Identification of a family of Fc receptor homologs with preferential B cell expression (original) (raw)
Abstract
Investigation of human genome sequences with a consensus sequence derived from receptors for the Fc region of Igs (FcR) led to the identification of a subfamily of five Ig superfamily members that we term the Fc receptor homologs (FcRHs). The closely linked_FcRH_ genes are located in a chromosome 1q21 region in the midst of previously recognized FcR genes. This report focuses on the FcRH1, FcRH2, and_FcRH3_ members of this gene family. Their cDNAs encode type I transmembrane glycoproteins with 3–6 Ig-like extracellular domains and cytoplasmic domains containing consensus immunoreceptor tyrosine-based activating and/or inhibitory signaling motifs. The five FcRH genes are structurally related, and their protein products share 28–60% extracellular identity with each other. They also share 15–31% identity with their closest FcR relatives. The_FcRH_ genes are expressed primarily, although not exclusively, by mature B lineage cells. Their conserved structural features, patterns of cellular expression, and the inhibitory and activating signaling potential of their transmembrane protein products suggest that the members of this FcRH multigene family may serve important regulatory roles in normal and neoplastic B cell development.
Receptors for the Fc region (FcRs) of Igs have broad tissue distribution patterns and can modulate cellular and humoral immunity by linking their antibody ligands with effector cells of the immune system (1, 2). These cellular receptors have the ability to sense humoral concentrations of antibody, initiate cellular responses in host defense, and participate in autoimmune disorders (3). Their diverse regulatory roles depend on the Ig isotype specificity and cellular distribution of the individual FcR. These Ig superfamily members share similarities in their ligand binding subunits, and they may have inhibitory or activating signaling motifs in their intracellular domains or instead pair with signal transducing subunits possessing activating signaling motifs.
Our view of the FcR family has been significantly extended with the characterization of FcαR homologs in mice, the paired Ig-like receptors (4, 5), and their relatives in humans the Ig-like transcripts/leucocyte Ig-like receptors (6, 7). This multigene family, which includes the FcαR (8) and the natural killer cell Ig-like receptors (9), is located in a human chromosome 19q13 region known as the leucocyte receptor complex (LRC) (10, 11). These Ig-like multigene families belong to a larger class of receptors characterized by their possession of common cytoplasmic tyrosine-based signaling motifs. These can be either immunoreceptor tyrosine-based activation motifs (ITAMs) containing two repeats of the consensus sequence Y-X-X-L/I spaced by 6–8 aa (E/D)-X-X-Y-X-X-(L/I)-X6–8-Y-X-X-(L/I) or immunoreceptor tyrosine-based inhibitory motifs (ITIMs) with a 6-aa consensus sequence (I/V/L/S)-X-Y-X-X-(L/V) (12–15). The phylogenetic conservation of these types of receptors in birds (16) and bony fish (17) is indicative of their biological value. After ligand binding of the activating receptor complexes, ITAM tyrosines are rapidly phosphorylated by Src family kinases to initiate a cascade of signaling events that trigger cellular activation. In the case of ITIM-bearing receptors, the tyrosines provide a docking site for phosphatases containing Src homology 2 domains that can abrogate cellular activation (18, 19). The balance in the utilization of these activating and inhibitory receptor pairs can serve to modulate cellular responses to a variety of stimuli.
The genes encoding the classical FcRs, FcγRI,FcγRII, FcγRIII, and FcɛRI, lie on the long arm of chromosome 1 (1q21–23) near the polymeric Ig receptor (pIgR) and Fcα/μR genes (1q32) (20–23). Members of this FcR subfamily have relatively low extracellular homology with the FcαR-related genes that reside in the LRC on chromosome 19. Like the FcγR- and FcɛR-activating receptors, the ligand binding chain of the FcαR coassociates with the ITAM containing FcR common γ-chain (24, 25). The concentration of genes in the 19q13.4 LRC locus that share a high degree of homology in their respective extracellular domains suggested the possibility that a similar cluster of FcγR and_FcɛR gene relatives might exist in the chromosome 1q21–23 region. In a search for new members of this FcR family, we derived a consensus amino acid motif through comparison of the FcγRI, FcγRII, FcγRIII, and pIgR extracellular regions. When this consensus motif was used in a GenBank protein database query (9/29/00), genomic clones were identified that were found to contain FcR_relatives that we term the Fc receptor homolog (FcRH) subfamily. Here we report the location, sequence, structure, and cellular expression patterns of three of the five functional_FcRH genes, FcRH1, FcRH2, and_FcRH3, which have not been previously described, and compare them with two recently reported members of this subfamily, Ig superfamily receptor translocation-associated genes 1 (IRTA1) (FcRH4) and 2 (IRTA2) (FcRH5) (26), and their FcR relatives.
Materials and Methods
Isolation of FcRH cDNA Clones.
Rapid amplification of cDNA ends (RACE)-PCR was performed by using a Marathon-Ready human lymph node cDNA library (CLONTECH). Gene-specific primers were as follows: FcRH3, forward 5′-GTGAGTCTCAGGGTCACAGTTCCG-3′ and reverse 5′-GCTCTTGAACTTGGATATTTAGGGGT-3′; FcRH2, forward 5′-CCAGTGTATGTCAATGTGGGCTCTG-3′ and reverse 5′-CGTTGAAAGAGCTCTTGGACTTTTATC-3′; and FcRH1, forward 5′-GCCTCAAAAGAAAAATAGGAAGACGTT-3′ and reverse 5′-AAGCTCACATCAGCGACAGGGAC-3′. RACE products were subjected to a second round of nested PCR and visualized by agarose gel electrophoresis and ethidium bromide staining.
Generation of Full-Length FcRH cDNAs.
Primers used in end-to-end amplification to generate full-length cDNAs were as follows: FcRH3, forward 5′-TCTTGGAGATAAGTCGGGCTTT-3′ and reverse 5′-ATCCTGCAGCCCAGCCTCGTAGGAG-3′; FcRH2, forward 5′-GGTCCTCATGCTGCTGTGGTCATT-3′ and reverse 5′-GCTGTTGATCTTCCCTTCTGATTC-3′; and FcRH1, forward 5′-ATGCTGCCGAGGCTGTTGCTGTTG3′ and reverse 5′-CATAGCATCTTCATAGTCCACATC-3′. Each amplification reaction underwent initial denaturation of 94°C for 30 s followed by 30 cycles of denaturation at 94°C for 5 s and annealing at 68°C for 4 min, and final extension at 72°C for 6 min.
Sequence Analysis.
PCR products were ligated into the pCR2.1 TOPO T/A vector (Invitrogen). Inserts were DNA-sequenced on both strands by the dideoxy chain termination method using Thermo Sequenase (Amersham Pharmacia) and an automated sequencer (Li-Cor, Lincoln, NE). Nucleotide and amino acid sequence alignment was analyzed with a dnastar(Madison, WI) software package, and homology searches were performed by using blast (27).
RNA Blot Analysis.
Northern blots (CLONTECH) were hybridized with α32P-dCTP-labeled probes: a 528-bp_Eco_RI fragment corresponding to the 5′ untranslated (UT)-EC1 regions of the FcRH3 cDNA, a 200-bp PCR product corresponding to a portion of the 3′ UT region of the FcRH2 cDNA, and a 257-bp PCR product corresponding to a portion of the 3′ UT region of the FcRH1 cDNA. Membranes were hybridized for 1 h at 65°C, washed, and exposed to x-ray film (4).
Reverse Transcription (RT)-PCR.
Human tonsillar cells, obtained with Institutional Review Board approval, were separated into CD19+ and CD19− subpopulations by magnetic cell sorting (Miltenyi Biotec, Auburn, CA). Viable CD19+ cells were stained with FITC-labeled anti-CD38 (Immunotech, Westbrook, ME) and phycoerythrin-labeled anti-IgD mAbs (Southern Biotechnology Associates) before sorting cells with a FACStarPlus instrument (Becton Dickinson) into Trizol reagent (Life Technologies, Grand Island, NY) for RNA isolation. Total cellular RNA was primed with random hexamers and oligo(dT) primers and reverse-transcribed with SuperScript II (Life Technologies) into single-stranded cDNA. RT-PCR was performed by using RNA from tonsillar B cells and cell lines, with GIBCO/BRL_Taq_ polymerase (Life Technologies). The following gene-specific primer pairs were used in the RT-PCR analysis of_FcRH1-5_ expresion in cell lines and tonsillar B cell subpopulations: FcRH1 forward, 5′-CTC AAC TTC ACA GTG CCT ACT GGG-3′ and reverse, 5′-TCC TGC AGA GTC ACT AAC CTT GAG-3′;FcRH2 forward, 5′-CCA GTG TAT GTC AAT GTG GGC TCT G and reverse, 5′-CAT TCT TCC CTC AAA TCT TTA CAC-3′; _FcRH3_forward, 5′-CAG CAC GTG GAT TCG AGT CAC-3′ and reverse, 5′-CAG ATC TGG GAA TAA ATC GGG TTG-3′ FcRH4 forward, 5′-TCT TCA GAG ATG GCG AGG TCA-3′ and reverse, 5′-TTT TGG GGT GTA CAT CAA CAT ACA AG-3′; and FcRH forward, 5′-TGT TGC CCT GTT TCT TCC AAT ACA-3′ and reverse, 5′-CAG AGT TGG CCG ACC TAC GC-3′. Each amplification reaction underwent initial denaturation at 94° for 5 min followed by 35 cycles of denaturation at 94° for 30 s, annealing at 60° for 30 s, extension at 72° for 1 min, and final extension at 72° for 7 min. Amplified products were visualized in 1% agarose gels containing ethidium bromide and documented with the Bio-Rad Fluor-S Imager.
Cell Lines.
Human cell lines included REH and Nalm 16 pro-B cell lines (28); 697, 207, and OB5 pre-B cell lines (29, 30); Ramos, Daudi, and Raji B cell lines (31–33); THP-1 and U937 monocytoid cell lines, HL-60 promyelocytic and KG-1 myelocytic cell lines, Jurkat T cell line and the K562 erythroid cell line (American Type Culture Collection).
Results
Identification of FcRH1 , FcRH2, and FcRH3.
A consensus sequence was generated that corresponds to the GenBank-derived amino terminal sequences of the second Ig-like domains of FcγR (FcγRI and FcγRII/III) and the third Ig-like domain of the polymeric Ig receptor: GEPIXLRCHSWKDKXLXKVTYXQNGKAXKFFH. A search (9/29/00) of the National Center for Biotechnology Information protein database with this sequence identified two overlapping human genomic bacterial artificial chromosome (BAC) clones, AL135929 andAL356276, which are located at 1q21.2–22. The second clone contained three putative Ig superfamily genes encoding complementary amino acid sequences that were designated FcRH1, FcRH2, and_FcRH3_ (Fig. 1). The predicted amino acid sequences of these gene segments shared 23–57% identity with each other and 14–28% identity with human FcγRI (CD64). Further analysis of the FcRH locus led to the identification of two additional genes (FcRH4, and FcRH5) and one pseudogene (FcRH4Ψ), immediately centromeric of_FcRH1–3_, two of which have recently been described as_IRTA1_ (FcRH4) and IRTA2(FcRH5) (26).
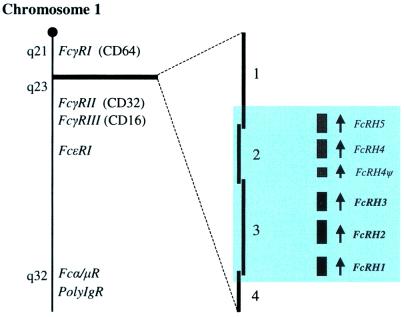
Figure 1.
Relative position of the FcRH locus within the_FcR_ cluster on chromosome 1. The cytogenetic location of the FcR genes is aapproximated from the GenBank Mapview database. The BAC clones (4, GenBank accession no. AL139409; 3, GenBank accession no. AL356276; 2, GenBank accession no. AL135929; and 1, GenBank accession no. AL353721) that span the locus are oriented in relation to their respective FcRH genes (shaded area).
To determine whether these genes are expressed by lymphocytes, the predicted amino acid sequences of their protein products were used to search the Lymphochip expressed sequence tag database with thetblastn algorithm (34). Two expressed sequence tags (AA505046 and AA282433) were identified that share complete identity over 23 aa in their translated ORFs with the N terminus of FcRH1. Lymphochip microarray data analysis indicated that these expressed sequence tags are expressed at relatively high levels in peripheral lymphoid tissues, including the lymph nodes, tonsils, resting peripheral B cells, and normal germinal center (GC) B cells. Among the different lymphoid malignancies, their expression proved to be highest in chronic lymphocytic leukemias, follicular lymphomas, and some diffuse large cell lymphomas of B lineage.
FcRH1, FcRH2, and FcRH3 cDNAs were isolated by RACE-PCR from a human lymph node cDNA library in both 5′ and 3′ directions. Full-length cDNAs of the coding regions for FcRH1, FcRH2, and_FcRH3_ were obtained by end-to-end PCR using unique primers generated from the cDNA sequences delineated for the 5′ UT and 3′UT regions. Southern blot analysis of human genomic DNA digested with_Bam_HI, _Eco_RI, or _Hin_dIII using cDNA probes specific for the 3′ UT regions of each cDNA revealed either one or two hybridizing fragments, suggesting that FcRH1, FcRH2, and FcRH3 are encoded by single genes. Analysis of full-length cDNA sequences indicated that FcRH1, FcRH2, and _FcRH3_have ORFs of 1,287 bp, 1,524 bp, and 2,202 bp, respectively, and encode type I transmembrane proteins of 429 aa, 508 aa, and 734 aa, respectively. Based on predicted consensus signal peptide cleavage sites (35, 36), the relative core peptide molecular masses were estimated as 45,158 for FcRH1, 53,407 for FcRH2, and 78,849 for FcRH3. These type I transmembrane proteins possess 3–6 extracellular C2 (37–39) type Ig-like domains with 3–7 potential N-linked glycosylation sites, uncharged transmembrane segments, and relatively long cytoplasmic tails containing consensus motifs for ITIMs and/or ITAMs (Fig. 2A).
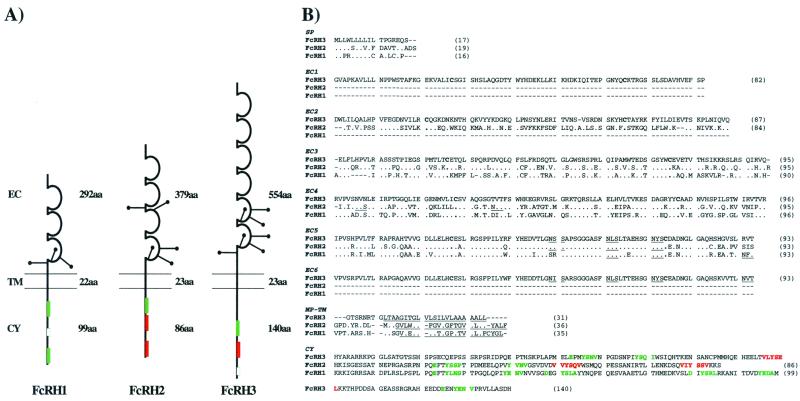
Figure 2.
FcRH1, FcRH2, and FcRH3 structural and sequence diversity. (A) Schematic representation of FcRH molecules. The three cDNAs encode type I transmembrane proteins with similar extracellular domains, but different cytoplasmic regions. The extracellular (EC) regions contain different numbers of C2-like Ig domains and potential sites of N-linked glycosylation (●—). The transmembrane (TM) domains are uncharged. The cytoplasmic (CY) region of FcRH1 contains two ITAMs (green boxes) and one ITAM-like region (small open box), whereas FcRH2 contains one ITAM and two ITIMs (red boxes). FcRH3 has a long cytoplasmic tail with one ITAM, one ITIM, and an ITAM-like region. The amino acid length of each region is indicated. (B) Multiple alignment comparison of FcRH1, FcRH2, and FcRH3 aa sequences (one-letter code) based on the_FcRH3_ sequence. Amino acid identity is represented by dots, and gaps are indicated by dashes. Predicted N-linked glycosylation sites and transmembrane domains are underlined in black. Consensus ITAM (green) and ITIM (red) motifs are indicated. Putative structural domains are labeled: SP, signal peptide; EC, extracellular domain; MP-TM, membrane proximal-transmembrane; and CY, cytoplasmic regions. Amino acid lengths are indicated in parentheses.
Multiple alignment analysis of the translated cDNAs, using FcRH3 as the index sequence of comparison, indicates that FcRH1, FcRH2, and FcRH3 have highly conserved hydrophobic signal peptides and corresponding Ig-like extracellular domains (Fig. 2B). Their hydrophobic, uncharged transmembrane domains (40) are also well conserved, but their cytoplasmic domains are not. FcRH1 has a long cytoplasmic tail containing three potential ITAMs, the first and third of which fit the consensus sequence (E/D)-X-X-Y-X-X-(L/I)-X6–8-Y-X-X-(L/I), whereas, the second has only one tyrosine residue. The shorter cytoplasmic domain of FcRH2 contains one potential ITAM and two ITIM consensus sequences (I/V/L/S)-X-Y-X-X-(L/V) separated by 22 aa. FcRH3 has the longest cytoplasmic tail. It contains one potential ITAM, one ITIM, and another potential ITAM that also has a single tyrosine residue.
Tissue Distribution of FcRH1, FcRH2, and_FcRH3_ Expression.
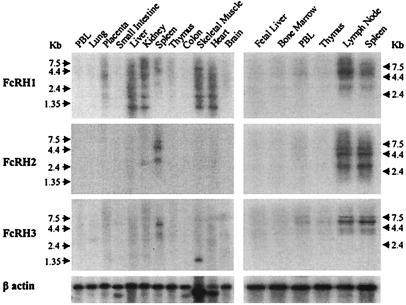
An RNA blot analysis with gene-specific probes was performed on 16 human tissues, including six primary or secondary lymphoid tissues. All three FcRH gene probes hybridized with transcripts in the secondary lymphoid organs, spleen and lymph node (Fig.3). An _FcRH1-_specific probe hybridized with spleen and lymph node transcripts of ≈3.5 kb and ≈6.0 kb. Additional hybridization bands of ≈0.7 kb and ≈1.5 kb were observed for heart, skeletal muscle, kidney, liver, and, in less abundance, placental tissue. Larger transcripts also were seen in skeletal muscle (≈6.0 kb) and in kidney and placenta (≈4.4 kb). An_FcRH2-_specific probe hybridized to ≈3.0-kb, ≈4.4-kb, and ≈5.5-kb transcripts most abundantly in spleen and lymph node. An ≈2.4-kb transcript was notable in the kidney. An FcRH3_probe hybridized with ≈3.5-kb, ≈5.5-kb, and ≈7.0-kb transcripts chiefly in spleen and lymph node. These also were seen, albeit in lesser abundance, in peripheral blood lymphocytes, thymus, and bone marrow samples. Additionally, a unique transcript of ≈1.35 kb was evident in skeletal muscle. These results indicated expression of_FcRH1, FcRH2, and FcRH3 in peripheral lymphoid organs, whereas tissue specific differences in alternative splicing or polyadenylation were suggested by the differential expression of transcripts with variable size in nonlymphoid tissues.
Figure 3.
Analysis of FcRH1, FcRH2, and_FcRH3_ expression in different tissues. RNA blots were analyzed with discriminating α32P-dCTP-labeled probes generated from the respective FcRH cDNAs. The following probes were used: (Top) a PCR-generated, 257-bp probe specific to the 3′ UT region of FcRH1; (Middle) a PCR-generated, 290-bp probe corresponding to the 3′ UT region of FcRH2; and (Bottom) a 528-bp _Eco_RI-digested fragment of the 5′ end of the FcRH3 cDNA corresponding to its 5′ UT region, S1, S2, and EC1 domains. Relative mRNA abundance is indicated by a β-actin probe.
When FcRH expression was examined by RT-PCR analysis of cell lines representing different hematopoietic lineages, FcRH1,FcRH2, and FcRH3 expression was found in every mature B cell line tested (Table 1).FcRH2 and FcRH3 expression was limited to the mature B cell lines and not seen in the other types of cells examined. In contrast, FcRH1 expression was seen in pro-B, T, and myeloid cell lines, although not in an erythroid cell line.
Table 1.
Expression of FcRH transcripts in human B cell lines
| Cell type | Cell line | FcRH1 | FcRH2 | FcRH3 |
|---|---|---|---|---|
| Pro-B | REH | + | − | − |
| Nalm16 | + | − | − | |
| Pre-B | 697 | − | − | − |
| 207 | − | − | − | |
| OB5 | − | − | − | |
| B | Ramos | + | + | + |
| Daudi | + | + | + | |
| Raji | + | + | + | |
| T | Jurkat | + | − | − |
| Monocytic | THP-1 | + | − | − |
| Myelomonocytic | U937 | + | − | − |
| Promyelocytic | HL-60 | + | − | − |
| Myelocytic | KG-1 | + | − | − |
| Erythroid | K562 | − | − | − |
RT-PCR analysis of sorted populations of peripheral blood cells indicated that FcRH1, FcRH2, FcRH3, and FcRH5 are expressed at relatively high levels in CD19+ B cells, whereas FcRH4 was expressed at only trace levels. FcRH3 expression was observed in CD3+ T cells whereas transcripts of_FcRH1_ were barely detectable. FcRH1 expression also was observed in circulating granulocytes.
Analysis of FcRH1, FcRH2, and_FcRH3_ Expression by Lymphoid Cells in the Tonsils.
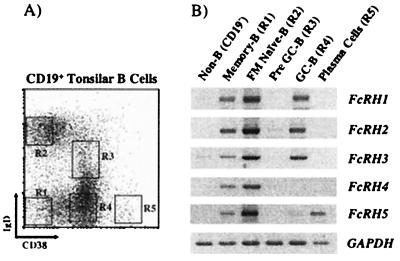
To refine the analysis of FcRH expression in secondary lymphoid tissues, tonsillar lymphocyte subpopulations were isolated. The five discrete subpopulations of B lineage cells that can be distinguished by their differential expression of cell surface IgD and CD38 (Fig. 4A) represent different stages in B cell differentiation: follicular mantle (IgD+CD38−), pre-GC (IgD+CD38+), GC (IgD−CD38+), memory (IgD−CD38−), and mature plasma cells (CD382+) (41). RT-PCR analysis indicated little or no expression of FcRH transcripts in the non-B lineage CD19− cells, most of which are T cells (Fig. 4B). However, CD19+subpopulations displayed coordinate expression of FcRH1,FcRH2, and FcRH3 transcripts in follicular mantle, naïve, GC, and memory B cell subpopulations, but yielded no evidence of FcRH transcripts in pre-GC B cells or plasma cells (Fig. 4B). In contrast, _FcRH4_transcripts were restricted to the follicular mantle and memory B cells, whereas FcRH5 expression extended to mature plasma cells.
Figure 4.
RT-PCR analysis of FcRH1–5 expression in tonsillar B cell subpopulations. (A) Viable cells were magnetically sorted into CD19− non-B cells and CD19+ B cells. The latter were stained with anti-IgD and anti-CD38 mAbs, and the five subpopulations indicated (CD38−IgD−, CD38− IgD+, CD38+ IgD+, CD38+IgD−, and CD382+) were sorted by flow cytometry. (B) RT-PCR analysis of _FcRH_transcripts in non-B cells and the B cell subpopulations. After cDNA preparation, PCR amplification was performed on the equivalent template of ≈10 k cells. Glyceraldehyde-3-phosphate dehydrogenase (GADPH) was amplified as a positive control.
Phylogenetic Analysis of the FcRH and FcR Families.
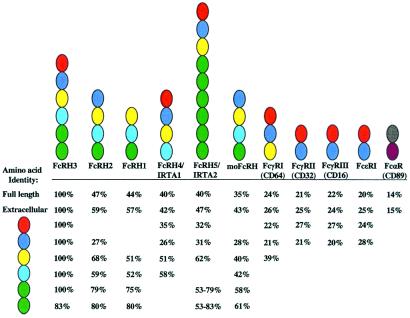
The relationship between the five FcRHs was examined by comparing their full-length, extracellular, and individual Ig-like domain amino acid sequences. This analysis, which included a recently identified mouse FcRH ortholog (moFcRH) and members of the FcR family, used theclustal method algorithm (42). Comparison of the full-length sequences of other FcRH family members with FcRH3 indicated 40–47% identity (Fig. 5). By comparison, the degree of FcRH3 homology with the moFcRH was found to be 35% and 21–24% with FcR members residing on chromosome 1, FcγRI, FcγRII, FcγRIII, and FcɛRI. A lower level of amino acid identity (14%) was observed for the chromosome 19 LRC member, FcαR. A slightly higher degree of extracellular homology was evident. Pairwise analysis of the individual Ig-like subunits indicated conservation in membrane-distal to membrane-proximal ordering of extracellular domain composition among family members (Fig. 5). Although similar Ig domain subunits were shared among family members, the individual receptors were found to be composed of unique domain combinations. The extracellular domain configuration of the moFcRH most closely resembled that of FcRH2, with which it has 46% identity. The extended pairwise comparison of the FcRH family with known FcRs suggested the conservation of these Ig-like domains to some degree throughout the greater family. The resemblance is particularly evident in the FcRH3 membrane-distal domains that correspond to the three FcγRI domains and the two domains of FcγRII, FcγRIII, and the FcɛR α-chain. This analysis suggests the ancestral occurrence of differential duplication and diversification of the individual Ig-like subunits in the respective FcRH family members. The data also indicate that the FcRHs are more similar to their FcR neighbors on chromosome 1 than to their FcαR relative on chromosome 19.
Figure 5.
Composite analysis of the extracellular homology among FcRH and FcR family members. Pairwise analysis of individual Ig-like subunits was performed with the clustal method algorithm using FcRH3 as the index of comparison. Individual homologous domains are color-coded to indicate relatedness. Percent amino acid identities for related domains are indicated and aligned in relation to the comparative FcRH3 subunit. The amino acid identity for the membrane proximal domains (green subunits) of FcRH5 are provided as the range of identity for all individually related domains. Comparisons that are not applicable are left blank. Amino acid sequences were derived from IRTA1 (GenBank accession no. AF343659), IRTA2 (GenBank accession no. AF34364), moFcRH (GenBank accession no. AAG28775) FcγRI (GenBank accession no.AAA35678), FcγRII (Swiss-Prot accession no. P31994), FcγRIII (Swiss-Prot accession no. P08637), FcɛRI (Swiss-Prot accession no.P12319), and FcαRI (Swiss-Prot accession no. P24071).
FcRH1, FcRH2, and FcRH3 Genomic Structure.
The genomic sequence analysis of relevant chromosome 1q21 BAC clones (Fig. 1) indicated that the entire FcRH locus spans ≈300 kb. The FcRH genes lie in the same transcriptional orientation toward the centomere. Exon-intron boundaries were characterized by sequence comparison of their respective cDNA clones and the AG/GT rule. The FcRH1 gene consists of 11 exons and 10 introns spanning ≈28 kb. The first exon, 5′ UT/S1, encodes the 5′ UT region, the ATG translation initiation codon, and the first half of a split signal peptide. S2, the second exon, is separated from 5′ UT/S1 by a long intron of ≈12.9 kb and, like the neighboring_FcRs_, is 21 bp in length (43–45). The extracellular region is encoded by three closely clustered exons, EC1–EC3, that code for the three Ig-like domains. The membrane-proximal, transmembrane, and the proximal portion of the cytoplasmic domain are encoded by a single sixth exon, TM. The cytoplasmic tail is encoded by five exons, CY1–CY5, and the CY5 also encodes the beginning of the 3′ UT region.
FcRH2 contains 12 exons and 11 introns that span ≈30 kb. It also contains two exons that encode a split signal peptide, the first of which, 5′UT/S1, includes the 5′ UT region, the ATG translation initiation codon, and first half of the signal peptide. The second exon, S2, is 21 bp in length. Exons 3–6 encode the four extracellular domains, EC1–EC4. The seventh exon encodes the membrane-proximal, transmembrane, and the proximal portion of the cytoplasmic domain. The FcRH2 cytoplasmic tail is encoded by five exons, CY1–CY5, the last exon of which includes the termination of the ORF and beginning of the 3′ UT region.
The FcRH3 gene consists of 16 exons and 15 introns that span ≈24 kb. Unlike FcRH1 and FcRH2, its 5′ UT region is encoded by two exons, 5′ UT1 and a second, 5′UT2/S1, that also encodes the ATG translation initiation codon and the beginning of the split signal peptide. The third exon, S2, is also 21 bp in length. Extracellular domains encoded by six exons, EC1–EC6, are followed by exon 10 that encodes the membrane-proximal, transmembrane, and the proximal portion of the cytoplasmic domain. The cytoplasmic tail is encoded by five exons, CY1–CY5; the last contains the beginning of the 3′ UT region.
Discussion
The FcRHs, identified here on the basis of sequence similarity with previously identified FcRs, are encoded by genes that reside near their closest FcR relatives on human chromosome 1 (q21–22). The FcRH family includes five functional genes and one pseudogene. Two of the FcRH members, FcRH4 and_FcRH5,_ are identical with the recently described_IRTA1_ and IRTA2, so named because of their discovery as genes that may be translocated in B cell malignancies (26). Three members of this family, FcRH1, FcRH2, and FcRH3, are described in this report. The FcRH family members share similar Ig-like domains, but possess distinctive cytoplasmic tails with consensus ITAMs and/or ITIMs. The signaling capacities of these transmembrane receptors thus are likely to differ. Our analysis of their tissue distribution indicates that the_FcRH1–3_ genes, like the previously described_FcRH4_ (IRTA1) and FcRH5(IRTA2), are expressed at relatively high levels in secondary lymphoid tissues, primarily by mature subsets of B cells.
The FcRH were identified on the basis of predicted amino acid sequence homology with the FcRγRs and the polymeric Ig receptor encoded by neighboring genes on chromosome 1. Initial bioinformatic screening using a consensus FcR sequence indicated the anticipated identity with FcR family members and, in addition, a lower correlative score was obtained for sequences found in chromosome 1 genomic BAC clones, one of which was remarkable for its inclusion of several putative Ig superfamily members. The BAC clones that proved to contain the_FcRH_ genes span the 1q21.2–1q22 region within the_FcR_ cluster. This genomic proximity, together with the amino acid homology of the predicted protein products of the _FcRH_genes, suggests a common phylogenetic ancestry. Analysis of their genomic structure further supports the relationship not only between the FcRH genes, but with their FcR neighbors as well. A conserved feature among the FcγRI, FcγRII,FcγRIII, and FcɛR genes is a signal peptide encoded by two separate exons, the second of which (S2) is a 21-bp “miniexon” (43–45). The conservation of the 21-bp S2 miniexon for all FcRH and FcR family members on chromosome 1 contrasts with the 36-bp S2 exon encoding the second half of the signal peptide for the FcαR gene on chromosome 19q13 (46).
The relationships between extracellular Ig-like domains of the FcRHs and those of the FcRs also suggest a common ancestry for the two subfamilies. This relationship is particularly evident for the individual Ig-like subunits, which display a conserved organizational pattern as an additional familial feature. Divergence of the_FcRHs_ from the FcRs is indicated by the duplication of novel membrane-proximal Ig-like domains in the FcRHs. However, the collective evidence of chromosomal proximity, genomic structural similarity, and Ig-like domain homology clearly imply that the FcRH genes are phylogenetic relatives of the_FcR_ family.
Like the previously identified (FcRH4) IRTA1 and (FcRH5) IRTA2 family members (ref. 26; our data), the FcRH1, FcRH2, and FcRH3 genes are preferentially expressed by B lineage cells in the spleen, tonsils, and lymph nodes. A possible alternatively spliced form of FcRH2, the recently described Src homology 2 domain-containing phosphatase anchor protein 1 (SPAP1), also has been shown to have a similar expression pattern (47). FcRH1 and FcRH5(IRTA2) transcripts were identified in nonlymphoid tissues also, suggesting a relatively broad biological potential for these receptors. Our analysis of FcRH1, FcRH2, and_FcRH3_ gene expression in cell lines representative of the different hematopoietic cell lineages indicates they are primarily expressed by mature B cells, suggesting a functional role in modulating the later stages of B cell maturation. In addition, _FcRH1_was found to be expressed by relatively immature hematopoietic cells of multiple cell lineages, including the T cell lineage. RT-PCR analysis of the expression of all five FcRH genes in primary lymphoid lineage cells in peripheral tissues indicated differential expression patterns for the different stages in the B cell differentiation pathway. Whereas transcripts for the FcRH1,FcRH2, and FcRH3 genes were expressed in naïve, GC, and memory B cell subsets, FcRH4_expression was limited to naïve and memory B cells and_FcRH5 expression persisted in mature plasma cells. These distinctive patterns of expression imply a highly coordinated regulation of FcRH gene expression during B cell activation and differentiation. In their in situ hybridization studies, Hatzivassiliou and coworkers (26) found IRTA1(FcRH4) expression to be topographically limited to marginal zone B cells, whereas IRTA2 (FcRH5) expression was also evident in GC B cells and immunoblasts. Their data also indicated a correlation between the IRTA2 overexpression and chromosomal 1q21 abnormalities in Burkitt lymphomas and multiple myelomas. Correspondingly, analysis of the Lymphochip data (34) indicates the differential expression of FcRH1 among the diffuse large B cell lymphomas. These observations have intriguing diagnostic and prognostic implications for these genes in lymphomagenesis.
The FcRH family members exhibit considerable diversity in their structure and signaling potential. Although their membrane-proximal Ig-like domains are relatively similar, the membrane-distal subunits display more limited identity, which may indicate diverse functional potential. The cytoplasmic tails of these receptors are also different. In contrast to the known FcRs that possess either activating or inhibitory properties, some of the FcRH have both types of motifs, a feature shared in common with the CD22 and CD31 (PECAM-1) transmembrane molecules (48–50). Whereas, the activating FcRs must associate with the FcR common γ chain via charged residues in their transmembrane region to transduce their signals (24, 25), the FcRHs have uncharged transmembrane domains and their own tyrosine-based signaling motifs. The capacity for both activating and inhibitory function in the cytoplasmic tails of individual FcRH members suggests the capacity for a dual role in regulation, or perhaps a fine-tuning modulation of the signaling threshold.
In addition to their structural differences and differential expression in B lineage cells, the individual FcRH expression in nonlymphoid tissues, apparently as splice variants, implies an additional degree of diversity for this family of receptors. The expression of a unique FcRH3 transcript of ≈1.3 kb in skeletal tissue, for example, suggests members of the _FcRH_family will have nonimmunological as well as immunological functions.
Acknowledgments
We thank Drs. Eric Vivier, Louis B. Justment, and Peter D. Burrows for critical comments on the manuscript, Marsha Flurry, Dottie Lang, and E. Ann Brookshire for help in manuscript preparation, Dr. Larry Gartland for help with flow cytometry, and Yoshiki Kubagawa for sequencing assistance. This work has been supported in part by National Institutes of Health Grants AI42127 and AI39816. M.D.C. is a Howard Hughes Medical Institute Investigator. R.S.D. was supported by the Walter B. Frommeyer, Jr. Fellowship in Investigative Medicine.
Abbreviations
FcR
Fc receptor
FcRH
FcR homolog
ITIM
immunoreceptor tyrosine-based inhibitory motif
ITAM
immunoreceptor tyrosine-based activation motif
LRC
leucocyte receptor complex
RACE
rapid amplification of cDNA ends
UT
untranslated
RT
reverse transcription
BAC
bacterial artificial chromosome
GC
germinal center
IRTA
Ig superfamily receptor translocation-associated gene
moFcRH
mouse FcRH
Footnotes
References
- 1.Ravetch J V, Kinet J-P. Annu Rev Immunol. 1991;9:457–492. doi: 10.1146/annurev.iy.09.040191.002325. [DOI] [PubMed] [Google Scholar]
- 2.Daeron M. Annu Rev Immunol. 1997;15:203–234. doi: 10.1146/annurev.immunol.15.1.203. [DOI] [PubMed] [Google Scholar]
- 3.Ravetch J V, Bolland S. Annu Rev Immunol. 2001;19:275–290. doi: 10.1146/annurev.immunol.19.1.275. [DOI] [PubMed] [Google Scholar]
- 4.Kubagawa H, Burrows P, Cooper M D. Proc Natl Acad Sci USA. 1997;94:5261–5266. doi: 10.1073/pnas.94.10.5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayami K, Fukuta D, Nishikawa Y, Yamashita Y, Inui M, Ohyama Y, Hikida M, Ohmori H, Takai T. J Biol Chem. 1997;272:7320–7327. doi: 10.1074/jbc.272.11.7320. [DOI] [PubMed] [Google Scholar]
- 6.Borges L, Hsu M-L, Fanger N, Kubin M, Cosman D. J Immunol. 1997;159:5192–5196. [PubMed] [Google Scholar]
- 7.Samaridis J, Colonna M. Eur J Immunol. 1997;27:660–665. doi: 10.1002/eji.1830270313. [DOI] [PubMed] [Google Scholar]
- 8.Kremer E J, Kalatzis V, Baker E, Callen D F, Sutherland G R, Maliszewski C R. Hum Genet. 1992;89:107–108. doi: 10.1007/BF00207054. [DOI] [PubMed] [Google Scholar]
- 9.Wagtmann N, Rojo S, Eichler E, Mohrenweiser H, Long E O. Curr Biol. 1997;7:615–618. doi: 10.1016/s0960-9822(06)00263-6. [DOI] [PubMed] [Google Scholar]
- 10.Wende H, Colonna M, Ziegler A, Volz A. Mamm Genome. 1999;10:154–160. doi: 10.1007/s003359900961. [DOI] [PubMed] [Google Scholar]
- 11.Wilson M J, Torkar M, Haude A, Milne S, Jones T, Sheer D, Beck S, Trowsdale J. Proc Natl Acad Sci USA. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. . (First Published April 18, 2000; 10.1073/pnas.080588597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reth M. Annu Rev Immunol. 1992;10:97–121. doi: 10.1146/annurev.iy.10.040192.000525. [DOI] [PubMed] [Google Scholar]
- 13.Vely F, Vivier E. J Immunol. 1997;159:2075–2077. [PubMed] [Google Scholar]
- 14.Ravetch J V, Lanier L L. Science. 2000;290:84–89. doi: 10.1126/science.290.5489.84. [DOI] [PubMed] [Google Scholar]
- 15.Gergely J, Pecht I, Sarmay G. Immunol Lett. 1999;68:3–15. doi: 10.1016/s0165-2478(99)00024-3. [DOI] [PubMed] [Google Scholar]
- 16.Dennis G, Jr, Kubagawa H, Cooper M D. Proc Natl Acad Sci USA. 2000;97:13245–13250. doi: 10.1073/pnas.230442897. . (First Published November 14, 2000; 10.1073/pnas.230442897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoder J A, Mueller M G, Wei S, Corliss B C, Prather D M, Willis T, Litman R T, Djeu J Y, Litman G W. Proc Natl Acad Sci USA. 2001;98:6771–6776. doi: 10.1073/pnas.121101598. . (First Published May 29, 2001; 10.1073/pnas.121101598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long E O. Annu Rev Immunol. 1999;17:875–904. doi: 10.1146/annurev.immunol.17.1.875. [DOI] [PubMed] [Google Scholar]
- 19.Unkeless J C, Jin J. Curr Opin Immunol. 1997;9:338–343. doi: 10.1016/s0952-7915(97)80079-9. [DOI] [PubMed] [Google Scholar]
- 20.Qui W Q, de Bruin D, Brownstein B H, Pearse R, Ravetch J V. Science. 1990;248:732–735. doi: 10.1126/science.2139735. [DOI] [PubMed] [Google Scholar]
- 21.Oakley R J, Howard T A, Hogarth P M, Tani K, Seldin M F. Immunogenetics. 1992;35:279–282. doi: 10.1007/BF00166834. [DOI] [PubMed] [Google Scholar]
- 22.Krajci P, Grzeschik K H, Geurts van Kessel A H, Olaisen B, Brandtzaeg P. Hum Genet. 1991;87:642–648. doi: 10.1007/BF00201717. [DOI] [PubMed] [Google Scholar]
- 23.Shibuya A, Sakamoto N, Shimizu Y, Shibuya K, Osawa M, Hiroyama T, Eyre H J, Sutherland G R, Endo Y, Fujita T, et al. Nat Immunol. 2000;1:441–446. doi: 10.1038/80886. [DOI] [PubMed] [Google Scholar]
- 24.Pfefferkorn L C, Yeaman G R. J Immunol. 1994;153:3228–3236. [PubMed] [Google Scholar]
- 25.Morton E C, Van den Herik-Oudijk I E, Vossebeld P, Snijders A, Verhoeven A J, Capel P J A, Van de Winkel J G J. J Biol Chem. 1995;270:29781–29787. doi: 10.1074/jbc.270.50.29781. [DOI] [PubMed] [Google Scholar]
- 26.Hatzivassiliou G, Miller I, Takizawa J, Palanisamy N, Rao P H, Iida S, Tagawa S, Taniwaki M, Russo J, Neri A, et al. Immunity. 2001;14:277–289. doi: 10.1016/s1074-7613(01)00109-1. [DOI] [PubMed] [Google Scholar]
- 27.Altschul S F, Gish W, Miller W, Meyers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Korsmeyer S J, Arnold A, Bakhshi A, Ravetch J V, Siebenlist U, Hieter P A, Sharrow S O, LeBien T W, Kersey J H, Poplack D, et al. J Clin Invest. 1983;71:301–313. doi: 10.1172/JCI110770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Findley H W, Cooper M D, Kim J H, Alvarado C, Ragab A H. Blood. 1982;60:1305–1309. [PubMed] [Google Scholar]
- 30.Martin D, Huang R, LeBien T W, Van Ness B. J Exp Med. 1991;173:639–645. doi: 10.1084/jem.173.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pulvertaft R J V. Lancet. 1964;1:238–240. doi: 10.1016/s0140-6736(64)92345-1. [DOI] [PubMed] [Google Scholar]
- 32.Klein E, Klein G, Nadkarni J S, Nadkarni J J, Wigzell H, Clifford P. Cancer Res. 1968;28:1300–1310. [PubMed] [Google Scholar]
- 33.Klein G, Giovinella B, Westman A, Stehlin J S, Mumford D. Intervirology. 1975;5:319–334. doi: 10.1159/000149930. [DOI] [PubMed] [Google Scholar]
- 34.Alizadeh A A, Eisen M B, Davis R E, Ma C, Lossos I S, Rosenwald A, Boldrick J C, Sabet H, Tran T, Yu X, et al. Nature (London) 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 35.Von Heijne G. Nucleic Acid Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 37.Williams A F, Barclay A N. Annu Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- 38.Bork P, Holm L, Sander C. J Mol Biol. 1994;242:309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- 39.Vaughn D E, Bjorkman P J. Neuron. 1996;16:261–273. doi: 10.1016/s0896-6273(00)80045-8. [DOI] [PubMed] [Google Scholar]
- 40.Sonnhammer E L L, von Heijne G, Krogh A. In: A Hidden Markov Model for Predicting Transmembrane Helices in Protein Sequences. Glasgow J, Littlejohn T, Major F, Lathrop R, Sankoff D, Sensen C, editors. Menlo Park, CA: Am. Assoc. for Artificial Intelligence; 1998. pp. 175–182. [PubMed] [Google Scholar]
- 41.Pascual V, Liu Y-J, Magalski A, de Bouteiller O, Bancereau J, Capra J D. J Exp Med. 1994;180:329–339. doi: 10.1084/jem.180.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Higgins D G, Sharp P M. Comput Appl Biosci. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 43.van de Winkel J G, Capel P J. Immunol Today. 1993;14:215–221. doi: 10.1016/0167-5699(93)90166-I. [DOI] [PubMed] [Google Scholar]
- 44.Kulczycki A, Jr, Webber J, Soares H A, Onken M D, Thompson J A, Chaplin D D, Loh D Y. Proc Natl Acad Sci USA. 1990;87:2856–2860. doi: 10.1073/pnas.87.7.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pang J, Taylor G R, Munroe D G, Ishaque A, Fung-Leung W P, Lau C Y, Liu F T, Zhou L. J Immunol. 1994;151:6166–6174. [PubMed] [Google Scholar]
- 46.de Wit T P M, Morton H C, Capel P J A, van de Winkel J G J. J Immunol. 1995;155:1203–1209. [PubMed] [Google Scholar]
- 47.Xu M, Zhao R, Zhao Z J. Biochem Biophys Res Commun. 2001;280:768–775. doi: 10.1006/bbrc.2000.4213. [DOI] [PubMed] [Google Scholar]
- 48.Newman P J, Berndt M C, Gorski J, White G C, Lyman S, Paddock C, Muller W A. Science. 1990;247:1219–1222. doi: 10.1126/science.1690453. [DOI] [PubMed] [Google Scholar]
- 49.Doody G M, Justement L B, Delibrias C C, Matthews R J, Lin J, Thomas M L, Fearon D T. Science. 1995;269:242–244. doi: 10.1126/science.7618087. [DOI] [PubMed] [Google Scholar]
- 50.Poe J C, Fujimoto M, Jansen P J, Miller A S, Tedder T F. J Biol Chem. 2000;275:17420–17427. doi: 10.1074/jbc.M001892200. [DOI] [PubMed] [Google Scholar]