux upstream of NLRP3 activation (Muñoz-Planillo et al., 2013), the exact molecular mechanism of NLRP3 activation still remains elusive.
A role for Nek7 in NLRP3 inflammasome activation
Very recently a new kinase Nek7 was identified as a central regulator of the NLRP3 inflammasome. Nek7 is a serine-threonine kinase previously known to be involved in mitosis. A genome-wide CRISPR/Cas9 screen in mouse macrophages found that the loss of Nek7 protected the cells from nigericin-induced pyroptosis (Schmid-Burgk et al., 2016). Similarly, a forward genetic screen employing _N_-ethyl-_N_-nitrosourea in C57BL/6J mice revealed that deficiency in Nek7 abolished nigericin-induced IL-1β secretion by macrophages (Shi et al., 2015a). Confirming these two genetic screens, a proteomic analysis of NLRP3-binding proteins using mass spectrometry identified Nek7 as an NLRP3-interacting protein (He et al., 2016). In response to NLRP3 activators such as nigericin and ATP, NEK7 interacts with the leucine-rich repeat domain of NLRP3 and forms a high molecular weight complex with NLRP3 facilitating ASC and caspase-1 oligomerization. However, the kinase activity of Nek7 was dispensable for the activation of NLRP3. In vivo, Nek7 promoted the NLRP3-dependent cellular inflammatory response to monosodium urate crystals and the development of experimental autoimmune encephalitis in mice (He et al., 2016; Shi et al., 2015a). These findings indicate that NEK7 represents a central hub upstream of NLRP3. Exactly how Nek7 is enlisted into the inflammasome pathway however is still an unresolved question.
Additional caspases and gasdermin D in inflammasome signaling
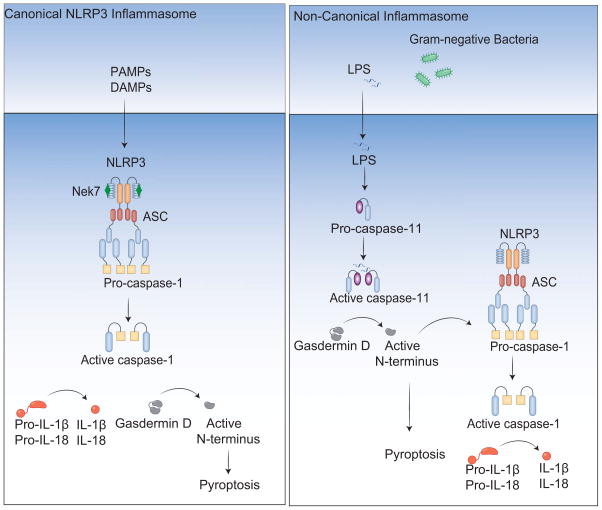
In addition to the canonical mode of inflammasome activation, additional caspases such as caspase-8 or caspase-11 (caspase 4/5 in humans), also contribute to inflammasome dependent control of IL-1β/IL18 processing and pyroptotic cell death. Caspase-8 is engaged downstream of several pathways including those triggered by dectin-1, Fas ligand, chemotherapeutic drugs, dsRNA engagement of TLR3 as well as certain classical activators of the NLRP3 inflammasome, where caspase-8 appears to serve a scaffolding function (Bossaller et al., 2012; Ganesan et al., 2014; Gurung et al., 2014; Karki et al., 2015; Lukens et al., 2014) Although caspase-8 can function as an IL-1β converting enzyme in certain contexts, in many of these cases, caspase-8 interacts with the NLRP3 inflammasome to modulate inflammasome effector functions (Man et al., 2013). Importantly, considerable progress in recent years has defined a non-canonical inflammasome pathway involving caspase-11, a caspase-1-like inflammatory protease (Kayagaki et al., 2011). Caspase-11 functions as a central regulator of non-canonical inflammasome activity during infection with Gram-negative bacterial pathogens that signal via NLRP3 (Rathinam et al., 2012b). Gram-negative bacterial pathogens require a licensing signal to facilitate NLRP3-dependent caspase-1 activation, cytokine maturation and pyroptotic cell death. This licensing signal involves transcriptional induction of caspase-11 and guanylate-binding proteins (GBPs) via TLR4-TRIF-type I IFN signaling (Broz et al., 2012; Gurung et al., 2012; Rathinam et al., 2012b). It is likely that additional interferon stimulated genes also contribute to these events. Studies over the last few years have also revealed that LPS from Gram-negative bacteria gains to access to the cytosol and signals independent of TLR4 by binding directly to caspase-11 (Hagar et al., 2013; Kayagaki et al., 2013; Shi et al., 2014). LPS binding to casapse-11 activates casapse-11 proteolytic activity leading to caspase-1-dependent IL-1β processing as well as a casapse-1-independent pyroptotic cell death. Latest developments in this area have shown that caspase-11 activation by intracellular LPS leads to cleavage of a new substrate gasdermin D. The resulting amino-terminal cleavage fragment of gasdermin D promotes NLRP3-dependent activation of caspase-1 and the demise of the cell via pyroptosis (He et al., 2015; Kayagaki et al., 2015; Shi et al., 2015). Collectively, all of these findings reveal different modes of inflammasome activation which in the case of NLRP3, the best-studied inflammasome has a canonical and non-canonical route of activation involving distinct inflammatory caspases. A schematic highlighting the advances in NLRP3 inflammasome signaling is shown in figure 1.
Figure 1. NLRP3 inflammasome activation: canonical and non-canonical modes of activation.
The canonical inflammasome pathway is triggered by multiple pathogens and inflammatory agents. Nek7 is recruited to the NLRP3 complex which recruits pro-caspase-1 monomers through the adaptor protein ASC, activating caspase-1. Caspase-1 processes the pro-inflammatory cytokine pro-IL-1β to generate mature IL-1β, which is presumably released by cell lysis during pyroptosis. Caspase-1 also initiates pyroptosis by cleaving gasdermin D. Murine caspase-11 (caspase-4 and caspase-5 in humans) oligomerizes upon binding with cytosolic LPS and becomes active. Active caspase-11 cleaves gasdermin D to drive pyroptosis and NLRP3 inflammasome-dependent cleavage of caspase-1 through an unknown mechanism.
Critical role of type I interferon signaling in inflammasome sensing of bacterial pathogens
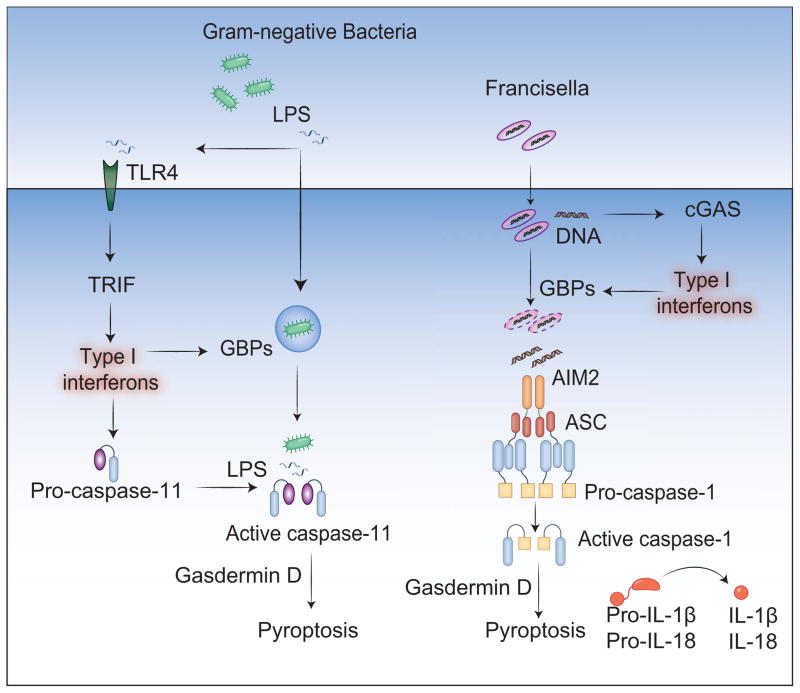
Type I interferons are integral to host innate and adaptive defenses against infectious organisms. Type I interferons are considered to inhibit NLRP3-dependent IL-1β response to DAMPs such as urate crystals and ATP primarily by inhibiting the transcription of pro-IL-1β (Guarda et al., 2011). However, a growing number of recent studies have unveiled a requirement for type I interferon signaling in enabling inflammasome activation by bacteria (Broz et al., 2012; Case et al., 2013; Casson et al., 2013; Henry et al., 2007; Jones et al., 2010; Man et al., 2015a; Meunier et al., 2014; 2015; Pilla et al., 2014; Rathinam et al., 2012b). During Gram-negative bacterial infections, type I interferon signaling sensitizes immune cells for cytosolic LPS and DNA sensing by enhancing the expression of the intracellular LPS receptor (caspase-11), GBPs, and potentially additional molecules (Figure 2) (Man et al., 2015a; Meunier et al., 2014; 2015; Rathinam et al., 2012b). Mechanistically, GBPs target the vacuolar and cytosolic bacteria and compromise the integrity of bacterial cells in a yet-to-be identified manner, thus exposing the microbial ligands LPS and DNA to caspase-11 and AIM2, respectively. GBPs are also considered to regulate yet-to-be identified event(s) subsequent to the entry of LPS into the cytosol (Pilla et al., 2014). In summary, emerging data point to the importance of type I interferon signaling in inflammasome surveillance of bacterial infections.
Figure 2. Essential roles of type I interferon signaling in inflammasome activation by bacteria.
Gram-negative bacterial infections lead to the activation of TLR4-TRIF-type I IFN pathway, which induces the expression of caspase-11 and GBPs. Type I interferon induction by Francisella via cGAS up regulates the expression of GBPs. Type I interferon-signaling aids in the cytosolic access of LPS and DNA leading to the activation of caspase-11 and AIM2, respectively.
Additional roles of Inflammasome complexes
While the ability of inflammasomes to control caspase-1 dependent maturation of IL-1β and IL-18 as well as cell death is well appreciated, several additional much less well-characterized effector functions of inflammasome complexes have also been described.
Unconventional Protein Secretion
Unlike most secretory proteins that contain aminoterminal or internal signal peptides to direct their sorting to the endoplasmic reticulum (ER) and subsequent release through the Golgi secretory pathway, IL-1β a leaderless cytoplasmic protein is secreted from cells by poorly defined mechanisms. This ER/Golgi-independent mechanism termed ‘unconventional protein secretion’ was shown to be dependent on caspase-1 activation. Although IL-1β cleavage and release are tightly coupled events whether caspase-1 plays a direct role in driving IL-1β release or if release occurs as a result of compromised membrane integrity which precedes cell death is still unclear. Early studies utilized mass spectrometry-based secretome analysis which demonstrated the caspase-1-mediated secretion of several leaderless proteins including caspase-1 itself as well as IL-1β, IL-1α, HMGB1 and fibroblast growth factor 2 (FGF2), several of which are not substrates of caspase-1 itself (Keller et al., 2008). The mechanisms and molecular components of this unconventional protein secretion pathway are still unclear. Multiple mechanisms have been proposed to facilitate the release of these leaderless proteins (Monteleone et al., 2015). IL-1β and caspase-1 are proposed to translocate into secretory lysosomes, a compartment that has features of both lysosomes and secretory granules. Upon activation of inflammasome complexes, secretory lysosomes then fuse with plasma membranes, resulting in the release of mature IL-1β and caspase-1 into the extracellular space. Alternative studies have suggested that IL-1β and caspase-1 are shed in microvesicles and released to the extracellular side of the plasma membrane or packed into multi vesicular bodies and released in exosomes (Nickel and Rabouille, 2009). More recent studies have defined the importance of autophagy in the unconventional secretion of IL-1β (Dupont et al., 2011). Autophagy induction cooperates with Golgi reassembly stacking protein (GRASP) proteins and Rab8a (a GTPase controlling post-Golgi polarized sorting and exocytosis) to regulate IL-1β secretion. Another Rab GTPase family member, Rab39a, itself a substrate of caspase-1 also plays a role in IL-1β secretion (Becker et al., 2009). All of these studies have analyzed secretory pathways downstream of caspase-1 in different cell types. It is possible that different mechanisms of unconventional secretion are operational in different cellular contexts. The recent discovery of gasdermin D as the executioner of pyroptotic cell death argues against a role for vesicle- and autophagy-dependent secretory pathways and instead favors a model whereby membrane rupture leads to IL-1β release. Indeed, gasdermin D-deficient cells fail to undergo membrane rupture and as a result cannot release IL-1β to the extracellular space despite having the ability to activate caspase-1 and induce maturation of IL-1β intracellularly (He et al., 2015; Kayagaki et al., 2015; Shi et al., 2015). Future studies are required to fully clarify and distinguish amongst these mechanisms.
Inflammasome induction of eicosanoids
An array of lipid molecules collectively known as lipid mediators are pivotal regulators of inflammatory responses (Shimizu, 2009). Eicosanoids, which comprise various lipid species including the well-studied prostaglandins, thromboxane, hydroxyeicosatetraenoic acid (HETEs) and leukotrienes, are bioactive molecules derived from membrane lipids. Eicosanoids play key roles in diverse homeostatic and pathological processes such as increasing vascular permeability and leukocyte recruitment (Dennis and Norris, 2015). Eicosanoid synthesis has been identified as a direct outcome of inflammasome activation (Moltke et al., 2012) (Figure 3); active caspase-1 generated by multiple inflammasomes such as NAIP/NLRC4 and NLRP1 stimulates rapid eicosanoid synthesis by the activation of cytosolic phospholipase A2. Surprisingly, this function of caspase-1 is independent of IL-1β, IL-18 and pyroptosis, the classic outcomes of inflammasome activation. Instead, active caspase-1 drives acute Ca2+ influx via membrane pore formation. Ca2+ influx in turn activates cytosolic phospholipase A2 (cPLA2) to generate arachidonic acid from membrane phospholipids. Arachidonic acid is ultimately converted to prostaglandins (PG) and thromboxanes by cyclooxygenases-1 (COX-1) and COX-2 and HETEs and leukotrienes by lipoxygenases (12/15-LOX and 5-LOX).
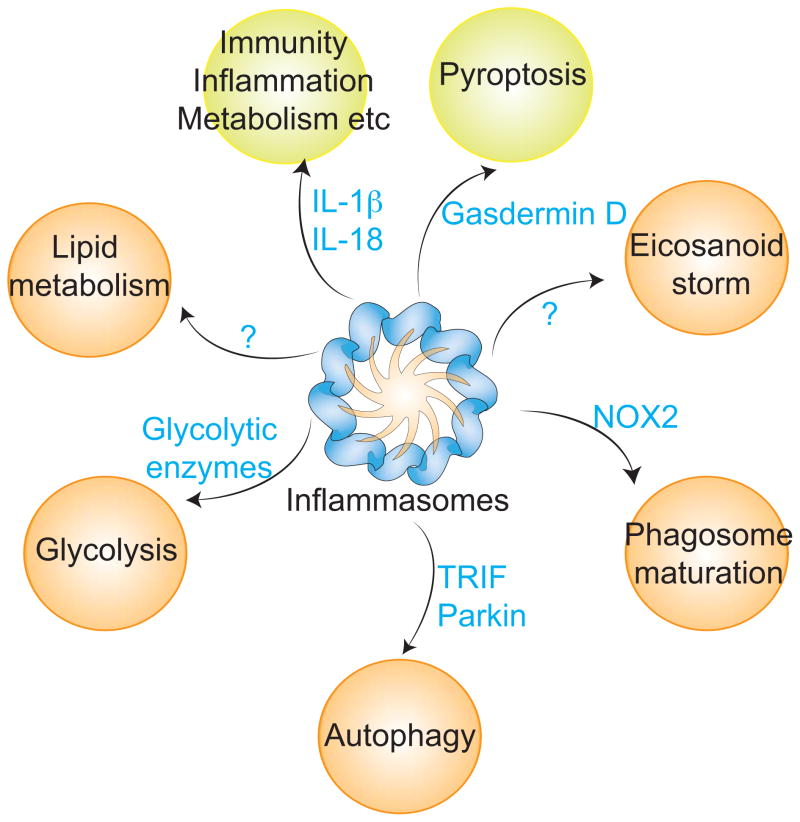
Figure 3. Novel functions of inflammasomes.
Inflammasome activation leads to the proteolytic activation of IL-1β, IL-18, and gasdermin D, the latter triggers inflammatory cell death, pyroptosis. In addition, caspase-1 activated by inflammasomes cleaves additional but partially known set of substrates such as Nox2, TRIF, parkin and thereby regulates a multitude of processes such as eicosanoid synthesis, phagosomal acidification, autophagy, glycolysis and lipid metabolism in an IL-1 cytokines- and pyroptosis-independent fashion. Green and orange circles represent the canonical and non-canonical functions of inflammasomes. Blue letters indicate caspase-1 substrates.
This caspase-1-mediated synthesis of eicosanoids, particularly PGE2, elicits a rapid and robust vasodilation, fluid loss from blood, hemoconcentration, diarrhea and hypothermia culminating in the death of mice. Consistently, mice lacking caspase-1, cPLA2, and Cox-1 are protected. Another unique feature of this ‘eicosanoid storm’ is its cellular origin; while inflammasome-mediated maturation of cytokines and cell death are common to multiple cell types only resident peritoneal macrophages but not thioglycollate elicited- or bone marrow-derived macrophages are equipped with the capability to synthesize and secrete substantial amounts of prostaglandins and leukotrienes in response to inflammasome triggers in vivo and ex vivo. It is believed that the high steady-state level expression of Cox1, Alox12/15 and Alox5 (enzymes involved in eicosanoid synthesis) in resident peritoneal macrophages likely accounts for these effects. Eicosanoid synthesis is also stimulated by the noncanonical caspase-11 inflammasome. The eicosanoid storm generated during caspase-11 mediated endotoxin shock contributes to host mortality, and the pharmacological inhibition of Cox-1 markedly improves host survival (Hagar et al., 2013). A localized, transient and controlled wave of eicosanoid production may be beneficial for the host to limit and eliminate foci of infection by recruiting phagocytic cells and complement components. On the other hand, sustained and systemic production of proinflammatory eicosanoids would be detrimental to the host. While the synthesis of proinflammatory eicosanoids as an output of inflammasome signaling is evident, whether caspase-1 activation can also stimulate the production of anti-inflammatory and proresolving lipid mediators is not yet known. Nonetheless, the eicosanoid storm downstream of inflammatory caspases expand the means by which inflammasomes can propagate inflammation and thus should be taken into consideration in developing new treatment measures for inflammatory disorders.
Autophagy and inflammasomes
Several recent studies have highlighted important cross talk between inflammasome activation and autophagy. Autophagy is a basic catabolic process that recycles cytosolic macromolecular structures and organelles (Ding et al., 2014). A multifaceted role for autophagy in the negative regulation of inflammasome activation is well established (Rathinam et al., 2012a). Autophagy of mitochondria, mitophagy, limits NLRP3 inflammasome activation by targeting damaged mitochondria for removal and deficiency of any of the key factors involved in this process such as LC3, Beclin, Ulk1, or Receptor interacting protein kinase 2 (RIP2) enhances caspase-1 activation (Levine et al., 2011; Lupfer et al., 2013; Nakahira et al., 2011; Rathinam et al., 2012a). Furthermore, autophagy mediates the removal of macromolecular inflammasome complexes to restrain caspase-1 activation (Shi et al., 2012). Interestingly, the antagonism between autophagy and inflammasomes is mutual; inflammasomes also display an inhibitory effect on autophagy itself. During Shigella flexneri infection of macrophages, NLRC4-mediated activation of caspase-1 suppresses autophagy and autophagosome formation (Suzuki et al., 2007). Similarly, TLR4- and TRIF-dependent stimulation of autophagy by Pseudomonas aeruginosa is also suppressed by the NLRC4 inflammasome (Jabir et al., 2014). Caspase-1 activated by the NLRC4 inflammasome cleaves TRIF at aspartic acid residues 286 (ILPDA) and 292 (AAPDT). Since TRIF is required for autophagy induction during Pseudomonas infection, caspase-1-mediated inactivation of TRIF down regulates autophagy. The suppression of autophagy by the inflammasome appears to favor P. aeruginosa replication in mice. Like the NLRC4 inflammasome, AIM2 and NLRP3 inflammasomes also negatively regulate autophagy. Active caspase-1 generated by the AIM2 and NLRP3 inflammasomes inactivates parkin, a key protein involved in mitophagy, by cleaving it at an aspartic acid residue (aa126) leading to the accumulation of damaged mitochondria (Yu et al., 2014). Autophagy is classically considered as a salvaging process whereas inflammasome activation invariably leads to the rapid demise of the cell. Therefore, the balance between autophagy and inflammasome activity during sterile inflammation and infection perhaps guides a cell’s decision to survive or die.
An exception to this suppressive effect of inflammasomes on autophagy is the NLRP6 inflammasome in the intestine. The NLRP6 inflammasome is important in maintaining intestinal homeostasis (Elinav et al., 2011). Microbial metabolites such as taurine regulate the activation of the NLRP6 inflammasome which in turn shapes the composition of the microbiome favoring a beneficial microbiome interface at the gut mucosa (Levy et al., 2015). An effector mechanism downstream of NLRP6 inflammasome that contribute to its homeostatic function is autophagy. Specifically, the NLRP6 inflammasome expressed in goblet cells promotes autophagy, which ensures optimal mucus secretion in the gut by facilitating goblet cell mucin granule exocytosis (Wlodarska et al., 2014). The precise molecular basis of NLRP6 inflammasome regulation of autophagy, particularly if it is direct, is yet to be determined. Nonetheless, these findings demonstrate that the relationship between inflammasomes and autophagy is not universally antagonistic but can be cell type-, tissue-, context-, and potentially inflammasome type-specific.
Regulation of phagosome maturation by inflammasomes
Phagocytosis is one of the most important anti-microbial defense mechanisms. The antimicrobial function of the phagosome—vesicles containing phagocytosed cargo—is conferred by the pH-sensitive degradative enzymes derived upon fusion with lysosomes. This process termed phagosome maturation relies on the acidification of phagosomes as they move from the plasma membrane towards lysosomes. The lowering of pH in the phagosomal lumen favors the activation of proteolytic and lipolytic enzymes. Phagosome acidification and maturation is a complex process involving delicate spatio-temporal interactions among several cellular proteins including small GTPases. A new addition to this growing list of regulators of phagosome maturation is inflammasomes (Sokolovska et al., 2013). The acidification of phagosomes with the cargo of Gram-positive bacteria such as Staphylococcus aureus and Group B streptococcus requires NLRP3 inflammasome activation. In this context, the activation of caspase-1 by the NLRP3 inflammasome occurs immediately upon phagocytosis of S. aureus. Such a rapid activation of NLRP3 seems to be driven by events associated with the phagocytic process itself including but not limited to the production of reactive oxygen species (ROS). Interestingly, the active caspase-1 molecule accumulates on phagosomes containing Gram-positive bacteria but not on phagosomes where the cargo is latex beads. This cargo-dependent accrual of active caspase-1 on phagosomes is important for subsequent lowering of the pH in the phagosomal lumen. Active caspase-1 targets the phagocyte NADPH oxidase (NOX2) complex, which assembles on the phagosomal membrane and generates superoxide anions in the phagosomal lumen causing alkalinization. Caspase-1 proteolytically inactivates one or more components of the transmembrane NOX2 complex such as gp91 and Rac1 and thus accelerates the rate of acidification of phagosomes. Consequently, the NLRP3 inflammasome facilitates antimicrobial activities of the phagosome leading to effective bacterial killing. Furthermore, the enhancement of the degradative capacity of phagosomes by the NLRP3-caspase-1 axis adversely impacts antigen cross presentation and CD8 T cell activation.
Inflammatory caspases regulate phagosome maturation at more than one stage. Caspase-11 in mice and its orthologues in humans (caspase-4 and caspase-5) mediate the fusion of _Legionella pneumophila_-containing phagocytic vacuoles with lysosomes by regulating the F-actin network assembly (Akhter et al., 2012). Surprisingly, caspase-11 deficiency does not impact the trafficking of phagosomes with nonpathogenic bacteria such as E. coli DH5α. L. pneumophila actively interferes with endosomal trafficking via its type IV secretion system effector proteins to prevent fusion of endosomes with lysosomes. Therefore it can be speculated that caspase-11 aids the host in overcoming such bacterial evasion mechanisms to ensure effective killing of intracellular bacteria.
Caspase-1 modulation of metabolism
An exciting theme emerging from the last decade of research is that inflammation and metabolism are more intimately linked than previously thought. While recent evidence indicates that the metabolic status of a cell is a critical determinant of the type and magnitude of the inflammatory response (O’Neill and Hardie, 2013), chronic inflammation is a risk factor underlying several metabolic disorders such as obesity and diabetes. Inflammasomes modulate carbohydrate and lipid metabolism via several mechanisms. The cytokine output of inflammasomes, specifically IL-1β, is well documented to play critical roles in diabetes and atherosclerosis. Nonetheless, inflammasomes control metabolic processes in an IL-1 cytokine-independent manner. An in vitro caspase-1 cleavage assay combined with the proteomic approach to identify substrates of caspase-1 revealed multiple proteins in the glycolysis pathway including aldolase, triose-phosphate isomerase, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), α-enolase, and pyruvate kinase as caspase-1 substrates (Shao et al., 2007). Indeed, several of these proteins were cleaved by caspase-1 during infection with Salmonella enterica serovar Typhimurium, which activates the NLRC4 inflammasome. As a result, the glycolytic rate was blunted in macrophages with active inflammasomes. This perhaps plays a role in making the intracellular microenvironment less hospitable for pathogens that persist inside the cells such as Salmonella. Even more importantly, the processing or degradation of GAPDH, aldolase and α-enolase was observed in diaphragm muscles of mice during endotoxic shock, which could compromise the contractibility of respiratory muscles and contribute to the respiratory failure in septic shock patients. These findings highlight a critical point from a therapeutic perspective that targeting the IL-1 family of cytokines alone during inflammasome-driven disorders such as sepsis might not be sufficient for a beneficial effect.
In addition to modulating glycolysis, inflammasomes also regulate lipid homeostasis. Caspase-1 activation by a bacterial pore forming toxin namely aerolysin in chinese hamster ovary (CHO) cells leads to the activation of sterol regulatory element binding proteins (SREBPs). SREBPs are endoplasmic reticulum (ER)-resident proteins that become active in cholesterol-poor conditions driving the transcription of cholesterol and fatty-acid biosynthetic genes (Gurcel et al., 2006). The stimulation of SREBPs by caspase-1 via an unknown mechanism leads to lipid biosynthesis, which eventually contributes to the repair of membrane damage induced by aerolysin. Triglyceride (TG) metabolism is another aspect of lipid homeostasis regulated by caspase-1. Caspase-1 was previously shown to play a role in TG absorption in the intestine and their production in the liver (van Diepen et al., 2013). However, recent evident suggest that caspase-1 facilitates the clearance of triglycerides from the plasma during fasting conditions as well as after the uptake of a lipid bolus (Kotas et al., 2013). As a result, lipid metabolism is substantially dysregulated in caspase-1-deficient mice. Although caspase-1 activation in these conditions is driven by the NLRP3 inflammasome, the effect of caspase-1 on TG metabolism is independent of IL-1 and IL-18 cytokines again implying the involvement of as yet-to-be identified mechanism(s).
Inflammasome complex-independent functions of NLR/ALR proteins
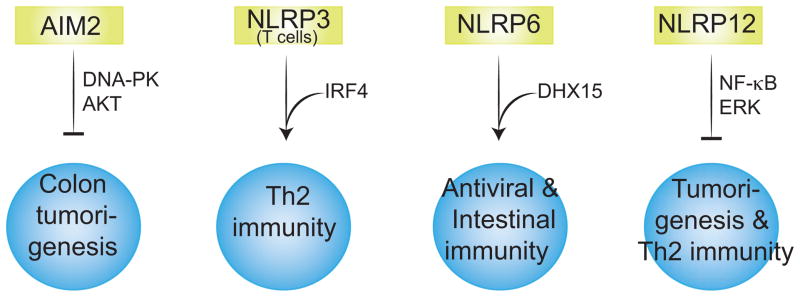
While all of the mechanisms described above are novel functions of inflammasome complexes, certain inflammasome components also appear to have biological functions that are independent of their capacity to form inflammasome complex and to activate caspase-1 and IL-1β/IL-18. Specifically, AIM2, NLRP3, NLRP6 and NLRP12 have all been linked to biological responses that appear to be independent of the ability of these proteins to engage ASC/Caspase-1 (Figure 4).
Figure 4. The inflammasome-independent biological roles of ALR and NLR proteins.
AIM2 controls intestinal stem cell proliferation and colon tumorigenesis by inhibiting DNA-PK-dependent AKT activation. NLRP3 interacts with IRF4 and acts as a transcriptional factor to regulate Th2 gene expression. NLRP6 functions both as a negative regulator of anti-bacterial immunity and a sensor of viral RNA. NLRP12 inhibits NF-κB and ERK signaling and thus colorectal tumor development and Th2 immunity.
AIM2 control of tumorigenesis
AIM2, which forms a caspase-1 activating inflammasome in response to cytosolic DNA, has recently been linked to prevention of colorectal cancer (Man et al., 2015b; Wilson et al., 2015). It was known that mutations in AIM2 were associated with colorectal cancer in humans. Two recent studies demonstrated that Aim2-deficient mice had increased susceptibility to colonic carcinogenesis. Unexpectedly, this tumor suppressive effect of AIM2 was independent of its inflammasome complex-forming capacity and inflammasome-associated cytokines. Rather, the mechanism driving AIM2-dependent protection against tumorigenesis was dependent on the ability of AIM2 to suppress proliferation of intestinal epithelial cells. Furthermore, AIM2 limits DNA-dependent protein kinase-mediated activation of Akt, a critical regulator of cell survival and proliferation (Wilson et al., 2015). In addition, the gut microbiota seems to also play a role in the hyper susceptibility of Aim2-deficient mice to colorectal tumorigenesis (Man et al., 2015b). Together, these findings reveal an important role for AIM2 in determining susceptibility to colorectal cancer and highlight inflammasome-independent roles for this DNA binding protein.
NLRP3, NLRP6 and NLRP12 regulation of innate and T cell immunity
An inflammasome-independent function has also recently been proposed for the best-studied NLRP3 inflammasome (Bruchard et al., 2015). NLRP3 was found to be expressed in differentiated T helper cells subsets and specifically control Th2 cell differentiation. Unlike myeloid cells where NLRP3 is cytosolic, NLRP3 is localized to the nucleus in T cells and lacks inflammasome-forming capabilities. Rather, NLRP3 interacts with the transcription factor IRF4 and binds to the promoter regions of Th2 cytokine genes to induce their expression. Consistent with a role for NLRP3 in coordinating Th2 cell target gene expression, NLRP3 regulates asthma and Th2 cell-dependent control of melanoma growth (Bruchard et al., 2015).
NLRs also regulate signaling pathways via inflammasome-independent means. For example, NLRP6 was shown to negatively regulate mitogen-activated protein kinase (MAPK) and the canonical NF-κB pathway in response to Toll-like receptor, but not NOD1/2 signaling. Accordingly, NLRP6 reduced the production of NF-κB- and MAPK-dependent cytokines and chemokines leading to enhanced susceptibility to infection with bacterial pathogens (Anand et al., 2012). Most recently, NLRP6 has also been shown to function as a sensor for enteric viruses (Wang et al., 2015). NLRP6 binds viral RNA via an RNA helicase DHX15 triggering the expression of type I/III interferons and interferon-stimulated genes (ISGs). Consequently, NLRP6 contributed to the control of encephalomyocarditis virus and norovirus infection of the gastrointestinal tract.
Several studies have also defined anti-inflammatory functions for NLRP12 that are independent of inflammasome functions. NLRP12 dampens NF-κB and ERK activation, in the absence of which mice are highly susceptible to colon inflammation and colorectal tumor development (Allen et al., 2012; Zaki et al., 2011). NLRP12 also limits noncanonical NF-κB signaling in osteoclast precursors and down regulates osteoclastogenesis thus altering bone homeostasis (Krauss et al., 2015). NLRP12 has also been shown to function in neutrophils as a negative regulator of cell migration in vitro (Zamoshnikova et al., 2016). Finally, an unexpected role for NLRP12 as a negative regulator of T-cell-mediated IL-4 production has also been defined (Lukens et al., 2015). Taken together, all of these emerging data reveal much broader roles for NLR and ALR members in different immune cells beyond inflammasome activation.
Conclusions and future directions
A solid body of evidence from in vitro and in vivo studies over the last several years has clearly positioned IL-1β, IL-18 as well as pyroptotic cell death as central mediators of inflammasome-driven biological responses. Recent studies, however, have also provided compelling evidence for the existence of additional effector mechanisms downstream of inflammasome complexes. In addition, new functions for some NLRs and AIM2 have been described which broaden our understanding of the functions of these proteins. These novel pathways as described in this Review play crucial roles not only in infection and sterile inflammatory conditions but also in controlling the cellular homeostasis. Such a diversification of inflammasome outcomes could be due to the many substrates available for the inflammatory caspases. The limited number of currently known substrates of caspase-1 and -11 could very well be the ‘tip of the iceberg’. Therefore, the identification and characterization of novel inflammasome substrates as well as further understanding of the inflammasome-independent roles of NLRs and ALRs would be of great biological as well as therapeutic interest as it would guide the design of the next generation of drugs to treat infectious and inflammatory diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akhter A, Caution K, Abu Khweek A, Tazi M, Abdulrahman BA, Abdelaziz DHA, Voss OH, Doseff AI, Hassan H, Azad AK, et al. Caspase-11 promotes the fusion of phagosomes harboring pathogenic bacteria with lysosomes by modulating actin polymerization. Immunity. 2012;37:35–47. doi: 10.1016/j.immuni.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand PK, Malireddi RKS, Lukens JR, Vogel P, Bertin J, Lamkanfi M, Kanneganti TD. NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature. 2012;488:389–393. doi: 10.1038/nature11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker CE, Creagh EM, O’Neill LAJ. Rab39a binds caspase-1 and is required for caspase-1-dependent interleukin-1beta secretion. J Biol Chem. 2009;284:34531–34537. doi: 10.1074/jbc.M109.046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossaller L, Chiang PI, Schmidt-Lauber C, Ganesan S, Kaiser WJ, Rathinam VAK, Mocarski ES, Subramanian D, Green DR, Silverman N, et al. Cutting edge: FAS (CD95) mediates noncanonical IL-1β and IL-18 maturation via caspase-8 in an RIP3-independent manner. The Journal of Immunology. 2012;189:5508–5512. doi: 10.4049/jimmunol.1202121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P, Ruby T, Belhocine K, Bouley DM, Kayagaki N. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature. 2012;490:288–291. doi: 10.1038/nature11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case CL, Kohler LJ, Lima JB, Strowig T, de Zoete MR, Flavell RA, Zamboni DS, Roy CR. Caspase-11 stimulates rapid flagellin-independent pyroptosis in response to Legionella pneumophila. Proceedings of the National Academy of Sciences. 2013;110:1851–1856. doi: 10.1073/pnas.1211521110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson CN, Copenhaver AM, Zwack EE, Nguyen HT, Strowig T, Javdan B, Bradley WP, Fung TC, Flavell RA, Brodsky IE, et al. Caspase-11 activation in response to bacterial secretion systems that access the host cytosol. PLoS Pathog. 2013;9:e1003400. doi: 10.1371/journal.ppat.1003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Liu S, Wang X, Dai Y, Khaidakov M, Deng X, Fan Y, Xiang D, Mehta JL. LOX-1, mtDNA damage, and NLRP3 inflammasome activation in macrophages: implications in atherogenesis. Cardiovasc Res. 2014:cvu114. doi: 10.1093/cvr/cvu114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. Embo J. 2011;30:4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, et al. NLRP6 Inflammasome Regulates Colonic Microbial Ecology and Risk for Colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan S, Rathinam VAK, Army K, Kaiser WJ, Mocarski ES, Dillon CP, Green DR, Mayadas TN, Levitz SM, Hise AG, et al. Caspase-8 Modulates Dectin-1 and Complement Receptor 3-Driven IL-1β Production in Response to β-Glucans and the Fungal Pathogen, Candida albicans. The Journal of Immunology. 2014;193:2519–2530. doi: 10.4049/jimmunol.1400276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarda G, Braun M, Staehli F, Tardivel A, Mattmann C, Förster I, Farlik M, Decker T, Pasquier Du RA, Romero P, et al. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34:213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Gurcel L, Abrami L, Girardin S, Tschopp J, van der Goot FG. Caspase-1 Activation of Lipid Metabolic Pathways in Response to Bacterial Pore-Forming Toxins Promotes Cell Survival. Cell. 2006;126:1135–1145. doi: 10.1016/j.cell.2006.07.033. [DOI] [PubMed] [Google Scholar]
- Gurung P, Anand PK, Malireddi RKS, Vande Walle L, Van Opdenbosch N, Dillon CP, Weinlich R, Green DR, Lamkanfi M, Kanneganti TD. FADD and caspase-8 mediate priming and activation of the canonical and noncanonical Nlrp3 inflammasomes. The Journal of Immunology. 2014;192:1835–1846. doi: 10.4049/jimmunol.1302839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung P, Malireddi RKS, Anand PK, Demon D, Vande Walle L, Liu Z, Vogel P, Lamkanfi M, Kanneganti TD. Toll or interleukin-1 receptor (TIR) domain-containing adaptor inducing interferon-β (TRIF)-mediated caspase-11 protease production integrates Toll-like receptor 4 (TLR4) protein- and Nlrp3 inflammasome-mediated host defense against enteropathogens. J Biol Chem. 2012;287:34474–34483. doi: 10.1074/jbc.M112.401406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science. 2013;341:1250–1253. doi: 10.1126/science.1240988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W-T, Wan H, Hu L, Chen P, Wang X, Huang Z, Yang Z-H, Zhong C-Q, Han J. Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res. 2015;25:1285–1298. doi: 10.1038/cr.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J Exp Med. 2007;204:987–994. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabir MS, Ritchie ND, Li D, Bayes HK, Tourlomousis P, Puleston D, Lupton A, Hopkins L, Simon AK, Bryant C, et al. Caspase-1 Cleavage of the TLR Adaptor TRIF Inhibits Autophagy and β-Interferon Production during Pseudomonas aeruginosa Infection. Cell Host & Microbe. 2014;15:214–227. doi: 10.1016/j.chom.2014.01.010. [DOI] [PubMed] [Google Scholar]
- Jones JW, Kayagaki N, Broz P, Henry T, Newton K, O’Rourke K, Chan S, Dong J, Qu Y, Roose-Girma M, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proceedings of the National Academy of Sciences. 2010;107:9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki R, Man SM, Malireddi RKS, Gurung P, Vogel P, Lamkanfi M, Kanneganti TD. Concerted activation of the AIM2 and NLRP3 inflammasomes orchestrates host protection against Aspergillus infection. Cell Host & Microbe. 2015;17:357–368. doi: 10.1016/j.chom.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signaling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, Miyake K, Zhang J, Lee WP, Muszynski A, et al. Noncanonical Inflammasome Activation by Intracellular LPS Independent of TLR4. Science. 2013;341:1246–1249. doi: 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]
- Keller M, Rüegg A, Werner S, Beer HD. Active Caspase-1 Is a Regulator of Unconventional Protein Secretion. Cell. 2008;132:818–831. doi: 10.1016/j.cell.2007.12.040. [DOI] [PubMed] [Google Scholar]
- Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. IFI16 Acts as a Nuclear Pathogen Sensor to Induce the Inflammasome in Response to Kaposi Sarcoma-Associated Herpesvirus Infection. Cell Host & Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477:592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotas ME, Jurczak MJ, Annicelli C, Gillum MP, Cline GW, Shulman GI, Medzhitov R. Role of caspase-1 in regulation of triglyceride metabolism. Proceedings of the National Academy of Sciences. 2013;110:4810–4815. doi: 10.1073/pnas.1301996110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nature Reviews Immunology. 2013;13:397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M, Thaiss CA, Zeevi D, Dohnalová L, Zilberman-Schapira G, Mahdi JA, David E, Savidor A, Korem T, Herzig Y, et al. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell. 2015;163:1428–1443. doi: 10.1016/j.cell.2015.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukens JR, Gurung P, Vogel P, Johnson GR, Carter RA, McGoldrick DJ, Bandi SR, Calabrese CR, Vande Walle L, Lamkanfi M, et al. Dietary modulation of the microbiome affects autoinflammatory disease. Nature. 2014;516:246–249. doi: 10.1038/nature13788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupfer C, Thomas PG, Anand PK, Vogel P, Milasta S, Martinez J, Huang G, Green M, Kundu M, Chi H, et al. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nature Immunology. 2013;14:480–488. doi: 10.1038/ni.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Karki R, Malireddi RKS, Neale G, Vogel P, Yamamoto M, Lamkanfi M, Kanneganti TD. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nature Immunology. 2015a;16:467–475. doi: 10.1038/ni.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Zhu Q, Zhu L, Liu Z, Karki R, Malik A, Sharma D, Li L, Malireddi RKS, Gurung P, et al. Critical Role for the DNA Sensor AIM2 in Stem Cell Proliferation and Cancer. Cell. 2015b;162:45–58. doi: 10.1016/j.cell.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- Meunier E, Dick MS, Dreier RF, Schürmann N, Kenzelmann Broz D, Warming S, Roose-Girma M, Bumann D, Kayagaki N, Takeda K, et al. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature. 2014;509:366–370. doi: 10.1038/nature13157. [DOI] [PubMed] [Google Scholar]
- Meunier E, Wallet P, Dreier RF, Costanzo S, Anton L, Rühl S, Dussurgey S, Dick MS, Kistner A, Rigard M, et al. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nature Immunology. 2015;16:476–484. doi: 10.1038/ni.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J, Ayres JS, Kofoed EM, Chavarría-Smith J, Vance RE. Recognition of bacteria by inflammasomes. Annu Rev Immunol. 2013;31:73–106. doi: 10.1146/annurev-immunol-032712-095944. [DOI] [PubMed] [Google Scholar]
- von Moltke J, Trinidad NJ, Moayeri M, Kintzer AF, Wang SB, van Rooijen N, Brown CR, Krantz BA, Leppla SH, Gronert K, et al. Rapid induction of inflammatory lipid mediators by the inflammasome in vivo. Nature. 2012;490:107–111. doi: 10.1038/nature11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe KM, Yang Z, Johnson JR, Geng X, Doitsh G, Krogan NJ, Greene WC. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. 2014;343:428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteleone M, Stow JL, Schroder K. Mechanisms of unconventional secretion of IL-1 family cytokines. Cytokine. 2015;74:213–218. doi: 10.1016/j.cyto.2015.03.022. [DOI] [PubMed] [Google Scholar]
- Muñoz-Planillo R, Kuffa P, Martínez-Colón G, Smith BL, Rajendiran TM, Núñez G. K+ Efflux Is the Common Trigger of NLRP3 Inflammasome Activation by Bacterial Toxins and Particulate Matter. Immunity. 2013;38:1142–1153. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VAK, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nature Immunology. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel W, Rabouille C. Mechanisms of regulated unconventional protein secretion. Nat Rev Mol Cell Biol. 2009;10:148–155. doi: 10.1038/nrm2617. [DOI] [PubMed] [Google Scholar]
- O’Neill LAJ, Hardie DG. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. 2013;493:346–355. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- Pilla DM, Hagar JA, Haldar AK, Mason AK, Degrandi D, Pfeffer K, Ernst RK, Yamamoto M, Miao EA, Coers J. Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proceedings of the National Academy of Sciences. 2014;111:6046–6051. doi: 10.1073/pnas.1321700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VAK, Vanaja SK, Fitzgerald KA. Regulation of inflammasome signaling. Nature Immunology. 2012a;13:333–332. doi: 10.1038/ni.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VAK, Vanaja SK, Waggoner L, Sokolovska A, Becker C, Stuart LM, Leong JM, Fitzgerald KA. TRIF Licenses Caspase-11-Dependent NLRP3 Inflammasome Activation by Gram-Negative Bacteria. Cell. 2012b;150:606–619. doi: 10.1016/j.cell.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, et al. HIN-200 Proteins Regulate Caspase Activation in Response to Foreign Cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- Shao W, Yeretssian G, Doiron K, Hussain SN, Saleh M. The caspase-1 digestome identifies the glycolysis pathway as a target during infection and septic shock. J Biol Chem. 2007;282:36321–36329. doi: 10.1074/jbc.M708182200. [DOI] [PubMed] [Google Scholar]
- Shi CS, Shenderov K, Huang NN, Kabat J. Activation of autophagy by inflammatory signals limits IL-1 [beta] production by targeting ubiquitinated inflammasomes for destruction. Nature. 2012;13:255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, Shao F. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–192. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- Sokolovska A, Becker CE, Ip WKE, Rathinam VAK, Brudner M, Paquette N, Tanne A, Vanaja SK, Moore KJ, Fitzgerald KA, et al. Activation of caspase-1 by the NLRP3 inflammasome regulates the NADPH oxidase NOX2 to control phagosome function. Nature Immunology. 2013;14:543–553. doi: 10.1038/ni.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Franchi L, Toma C, Ashida H, Ogawa M, Yoshikawa Y, Mimuro H, Inohara N, Sasakawa C, Núñez G. Differential Regulation of Caspase-1 Activation, Pyroptosis, and Autophagy via Ipaf and ASC in Shigella-Infected Macrophages. PLoS Pathog. 2007;3:e111. doi: 10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Diepen JA, Stienstra R, Vroegrijk IOCM, van den Berg SAA, Salvatori D, Hooiveld GJ, Kersten S, Tack CJ, Netea MG, Smit JWA, et al. Caspase-1 deficiency in mice reduces intestinal triglyceride absorption and hepatic triglyceride secretion. J Lipid Res. 2013;54:448–456. doi: 10.1194/jlr.M031963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Petrucelli AS, Chen L, Koblansky AA, Truax AD, Oyama Y, Rogers AB, Brickey WJ, Wang Y, Schneider M, et al. Inflammasome-independent role of AIM2 in suppressing colon tumorigenesis via DNA-PK and Akt. Nat Med. 2015;21:906–913. doi: 10.1038/nm.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M, Thaiss CA, Nowarski R, Henao-Mejia J, Zhang JP, Brown EM, Frankel G, Levy M, Katz MN, Philbrick WM, et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 2014;156:1045–1059. doi: 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Yang J, Gao W, Li L, Li P, Zhang L, Gong YN, Peng X, Xi JJ, Chen S, et al. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature. 2014;513:237–241. doi: 10.1038/nature13449. [DOI] [PubMed] [Google Scholar]
- Yu J, Nagasu H, Murakami T, Hoang H, Broderick L, Hoffman HM, Horng T. Inflammasome activation leads to Caspase-1-dependent mitochondrial damage and block of mitophagy. Proceedings of the National Academy of Sciences. 2014;111:15514–15519. doi: 10.1073/pnas.1414859111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H, Liu L, Shao F. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]