Long non-coding RNAs, ASAP1-IT1, FAM215A, and LINC00472, in epithelial ovarian cancer (original) (raw)
. Author manuscript; available in PMC: 2017 Dec 1.
Published in final edited form as: Gynecol Oncol. 2016 Sep 23;143(3):642–649. doi: 10.1016/j.ygyno.2016.09.021
Abstract
Objective
Long non-coding RNAs (lncRNAs) are a class of non-protein coding transcripts that has gained significant attention lately due to their important biological actions and potential involvement in cancer. Ovarian cancer is a devastating disease with poor prognosis, and our understanding of lncRNA's involvement in the malignancy is limited. To further our knowledge, we measured the expression of three lncRNAs, ASAP1-IT1, FAM215A, and LINC00472, in tumor samples, and analyzed their associations with disease characteristics and patient survival.
Methods
Two hundred sixty-six patients diagnosed with primary epithelial ovarian cancers were recruited for the study. Fresh-frozen tumor samples were obtained from the patients at tumor resection and analyzed by RT-qPCR for expression of ASAP1-IT1, FAM215A, and LINC00472. Associations of lncRNA expression with patient survival were determined using Cox proportional hazards regression models.
Results
We observed high expression of ASAP1-IT1, FAM215A and LINC00472 more frequently in low grade tumors and early stage disease compared to high grade tumors and late stage disease, respectively. High expression of ASAP1-IT1 and FAM215A were associated with favorable overall survival, and the survival association with ASAP1-IT1 was independent of tumor grade and disease stage. Analyses of online data also demonstrated similar survival associations with ASAP1-IT1 and FAM215A, suggesting that these lncRNAs may be involved in ovarian cancer progression.
Conclusions
LncRNAs may play appreciable roles in ovarian cancer and more research is needed to elucidate their biological mechanisms and clinical implications in tumor characterization as well as disease prognosis and treatment.
Keywords: ASAP1-IT1, FAM215A, LNC00472, ASAP1, Ovarian cancer, Prognosis
1 Introduction
Ovarian cancer is the most lethal gynecological malignancy, attributable to 5% of female cancer deaths in the US [1]. >50% of ovarian cancer patients succumb to the tumor within 5 years of diagnosis. It is believed that ovarian cancer survival may be significantly improved if the disease can be detected early when the tumors are still confined to the ovaries [1]. Since only a small percentage of patients are diagnosed with localized disease, a means that allows detection of ovarian cancer at early stages is urgently needed. Further elucidating the molecular features of ovarian cancer may help to achieve this goal. Previous knowledge of ovarian cancer biology has been largely centered on proteins and their coding genes. Since only 2% of the human genome encodes proteins [2], our understanding of ovarian cancer from the genome perspective is quite limited. It is now known that >90% of the genome is transcribed into RNAs, and a majority of them are non-coding RNAs. Many of these non-coding RNAs are biologically functional, and are involved in regulation of cell activities and functions. Dysregulation of non-coding RNAs may play an important role in various pathogenic processes of human diseases including cancer [3–5].
Non-coding RNAs with sequences of 200 nucleotides or more are called long non-coding RNAs (lncRNAs) [6]. In ovarian cancer initiation and progression, little is known about the role of lncRNAs. In this report, we studied the expression of three lncRNAs, ASAP1-IT1, FAM215A and LINC00472, in primary epithelial ovarian cancer, and analyzed their relationships with tumor characteristics and disease outcomes. These lncRNAs were selected for study either because our previous investigations provided evidence of its potential involvement in cancer (LINC00472) or our analyses using the Kaplan-Meier Plotter (http://kmplot.com/analysis/) of online databases suggested their possible associations with ovarian cancer survival (ASAP1-IT1, FAM215A) [7]. LINC00472 is a long intergenic non-coding RNA located on chromosome 6q13. Our recent studies revealed that this lincRNA may be associated with tumor suppression in breast cancer [8, 9]. ASAP1-IT1 is an intronic transcript of the ASAP1 (AMAP1; DDEF1) gene which encodes ASAP1, an ADP-ribosylation factor (ARF) GTPase-activating protein involved in membrane trafficking and cytoskeleton remodeling [10, 11]. Reports have suggested that ASAP1 is associated with tumor metastasis and poor cancer survival [12–14]. LncRNA ASAP1-IT1 may antagonize the function of ASAP1, and our analysis of online data showed that high expression of ASAP1-IT1 was associated with favorable survival in ovarian cancer. A similar association was also observed in the public database for another lncRNA, FAM215A (family with sequence similarity 215 member A, or C17orf88, LINC00530). Currently, little is known about FAM215A with regard to its biologic activities and associations with cancer outcomes.
2 Materials and methods
2.1 Patient information
Patients with epithelial ovarian cancer were recruited from two hospitals affiliated with the University of Turin in Turin, Italy. Patient enrollment occurred between October 1991 and February 2000 in one hospital (group one: n = 191), and between April 1997 and January 2013 in the other (group two: n = 75). All patients enrolled in the study underwent cytoreduction surgery for primary ovarian cancer, and 208 (78%) of these patients received standard post-operative platinum-based chemotherapy after surgery, which included cisplatin and cyclophosphamide between 1991 and 1995 (n = 63) and carboplatin and paclitaxel after 1995 (n = 145). Fresh tumor samples were collected from the patients during surgery. The specimens were snap-frozen in liquid nitrogen immediately after resection and then transferred to − 80°C freezers for storage. Patient information on age at surgery, disease stage, tumor grade and histology was obtained from medical records and pathology reports. Disease stage and tumor grade were categorized based on the International Federation of Gynecology and Obstetrics (FIGO) Classification and the WHO Guidelines, respectively [15, 16]. Patients were followed for disease progression and survival outcomes from surgery through June 2005 (group one: n = 180) or through March 2015 (group two: n = 68). The median follow-up time was 29.9 months (range: 0.6–114.1) for the former group and 40.5 months (range: 3.4–165.5) for the latter, respectively. Information on treatment response was available for 179 patients, including 128 who had complete response and 51 who did not. Treatment response was assessed one month after chemotherapy, and complete response was defined as resolution of all evidence of the disease for at least a month. The study was approved by ethics review committees at the hospitals, and informed consent was obtained from each patient who participated in the study.
2.2 Tumor analysis
Frozen tumor samples collected for study were evaluated by two independent pathologists to confirm that tumor cells were present in > 80% of each tumor specimen. The tissue samples were pulverized using a tissue homogenizer. Samples of approximately 30 mg of pulverized tissue powder were used for total RNA extraction, which was performed using the AllPrep DNA/RNA Mini Kit (Qiagen). The extracted total RNAs were treated with RNase-free DNase to remove DNA contamination. The quality of the RNA samples was assessed by measuring light absorbance and RNA Integrity Number (RIN) using the NanoDrop spectrophotometer (NanoDrop 2000, Thermo Fisher) and Agilent 2100 Bioanalyzer System, respectively. The assessment showed a 260/280 ratio of 1.8 or higher and an average RIN number of 5.63 based on the 28s:18s rRNA ratio. High Capacity cDNA Reverse Transcription Kit was used to convert total RNA to cDNA (Applied Biosystems). The cDNA samples were analyzed for lncRNA and ASAP1 expression using the SYBR green-based real-time PCR (qPCR). The PCR reaction was performed in a LightCycler 480 instrument (Roche) using LightCycler 480 SYBR Green I Master with UDG (Roche). In the PCR reaction (10 µl), 1 µl cDNA template was mixed with 200 nM primers and 5 µl SYBR PCR master mix (LifeTech). The PCR reaction conditions included incubation at 50 °C for 2 min to activate UDG, 95 °C for 2 min to activate Taq polymerase, and 40 cycles of 95 °C for 15s and 60°C for 1 min. Melting curves were generated after each PCR run to evaluate the size of PCR products. Each sample was tested in triplicate, and the mean value of three reactions was used for analysis if the coefficient of variation was <10%. If not, the mean of two closest reactions was used. As an internal reference, GAPDH expression was also measured simultaneously with lncRNAs and ASAP1 in all of the tumor samples. Primer sequences for the PCR reactions are provided in Supplementary Table 1.
2.3 Statistical analysis
Expression index (EI) was calculated as levels of RNA expression for ASAP1-IT1, FAM215A, and LINC00472 after adjusting for GAPDH, and calculation was based on the formula 1000 × 2(−ΔCt), where Ct is the cycle threshold and Δ Ct is the difference between CtlncRNA and CtGAPDH. After calculation, EI values were grouped into 3 categories, low, medium and high, based on the tertile distribution of each lncRNA among the patients. The ordinal values were then analyzed for their associations with clinical and pathological variables, using the Chi-square test or Mann-Whitney U statistics where appropriate. Survival analyses were performed using the Cox proportional hazards regression model at both univariate and multivariate levels. In the multivariate analyses, age at surgery, disease stage, tumor grade and histological type were included for adjustment. The log-rank test with two degree of freedom was used for comparison of three Kaplan-Meier survival curves. Two survival endpoints, progression-free survival and overall survival, were studied as disease outcomes. Progression-free survival was the time interval from the date of surgery to the date of disease progression or last follow-up; overall survival was the time between surgery and last follow-up or death. SAS (version 9.4) and R (version 3.0.2) were used for statistical analyses. All p-values were two-sided.
3 Results
In total, 266 patients were included in the study. The median age of patients at surgery was 59.1 years, ranging from 24.4 to 82.1 years. Of the patients, 73 (28%) had stage I or II disease, and 191 (72%) had stage III or IV disease. Twenty-nine patients (11%) had grade 1 tumors, and 235 (89%) had grade 2 or 3 tumors. Two patients had no information either on tumor grade or disease stage. Forty-five percent of the patients (n = 121) were diagnosed with serous tumors, and 55% (n = 145) had other histotypes, including endometrioid, mucinous, clear cell and undifferentiated. Median levels of lncRNA expression were 3.57 EI (range: 0.03–83.33) for ASAP1-IT1, 0.62 EI (range: 0.02–23.85) for FAM215A, and 8.44 EI (range: 0.03–289.84) for LINC00472. The ranges of each tertile group were: 0.03–2.36 (low), 2.36–5.43 (mid) and 5.43–83.33(high) for ASAP1-IT1; 0.018–0.39 (low), 0.39–0.92 (mid) and 0.92–23.85 (high) for FAM215A; and 0.03–5.5 (low), 5.5–13.67 (mid) and 13.67–289.84 (high) for LINC00472. Table 1 shows the associations of lncRNA expression with clinical and pathological variables. No correlations were observed between lncRNA expression and patient age at surgery, except for FAM215A. Disease stage and tumor grade were associated with expression of LINC00472 and FAM215A. High expression of these lncRNAs correlated with lower tumor grades and earlier disease stages. For FAM215A, 45.21% of patients with stage I or II disease had high expression compared to 27.89% of those with stage III or IV patients (p = 0.0072), and 51.72% of patients with grade 1 tumors had high expression compared to 30.34% of patients with grade 2 or 3 tumors (p = 0.004). Patients with high expression of FAM215A were more likely to respond to chemotherapy compared to those with low expression (p = 0.017). For LINC00472, the corresponding numbers were 45.21% versus 28.04% when comparing disease stage (p = 0.024), and 51.72% versus 30.47% when comparing tumor grade (p = 0.004). ASAP1-IT1 expression appeared to differ by disease stage (p = 0.025), but no significant trend was observed with regard to the difference by tumor grade. Expression of LINC00472, FAM215A and ASAP1-IT1 did not show appreciable differences between serous and non-serous tumors.
Table 1.
Associations of lncRNAs and ASAP1 with clinical and pathological variables.
| Variables | Total no. (%) | Low no. (%) | Mid no. (%) | High no. (%) | p value |
|---|---|---|---|---|---|
| ASAP1-IT1 | |||||
| Age | 0.19 | ||||
| ≥ 59.08 | 121 (46.18) | 41 (33.88) | 35 (28.93) | 45 (37.19) | |
| < 59.08 | 141 (53.82) | 45 (31.91) | 55 (39.01) | 41 (29.08) | |
| Disease stage | 0.025 | ||||
| I–II | 73 (27.65) | 22 (30.14) | 18 (24.66) | 33 (45.21) | |
| III–IV | 191 (72.35) | 66 (34.55) | 71 (37.17) | 54 (28.27) | |
| Tumor grade | 0.55 | ||||
| 1 | 29 (10.98) | 8 (27.59) | 9 (31.03) | 12 (41.38) | |
| 2–3 | 235 (89.02) | 80 (34.04) | 81 (34.47) | 74 (31.49) | |
| Histologic type | 0.79 | ||||
| Non-serous | 145 (54.51) | 49 (33.79) | 47 (32.41) | 49 (33.79) | |
| Serous | 121 (45.49) | 39 (32.23) | 44 (36.36) | 38 (31.40) | |
| Treatment responsea | 0.59 | ||||
| Yes | 128 (71.51) | 40 (31.25) | 42 (32.81) | 46 (35.94) | |
| No | 51 (28.49) | 20 (39.22) | 15 (29.41) | 16 (31.37) | |
| FAM215A | |||||
| Age | 0.028 | ||||
| ≥ 59.08 | 120 (45.98) | 46 (38.33) | 31 (25.83) | 43 (35.83) | |
| < 59.08 | 141 (54.02) | 39 (27.66) | 58 (41.13) | 44 (31.21) | |
| Disease stage | 0.0072 | ||||
| I–II | 73 (27.76) | 15 (20.55) | 25 (34.25) | 33 (45.21) | |
| III–IV | 190 (72.24) | 73 (38.42) | 64 (33.68) | 53 (27.89) | |
| Tumor grade | 0.004 | ||||
| 1 | 29 (11.03) | 2 (6.90) | 12 (41.38) | 15 (51.72) | |
| 2–3 | 234 (88.97) | 86 (36.75) | 77 (32.91) | 71 (30.34) | |
| Histologic type | 0.85 | ||||
| Non-serous | 144 (54.34) | 47 (32.64) | 47 (32.64) | 50 (34.72) | |
| Serous | 121 (45.66) | 41 (33.88) | 42 (34.71) | 38 (31.40) | |
| Treatment responsea | 0.017 | ||||
| Yes | 128 (71.91) | 38 (29.69) | 41 (32.03) | 49 (38.28) | |
| No | 50 (28.09) | 20 (40.00) | 22 (44.00) | 8 (16.00) | |
| LINC00472 | |||||
| Age | 0.65 | ||||
| ≥ 59.08 | 121 (46.54) | 41 (33.88) | 37 (30.58) | 43 (35.54) | |
| < 59.08 | 139 (53.46) | 44 (31.65) | 50 (35.97) | 45 (32.37) | |
| Disease stage | 0.024 | ||||
| I–II | 73 (27.86) | 18 (24.66) | 22 (30.14) | 33 (45.21) | |
| III–IV | 189 (72.14) | 70 (37.04) | 66 (34.92) | 53 (28.04) | |
| Tumor grade | 0.004 | ||||
| 1 | 29 (11.07) | 2 (6.90) | 12 (41.38) | 15 (51.72) | |
| 2–3 | 233 (88.93) | 86 (36.91) | 76 (32.62) | 71 (30.47) | |
| Histologic type | 0.21 | ||||
| Non-serous | 145 (54.92) | 55 (37.93) | 46 (31.72) | 44 (30.34) | |
| Serous | 119 (45.08) | 33 (27.73) | 42 (35.29) | 44 (36.97) | |
| Treatment responsea | 0.096 | ||||
| Yes | 128 (71.51) | 40 (31.25) | 39 (30.47) | 49 (38.28) | |
| No | 51 (28.49) | 19 (37.25) | 21 (41.18) | 11 (21.57) | |
| ASAP1 | |||||
| Age | 0.40 | ||||
| ≥ 59.08 | 119 (46.12) | 40 (33.61) | 36 (30.25) | 43 (36.13) | |
| < 59.08 | 139 (53.88) | 43 (30.94) | 53 (38.13) | 43 (30.94) | |
| Disease stage | 0.051 | ||||
| I–II | 71 (27.31) | 28 (39.44) | 16 (22.54) | 27 (38.03) | |
| III–IV | 189 (72.69) | 58 (30.69) | 73 (38.62) | 58 (30.69) | |
| Tumor grade | 0.048 | ||||
| 1 | 28 (10.77) | 15 (53.57) | 6 (21.43) | 7 (25.00) | |
| 2–3 | 232 (89.23) | 71 (30.60) | 83 (35.78) | 78 (33.62) | |
| Histologic type | 0.40 | ||||
| Non-serous | 143 (54.58) | 52 (36.36) | 46 (32.17) | 45 (31.47) | |
| Serous | 119 (45.42) | 34 (28.57) | 44 (36.97) | 41 (34.45) | |
| Treatment responsea | 0.75 | ||||
| Yes | 124 (70.86) | 43 (34.68) | 41 (33.06) | 40 (32.26) | |
| No | 51 (29.14) | 15 (29.41) | 17 (33.33) | 19 (37.25) |
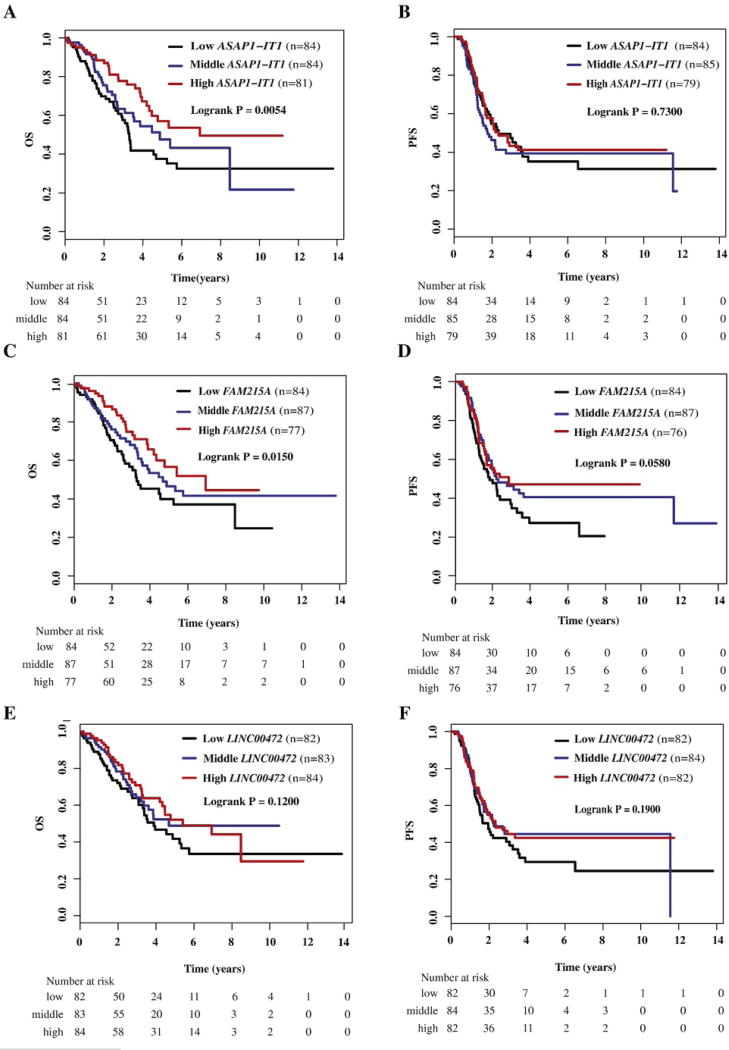
The results of survival analyses are shown in Table 2. ASAP1-IT1 expression was associated with overall survival, but not with progression-free survival (Fig. 1A and B). Patients with high expression had better overall survival compared to those with low expression, and the survival association remained significant after adjustment for patient age, disease stage, tumor grade and histologic type. FAM215A expression was associated with overall survival and possibly with progression-free survival, but these associations were observed only in univariate analysis (Fig. 1C and D). After adjusting for disease stage, tumor grade and histology, the survival associations with FAM215A were no longer significant. LINC00472 expression was not associated with either progression-free or overall survival (Fig. 1E and F). Since tumor grade and disease stage were associated with some of the lncRNA expression, we also performed survival analyses among patients stratified by their tumor grade or disease stage. These additional analyses did not change the study results substantially (data not shown).
Table 2.
Associations of lncRNAs and ASAP1 with ovarian cancer survival.
| Expression | HR for progression | 95% CI | p value | HR for death | 95% CI | p value |
|---|---|---|---|---|---|---|
| ASAP1-IT1 | ||||||
| Unadjusted Cox regression model | ||||||
| Low | 1 | 1 | ||||
| Mid | 1.12 | 0.74–1.70 | 0.59 | 0.77 | 0.49–1.20 | 0.24 |
| High | 0.93 | 0.60–1.42 | 0.72 | 0.51 | 0.32–0.82 | 0.0057 |
| Continuous | 0.96 | 0.78–1.19 | 0.72 | 0.72 | 0.57–0.91 | 0.0055 |
| Adjusted Cox regression modela | ||||||
| Low | 1 | 1 | ||||
| Mid | 0.97 | 0.63–1.47 | 0.87 | 0.69 | 0.44–1.08 | 0.11 |
| High | 1.09 | 0.71–1.67 | 0.70 | 0.56 | 0.35–0.90 | 0.017 |
| Continuous | 1.04 | 0.84–1.30 | 0.72 | 0.74 | 0.58–0.94 | 0.015 |
| FAM215A | ||||||
| Unadjusted Cox regression model | ||||||
| Low | 1 | 1 | ||||
| Mid | 0.72 | 0.48–1.08 | 0.11 | 0.76 | 0.49–1.18 | 0.22 |
| High | 0.67 | 0.44–1.02 | 0.063 | 0.55 | 0.34–0.90 | 0.017 |
| Continuous | 0.81 | 0.65–1.01 | 0.059 | 0.74 | 0.59–0.95 | 0.016 |
| Adjusted Cox regression modela | ||||||
| Low | 1 | 1 | ||||
| Mid | 0.74 | 0.49–1.13 | 0.17 | 0.83 | 0.53–1.31 | 0.43 |
| High | 0.92 | 0.60–1.41 | 0.71 | 0.74 | 0.45–1.20 | 0.22 |
| Continuous | 0.95 | 0.76–1.18 | 0.63 | 0.86 | 0.67–1.09 | 0.21 |
| LINC00472 | ||||||
| Unadjusted Cox regression model | ||||||
| Low | 1 | 1 | ||||
| Mid | 0.78 | 0.52–1.18 | 0.24 | 0.76 | 0.48–1.21 | 0.25 |
| High | 0.76 | 0.50–1.15 | 0.19 | 0.70 | 0.44–1.11 | 0.13 |
| Continuous | 0.87 | 0.70–1.07 | 0.19 | 0.83 | 0.66–1.05 | 0.12 |
| Adjusted Cox regression modela | ||||||
| Low | 1 | 1 | ||||
| Mid | 0.87 | 0.58–1.33 | 0.53 | 0.89 | 0.55–1.42 | 0.62 |
| High | 0.92 | 0.60–1.41 | 0.71 | 0.88 | 0.55–1.42 | 0.61 |
| Continuous | 0.96 | 0.77–1.19 | 0.70 | 0.94 | 0.74–1.19 | 0.60 |
| ASAP1 | ||||||
| Unadjusted Cox regression model | ||||||
| Low | 1 | 1 | ||||
| Mid | 1.81 | 1.17–2.82 | 0.0083 | 1.11 | 0.70–1.76 | 0.66 |
| High | 1.59 | 1.02–2.49 | 0.042 | 0.79 | 0.49–1.27 | 0.32 |
| Continuous | 1.24 | 1.00–1.52 | 0.048 | 0.89 | 0.71–1.12 | 0.31 |
| Adjusted cox regression modela | ||||||
| Low | 1 | 1 | ||||
| Mid | 1.54 | 0.98–2.39 | 0.059 | 0.94 | 0.59–1.50 | 0.79 |
| High | 1.63 | 1.04–2.56 | 0.034 | 0.75 | 0.47–1.21 | 0.24 |
| Continuous | 1.26 | 1.02–1.57 | 0.036 | 0.87 | 0.69–1.10 | 0.24 |
Fig. 1.
Kaplan-Meier survival by tertiles of ASAP1-IT1, FAM215A and LINC00472 expression in ovarian cancer patients. A) Overall survival (OS) by low, middle, and high ASAP1-IT1; B) progression-free survival (PFS) by low, middle, and high ASAP1-IT1; C) overall survival (OS) by low, middle, and high FAM215A; D) progression-free survival (PFS) by low, middle, and high FAM215A; E) overall survival (OS) by low, middle, and high LINC00472; F) progression-free survival (PFS) by low, middle, and high LINC00472.
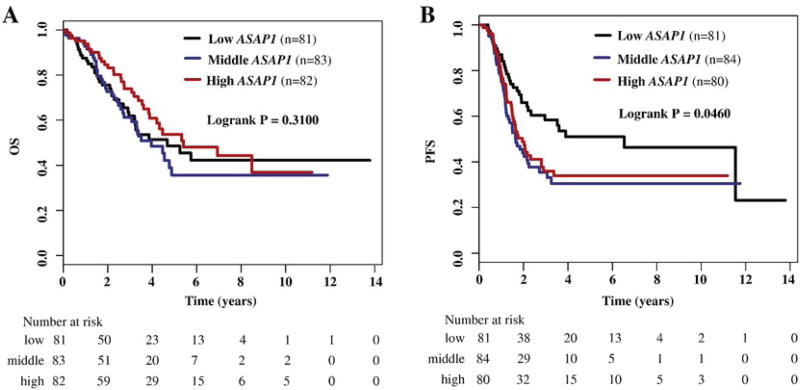
ASAP1-IT1 is transcribed from an intron of the ASAP1 gene, and may regulate the activity and function of ASAP1. We measured the mRNA expression of ASAP1 in ovarian cancer and analyzed its association with the disease characteristics and ASAP1-IT1 expression. ASAP1 expression appeared to be higher in high grade than in low grade tumors (p = 0.048) (Table 1), and high expression was associated with increased risk of tumor progression, although no association was observed for overall survival (Table 2, Fig. 2A and B). Expression of ASAP1-IT1 and ASAP1 were positively correlated in ovarian tumors (Spearman r = 0.645; p <0.001).
Fig. 2.
Kaplan-Meier survival by tertiles of ASAP1 expression in ovarian cancer patients. A) Overall survival (OS) by low, middle, and high ASAP1; B) progression-free survival (PFS) by low, middle, and high ASAP1.
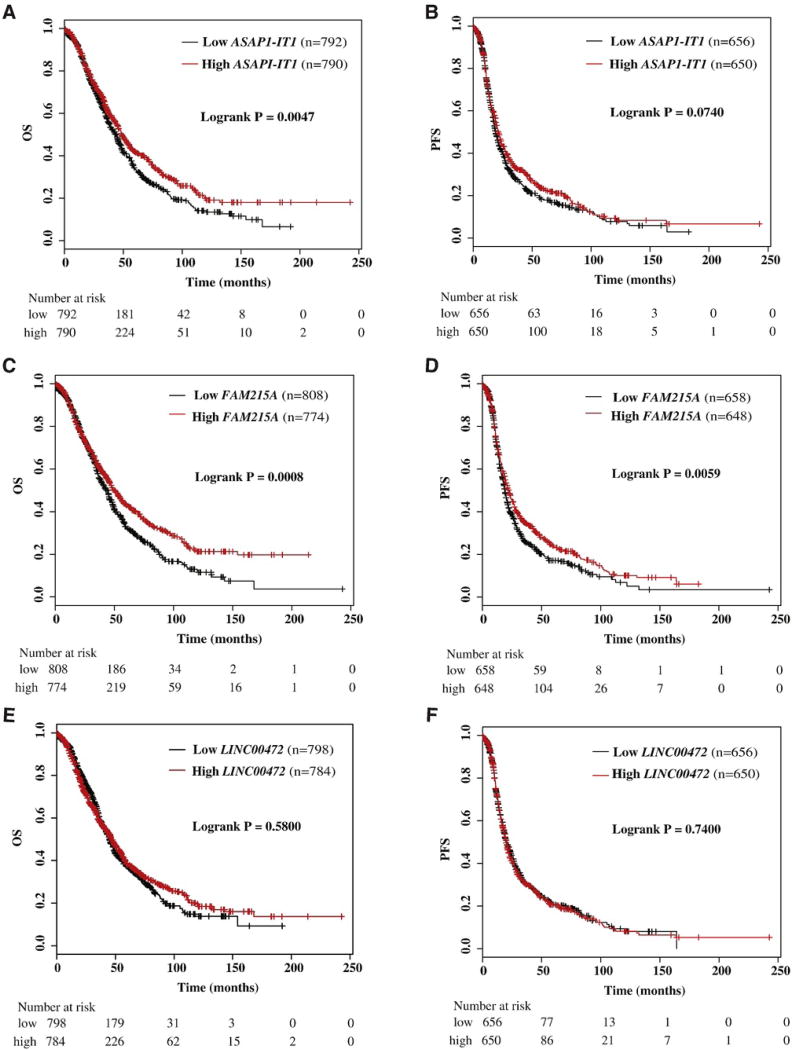
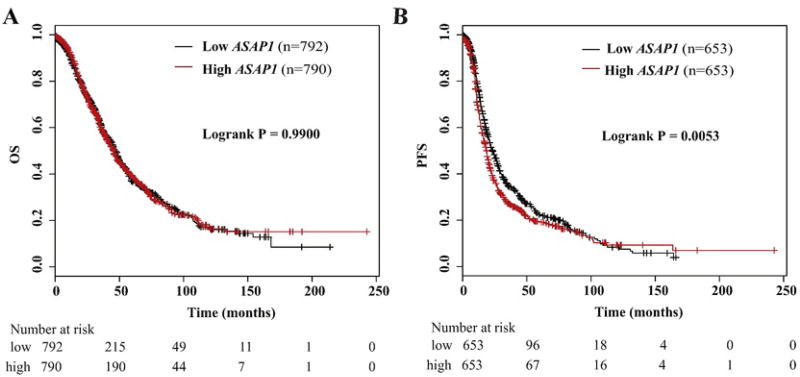
Using the Kaplan-Meier Plotter (http://kmplot.com/analysis/), we found that high expression of ASAP1-IT1 was also associated with favorable overall survival of ovarian cancer patients (p = 0.0047) (Fig. 3A), but no association with progression-free survival (Fig. 3B). Similarly, analyses of online data showed that high expression of FAM215A was significantly associated with favorable overall survival (p = 0.00080) (Fig. 3C) and progression-free survival (p = 0.0059) (Fig. 3D). LINC00472 was not associated with either overall survival (Fig. 3E) or progression-free survival (Fig. 3F). We also checked the association of ASAP1 with patient survival using the online data, and found high expression associated with poor progression-free survival, but not with overall survival (Fig. 4A and B).
Fig. 3.
Kaplan-Meier survival by expression tertiles of ASAP1-IT1, FAM215A and LINC00472 in an online database analyzed by Kaplan-Meier Plotter. A) Overall survival (OS) by low and high ASAP1-IT1; B) progression-free survival (PFS) by low and high ASAP1-IT1; C) overall survival (OS) by low and high FAM215A; D) progression-free survival (PFS) by low and high FAM215A; E) overall survival (OS) by low and high LINC00472; F) progression-free survival (PFS) by low and high LINC00472.
Fig. 4.
Kaplan-Meier survival by expression tertiles of ASAP1 in an online database analyzed by Kaplan-Meier Plotter. A) Overall survival (OS) by low and high ASAP1; B) progression-free survival (PFS) by low and high ASAP1.
4 Discussion
Our study showed that tumor expression of two lncRNAs, LINC00472 and FAM215A, differed significantly by tumor grade and disease stage: high expression associated with low tumor grades and early disease stages. ASAP1-IT1 had a similar trend in association with disease stage, but not tumor grade. High expression of ASAP1-IT1 and FAM215A were also associated with more favorable overall survival of ovarian cancer. Similar survival associations were further observed in an online dataset [7], indicating that the findings were somehow consistent across different studies. We also analyzed the expression of ASAP1 and found high expression associated with poor progression-free survival. Furthermore, ASAP1 expression was positively correlated with ASAP1-IT1 expression, suggesting that the effects of ASAP1-IT1 on ASAP1, if any, may not be achieved through its down-regulation of ASAP1 expression.
Based on their locations in or relative to the coding genes, lncRNAs are classified into intronic, intergenic, and overlapping (either in sense or antisense orientation) transcripts [17]. Several studies have reported that some intronic non-coding RNAs are positively correlated with expression of their corresponding protein-coding genes, whereas others are inversely correlated [18–21]. These observations indicate potentially complex regulations between intronic lncRNAs and their surrounding genes. Non-coding RNAs have been found to act on their targets either at the transcriptional level or post-transcriptionally. The mechanisms that determine the pre- and post-transcription regulation remain to be elucidated. ASAP1-IT1 is located in an intron of the ASAP1 gene, and the biological functions of ASAP1-IT1 are still unknown despite the fact that the ASAP1 gene has been well characterized. Evidence suggests that ASAP1 may be an oncogene as ASAP1 expression is highly up-regulated in a variety of tumors in comparison with normal tissue, and in colorectal cancer, the expression correlates with poor prognosis. ASAP1 enhances metastasis in vivo, and stimulates tumor cell migration, invasion, and adhesion in vitro [12]. Studies have indicated that ASAP1 is highly expressed in primary prostate cancer and metastatic prostate tumors compared to benign prostate tissue. Down-regulation of ASAP1 in PC-3 markedly inhibits cell migration and invasion [14]. ASAP1 also enhances the invasion of breast cancer cells [22]. Recently, Hou et al. found that ASAP1 expression was higher in epithelial ovarian cancer than in normal ovarian tissue, and high expression was associated with poor progression-free and overall survivals [13]. In our study, we found a similar association between ASAP1 expression and progression-free survival, though not with overall survival. We also found that ASAP1 expression was positively associated with tumor grade. How ASAP1 may be regulated by lncRNA ASAP1-IT1 remains unknown. The finding in our study suggests that ASAP1-IT1 may not antagonize the action of ASAP1 through suppressing its expression. Previous studies have found positive correlations between the expression of host genes and their associated intronic lncRNAs [19, 20]. Sometimes, lncRNAs are not simply co-expressed with their host genes, and their expression may be independent of the host genes. Intronic lncRNAs can modulate the biological pathways of their host genes. For example, SPRY4 is an inhibitor of the receptor-transduced mitogen-activated protein kinase (MAPK) signaling pathway. SPRY4-IT1, which is transcribed from an intron of the SPRY4 gene, can affect the MAPK signaling pathway through its interaction with Raf1, B-Raf, MEK1/2, TESK1, MARKK, and MARK2 [23]. Intronic lncRNAs can also negatively regulate their host genes. NPTN is overexpressed in breast cancer cells resulting in significant tumor growth, but its intronic lncRNA, lnc-LET, is a tumor suppressor that appears to inhibit the metastasis of hepatocellular carcinoma [5].
Currently, few studies have evaluated the function of FAM215A. One report showed that suppression of FAM215A expression by siRNA increased the death of A375 melanoma cells induced by MLN4924 [24]. Although our study and the data from online sources indicated a possible association with ovarian cancer survival, the biological relevance of FAM215A in ovarian cancer is still unclear.
LINC00472 has been observed by our group to be involved in breast cancer. In our previous studies, we found that high expression of LINC00472 was associated with favorable overall survival of breast cancer patients in our study as well as in >2 dozen clinical data sets available online [8, 9]. This finding was consistent across study populations and with different analytical technologies, as well as supported by in vitro experiments. Using breast cancer cell lines to manipulate the expression of LINC00472, we demonstrated that increasing LINC00472 expression was associated with reduced cell proliferation and migration. Since ovarian cancer shares certain aspects in etiology with breast cancer as suggested by BRCA1 and BRCA2 mutations, we analyzed LINC00472 expression in epithelial ovarian cancer, the most common form, and its associations with disease characteristics and patient survival [25]. In our analysis, although we did not find any associations between the lncRNA expression and disease outcome, we did observe that high expression of LINC00472 was associated with low grade tumors and early stage disease. These findings are consistent with what we have seen in breast cancer, offering more evidence in support of the speculation that LINC00472 may play a role in cancer as a tumor suppressor.
In summary, we investigated the clinical significance of three lncRNAs, LINC00472, ASAP1-IT1 and FAM215A, in ovarian cancer, and found that two of the lncRNAs, LINC00472 and FAM215A, were associated with tumor grade and disease stage. Furthermore, expression of FAM215A and ASAP1-IT1 were associated with disease outcomes, and these associations were also seen in other datasets. Our findings suggest that lncRNAs may play appreciable roles in cancer and that more research is needed to elucidate the biological mechanisms of lncRNAs involved in tumorigenesis and disease progression, as well as their clinical implications in tumor characterization and patient management.
Supplementary Material
Supplementary Table S1
Highlights.
- ASAP1-IT1, FAM215A and LINC00472 were highly expressed in early stage disease of EOC.
- ASAP1-IT1, FAM215A and LINC00472 were also highly expressed in low grade tumors.
- High expression of ASAP1-IT1 and FAM215A was associated with favorable OS.
- Large online database showed similar survival associations with ASAP1-IT1 and FAM215A.
Footnotes
Conflict of interest statement
The authors declare that there are no conflicts of interest.
E-Extra
Frozen tumor samples collected for study were evaluated by two independent pathologists to confirm that tumor cells were present in >80% of each tumor specimen. The tissue samples were pulverized using a tissue homogenizer. Samples of approximately 30 mg of pulverized tissue powder were used for total RNA extraction, which was performed using the AllPrep DNA/RNA Mini Kit (Qiagen). The extracted total RNAs were treated with RNase-free DNase to remove DNA contamination. The quality of the RNA samples was assessed by measuring light absorbance and RNA Integrity Number (RIN) using the NanoDrop spectrophotometer (NanoDrop 2000, Thermo Fisher) and Agilent 2100 Bioanalyzer System, respectively. The assessment showed a 260/280 ratio of 1.8 or higher and an average RIN number of 5.63 based on the 28s:18s rRNA ratio. High Capacity cDNA Reverse Transcription Kit was used to convert total RNA to cDNA (Applied Biosystems). The cDNA samples were analyzed for lncRNA and ASAP1 expression using the SYBR green-based real-time PCR (qPCR). The PCR reaction was performed in a LightCycler 480 instrument (Roche) using LightCycler 480 SYBR Green I Master with UDG (Roche). In the PCR reaction (10 µl), 1 µl cDNA template was mixed with 200 nM primers and 5 µl SYBR PCR master mix (LifeTech). The PCR reaction conditions included incubation at 50 °C for 2 min to activate UDG, 95 °C for 2 min to activate Taq polymerase, and 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Melting curves were generated after each PCR run to evaluate the size of PCR products. Each sample was tested in triplicate, and the mean value of three reactions was used for analysis if the coefficient of variation was <10%. If not, the mean of two closest reactions was used. As an internal reference, GAPDH expression was also measured simultaneously with lncRNAs and ASAP1 in all of the tumor samples. Primer sequences for the PCR reactions are provided in Supplementary Table 1.
E-component
The following are the supplementary data related to this article.
References
- 1.Cancer Facts & Figures 2015. American Cancer Society; Atlanta: 2015. [Google Scholar]
- 2.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 3.Li L, Liu B, Wapinski OL, Tsai MC, Qu K, Zhang J, et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5:3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 5.Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gyorffy B, Lanczky A, Szallasi Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr. Relat. Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 8.Shen Y, Katsaros D, Loo LW, Hernandez BY, Chong C, Canuto EM, et al. Prognostic and predictive values of long non-coding RNA LINC00472 in breast cancer. Oncotarget. 2015;6:8579–8592. doi: 10.18632/oncotarget.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen Y, Wang Z, Loo LW, Ni Y, Jia W, Fei P, et al. LINC00472 expression is regulated by promoter methylation and associated with disease-free survival in patients with grade 2 breast cancer. Breast Cancer Res. Treat. 2015;154:473–482. doi: 10.1007/s10549-015-3632-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 11.Gillingham AK, Munro S. The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 2007;23:579–611. doi: 10.1146/annurev.cellbio.23.090506.123209. [DOI] [PubMed] [Google Scholar]
- 12.Muller T, Stein U, Poletti A, Garzia L, Rothley M, Plaumann D, et al. ASAP1 promotes tumor cell motility and invasiveness, stimulates metastasis formation in vivo, and correlates with poor survival in colorectal cancer patients. Oncogene. 2010;29:2393–2403. doi: 10.1038/onc.2010.6. [DOI] [PubMed] [Google Scholar]
- 13.Hou T, Yang C, Tong C, Zhang H, Xiao J, Li J. Overexpression of ASAP1 is associated with poor prognosis in epithelial ovarian cancer. Int. J. Clin. Exp. Pathol. 2014;7:280–287. [PMC free article] [PubMed] [Google Scholar]
- 14.Lin D, Watahiki A, Bayani J, Zhang F, Liu L, Ling V, et al. ASAP1, a gene at 8q24, is associated with prostate cancer metastasis. Cancer Res. 2008;68:4352–4359. doi: 10.1158/0008-5472.CAN-07-5237. [DOI] [PubMed] [Google Scholar]
- 15.Shepherd JH. Revised FIGO staging for gynaecological cancer. Br. J. Obstet. Gynaecol. 1989;96:889–892. doi: 10.1111/j.1471-0528.1989.tb03341.x. [DOI] [PubMed] [Google Scholar]
- 16.Scully RE, Sobin LH. Histologic typing of ovarian tumors. Arch. Pathol. Lab. Med. 1987;111:794–795. [PubMed] [Google Scholar]
- 17.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakaya HI, Amaral PP, Louro R, Lopes A, Fachel AA, Moreira YB, et al. Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome Biol. 2007;8:R43. doi: 10.1186/gb-2007-8-3-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. U. S. A. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 22.Onodera Y, Hashimoto S, Hashimoto A, Morishige M, Mazaki Y, Yamada A, et al. Expression of AMAP1, an ArfGAP, provides novel targets to inhibit breast cancer invasive activities. EMBO J. 2005;24:963–973. doi: 10.1038/sj.emboj.7600588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, et al. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852–3862. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- 24.Blank JL, Liu XJ, Cosmopoulos K, Bouck DC, Garcia K, Bernard H, et al. Novel DNA damage checkpoints mediating cell death induced by the NEDD8-activating enzyme inhibitor MLN4924. Cancer Res. 2013;73:225–234. doi: 10.1158/0008-5472.CAN-12-1729. [DOI] [PubMed] [Google Scholar]
- 25.King MC, Marks JH, Mandell JB New York Breast Cancer Study G. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1