Transgene analysis proves mRNA trans-splicing at the complex mod(mdg4) locus in Drosophila (original) (raw)
Abstract
The Drosophila BTB domain containing gene_mod(mdg4)_ produces a large number of protein isoforms combining a common N-terminal region of 402 aa with different C termini. We have deduced the genomic structure of this complex locus and found that at least seven of the mod(mdg4) isoforms are encoded on both of its antiparallel DNA strands, suggesting the generation of mature mRNAs by trans-splicing. In transgenic assays, we demonstrate the ability of Drosophila to produce_mod(mdg4)_ mRNAs by trans-splicing of pre-mRNAs generated from transgenes inserted at distant chromosomal positions. Furthermore, evidence is presented for occurring of trans-splicing of_mod(mdg4)_-specific exons encoded by the parallel DNA strand. The mod(mdg4) locus represents a new type of comlpex gene structure in which genetic complexity is resolved by extensive trans-splicing, giving important implications for genome sequencing projects. Demonstration of naturally occurring trans-splicing in the model organism Drosophila opens new experimental approaches toward an analysis of the underlying mechanisms.
Trans-splicing is a process that produces mature transcripts by combining exons of independent pre-mRNA molecules and was first reported in trypanosomes (1–3). The capability of mammalian cells for mRNA trans-splicing was first shown by Eul et al. (4). Recently, additional reports demonstrated trans-splicing in higher eukaryotes. Li et al. (5) identified a splice variant of the human acyltransferase ACAT-1 that contains a 5′-untranslated exon encoded from chromosome 7 connected with exons 1–16 encoded from chromosome 1. In rat liver cells, carnitine octanoyltransferase mRNA variants with duplications of coding exons 2 and 3 not present in the genomic complement have been identified (6). In this case, different proteins are encoded by the splice variants. However, the physiological significance of these trans-splicing events remains largely unknown.
So far no experimental proof for naturally occurring trans-splicing in insects has been documented. Recently, Labrador_et al._ (7) have shown that the mod(mdg4)-67.2 isoform in Drosophila is encoded on both DNA strands and suggested formation of the mature mRNA by trans-splicing of two independent transcripts.
Sequence analysis of a large number of mod(mdg4) cDNA clones (8–12) and the available genomic sequence of the mod(mdg4) locus (13) indicated that usage of both DNA strands as coding strands is a general property of the mod(mdg4) locus. The common exons 1–4, which are found in all identified transcripts, are located at the 5′ end of the locus whereas the alternatively spliced 3′-specific exons are organized in five groups on both DNA strands. In transgenic assays, we provide evidence that trans-splicing is the basic mechanism responsible for production of multiple mod(mdg4) isoforms.
Materials and Methods
Constructs and P Element-Mediated Germline Transformation.
The transgene containing the tagged specific exons_mod(mdg4)-55.1_ and mod(mdg4)-53.1 has been constructed as follows. An enhanced green fluorescent protein (EGFP) (CLONTECH) coding sequence (tag1) has been inserted upstream the stop codon of mod(mdg4)-55.1 and the Flag tag sequence (tag2) upstream the stop codon of mod(mdg4)-53.1 via introducing restriction sites by PCR. The final construct containing 2-kb upstream of the 5′ splice site of mod(mdg4)-55.1 exon 5 and 1.2-kb upstream of mod(mdg4)-53.1 exon 5 was cloned as a 5.2-kb_Not_I fragment into the upstream activating sequence containing transformation vector pUAST (14) in both orientations. The construct was partially sequenced and used for germline mediated transformation. However, by sequencing the transgene specific reverse transcription (RT)-PCR product, we found a frame shift mutation at amino acid position 499 in mod(mdg4)-55.1, preventing the expression of the EGFP-fusion protein.
Germline transformation was performed as described in Rubin and Spradling (15) with the w1118 strain as a host. Four transgenic lines have been obtained for the construct [pUAST-55.1-tag1-tag2–53.1], two lines on the second chromosome (TG-1 and -2) and two lines on the third chromosome (TG-3 and -4). In the case of the [pUAST-53.1-tag2-tag1–55.1] construct, we obtained two independent lines, both on the third chromosome, TG-A and B). As the GAL4-driver strain, we used the third chromosomal nanos-GAL4-driver line 4937 (Stock Center, Bloomington, IN).
Reverse Transcription–PCR (RT-PCR).
For transgenic assays, total RNA was isolated from females of the indicated genotypes with Trizol (Invitrogen). Total RNA (1 μg) was used for RT with Moloney murine leukemia virus reverse transcriptase and random hexamer primer (Promega) according to the manufacturer's protocol. Usually one-fifth of the product was used for PCR amplification. PCR was performed in a volume of 30 μl with 8 pmol of each primer, 0.15 mM dNTPs, and 1.65 mM MgCl2. For PCR conditions, we used 95°C for 3 min, 55°C for 40 sec, and 72°C for 40 sec for 35 cycles. The reaction (5–10 μl) was separated in agarose gels. In all experiments, the forward primer E4-F (5′-CGCAAATGTTATGGACCCTCTC-3′) annealing to exon 4 was used in combination with specific backward primers: tag1-back, used for specific detection of tagged exon 5 of mod(mdg4)-55.1 (5′-CTTGTGGCCGTTTACGTC-3′), tag2-back, used for specific detection of tagged exon 5 of mod(mdg4)-53.1 (5′-CGTCATCGTCCTTGTAGTC-3′), and mod(mdg4)-55.1-RT-B (5′-CCCGTCCTGTTCTTTTTGAGG-3′).
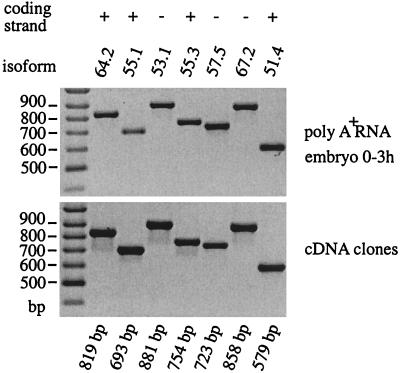
For RT-PCR experiments (shown in Fig. 3), 1 μg of poly(A)+ RNA isolated from 0- to 3-h embryos of_Drosophila melanogaster_ was used. The specific backward primers are mod(mdg4)-64.2-RT-B (5′-CAACTTGCAGTCCTTGCCGTC-3′), -53.1-RT-B (5′-GGGTTGGCTGGAAAATTGATTG-3′), -55.3-RT-B (5′-CCAAGGCATCTTTAGGCTTTGTTG-3′), -67.2-RT-B (5′-ATATGACTCCCGATTCGCCAGG-3′), -57.4-RT-B (5′-TTGGCCGCCTCAATACGC-3′), and -51.4-RT-B (5′-CAAGACCAATAAGTTTTCAATCCCG-3′).
Figure 3.
Results of RT-PCR experiments demonstrating the existence of selected mature mRNAs in poly(A)+ RNA isolated from early embryos (0–3 h). A primer deduced from common exon 4 (E4-F) was used as forward primer and specific backward primers deduced from the alternative exons 5 indicated above the Upper panel were used for PCR. Specific exons encoded by the same DNA strand as common exons 1–4 are indicated by (+) and those encoded by the antiparallel DNA strand are indicated by (−). PCR-fragments obtained with the same pairs of primers and the corresponding cDNA clones as DNA template are shown in the Lower panel. The expected fragment size for each isoform is indicated at the bottom.
Isolation and Characterization of cDNA Clones.
Two new mod(mdg4) cDNA clones [mod(mdg4)-52.2 and _mod(mdg4)-54.1_] have been isolated from a 2- to 12-h embryonic cDNA library in λZAPII (Stratagene), as described previously (11). The 0.5-kb genomic _Sal_I fragment contained in exon 4 was used as a radioactively labeled probe. The two transcripts mod(mdg4)-53.6 and mod(mdg4)-54.7 have been identified by RT-PCR on poly(A)+ RNA from 0- to 3-h embryos with the primer E4-F and the specific primers mod(mdg4)-53.6–1-RT-B (5′-GGCTTAAAGGCATCCGCCGGATG-3′) and mod(mdg4)-54.7-RT-B (5′-GGATTTGGTCACCACACGGGCGGAGC-3′). The missing 3′ end of isoform mod(mdg4)58.8 was identified by RT-PCR with primer E4-F and primer 58.8-RT-B (5′-TACACTTGGACTTAAGACTAG-3′).
Results
All mod(mdg4) Isoforms Contain Identical N Termini but Variable C Termini.
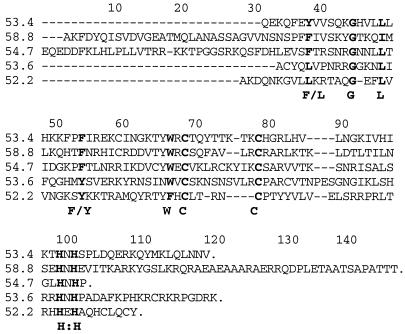
During the molecular analysis of the mod(mdg4) locus, we identified 26 different classes of transcripts all containing a common 5′ sequence (exons 1–4) but different 3′ regions. The deduced proteins contain an N-terminal (BTB) domain (17). Furthermore, in most of the isoforms a conserved C-terminal C2H2-containing protein motif is found (11). Two new cDNA clones representing isoforms_mod(mdg4)-52.2_ and mod(mdg4)-54.1 were isolated by screening an embryonic cDNA library (see Materials and Methods). Two additional putative isoforms,mod(mdg4)-53.6 and mod(mdg4)-54.7 have been detected by searching the genomic region of mod(mdg4) for ORFs containing the C-terminal C2H2 consensus sequence.Mod(mdg4)-58.8 represents another new isoform, which was so far identified by a 3′-truncated cDNA clone. RT-PCR experiments revealed the existence of mod(mdg4)-53.6, -54.7, and -58.8 transcripts in early embryos. By sequencing the resulting PCR products, we deduced the putative proteins (Fig.1). All of them contain the conserved C-terminal protein consensus sequence. Altogether 20 of 26 identified_mod(mdg4)_ isoforms combine both the N-terminal BTB domain and the C-terminal consensus sequence, which might be of functional significance.
Figure 1.
Sequence comparison of new Mod(mdg4) protein isoforms. All protein sequences start with amino acid position 403. The most N-terminal 402 aa common to all Mod(mdg4) isoforms are not listed. The conserved C-terminal C2H2 consensus sequence is present in all newly identified isoforms. Strongly conserved amino acid positions are shown in bold. Alignment was performed with the clustal x program (16).
Genomic Structure of the mod(mdg4) Locus.
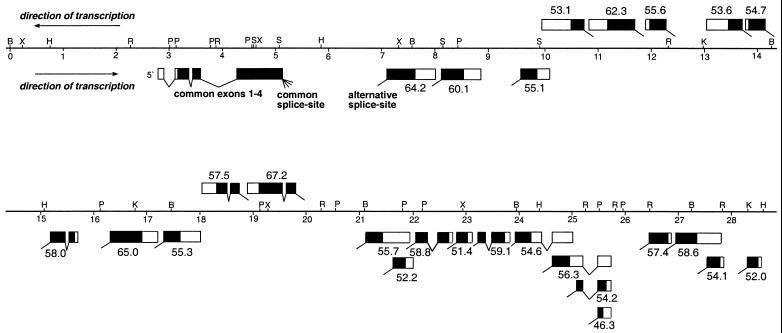
On the basis of our extensive cDNA sequence data and the available sequence of the mod(mdg4) region, we deduced the exon/intron structure of mod(mdg4) (Fig.2). Interestingly, seven of the transcripts are not colinearly located within the locus. Relative to the common exons 1–4, exon 5 of isoforms mod(mdg4)-53.1,-62.3, -55.6, -53.6, -54.7,-57.4, and -67.2 are encoded by the antiparallel DNA strand. Beside this finding, the exon/intron structure of_mod(mdg4)_ suggests differential splicing within isoform-specific exons (isoforms mod(mdg4)-55.7 and_-52.2_; mod(mdg4)-54.6, -56.3,-54.2, and -46.3; mod(mdg4)-58.6 and_-54.1_). According to its genomic structure, the genetic density at the mod(mdg4) complex is unusually high. Altogether 26 independent transcripts of an average size of 2 kb are encoded in a genomic region of 28 kb.
Figure 2.
Genomic region of the mod(mdg4) complex and the exon/intron structure of identified mRNAs. The alternative 3′-splice site at the exon 4 boundary is used to generate all mature mRNAs indicated by the molecular weight of the deduced proteins. Exons shown below the physical map are encoded by the same DNA strand, whereas those encoded by the antiparallel DNA strand are shown above the physical map. Direction of transcription is indicated by arrows. Untranslated regions are shown as open boxes and translated regions as black boxes. Note the overlap of specific exons five of isoforms_mod(mdg4)-55.1_ and -53.1 within their 3′-untranslated region and the differential splicing within the specific exons 5 of mod(mdg4)-55.7 and_-52.2_; -54.6, -56.3,-54.2 and -46.3; -58.6 and_-54.1_. Restriction sites indicated in the physical map are B, _Bam_HI; X, _Xho_I; H,_Hin_dIII; R, _Eco_RI; P,_Pst_I; S, _Sal_I; and K,_Kpn_I.
For most of the mRNAs, more than two independent cDNA clones have been isolated (11). In addition we proved the existence of mRNAs in early embryos for a subset of seven isoforms by RT-PCR. Representative data with the forward primer located within the common exon 4 (primer E4-F) and specific backward primers located within the appropriate specific exons of isoforms mod(mdg4)-64.2, -55.1,-53.1, -55.3, -57.4, -67.2, and -51.4 are shown in Fig. 3. All amplicons correspond to the expected size deduced from the cDNA clones and sequencing of the RT-PCR products confirmed the sequence of corresponding cDNA clones. These results support the generation of independent mRNAs from both antiparallel DNA strands in case of isoforms mod(mdg4)-53.1, -62.3, -55.6,-53.6, -54.7, -57.4, and_-67.2_. This is in agreement with RNase protection experiments performed on isoform mod(mdg4)-67.2 (ref. 7).
Furthermore, we compared all 3′ intron sequences upstream of the specific exons 5. In all cases, the conserved sequence elements known to be involved in mRNA splicing, the putative branch point, a pyrimidine-rich tract, and the dinucleotide AG at the 3′ intron border are present (Fig. 4). These data do not indicate significant sequence differences allowing the discrimination between putative cis-splicing of specific exons located colinearly to exons 1–4 and those 3′ exons encoded by the antiparallel DNA strand, which are expected to be trans-spliced.
Figure 4.
Alignment of 3′-intron sequences upstream of the specific exons 5 of all identified alternatively spliced mod(mdg4) mRNAs. All sequences contain the conserved dinucleotide AG at the 3′-boundary. Pyrimidine-rich tracts are underlined and the putative branch point is indicated by a small bold letter. Specific exons encoded by the same DNA strand as exons 1–4 are indicated by (+) and those encoded by the cDNA strand by (−).
Specific Exons Transcribed from Transgenes at Different Chromosomes Are Templates for Trans-Splicing.
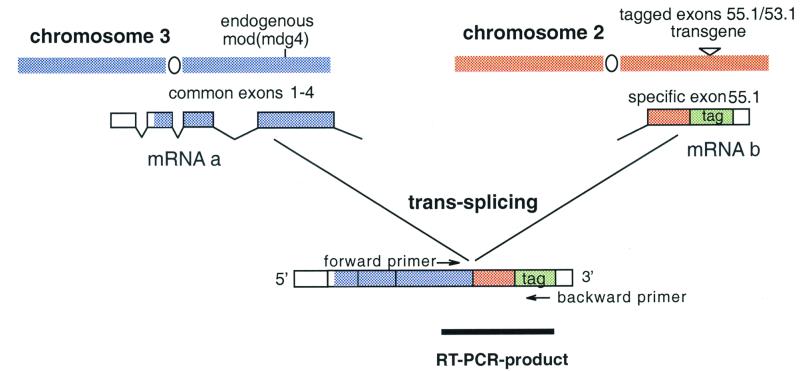
To demonstrate trans-splicing, we have separated specific exons from the common exons 1–4 to different chromosomes via transgene insertions (Fig. 5). In this assay, we have chosen the specific exons mod(mdg4)-55.1 and_mod(mdg4)-53.1_ (cf. Fig. 1), which are encoded by antiparallel DNA strands. Notably, the 3′-untranslated regions of both isoforms are antisense within a region of 149 nt. This experiment should also test for putative trans-splicing of isoform_mod(mdg4)-55.1_, which is colinearly located with respect to the common exons 1–4. In the transgenes, both specific exons have been sequence-tagged via PCR and the resulting fragment containing genomic sequences of 2.0-kb and 1.2-kb upstream of the splice sites of_mod(mdg4)-55.1_ and mod(mdg4)-53.1, respectively, was cloned in both orientations into the Drosophila transformation vector pUAST (14). Expression of the inserted sequences is induced by the yeast transcriptional activator GAL4, expressed from an independent driver element.
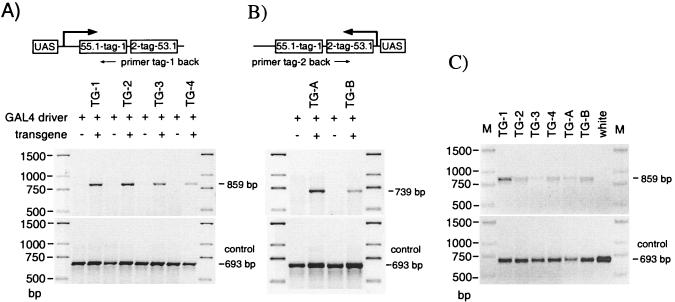
Figure 5.
Transgenic assay to demonstrate the existence of trans-splicing at_mod(mdg4)_. Schematic representation of the trans-splicing assay in case of a second chromosomal transgene. Common exons of mod(mdg4) are expressed from the endogenous locus producing mRNA a. The sequence-tagged specific exon mod(mdg4)-55.1 is expressed under control of GAL4 binding to the UAS upstream of the transgene, producing mRNA b. The mature trans-spliced mRNA is detected by RT-PCR with the indicated pair of primers.
Four transformants containing the tagged specific exon 5 of_mod(mdg4)-55.1_ (transgenes TG-1 to 4) and two transformants carrying the tagged specific exon 5 of mod(mdg4)-53.1 (transgenes TG-A and B) under control of the upstream activating sequence (UAS) element have been obtained. For expression of the transgene, all transformants have been crossed to GAL4-driver lines expressing GAL4 either under hsp70 control or the promoter of the maternal gene nanos. To test for trans-splicing events, total RNA was isolated from progeny females containing the GAL4-driver element and the transgene construct and from females containing the GAL4-driver element but no transgene, respectively. RT-PCR was performed with the forward primer E4-F, which is complementary to common exon 4, and backward primers specific for the two sequence tags. Fig.6 represents the RT-PCR results obtained from crosses with the nanos driver element. In all females containing the transgene construct and the driver element, an amplicon of the expected size is found, whereas in females not containing the transgene construct no fragment was amplified. Fig. 6A represents the results obtained for transgenic lines TG-1 to 4 containing the sequence tagged exon 5 of isoform_mod(mdg4)-55.1_ under control of UAS/GAL4. RT-PCR results with the transgenic lines expressing the sequence tagged exon_mod(mdg4)-53.1_ under control of GAL4 are shown in Fig.6B. As a control for all genotypes, RT-PCR was performed with the forward primer E4-F and the backward primer 55.1-RT-B annealing upstream to the sequence tag of isoform_mod(mdg4)-55.1_ (Fig. 6 A and B Lower). The expected control PCR fragment of 693 bp was obtained in all genotypes.
Figure 6.
RT-PCR demonstrating trans-splicing of endogenous_mod(mdg4)_ common exons 1–4 and tagged-specific_mod(mdg4)_ exons expressed from a transgene. (A) Expression of the tagged exon_mod(mdg4)-55.1_ is driven by crossing heterozygous transgenic lines to homozygous nanos_-GAL4-producing driver lines. The amplicon of the expected size is only obtained in case of trans-splicing of endogenous exon 4 and tagged exon 5 expressed from the transgene. The primer E4-F and tag1-back have been used for PCR. As a control for RT-PCR a backward primer complementary to exon 5 upstream of the sequence tag was used in combination with E4-F (Lower). (B) RT-PCR results obtained with transgenic lines expressing the tagged-specific exon_mod(mdg4)-53.1 under control of the nanos GAL4-driver line. The tag2-back primer has been used in combination with primer E4-F. Only in case of correct trans-splicing of the two independent transcripts, the expected amplicon is obtained. No amplicon is obtained in absence of the transgene. For control of RT-PCR the same primer pair as in A was used (Lower). (C) Demonstration of a sequence element with putative promoter activity upstream of specific exon 5 of_mod(mdg4)-55.1_ contained in the transgene. RT was performed with total RNA from females containing the transgenes TG-1 to 4 (pUAST-55.1-tag1-tag2–53.1) and TG-A and -B (pUAST-53.1-tag2-tag1–55.1) in the absence of a GAL4-driver element. For PCR the forward primer E4-F and the back primer tag1-back were used. In all transgenic lines, a PCR fragment of the expected size was obtained. Quantitative differences among the transgenic lines could be because of position effects. No fragment was amplified with total RNA isolated from the strain w 1118. As a control for RT-PCR, the backward primer deduced from the specific exon_mod(mdg4)-55.1_ upstream of the sequence tag was used in combination with the E4-F primer.
Sequencing of the RT-PCR fragments proved exact trans-splicing of endogenous common exon 4 to the tagged specific exon 5 transcribed from the transgene. In the case of the second chromosomal transgenic lines TG-1 and 2, the results demonstrate trans-splicing of mRNAs generated from different chromosomes. Our results furthermore show that the specific exon mod(mdg4)-55.1, which is colinearly located with respect to exons 1–4 within the endogenous locus, is trans-spliced in our assay. The latter result also suggests that 3′-specific exons on both strands are joined to the common exons 1–4 by trans-splicing.
Detection of Isoform-Specific Promoter Elements.
One of the important criteria for efficient mRNA trans-splicing is the presence of a 3′ splice site in absence of a functional 5′ splice site as represented in outrons (2). To meet this criteria, we would expect the production of independent transcript(s) containing one or several of the endogenous-specific mod(mdg4) exons. Searching for putative promoter elements within the_mod(mdg4)_ complex, several TATA-box-containing elements were found. One of these is located upstream of the specific exon_mod(mdg4)-55.1_ and is contained within the transgene construct used for the trans-splicing assay. To prove its function_in vivo_, all transgenic lines have been tested for expression of the transgene in absence of any GAL4-driver element (Fig.6C). Independent of the insertion site of the transgene and its orientation relative to the UAS sequence, trans-splicing of the tagged specific exon mod(mdg4)-55.1 could be demonstrated. For PCR primers, again the forward primer E4-F and the primer 55.1-tag1-back have been used. However, the level of expression and/or the efficiency of trans-splicing is variable in different transformants. This could be because of chromosomal position effects, depending on the insertion site of the transgene. We conclude from these results that independent mRNAs containing the common exons 1–4 on one hand and mRNAs containing the specific exon_mod(mdg4)-55.1_ on the other hand are produced endogenously.
Trans-Splicing of the Common Exons 1–4 Is Independent of Their Proximity to the 3′-Specific Exons.
If trans-splicing is a general property of mod(mdg4), we would expect that separation of common exons 1–4 and 3′-specific exons via transgene insertion should result in the generation of mRNAs containing endogenous specific exons. To test this we made use of a second chromosomal transgene containing the 7.5 kb-Bam_HI genomic fragment (at position 0–7.5 according to the physical map, Fig. 2). This transgene was shown to partially rescue the recessive lethality of the_mod(mdg4)neo129 mutation. This mutation is caused by insertion of the pUChsneory+ transposon within the third intron (11). Sequence analysis revealed a DNA polymorphism at nucleotide position 555 in exon 4 (C in transgene, T in mutant chromosome), which allows detection of mRNAs originated from the transgene. Trans-splicing of transgene encoded common exons 1–4 and endogenous 3′-specific exons was assayed by RT-PCR with total RNA isolated from females containing the transgene and which are homozygous for the mod(mdg4)neo129 mutation. To prove that the specific exon mod(mdg4)-55.1 is correctly spliced to exon 4, PCR was performed with primer E4-F and primer 55.1-RT-B. Sequencing of four cloned PCR fragments obtained in two independent RT-PCR experiments indicated that one clone contains the transgene-encoded sequence polymorphism. This result demonstrates correct trans-splicing of common exon 4 to the endogenous 3′ exon of isoform mod(mdg4)-55.1 independent of their chromosomal location.
Discussion
In this study, we provide experimental proof for the occurrence of mRNA trans-splicing in Drosophila. The mod(mdg4) locus represents an unusual type of gene structure. Both DNA strands within the locus are used to encode a large number of protein isoforms. Our transgenic approach clearly demonstrates that both the colinearly located specific exons (demonstrated for exon_mod(mdg4)-55.1)_ and those encoded by the antiparallel DNA strand (shown for exon mod(mdg4)-53.1) are substrates for trans-splicing. This result also suggests that all other_mod(mdg4)_ isoforms might be generated by trans-splicing, implying the initiation of independent pre-mRNAs at several promoter elements within the mod(mdg4) complex. We indeed found multiple TATA-box-containing elements throughout the locus. One of these is located upstream of the mod(mdg4)-55.1 isoform and is contained in the transgene. Expression of the transgene independent of the inducible promoter element in six independent transgenic lines indicates a putative promoter function. However, further experiments should demonstrate the existence of multiple promoters at the_mod(mdg4)_ locus. Moreover, our results demonstrate that trans-splicing occurs within mod(mdg4), independent of the chromosomal context of the common exons 1–4 and the 3′-specific exons. This also raises the question about the special requirements for initiating trans-splicing at mod(mdg4). Further experiments should clarify whether RNA recognition or nuclear compartmentalization or both plays a role in the initiation of trans-splicing.
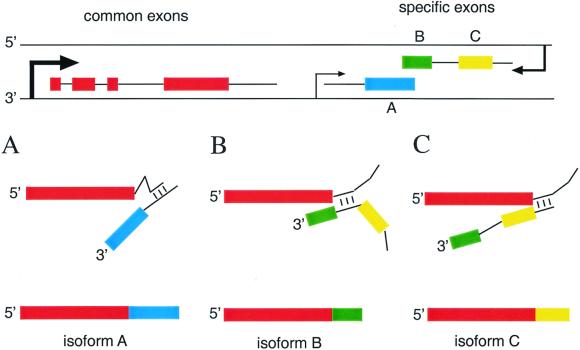
Our data suggest that trans-splicing is a general property of the_mod(mdg4)_ locus. Three possible types of trans-splicing events are shown by the model presented in Fig.7. The transcript containing common exons 1–4 (shown in red) is produced in large quantities and contains putative interaction sites with upstream regions of pre-mRNAs containing the specific mod(mdg4) exon(s). The specific exons are transcribed as mono-exonic, di-exonic, or polyexonic mRNAs from both cDNA strands. Depending on the site of trans-splicing, three different protein isoforms (A, B, and C) are produced. The expression of the alternatively spliced mod(mdg4) isoforms could be regulated at several levels: (i) differential spatial and temporal expression of pre-mRNAs containing one (or groups of) alternatively spliced specific exons; (ii) differences in selectivity and efficiency of trans-splicing to generate different quantities of mature mRNAs, and (iii) variable stability of the isoform specific transcripts. As a result, >20 different Mod(mdg4) protein isoforms are produced that all contain a common region of 402 aa, including the N-terminal BTB domain implicated in dimerization/oligomerization (18, 19) and variable C termini with the conserved C2H2 motif (11). The variable C termini are implicated to specify the function of individual isoforms in different processes like chromatin insulator function (9, 20), programmed cell death (10), or modification of gene silencing (8).
Figure 7.
The model demonstrating different mRNA trans-splicing events at the_mod(mdg4)_ locus. Common exons 1–4 located at the 5′ end of the locus are supposed to be strongly transcribed as a unique transcription unit. Additional transcription units containing one (A) two (B and C) or even more (not shown) isoform-specific exons are initiated at the same coding strand (A) or at the antiparallel DNA strand (B/C). In case of mRNAs containing two or more specific exons, different 5′ splice sites can be used, resulting in the generation of different mRNA variants (B vs. C). The relative amount of single_mod(mdg4)_ protein isoforms could be regulated by the rate of transcription initiation, by the efficiency of trans-splicing, and by the stability of the resulting mRNA.
Our results demonstrate the existence of trans-splicing in_Drosophila_ and provide a new basis for a genetic and molecular analysis of key elements involved in this process.
Acknowledgments
We thank M. Kube and K. Kittlaus for technical assistance. This work was supported by a grant from the Deutsche Forschungsgemeinschaft to R.D.
Abbreviations
RT-PCR
reverse transcription–PCR
UAS
upstream activating sequence
BTB
_B_R-C,_t_tk,_b_ab
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the European Molecular Biology Laboratory (EMBL) database (accession nos. AJ320161 and AJ320165).
References
- 1.Murphy W J, Watkins K P, Agabian N. Cell. 1986;47:517–527. doi: 10.1016/0092-8674(86)90616-1. [DOI] [PubMed] [Google Scholar]
- 2.Blumenthal T. Trends Genet. 1995;11:132–136. doi: 10.1016/s0168-9525(00)89026-5. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Blanco M A, Puttaraju M, Mansfield S G, Mitchell L G. Gene Ther Regul. 2000;1:141–163. doi: 10.1038/sj.gt.3301307. [DOI] [PubMed] [Google Scholar]
- 4.Eul J, Graessmann M, Graessmann A. EMBO J. 1995;14:3226–3235. doi: 10.1002/j.1460-2075.1995.tb07325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li B-L, Li X-L, Duan Z-J, Lee O, Lin S, Ma Z-M, Chang C C Y, Yang X-Y, Park J P, Mohandas T K, et al. J Biol Chem. 1999;274:11060–11071. doi: 10.1074/jbc.274.16.11060. [DOI] [PubMed] [Google Scholar]
- 6.Caudevilla C, Serra D, Miliar A, Codony C, Asins G, Bach M, Hegardt F G. Proc Natl Acad Sci USA. 1998;95:12185–12190. doi: 10.1073/pnas.95.21.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Labrador M, Mongelard F, Plata-Rengifo P, Baxter E M, Corces V G, Gerasimova T I. Nature (London) 2001;409:1000. doi: 10.1038/35059000. [DOI] [PubMed] [Google Scholar]
- 8.Dorn R, Krauss V, Reuter G, Saumweber H. Proc Natl Acad Sci USA. 1993;90:11376–11380. doi: 10.1073/pnas.90.23.11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerasimova T I, Gdula D A, Gerasimov D V, Simonova O, Corces V G. Cell. 1995;82:587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- 10.Harvey A J, Bidwai A P, Miller L K. Mol Cell Biol. 1997;5:2835–2843. doi: 10.1128/mcb.17.5.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Büchner K, Roth P, Schotta G, Krauss V, Saumweber H, Reuter G, Dorn R. Genetics. 2000;155:141–157. doi: 10.1093/genetics/155.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Read D, Butte M J, Dernburg A F, Frasch M, Kornberg T B. Nucleic Acids Res. 2000;28:3864–3870. doi: 10.1093/nar/28.20.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 14.Brand A, Rerrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 15.Rubin G M, Spradling A C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 16.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zollmann S, Godt D, Prive G G, Couderc J L, Laski F A. Proc Natl Acad Sci USA. 1994;91:10717–10721. doi: 10.1073/pnas.91.22.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmad K F, Engel C K, Prive G G. Proc Natl Acad Sci USA. 1998;95:12123–12128. doi: 10.1073/pnas.95.21.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bardwell V J, Treisman R. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 20.Bell A C, West A G, Felsenfeld G. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]