Human Splicing Factor SF3a, but Not SF1, Is Essential for Pre-mRNA Splicing In Vivo (original) (raw)
Abstract
The three subunits of human splicing factor SF3a are essential for the formation of the functional 17S U2 snRNP and prespliceosome assembly in vitro. RNAi-mediated depletion indicates that each subunit is essential for viability of human cells. Knockdown of single subunits results in a general block in splicing strongly suggesting that SF3a is a constitutive splicing factor in vivo. In contrast, splicing of several endogenous and reporter pre-mRNAs is not affected after knockdown of SF1, which functions at the onset of spliceosome assembly in vitro and is essential for cell viability. Thus, SF1 may only be required for the splicing of a subset of pre-mRNAs. We also observe a reorganization of U2 snRNP components in SF3a-depleted cells, where U2 snRNA and U2-B″ are significantly reduced in nuclear speckles and the nucleoplasm, but still present in Cajal bodies. Together with the observation that the 17S U2 snRNP cannot be detected in extracts from SF3a-depleted cells, our results provide further evidence for a function of Cajal bodies in U2 snRNP biogenesis.
INTRODUCTION
Introns are removed from nuclear pre-mRNA by the spliceosome, a macromolecular complex composed of the U1, U2, U4, U5, and U6 small nuclear ribonucleoprotein particles (snRNPs) and many non-snRNP proteins (reviewed in Krämer, 1996; Burge et al., 1999; Jurica and Moore, 2003). SnRNPs play key roles in the recognition of conserved sequence elements that define the exon/intron junctions and juxtapose the splice sites in the catalytic center of the spliceosome for intron excision (reviewed in Collins and Guthrie, 2000; Brow, 2002; Nilsen, 2003). The U1 snRNP interacts with the 5′ splice site and the first splicing-specific complex (E) is formed. Binding of the U2 snRNP to the intron branch site leads to the formation of the prespliceosomal complex A. Addition of the U4/U6.U5 tri-snRNP results in the assembly of complex B and after conformational rearrangements the intron is removed in complex C.
Despite considerable evidence that splicing is RNA-catalyzed (Collins and Guthrie, 2000; Nilsen, 2003), it requires the action of snRNP-associated and non-snRNP proteins which, for example, aid splice site recognition, drive conformational changes or regulate the activity of splicing factors. Splicing factors SF1 and SF3a function in 3′ splice site recognition at early stages of spliceosome assembly. During the formation of complex E, SF1 interacts with U2AF65 followed by cooperative binding to the intron branch site and the neighboring polypyrimidine tract, respectively (Berglund et al., 1997, 1998; Rain et al., 1998; Liu et al., 2001; Selenko et al., 2003). Although SF1 was initially isolated as a factor necessary for prespliceosome formation in vitro (Krämer, 1992), later studies suggested a kinetic rather than essential role in splicing (Rutz and Séraphin, 1999; Guth and Valcárcel, 2000). Nevertheless, in Saccharomyces cerevisiae, SF1 has been implicated in the removal of introns with suboptimal splice sites and appears to play a role in nuclear pre-mRNA retention (Rutz and Séraphin, 2000; Galy et al., 2004).
SF3a is composed of three subunits of 60, 66, and 120 kDa (Brosi et al., 1993b). Together with SF3b it binds to the 12S U2 snRNP comprising seven Sm proteins common to the spliceosomal snRNPs and the U2-specific proteins U2-A′ and U2-B″ (Will and Lührmann, 2001). Initial binding of SF3b results in the formation of an intermediate 15S particle, which is converted into the active 17S U2 snRNP after association of SF3a (Brosi et al., 1993a; Krämer et al., 1999). During the assembly of presplicing complex A, U2 snRNA base pairs with the branch site and several subunits of SF3a and SF3b interact with surrounding sequences and the branch site adenosine, suggesting roles for SF3a and SF3b in facilitating branch site recognition by U2 snRNA and tethering the U2 snRNP to the pre-mRNA (Gozani et al., 1996, 1998; Query et al., 1996; Will et al., 2001).
All SF3a subunits are necessary for prespliceosome assembly in HeLa cell extracts (Nesic and Krämer, 2001). Genetic studies have demonstrated essential functions of S. cerevisiae, Drosophila, and Caenorhabditis elegans orthologues in vivo (Krämer, 1996; Meyer et al., 1998; Kamath et al., 2003; Simmer et al., 2003; Boutros et al., 2004). Similarly, SF1 orthologues in yeast and C. elegans are essential for viability (Abovich and Rosbash, 1997; Fromont-Racine et al., 1997; Mazroui et al., 1999). Here we have used RNA interference (RNAi) to study the consequences of SF1 and SF3a depletion on viability and pre-mRNA splicing in human cells.
In addition, we have combined RNAi and immunolocalization studies to test how SF3a depletion influences the maturation of the U2 snRNP. Newly synthesized snRNAs, except for U6, are exported to the cytoplasm and assemble with the Sm proteins. After cap hypermethylation and 3′ end trimming, the core snRNPs are reimported into the nucleus (Will and Lührmann, 2001) and pass through Cajal bodies (CBs) before they reach sites of storage and splicing (Sleeman and Lamond, 1999a). CBs are dynamic structures present in varying number in many different cell types (Matera, 1999; Gall, 2000; Ogg and Lamond, 2002). Based on the association of CBs with nucleolar factors, components of the transcription and cell cycle machineries and specific gene loci, such as histone and U2 snRNP gene clusters, roles in the assembly of macromolecular complexes have been proposed. Several recent studies provide evidence for functions of CBs in snRNP biogenesis. First, upon import from the cytoplasm, newly synthesized snRNPs pass through CBs, before they move to sites of splicing and storage (Ferreira et al., 1994; Sleeman and Lamond, 1999a; Sleeman et al., 2001; Ogg and Lamond, 2002). Second, small RNAs that guide snRNA base modifications localize specifically to CBs (Kiss et al., 2002; Jády et al., 2003). Third, proteins required for the assembly of the U4/U6 and U4/U6.U5 snRNPs are enriched in CBs (Makarova et al., 2002; Stanek et al., 2003). Moreover, RNAi-mediated knockdown of U4/U6 or U5-specific proteins inhibits the formation of the U4/U6.U5 tri-snRNP and results in an accumulation of U4/U6 snRNPs and p110 in CBs (Schaffert et al., 2004), suggesting that CBs are sites of snRNP recycling.
Under steady-state conditions, snRNPs (including 12S U2 snRNP components) are detected in CBs and nuclear speckles (or interchromatin granule clusters), which are most likely storage sites for splicing components (Lamond and Spector, 2003). The speckled pattern is superimposed onto a diffuse nucleoplasmic component, thought to represent sites of cotranscriptional splicing. Although the SF3a subunits are part of the mature 17S U2 snRNP, they have not been detected in CBs (Nesic et al., 2004, and references therein), suggesting that the U2 population in CBs represents immature and nonfunctional particles. In contrast, transiently expressed mutant SF3a60 and SF3a66 impaired in binding to the U2 snRNP accumulate in CBs and are otherwise diffusely distributed in the nucleus (Nesic et al., 2004) similar to snRNPs shortly after nuclear import (Sleeman and Lamond, 1999a). On the basis of these observations we proposed that the binding of U2-specific proteins takes place in CBs (Nesic et al., 2004).
In this study we show that SF3a is essential for the splicing of many if not all U2-type introns in human cells. As a consequence of a block in splicing other steps in gene expression are affected, resulting in cell death. Cells depleted of SF3a lack the 17S U2 snRNP, whereas the 12S U2 snRNP accumulates. In addition, components of the U2 snRNP are highly diminished in speckles, but still detected in CBs, further supporting a role for CBs in U2 snRNP biogenesis. In contrast, although SF1 is also essential for cell viability, its depletion does not reduce general splicing, suggesting that SF1 is either only required for the splicing of certain pre-mRNAs or fulfils another essential function in vivo.
MATERIALS AND METHODS
Cell Culture, RNAi, and Transfection Procedures
HeLa cells were propagated in DMEM high glucose (Invitrogen, Basel, Switzerland) supplemented with 3.7 g/l NaHCO3 (Invitrogen) and 10% fetal calf serum (Sigma-Aldrich, Deisenhofen, Germany). Cells (2 × 105) were transfected with 0.2 μM chemically synthesized and annealed siRNAs (Dharmacon, Boulder, CO) in the presence of oligofectamine (Invitrogen) according to the instructions supplied by Dharmacon. For triple transfections, individual siRNAs were used at a concentration of 0.07 μM. Control transfections were performed in the absence of siRNA. The sequences of the top strand of the siRNAs were as follows: 60/1, UUCUGAUCACCGCACUCGGdTdT (nts 114–132 of the coding sequence); 60/2, GGAGGAGCUCAAUGCCAUUdTdT (nts 207–225); 66/1, CAAGGACCCGUACUUCAUGdTdT (nts 123–141); 66/2, UGAGGGGAGCUACCUGGCAdTdT (nts 195–213); 120/1, GGAGGAUUCUGCACCUUCUdTdT (nts 89–107); 120/2, GCUAGGAUCCGACAGAACGdTdT (nts 204–222); SF1/1, GACCUGACUCGUAAACUGCdTdT (nts 178–196). The GL2 siRNA targeting the firefly (Photinus pyralis) luciferase mRNA served as a control where indicated (Elbashir et al., 2002).
Cell viability was followed for 3 d after transfection. At 24-h intervals attached and detached cells were collected by centrifugation, washed with phosphate-buffered saline (PBS), stained with trypan blue, and counted.
To monitor splicing of a reporter pre-mRNA, HeLa cells were transfected with plasmid pAdCMV-glob (Estmer Nilsson et al., 2001) by calcium phosphate precipitation (Jordan et al., 1996) 24 h after treatment with siRNAs and grown for an additional 48 h before RT-PCR.
Isolation of RNA, RT-PCR, and Northern Blotting
Cells were collected 60 or 72 h posttransfection and total or cytoplasmic RNA was isolated with the RNeasy Mini kit (Qiagen, Basel, Switzerland).
Total cellular RNA was treated with 2 U RQ-DNase (Promega, Madison, WI) for 45 min at 37°C and extracted with phenol-chloroform. RT reactions were done in the presence of 1 μg total RNA, 2 μg oligo dT(12–18) (Sigma-Aldrich), 40 U rRNasin (Promega), and 200 U MMLV-RT (Invitrogen) for 2 h at 37°C. PCR reactions were performed with the Expand high-fidelity PCR system (Roche, Rotkreuz, Switzerland) for 30 cycles. Primers were complementary to exons 5 and 6 of SF3a60, exons 6 and 7 of SF3a66, exons 6 and 7 of SF3a120, exons 5 and 6 of SF1, exons 1 and 2 of β-globin, nts 150–166 and 423–440 of histone H3F1 mRNA, exons 2 and 3 of p80-coilin and exons 1 and 2 of SC35. PCR products were separated in agarose gels.
Northern blot analysis of snRNAs was performed according to Utans et al. (1992). The blot was hybridized with 32P-UTP–labeled antisense snRNA transcripts.
Metabolic Labeling
HeLa cells (6 × 105) were transfected with siRNAs and incubated 36 h later for 15 min at 37°C in DMEM high glucose, without methionine and cysteine, followed by a 1-h incubation at 37°C in the presence of 0.1 mCi/ml [35S]methionine and 35S-cysteine (Promix; Amersham, Zürich, Switzerland). Cells were washed in PBS, collected by centrifugation, and lysed in 2× SDS gel loading buffer. Equal aliquots of the lysates were separated by SDS-PAGE, and proteins were visualized by autoradiography.
In Vitro Translation
Cytoplasmic RNA (1 μg) isolated 60 h after siRNA transfection was translated in vitro with the Rabbit Reticulocyte Lysate System (Promega). Translation products were separated by SDS-PAGE and visualized by autoradiography.
Western Blotting
Total cells were collected 48 or 72 h posttransfection, washed with PBS, lysed in 2× SDS gel loading buffer and sonicated briefly. Proteins were separated by SDS-PAGE and transferred to nitrocellulose. Membranes were stained with Ponceau S, blocked with 5% dry milk in 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.05% Tween 20 and probed with primary and secondary antibodies diluted in blocking buffer. Primary antibodies were as follows: rabbit antibodies against SF3a60 (pAb60; Krämer et al., 1994), SF3a120 (pAb120; Krämer et al., 1995), and p80-coilin (Bohmann et al., 1995), and mouse monoclonal antibodies against SF3a66 (mAb66; Brosi et al., 1993b), SF1 (mAbSF1; Z. Rafi and A. Krämer, unpublished data), RNA pol II (ProGen, Heidelberg, Germany), antitubulin (Sigma-Aldrich), U2-B″ (Habets et al., 1989), U2AF65 (Gama-Carvalho et al., 1997), WT1 (Santa Cruz Biotechnology, Santa Cruz, CA), SC35 (Sigma-Aldrich), and SMN (BD Transduction Laboratories, Lexington, KY). Secondary horseradish peroxydase–conjugated rabbit anti-mouse and swine anti-rabbit antibodies (DAKO, Zug, Switzerland) were detected with the SuperSignal kit (Pierce, Rockford, IL).
Detection of U2 snRNP Complexes
Small scale nuclear extracts (Krämer and Keller, 1990) were prepared from mock-treated or siRNA-treated HeLa cells and incubated with a 5′ end-labeled, 2′-_O_-methyl oligoribonucleotide complementary to the 5′ end of U2 snRNA as described (Brosi et al., 1993a). Control reactions were performed in the presence of a partially purified fraction enriched in 15S and 17S U2 snRNPs (Krämer et al., 1999). Reaction products were resolved in a native 4% polyacrylamide gel and visualized by autoradiography (Krämer, 1988).
Indirect Immunofluorescence
HeLa cells were grown on coverslips, transfected with siRNAs, and washed twice with PBS. For indirect immunofluorescence cells were fixed with methanol for 5 min at -20°C and washed in PBS for 5 min at room temperature before incubation with primary and secondary antibodies for 30 min at room temperature each. Antibodies were diluted in PBS containing 0.2% Nonidet P40. In addition to the primary antibodies mentioned above mouse monoclonal anti-p80-coilin 5P10 (Almeida et al., 1998) was used. Secondary antibodies were as follows: fluorescein (FITC)-conjugated donkey anti-rabbit and anti-mouse, and Texas Red-conjugated donkey anti-rabbit and anti-mouse (all from Jackson ImmunoResearch Europe, Cambridgeshire, United Kingdom).
Transcription Analysis
Sixty hours after transfection with siRNA 60/2, HeLa cells were pulse-labeled with 2 mM 5-fluoro-uridine (FU) (Sigma) for 45 min, washed twice in PBS, fixed with 2% paraformaldehyde in PBS for 10 min at room temperature, and extracted with 0.1% Triton X-100 in PBS for 10 min at room temperature. FU incorporation was detected with mouse anti-bromodesoxy-uridine (BrdU) (Sigma-Aldrich) and FITC-donkey anti-mouse.
Fluorescence In Situ Hybridization
Cells were fixed with 3% paraformaldehyde in 100 mM KCl, 3 mM MgCl2, 1 mM CaCl2, 200 mM sucrose, and 10 mM HEPES-KOH, pH 7.1, for 5 min on ice and 15 min at room temperature. Extraction was performed in PBS, 0.5% bovine serum albumin, 20 mM glycine, 0.1% saponin, and 0.1% NaN3 for 15 min at room temperature followed by washing with PBS for 5 min at room temperature. Fluorescein (FAM) or Cy3-conjugated oligonucleotides complementary to nts 4–44 of U2 snRNA, and a Cy3-conjugated oligonucleotide complementary to nts 1–18 of U4 snRNA were used for hybridization according to Taneja et al. (1992). Samples were stained for immunofluorescence with pAb60 and 7-amino-4-methylcoumarin-3-acetic acid (AMCA)-conjugated goat anti-rabbit (Vector Laboratories, Burlingame, CA).
Fluorescence Microscopy
Microscopy was performed on an inverted fluorescence microscope (Axiovert TV135; Zeiss, Jena, Germany) with a 100× oil objective at standard wavelengths and filters for the fluorophores mentioned above. Images were recorded with a CCD camera (CH250, Photometrics, Tucson, AZ), using the software package IPLab spectrum V2.3 (Scanalytics, Billerica, MA). Images were recorded individually for each filter-channel and subsequently pseudocolored and superimposed in Adobe Photoshop 7.0 (Adobe Systems, San Jose, CA).
The analysis of transcription inhibition (see Figure 5) and the comparison of U2-B″ and U2 snRNA staining after SF3a60 depletion (see Figure 6, B and C) were performed with a filter-free confocal microscope (TCS SP2 AOBS, Leica, Heerbrugg, Switzerland) equipped with a HCX PL APO Ibd. BL 63× 1.4 oil objective. Images were recorded sequentially for AMCA-emission (detection of SF3a60), FITC-emission (U2-B″), and Cy3-emission (U2 snRNA). Stacks of 10 _z_-sections with 285-nm _z_-step were collected. Projections of all optical sections are shown.
Figure 5.
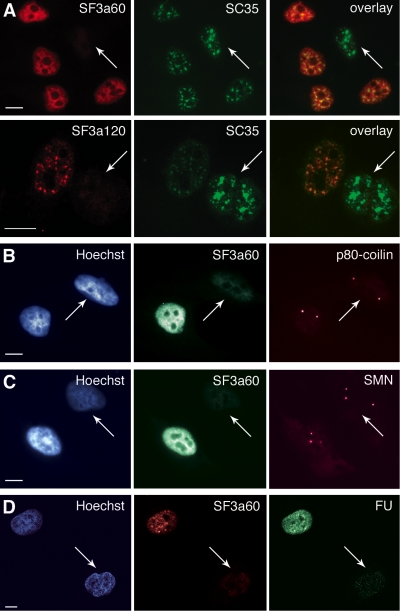
Effects of SF3a depletion on nuclear structures and transcription. (A) HeLa cells were transfected with siRNAs 60/2 or 120/2, fixed 60 h posttransfection, and immunostained with pAb60 or pAb120 and Texas Red-anti-rabbit, and anti-SC35 and FITC-anti-mouse as indicated. The right panels show computer-generated overlays of the images. Arrows indicate cells depleted of SF3a. (B) and (C) HeLa cells transfected with siRNA 60/2 were fixed 60 h posttransfection and immunostained with pAb60 and FITC-anti-rabbit, and anti-p80-coilin (B) or anti-SMN (C) and Texas Red-anti-mouse as indicated. Nuclei were visualized by Hoechst staining (left panels). (D) HeLa cells transfected with siRNA 60/2 were pulse-labeled 60 h posttransfection with FU for 45 min. Incorporation of FU into newly synthesized RNA was detected by confocal microscopy after staining with anti-BrdU and FITC-anti-mouse (right). SF3a60 was visualized with pAb60 and Texas Red anti-rabbit (middle). Nuclei were visualized by Hoechst staining (left). Scale bars, 10 μm.
Figure 6.
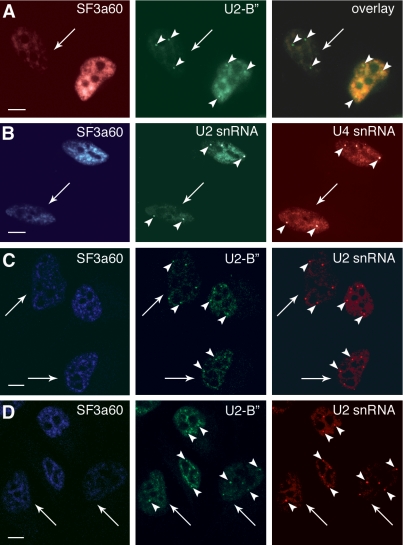
Depletion of U2 snRNA and U2-B″ from speckles, but not from CBs after knockdown of SF360. (A) HeLa cells transfected with siRNA 60/2 were fixed 60 h posttransfection. Cells were stained with pAb60 and Texas Red-anti-rabbit, and anti-U2-B″ and FITC-anti-mouse as indicated. The right panel shows a computer-generated overlay of the left and middle images. (B) Cells transfected and fixed as in A were incubated with pAb60 and AMCA-anti-rabbit and hybridized with FAM- and Cy3-conjugated oligonucleotides complementary to U2 and U4 snRNAs, respectively, as indicated. (C) and (D) Confocal images of cells transfected with siRNA 60/2 and stained for U2-B″ and U2 snRNA as in panels A and B 60 h posttransfection. Arrows mark SF3a60-depleted cells; arrowheads indicate CBs. Scale bars, 10 μm.
RESULTS
The SF3a Subunits and SF1 Are Required for Cell Viability
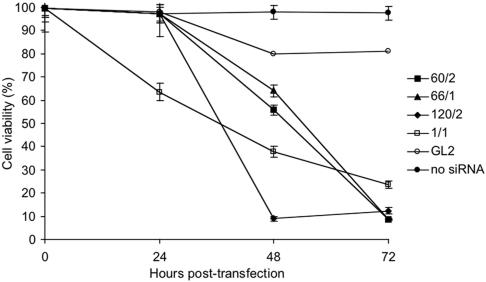
Single SF3a subunits were depleted from HeLa cells by transfection with synthetic small interfering (si) RNAs. Effects on cell proliferation were apparent in cells treated with siRNAs 60/2, 66/1, and 120/2 48 h later (Figure 1). At 72 h posttransfection, cell viability was reduced to ∼10% in all cases and the levels of the targeted SF3a subunits were highly decreased (see Figure 3A). Low amounts of SF3a60 and SF3a66 were still present, but apparently insufficient for cell survival. Depletion of the SF3a subunits by a second set of siRNAs was less efficient and consequently fewer cells died (see Figure 3A; unpublished data). Transfection of a control siRNA (GL2) targeting the firefly luciferase mRNA resulted in <20% cell death after 72 h, and mock-transfection in the absence of siRNA had no effect (Figure 1). None of the SF3a subunits nor SF1 were depleted in these experiments (see Figure 3, A and B; unpublished data). Down-regulation of SF3a caused necrotic cell death, as shown by positive propidium-iodide staining and a nuclear morphology typical for necrotic cells (unpublished data). Staining of SF3a-depleted cells with a marker for activated caspases (FITC-Val-Ala-DL-Asp(O-methyl)-fluoro-methylketone) was negative and DNA-laddering typical for apoptotic cells was not detected.
Figure 1.
Effects of SF3a and SF1 depletion on viability of HeLa cells. HeLa cells were transfected with siRNAs 60/2, 66/1, 120/2, SF1/1, and GL2, or mock-transfected in the absence of siRNA. Cell viability was monitored 24, 48, and 72 h posttransfection. The average of three independent experiments (except for GL2) is shown.
Figure 3.
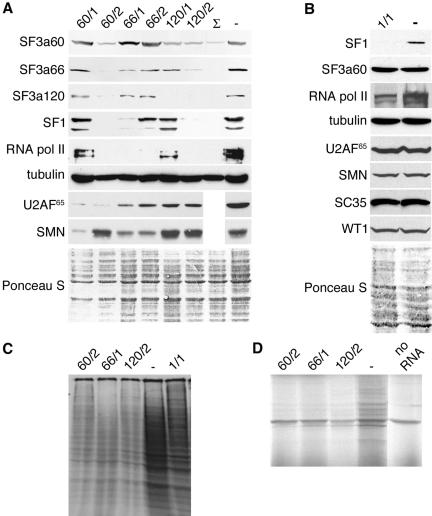
Effects of SF3a and SF1 depletion on protein expression. (A) HeLa cells were transfected with siRNAs indicated above the figure or oligofectamine alone (-). A triple transfection (Σ) was performed with siRNAs 60/2, 66/1, and 120/2. Total cell lysates were prepared 72 h posttransfection, separated by 7.5% SDS-PAGE, and transferred to nitrocellulose. Membranes were incubated with the antibodies indicated on the left. The bottom panel shows a representative Ponceau S–stained membrane as a loading control. (B) Total lysates of cells transfected with siRNA SF1/1 or oligofectamine alone (-) were prepared 48 h posttransfection and analyzed as in A. (C) HeLa cells transfected with siRNAs 60/2, 66/1, 120/2, SF1/1, or oligofectamine alone (-) were metabolically labeled with [35S]methionine and 35S-cysteine. Cell lysates were separated by 10% SDS-PAGE, and translation products were visualized by autoradiography. (D) Cytoplasmic RNA was isolated from HeLa cells transfected with siRNAs 60/2, 66/1, 120/2, or oligofectamine alone (-) and translated in vitro. A control reaction was performed in the absence of RNA (no RNA). Translation products were separated by 12% SDS-PAGE and visualized by autoradiography.
SF1 exists in several isoforms that share N-terminal sequences but differ in their C termini (Krämer et al., 1998 and references therein). To deplete all known SF1 isoforms, two siRNAs targeting the common 5′ region of the mRNAs were used. Transfection of HeLa cells with either siRNA resulted in SF1 depletion and reduced cell viability to ∼25% within 72 h (see Figures 1 and 3B; unpublished data). Hoechst staining of SF1-depleted cells revealed fragmented nuclei typical for apoptotic cells (unpublished data). In control experiments apoptosis was delayed after treatment with the caspase inhibitor Z-Val-Ala-DL-Asp(_O_-methyl)-fluoro-methylketone. Moreover, Western blot analysis indicated a time-dependent cleavage of procaspase 3 and accumulation of caspase 3, which correlated with the degree of SF1 depletion.
Knockdown of the SF3a Subunits but Not SF1 Leads to an Accumulation of Unspliced pre-mRNA
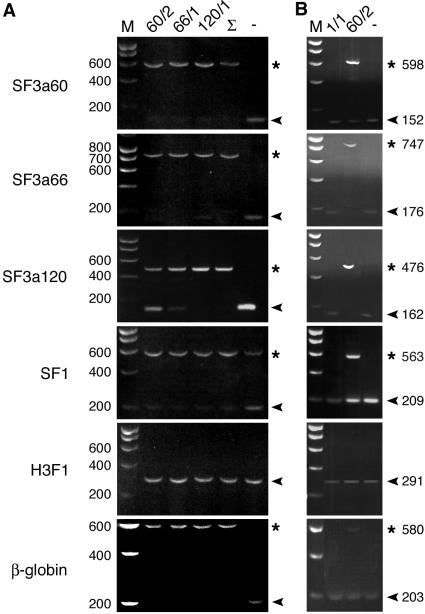
To determine whether depletion of SF3a and SF1 caused a splicing defect in vivo, total RNA was isolated from siRNA-treated cells 60 h posttransfection, and splicing of endogenous pre-mRNAs encoding the SF3a subunits and SF1 was analyzed by RT-PCR with primers designed to neighboring exons. As expected, after transfection of SF3a siRNAs the targeted mRNAs were no longer detectable (Figure 2A). In addition, mRNA levels of the nontargeted SF3a subunits and SF1 were highly reduced. At the same time the corresponding pre-mRNAs accumulated, consistent with a block in splicing. In time-course experiments pre-mRNAs were observed as early as 48 h (unpublished data). mRNA stability did not appear to be affected, because the levels of histone H3F1 mRNA, which is derived from an intron-less gene, were comparable in siRNA-treated and control cells. Moreover, neither mRNAs nor pre-mRNAs were aberrantly degraded. Similar to the results obtained with endogenous pre-mRNAs, a β-globin pre-mRNA transcribed from a plasmid transfected 24 h after siRNA transfection accumulated in siRNA- but not mock-transfected cells. Thus, in agreement with in vitro results (Nesic and Krämer, 2001), each SF3a subunit is essential for the splicing of at least a subset of pre-mRNAs in vivo.
Figure 2.
Effects of SF3a and SF1 depletion on pre-mRNA accumulation. (A) HeLa cells were transfected with siRNAs 60/2, 66/1, 120/2, all three (Σ) or oligofectamine alone (-). Total RNA isolated 60 h posttransfection was used for RT-PCR with primers specific for endogenous SF3a60, SF3a66, SF3a120, SF1 and histone H3F1 RNAs, and RNA transcribed from the transiently transfected β-globin reporter plasmid pAdCMV-glob. (B) HeLa cells were transfected with siRNAs SF1/2, 60/2 or oligofectamine alone (-). RNA isolation and RT-PCR was performed as in A. Reaction products were separated in agarose gels. RT-PCR products derived from pre-mRNAs and mRNAs are marked by asterisks and arrowheads, respectively. The sizes of DNA markers (M) are shown on the left.
RT-PCR of total RNA isolated from cells transfected with SF1 siRNAs indicated reduced amounts of SF1 mRNA as expected (Figure 2B). However, no accumulation of the corresponding pre-mRNA was evident, which is in contrast to the results obtained after depletion of SF3a60 or the other subunits. Furthermore, no decrease in the amount of mRNAs of the SF3a subunits was apparent and splicing of the β-globin reporter pre-mRNA was unaffected. Again, the intron-less H3F1 mRNA was stable in SF1-depleted cells. These results suggest that SF1 is not essential for the splicing of the pre-mRNAs tested.
RNAi of Single SF3a Subunits Severely Reduces Protein Expression
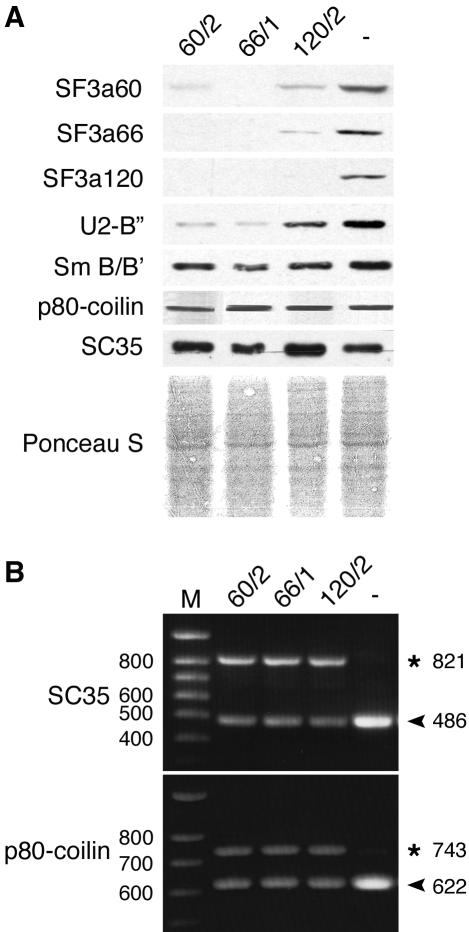
Given that depletion of individual SF3a subunits blocked splicing of selected pre-mRNAs, we examined whether protein levels were affected as well. To this end, HeLa cells were transfected with both sets of siRNAs, total cell lysates were prepared 72 h posttransfection and analyzed by Western blotting. In cells transfected with siRNA 60/2 not only SF3a60, but also SF3a66 and SF3a120 were highly reduced when compared with mock-transfected cells (Figure 3A), consistent with the observation that the splicing of the corresponding pre-mRNAs was inhibited (Figure 2A). Similarly, transfection of siRNAs 66/1 and 120/2 resulted in decreased levels of all subunits. Western blotting with additional antibodies indicated that at least two SF1 isoforms and differentially phosphorylated forms of the largest subunit of RNA polymerase II (RNA pol II) were highly diminished (Figure 3A). Moreover, variable effects on the expression of U2AF65 and the survival of motor neuron (SMN) protein were evident. In contrast, tubulin levels, tested as a control, were not significantly changed. As mentioned above, effects on cell viability of siRNAs 60/1, 66/2, and 120/1 were less pronounced compared with the other siRNAs. Accordingly, transfection of cells with these siRNAs resulted in reduced depletion of both targeted and nontargeted proteins. A triple transfection of HeLa cells with siRNAs 60/2, 66/1, and 120/2 had the strongest effect on protein levels. Thus, the degree of codepletion correlates with the level of depletion of the targeted protein. These results indicate that depletion of single SF3a subunits causes a concomitant depletion of other proteins as a consequence of a block in splicing.
In contrast to the results obtained after SF3a depletion, knockdown of SF1 did not result in codepletion of SF3a60, RNA pol II, U2AF65, or SMN (Figure 3B). Moreover, the relative abundance of several proteins encoded by intron-containing genes, such as splicing factor SC35, Wilms' tumor protein 1 (WT1), or SMN, was not visibly reduced. We cannot exclude that some of these proteins or their mRNAs have a relatively long half-life (see below). However, the observation that SF1 depletion does not affect SF3a60, U2AF65, or RNA pol II levels, which are clearly reduced after RNAi of SF3a, strengthens the idea that SF1 is not involved in the splicing of the corresponding pre-mRNAs.
As an indirect means to examine whether knockdown of the SF3a subunits caused a more general splicing defect, we metabolically labeled siRNA-transfected cells. Figure 3C shows that depletion of each SF3a subunit significantly reduced protein synthesis as early as 36 h posttransfection. To rule out that this effect was due to a defect in translation, cytoplasmic RNA isolated from siRNA- and mock-treated cells 60 h posttransfection was translated in a rabbit reticulocyte lysate. In contrast to reactions carried out with cytoplasmic RNA from mock-transfected cells, the incorporation of [35S]methionine into newly synthesized proteins was almost completely abolished in the presence of RNA from siRNA-treated cells and was comparable to the background translation observed in the absence of added RNA (Figure 3D). Thus, the severe reduction in protein synthesis following depletion of SF3a is caused by a reduction in the amount of translatable cytoplasmic RNA. Depletion of SF1 only marginally reduced overall protein synthesis in metabolically labeled HeLa cells (Figure 3C).
During the course of our experiments we observed that protein levels of SC35, p80-coilin, a protein specifically associated with CBs (Raska et al., 1991), and the snRNP-associated proteins SmB/B′ were not or only partially reduced after SF3a depletion (Figure 4A). Because these proteins are encoded by intron-containing genes, we performed RT-PCR to test whether SF3a depletion affected splicing of the corresponding pre-mRNAs. Unspliced RNA of SC35 and p80-coilin had accumulated 72 h after transfection of SF3a siRNAs (Figure 4B). At the same time mRNA was still detectable in siRNA-treated cells, however, at reduced levels. Control RT-PCR indicated a complete block in splicing of the SF3a pre-mRNAs similar to the results presented above (unpublished data). Thus it is likely that the splicing of SC35 and p80-coilin pre-mRNAs also depends on SF3a. The persistence of SC35 and p80-coilin (and probably also that of SmB/B') in SF3a-depleted cells could be explained by a longer half-life of the proteins and/or the mRNAs.
Figure 4.
Effects of SF3a depletion on U2-B″, SmB/B′, p80-coilin, and SC35 expression. (A) Cell lysates prepared from HeLa cells transfected with siRNAs 60/2, 66/1, 120/2, or oligofectamine alone (-) were prepared 60 h later and separated by 12% SDS-PAGE followed by detection of the proteins indicated on the left by Western blotting. The bottom panel shows the Ponceau S–stained membrane as a loading control. (B) HeLa cells were transfected with siRNAs 60/2, 66/1, 120/2, or oligofectamine alone (-). Total RNA isolated 60 h posttransfection was used for RT-PCR with primers specific for SC35 and p80-coilin RNAs. Reaction products were analyzed as in Figure 2.
Together these data demonstrate that down-regulation of single SF3a subunits results in global effects on gene expression rather than compromising the expression of only a subset of intron-containing genes. Thus, each SF3a subunit is most likely required for general splicing in vivo. In contrast, SF1 does not appear to classify as a constitutive splicing factor, because its depletion does not inhibit the splicing of several pre-mRNAs tested nor overall translation.
Depletion of SF3a Results in an Enlargement of Nuclear Speckles and a Reduction in Transcriptional Activity
In light of the marked effects of SF3a depletion on splicing and protein expression, we asked whether the intracellular distribution of splicing components and/or nuclear morphology was affected by SF3a down-regulation. HeLa cells were stained with an antibody against SC35, which exhibits a characteristic speckled localization (Fu and Maniatis, 1990). SC35 was detected in speckles in nontransfected cells, but accumulated in larger bright foci in cells depleted of SF3a60 or SF3a120 (Figure 5A). These foci resemble enlarged speckles observed after inhibition of splicing or transcription (Carmo-Fonseca et al., 1992; O'Keefe et al., 1994). Thus, in agreement with the block in splicing demonstrated by RT-PCR (Figures 2A and 4B), SF3a depletion causes changes in nuclear morphology typical for splicing inhibition.
SnRNP components are not only associated with speckles but also found in CBs. The fluorescence intensity of these structures was unchanged in cells depleted for SF3a60 or the other subunits as revealed by immunofluorescence with anti-p80-coilin (Figure 5B; unpublished data). Moreover, the number of CBs remained constant with more than 70% of transfected and mock-treated cells containing 2–4 CBs (unpublished data). Similarly, we observed no changes in the morphology of gems (Figure 5C), which contain components of the SMN complex and are related to and often found associated with CBs (Sleeman and Lamond, 1999b).
The largest subunit of RNA pol II was at least partially depleted after RNAi of SF3a (Figure 3A). Thus, to test whether transcription was inhibited, cells transfected with siRNA 60/2 were pulse-labeled for 45 min with FU 60 h posttransfection. Immunofluorescence with anti-BrdU antibodies indicated that depletion of SF3a60 caused a severe reduction of FU incorporation into newly transcribed RNA (Figure 5D), demonstrating that SF3a depletion not only inhibits splicing but, probably as a consequence, also blocks overall transcription.
It has been reported that siRNAs can nonspecifically stimulate or repress gene expression in a concentration-dependent manner (Semizarov et al., 2003; Persengiev et al., 2004). For the following reasons we believe that the effects seen after SF3a depletion are specific. First, in database searches no significant homology between the siRNAs used and sequences other than the target sequences were found. Second, knockdown with two different siRNAs for each subunit resulted in similar effects on splicing and other aspects of gene expression. Moreover, residual cell viability and decrease in the concentration of other proteins correlated with the degree of depletion of each SF3a subunit by the two siRNAs used. Third, as expected from proteins that function in a tight complex, depletion of each subunit generated highly related in vivo phenotypes. Finally, depletion of SF1 with the same protocol resulted in different, less pronounced, or no effects.
Down-regulation of SF3a Causes Depletion of Other U2 snRNP Components from the Nucleoplasm and Speckles, but Not from CBs
Recent evidence suggests that the incorporation of SF3a into the U2 snRNP, resulting in the formation of the mature 17S U2 snRNP, takes place in CBs (Nesic et al., 2004). The observation that constituents of the 12S U2 snRNP, e.g., U2 snRNA, U2-B″ and Sm proteins, are distributed in speckles and CBs under steady state conditions (Carmo-Fonseca et al., 1991, 1992; Matera and Ward, 1993), whereas SF3a is only detected in speckles (Nesic et al., 2004; Figure 6), could be explained by a rapid movement of the U2 snRNP from CBs to sites of splicing and storage once its maturation is completed. We therefore reasoned that SF3a depletion could block U2 snRNP maturation and consequently lead to an accumulation of immature U2 particles in CBs. Consistent with this prediction, staining of SF3a60-depleted cells with anti-U2-B″ showed a reduction of U2-B″ in the nucleoplasm, but the protein was still detected in CBs (Figure 6A). The same observation was made for U2 snRNA, visualized by fluorescence in situ hybridization (FISH; Figure 6B). Figures 6, C and D, show confocal images of cells depleted of SF3a60 to varying degrees. Although the nucleoplasmic staining of U2-B″ and U2 snRNA varies in these cells, CBs are stained in all nuclei. Quantification of the fluorescence intensity in nuclei and CBs of a large number of confocal images indicated that SF3a60 depletion caused a significant increase of both U2 snRNA and U2-B″ in CBs compared with the remaining nucleoplasm (unpublished data), suggesting that SF3a depletion mostly affects the nucleoplasmic pool of the U2 snRNP. Changes in the localization of U4 snRNA, analyzed as a control, were not apparent (Figure 6B). In addition, we did not observe significant changes in the localization or abundance of Sm proteins, which are common to all snRNPs (unpublished data). To analyze the distribution of SF3b, which is part of the mature U2 snRNP, we used antibodies against SF3b155. The protein could not be detected in SF3a-depleted cells, because it was efficiently codepleted with SF3a (unpublished data).
The Mature 17S U2 snRNP Cannot Be Detected in Extracts of SF3a60-depleted Cells
In HeLa nuclear extracts, the majority of the U2 snRNP is present in the 17S form (Behrens et al., 1993; Brosi et al., 1993a; Krämer et al., 1999). To test whether SF3a depletion affected the integrity of the particle, nuclear extract from SF3a60-depleted cells was incubated with a radiolabeled oligoribonucleotide complementary to the 5′ end of U2 snRNA, followed by native PAGE and visualization of U2 snRNP complexes by autoradiography. Consistent with previous results (Brosi et al., 1993a) most of the U2 snRNP in an extract from mock-treated cells migrated at the position of the 17S U2 snRNP (Figure 7A, cf. lanes 1 and 2). In contrast, the 17S U2 snRNP was not visible in the SF3a60-depleted extract (lane 3). Instead, a smear of radioactivity below the migration of the 15S U2 snRNP was observed in addition to an accumulation of particles migrating at the position of the 12S U2 snRNP (cf. lanes 1 and 3). Partially purified 17S and 15S U2 snRNPs (lane 1) used to complement the extract from SF3a60-depleted cells were not affected in their migration and appeared stable in the extract (lane 4), ruling out nonspecific degradation of larger U2 particles. Thus, depletion of SF3a60 from HeLa cells causes a disappearance of the 17S U2 snRNP, as expected if one or more SF3a subunits are reduced in amount or absent. The integrity of the 15S U2 snRNP was also affected, consistent with the codepletion of SF3b155 and presumably other SF3b subunits. In contrast, considerable amounts of the 12S U2 snRNP remained after SF3a60 depletion, suggesting only minor effects on the integrity of this particle. Furthermore, U2 snRNA or other snRNAs were not reduced in abundance in SF3a-depleted cells as revealed by Northern blotting of total RNA (Figure 7B), consistent with the high metabolic stability of snRNAs (Fury and Zieve, 1996).
Figure 7.
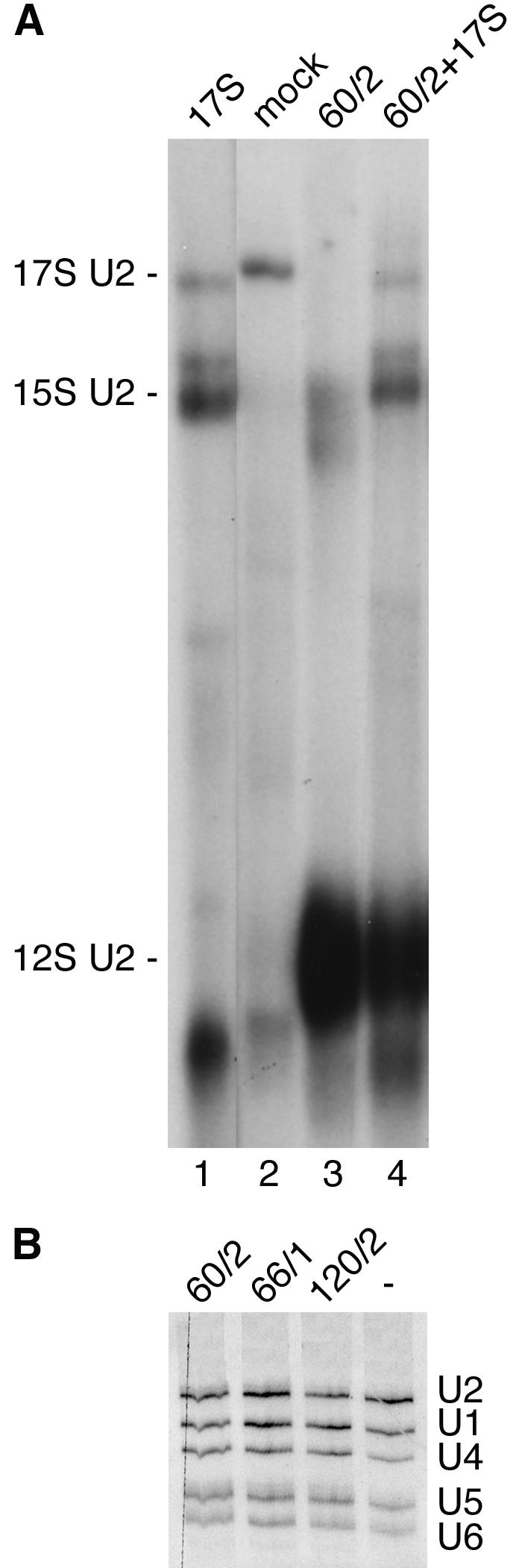
Analysis of U2 snRNPs and snRNAs in SF3a60-depleted cells. (A) HeLa cells were mock-treated or transfected with siRNA 60/2. Aliquots of small-scale nuclear extracts (normalized for protein concentration) prepared 72 h posttransfection from mock-treated (lane 2) and siRNA-transfected (lane 3) cells were incubated with a labeled oligoribonucleotide complementary to the 5′ end of U2 snRNA. Partially purified 17S and 15S U2 snRNPs (lane 1) were used as markers for the migration of U2 particles. The extract from siRNA-treated cells was supplemented with the U2 snRNP fraction (lane 4). SnRNP complexes were separated by native 4% PAGE and visualized by autoradiography. The migration of U2 complexes is indicated on the left. (B) SnRNAs present in total RNA isolated from cells 60 h after transfection with siRNAs 60/2, 66/1, and 120/2 or oligofectamine alone (-) were analyzed by Northern blotting. The snRNAs are indicated on the right.
DISCUSSION
Proteomic approaches suggest that more than 100 proteins participate in splicing in addition to five snRNAs (Jurica and Moore, 2003; Nilsen, 2003). Many of these are essential for growth and splicing in S. cerevisiae (Hodges et al., 1997). Although genetic methods and RNAi revealed a requirement for viability of splicing factors also in metazoan cells, information regarding their role in splicing in vivo is limited (for examples see Rudner et al., 1996; Wang et al., 1996; Jumaa et al., 1999; Longman et al., 2000; Wang et al., 2001; Piano et al., 2002; Kamath et al., 2003; Simmer et al., 2003; Boutros et al., 2004; Ding et al., 2004; Schaffert et al., 2004). Here we have examined the in vivo function of two splicing factors after RNAi-mediated knockdown in human cells.
SF3a Is a Constitutive Splicing Factor
SF3a is essential for splicing in vitro and each subunit is necessary for function (Brosi et al., 1993b; Nesic and Krämer, 2001). We have shown that the SF3a subunits are also essential for viability of human cells, similar to their orthologues in other organisms (Krämer, 1996; Meyer et al., 1998; Piano et al., 2002; Kamath et al., 2003; Simmer et al., 2003; Boutros et al., 2004). Knockdown of each subunit blocked splicing of selected pre-mRNAs. In addition, several proteins were codepleted with SF3a. Levels of other proteins (p80-coilin, Sm B/B′ and SC35) were only marginally affected in the time frame analyzed. Pre-mRNAs also accumulated in these cases, indicating that their splicing was compromised and suggesting that either the corresponding mRNAs or proteins have relatively long half-lives. The decrease in overall protein synthesis after SF3a knockdown can at least in part be attributed to a reduction in the amount of translatable mRNA. In addition, defects in transcription, mRNA export and translation, most likely caused by depletion of proteins involved in these processes due to a failure in splicing, can contribute to the observed reduction in protein levels. Together, these results are consistent with a function of SF3a in the splicing of many, if not all U2-type pre-mRNAs. Thus, SF3a represents an essential, most likely constitutive splicing factor in vivo.
In addition to its association with SF3a, SF3a120 was found in a nuclear receptor corepressor complex involved in transcriptional silencing (Underhill et al., 2000). Hence, SF3a120 depletion may also compromise cell viability by a direct influence on transcription. SF3a66 has recently been described as a microtubule-binding and bundling protein in neuronal cells (Takenaka et al., 2004). As SF3a66 is not detected in the cytoplasm of HeLa cells by immunofluorescence (Nesic et al., 2004), direct effects of SF3a66 depletion on microtubule dynamics in HeLa cells can probably be ruled out.
SF1 Is Essential for Cell Viability but Not for General Splicing
Knockdown of SF1 did not block splicing of any pre-mRNA tested or decrease the concentration of specific proteins or overall translation. Thus, by these criteria SF1 does not classify as a constitutive splicing factor. Nevertheless, SF1 depletion interferes with viability in human cells, indicating that it fulfils an essential function in vivo. Genetic or biochemical depletion of SF1 from S. cerevisiae and human cells did not affect the final outcome of splicing; however, the kinetics of early steps of spliceosome assembly were slowed down (Rutz and Séraphin, 1999; Guth and Valcárcel, 2000). In S. cerevisiae, SF1 dissociates from the spliceosome upon U2 snRNP binding to the branch site and is presumably recycled to participate in new rounds of spliceosome assembly, suggesting that SF1 acts catalytically (Rutz and Séraphin, 2000). Thus, low amounts of SF1 remaining after RNAi could be sufficient for splicing. Although we cannot formally exclude this possibility, it is more likely that residual SF1 detected in extracts from siRNA-treated cells is derived from nontransfected cells. Despite the fact that we failed to detect an inhibition of splicing, SF1 may be required for the splicing of only certain pre-mRNAs, for example, those with suboptimal splice sites, as suggested from studies in S. cerevisiae (Rutz and Séraphin, 2000).
In yeast, SF1 has also been implicated in nuclear pre-mRNA retention (Rutz and Séraphin, 2000; Galy et al., 2004). Given the deleterious effects of pre-mRNA export on cell metabolism, a function of SF1 in pre-mRNA retention could be essential for viability in mammalian cells. In addition, roles for SF1 in transcriptional repression have been reported (Zhang and Childs, 1998; Zhang et al., 1998; Goldstrohm et al., 2001). SF1 depletion did not reduce FU incorporation into nascent transcripts nor reverse the repression of transcription elongation mediated by CA150 (Goldstrohm et al., 2001; Goldstrohm et al., unpublished data). Thus, further experiments are required to solve the question of why SF1 is essential for viability in human cells.
SF3a Depletion Causes a Redistribution of U2 snRNP Components in the Nucleus
To provide further support for our proposal that CBs represent sites of U2 snRNP maturation (Nesic et al., 2004), we analyzed the localization of U2 snRNP components after SF3a knockdown. Depending on the degree of SF3a depletion, U2 snRNA and U2-B″ were more or less reduced or no longer visible in the nucleoplasm, but both components remained concentrated in CBs. This pattern may be expected if mature U2 snRNPs turned over in the nucleoplasm, whereas newly imported, immature U2 snRNPs are blocked in their movement from CBs to sites of splicing and/or storage because of decreased SF3a levels. U2-B″ was reduced in abundance after SF3a knockdown, suggesting that at least a portion of the protein turned over. In contrast, U2 snRNA levels were not visibly affected, consistent with the stability of snRNAs (Fury and Zieve, 1996). However, 72 h after SF3a depletion U2 snRNA was no longer associated with 17S particles, but accumulated as 12S particles. Thus, the discrepancy between unchanged U2 snRNA levels and decreased nucleoplasmic staining could be explained by a weakened association of partially or completely disassembled U2 snRNPs with speckles and a consequent loss during the extraction and fixation procedures. On the other hand, newly imported, immature U2 snRNPs may be tightly anchored in CBs until fully matured. Given the severe effects of SF3a depletion on gene expression, it is also possible that the reimport of core U2 snRNPs into the nucleus is slowed down and the cytoplasmic fraction of the U2 snRNP may be lost during sample preparation.
It is unlikely that the changes in U2 snRNA and U2-B″ localization after SF3a depletion are merely a consequence of the observed inhibition of splicing or transcription. When these processes are inhibited by drug treatment or injection of snRNA antisense oligonucleotides or anti-snRNP antibodies, U2 snRNA and U2-B″ accumulate in enlarged speckles and are no longer detected in CBs (Carmo-Fonseca et al., 1992; O'Keefe et al., 1994). We therefore conclude that after SF3a depletion partially assembled U2 snRNPs remain in CBs because of the lack of SF3a and are inhibited from further movement, whereas fully assembled U2 snRNPs turn over in the nucleoplasm, and the decrease in functional U2 snRNPs ultimately results in an inhibition of splicing.
The concentration of U2 snRNA and U2-B″ in CBs and their depletion from the nucleoplasm after SF3a knockdown is paralleled by findings of Schaffert et al. (2004) that knockdown of U4/U6- or U5-specific proteins leads to an accumulation of U4/U6 snRNPs in CBs and a decrease of U4/U6.U5 snRNPs levels in cell extracts. In this case CBs were suggested to function in snRNP recycling between rounds of spliceosome assembly. Thus far, there is no indication that U2 snRNPs disassemble after dissociation from spliced introns, which could explain why we did not observe an increase in U2 snRNA and U2-B″ levels in CBs after SF3a depletion.
In summary, we have used RNAi to determine whether two proteins implicated in splicing in vitro are also essential for this process in vivo. Although this is the case for SF3a, SF1 does not appear to be a constitutive splicing factor and further studies are required to elucidate its in vivo function. Furthermore, our results indicate that RNAi-mediated knockdown of splicing proteins is a useful approach to gain insight into the nuclear dynamics of splicing components.
Acknowledgments
We thank Maria Carmo-Fonseca, Angus Lamond, and Walther Van Venrooij for antibodies. This work was supported by an exchange fellowship of the Swiss Confederation to G.T. and the Schweizerischer Nationalfonds, the Fondation Medic, and the Canton of Geneva to A.K.
Abbreviations used: AMCA, 7-amino-4-methylcoumarin-3-acetic acid; BrdU, bromo-desoxy-uridine; CB, Cajal body; FISH, fluorescence in situ hybridization; FU, fluoro-uridine; RNAi, RNA interference; RNA pol II, RNA polymerase II; siRNA, small interfering RNA; SMN, survival of motor neurons; snRNA, small nuclear RNA; snRNP, small nuclear ribonucleoprotein particle; WT1, Wilms' tumor 1.
References
- Abovich, N., and Rosbash, M. (1997). Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89, 403-412. [DOI] [PubMed] [Google Scholar]
- Almeida, F., Saffrich, R., Ansorge, W., and Carmo-Fonseca, M. (1998). Microinjection of anti-coilin antibodies affects the structure of coiled bodies. J. Cell Biol. 142, 899-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens, S. E., Galisson, F., Legrain, P., and Lührmann, R. (1993). Evidence that the 60-kDa protein of 17S U2 small nuclear ribonucleoprotein is immunologically and functionally related to the yeast PRP9 splicing factor and is required for the efficient formation of prespliceosomes. Proc. Natl. Acad. Sci. USA 90, 8229-8233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund, J. A., Chua, K., Abovich, N., Reed, R., and Rosbash, M. (1997). The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell 89, 781-787. [DOI] [PubMed] [Google Scholar]
- Berglund, J. A., Abovich, N., and Rosbash, M. (1998). A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 12, 858-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann, K., Ferreira, J. A., and Lamond, A. I. (1995). Mutational analysis of p80 coilin indicates a functional interaction between coiled bodies and the nucleolus. J. Cell Biol. 131, 817-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros, M. et al. (2004). Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303, 832-835. [DOI] [PubMed] [Google Scholar]
- Brosi, R., Gröning, K., Behrens, S.-E., Lührmann, R., and Krämer, A. (1993a). Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science 262, 102-105. [DOI] [PubMed] [Google Scholar]
- Brosi, R., Hauri, H. P., and Krämer, A. (1993b). Separation of splicing factor SF3 into two components and purification of SF3a activity. J. Biol. Chem. 268, 17640-17646. [PubMed] [Google Scholar]
- Brow, D. A. (2002). Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36, 333-360. [DOI] [PubMed] [Google Scholar]
- Burge, C., Tuschl, T., and Sharp, P. (1999). Splicing of precursors to mRNA by the spliceosome. In: The RNA World, 2nd ed., ed. R. Gesteland, T. Cech, and J. Atkins, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 525-560.
- Carmo-Fonseca, M., Pepperkok, R., Sproat, B. S., Ansorge, W., Swanson, M. S., and Lamond, A. I. (1991). In vivo detection of snRNP-rich organelles in the nuclei of mammalian cells. EMBO J. 10, 1863-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca, M., Pepperkok, R., Carvalho, M. T., and Lamond, A. I. (1992). Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J. Cell Biol. 117, 1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, C. A., and Guthrie, C. (2000). The question remains: is the spliceosome a ribozyme? Nat. Struct. Biol. 7, 850-854. [DOI] [PubMed] [Google Scholar]
- Ding, J. H. et al. (2004). Dilated cardiomyopathy caused by tissue-specific ablation of SC35 in the heart. EMBO J. 23, 885-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S. M., Harborth, J., Weber, K., and Tuschl, T. (2002). Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26, 199-213. [DOI] [PubMed] [Google Scholar]
- Estmer Nilsson, C., Petersen-Mahrt, S., Durot, C., Shtrichman, R., Krainer, A. R., Kleinberger, T., and Akusjärvi, G. (2001). The adenovirus E4-ORF4 splicing enhancer protein interacts with a subset of phosphorylated SR proteins. EMBO J. 20, 864-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, J. A., Carmo-Fonseca, M., and Lamond, A. I. (1994). Differential interaction of splicing snRNPs with coiled bodies and interchromatin granules during mitosis and assembly of daughter cell nuclei. J. Cell Biol. 126, 11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine, M., Rain, J.-C., and Legrain, P. (1997). Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16, 277-282. [DOI] [PubMed] [Google Scholar]
- Fu, X.-D., and Maniatis, T. (1990). Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature 343, 437-441. [DOI] [PubMed] [Google Scholar]
- Fury, M. G., and Zieve, G. W. (1996). U6 snRNA maturation and stability. Exp. Cell Res. 228, 160-163. [DOI] [PubMed] [Google Scholar]
- Gall, J. G. (2000). Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol. 16, 273-300. [DOI] [PubMed] [Google Scholar]
- Galy, V., Gadal, O., Fromont-Racine, M., Romano, A., Jacquier, A., and Nehrbass, U. (2004). Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116, 63-73. [DOI] [PubMed] [Google Scholar]
- Gama-Carvalho, M., Krauss, R. D., Chiang, L., Valcárcel, J., Green, M. R., and Carmo-Fonseca, M. (1997). Targeting of U2AF65 to sites of active splicing in the nucleus. J. Cell Biol. 137, 975-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm, A. C., Albrecht, T. R., Sune, C., Bedford, M. T., and Garcia-Blanco, M. A. (2001). The transcription elongation factor CA150 interacts with RNA polymerase II and the pre-mRNA splicing factor SF1. Mol. Cell. Biol. 21, 7617-7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani, O., Feld, R., and Reed, R. (1996). Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 10, 233-243. [DOI] [PubMed] [Google Scholar]
- Gozani, O., Potashkin, J., and Reed, R. (1998). A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol. 18, 4752-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guth, S., and Valcárcel, J. (2000). Kinetic role for mammalian SF1/BBP in spliceosome assembly and function after polypyrimidine tract recognition by U2AF. J. Biol. Chem. 275, 38059-38066. [DOI] [PubMed] [Google Scholar]
- Habets, W. J., Hoet, M. H., De Jong, B. A., Van der Kemp, A., and Van Venrooij, W. J. (1989). Mapping of B cell epitopes on small nuclear ribonucleoproteins that react with human autoantibodies as well as with experimentally-induced mouse monoclonal antibodies. J. Immunol. 143, 2560-2566. [PubMed] [Google Scholar]
- Hodges, P., Plumpton, M., and Beggs, J. (1997). Pre-mRNA splicing factors in the yeast Saccharomyces cerevisiae. In: Eukaryotic mRNA Processing, ed. A. Krainer, Oxford: IRL Press, 213-241.
- Jády, B. E., Darzacq, X., Tucker, K. E., Matera, A. G., Bertrand, E., and Kiss, T. (2003). Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J. 22, 1878-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, M., Schallhorn, A., and Wurm, F. M. (1996). Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 24, 596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumaa, H., Wei, G., and Nielsen, P. J. (1999). Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Curr. Biol. 9, 899-902. [DOI] [PubMed] [Google Scholar]
- Jurica, M. S., and Moore, M. J. (2003). Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12, 5-14. [DOI] [PubMed] [Google Scholar]
- Kamath, R. S. et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231-237. [DOI] [PubMed] [Google Scholar]
- Kiss, A. M., Jady, B. E., Darzacq, X., Verheggen, C., Bertrand, E., and Kiss, T. (2002). A Cajal body-specific pseudouridylation guide RNA is composed of two box H/ACA snoRNA-like domains. Nucleic Acids Res. 30, 4643-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer, A. (1988). Pre-splicing complex formation requires two proteins and U2 snRNP. Genes Dev. 2, 1155-1167. [DOI] [PubMed] [Google Scholar]
- Krämer, A., and Keller, W. (1990). Preparation and fractionation of mammalian extracts active in pre-mRNA splicing. In: Methods Enzymology, Vol. 181, ed. J. E. Dahlberg and J. N. Abelson, New York: Academic Press, 3-19. [DOI] [PubMed] [Google Scholar]
- Krämer, A. (1992). Purification of splicing factor SF1, a heat-stable protein that functions in the assembly of a pre-splicing complex. Mol. Cell. Biol. 12, 4545-4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer, A., Legrain, P., Mulhauser, F., Gröning, K., Brosi, R., and Bilbe, G. (1994). Splicing factor SF3a60 is the mammalian homologue of PRP9 of S. cerevisiae: the conserved zinc finger-like motif is functionally exchangeable in vivo. Nucleic Acids Res. 22, 5223-5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer, A., Mulhauser, F., Wersig, C., Gröning, K., and Bilbe, G. (1995). Mammalian splicing factor SF3a120 represents a new member of the SURP family of proteins and is homologous to the essential splicing factor PRP21p of S. cerevisiae. RNA 1, 260-272. [PMC free article] [PubMed] [Google Scholar]
- Krämer, A. (1996). The structure and function of proteins involved in nuclear pre-mRNA splicing. Annu. Rev. Biochem. 65, 367-409. [DOI] [PubMed] [Google Scholar]
- Krämer, A., Quentin, M., and Mulhauser, F. (1998). Diverse modes of alternative splicing of human splicing factor SF1 deduced from the exon-intron structure of the gene. Gene 211, 29-37. [DOI] [PubMed] [Google Scholar]
- Krämer, A., Grüter, P., Gröning, K., and Kastner, B. (1999). Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J. Cell Biol. 145, 1355-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond, A. I., and Spector, D. L. (2003). Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell. Biol. 4, 605-612. [DOI] [PubMed] [Google Scholar]
- Liu, Z., Luyten, I., Bottomley, M., Messias, A., Houngninou-Molango, S., Sprangers, R., Zanier, K., Krämer, A., and Sattler, M. (2001). Structural basis for recognition of the intron branch site by splicing factor 1. Science 294, 1098-1102. [DOI] [PubMed] [Google Scholar]
- Longman, D., Johnstone, I. L., and Cáceres, J. F. (2000). Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 19, 1625-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova, O. V., Makarov, E. M., Liu, S., Vornlocher, H. P., and Lührmann, R. (2002). Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6.U5 tri-snRNP formation and pre-mRNA splicing. EMBO J. 21, 1148-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera, A. G., and Ward, D. C. (1993). Nucleoplasmic organization of small nuclear ribonucleoproteins in cultured human cells. J. Cell Biol. 121, 715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera, A. G. (1999). Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol. 9, 302-309. [DOI] [PubMed] [Google Scholar]
- Mazroui, R., Puoti, A., and Krämer, A. (1999). Splicing factor SF1 from Drosophila and Caenorhabditis: presence of an N-terminal RS domain and requirement for viability. RNA 5, 1615-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, V., Oliver, B., and Pauli, D. (1998). Multiple developmental requirements of NOISETTE, the Drosophila homolog of the U2 snRNP-associated polypeptide SF3a60. Mol. Cell. Biol. 18, 1835-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesic, D., and Krämer, A. (2001). Domains in human splicing factors SF3a60 and SF3a66 required for binding to SF3a120, assembly of the 17S U2 snRNP, and prespliceosome formation. Mol. Cell. Biol. 21, 6406-6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesic, D., Tanackovic, G., and Krämer, A. (2004). A role for Cajal bodies in the final steps of U2 snRNP biogenesis. J. Cell Sci. 117, 4423-4433. [DOI] [PubMed] [Google Scholar]
- Nilsen, T. W. (2003). The spliceosome: the most complex macromolecular machine in the cell? Bioessays 25, 1147-1149. [DOI] [PubMed] [Google Scholar]
- O'Keefe, R. T., Mayeda, A., Sadowski, C. L., Krainer, A. R., and Spector, D. L. (1994). Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J. Cell Biol. 124, 249-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg, S. C., and Lamond, A. I. (2002). Cajal bodies and coilin—moving towards function. J. Cell Biol. 159, 17-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persengiev, S. P., Zhu, X., and Green, M. R. (2004). Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs). RNA 10, 12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piano, F., Schetter, A. J., Morton, D. G., Gunsalus, K. C., Reinke, V., Kim, S. K., and Kemphues, K. J. (2002). Gene clustering based on RNAi phenotypes of ovary-enriched genes in C. elegans. Curr. Biol. 12, 1959-1964. [DOI] [PubMed] [Google Scholar]
- Query, C. C., Strobel, S.A., and Sharp, P. A. (1996). Three recognition events at the branch-site adenine. EMBO J. 15, 1392-1402. [PMC free article] [PubMed] [Google Scholar]
- Rain, J.-C., Rafi, Z., Rhani, Z., Legrain, P., and Krämer, A. (1998). Conservation of functional domains involved in RNA binding and protein-protein interactions in human and Saccharomyces cerevisiae pre-mRNA splicing factor SF1. RNA 4, 551-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska, I., Andrade, L. E., Ochs, R. L., Chan, E. K., Chang, C. M., Roos, G., and Tan, E. M. (1991). Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp. Cell Res. 195, 27-37. [DOI] [PubMed] [Google Scholar]
- Rudner, D. Z., Kanaar, R., Breger, K. S., and Rio, D. C. (1996). Mutations in the small subunit of the Drosophila U2AF splicing factor cause lethality and developmental defects. Proc. Natl. Acad. Sci. USA 93, 10333-10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz, B., and Séraphin, B. (1999). Transient interaction of BBP/ScSF1 and Mud2 with the splicing machinery affects the kinetics of spliceosome assembly. RNA 5, 819-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz, B., and Séraphin, B. (2000). A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J. 19, 1873-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffert, N., Hossbach, M., Heintzmann, R., Achsel, T., and Lührmann, R. (2004). RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J. 23, 3000-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selenko, P., Gregorovic, G., Sprangers, R., Stier, G., Rhani, Z., Krämer, A., and Sattler, M. (2003). Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol. Cell 11, 965-976. [DOI] [PubMed] [Google Scholar]
- Semizarov, D., Frost, L., Sarthy, A., Kroeger, P., Halbert, D. N., and Fesik, S. W. (2003). Specificity of short interfering RNA determined through gene expression signatures. Proc. Natl. Acad. Sci. USA 100, 6347-6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer, F., Moorman, C., Van Der Linden, A. M., Kuijk, E., Van Den Berghe, P. V., Kamath, R., Fraser, A. G., Ahringer, J., and Plasterk, R. H. (2003). Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1, 77-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman, J. E., and Lamond, A. I. (1999a). Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol. 9, 1065-1074. [DOI] [PubMed] [Google Scholar]
- Sleeman, J. E., and Lamond, A. I. (1999b). Nuclear organization of pre-mRNA splicing factors. Curr. Opin. Cell Biol. 11, 372-377. [DOI] [PubMed] [Google Scholar]
- Sleeman, J. E., Ajuh, P., and Lamond, A. I. (2001). snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J. Cell Sci. 114, 4407-4419. [DOI] [PubMed] [Google Scholar]
- Stanek, D., Rader, S. D., Klingauf, M., and Neugebauer, K. M. (2003). Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J. Cell Biol. 160, 505-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka, K., Nakagawa, H., Miyamoto, S., and Miki, H. (2004). The pre-mRNA-splicing factor SF3a66 functions as a microtubule-binding and -bundling protein. Biochem. J. 382, 223-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja, K. L., Lifshitz, L. M., Fay, F. S., and Singer, R. H. (1992). Poly(A) RNA codistribution with microfilaments: evaluation by in situ hybridization and quantitative digital imaging microscopy. J. Cell Biol. 119, 1245-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill, C., Qutob, M. S., Yee, S. P., and Torchia, J. (2000). A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem. 275, 40463-40470. [DOI] [PubMed] [Google Scholar]
- Utans, U., Behrens, S.-E., Lührmann, R., Kole, R., and Krämer, A. (1992). A splicing factor that is inactivated during in vivo heat shock is functionally equivalent to the [U4/U6.U5] triple snRNP-specific proteins. Genes Dev. 6, 631-641. [DOI] [PubMed] [Google Scholar]
- Wang, H. Y., Xu, X., Ding, J. H., Bermingham, J. R., Jr., and Fu, X. D. (2001). SC35 plays a role in T cell development and alternative splicing of CD45. Mol. Cell 7, 331-342. [DOI] [PubMed] [Google Scholar]
- Wang, J., Takagaki, Y., and Manley, J. L. (1996). Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes Dev. 10, 2588-2599. [DOI] [PubMed] [Google Scholar]
- Will, C. L., and Lührmann, R. (2001). Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13, 290-301. [DOI] [PubMed] [Google Scholar]
- Will, C. L., Schneider, C., MacMillan, A. M., Katopodis, N. F., Neubauer, G., Wilm, M., Lührmann, R., and Query, C. C. (2001). A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J. 20, 4536-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D., and Childs, G. (1998). Human ZFM1 protein is a transcriptional repressor that interacts with the transcription activation domain of stage-specific activator protein. J. Biol. Chem. 273, 6868-6877. [DOI] [PubMed] [Google Scholar]
- Zhang, D., Paley, A. J., and Childs, G. (1998). The transcriptional repressor ZFM1 interacts with and modulates the ability of EWS to activate transcription. J. Biol. Chem. 273, 18086-18091. [DOI] [PubMed] [Google Scholar]