Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera (original) (raw)
Abstract
Aphids possess bacteriocytes, cells specifically differentiated to harbor obligatory mutualistic bacteria of the genus Buchnera, which have lost many genes that are essential for common bacterial functions. To understand the host's role in maintaining the symbiotic relationship, bacteriocytes were isolated from the pea aphid, Acyrthosiphon pisum, and the host transcriptome was investigated by using EST analysis and real-time quantitative RT-PCR. A number of genes were highly expressed specifically in the bacteriocyte, including (i) genes for amino acid metabolism, including those for biosynthesis of amino acids that Buchnera cannot produce, and those for utilization of amino acids that Buchnera can synthesize; (ii) genes related to transport, including genes for mitochondrial transporters and a gene encoding Rab, a G protein that regulates vesicular transport; and (iii) genes for putative lysozymes that degrade bacterial cell walls. Significant up-regulation of i clearly indicated that the bacteriocyte is involved in the exchange of amino acids between the host aphid and Buchnera, the key metabolic process in the symbiotic system. Conspicuously high expression of ii and iii shed light on previously unknown aspects of the host–Buchnera interactions in the symbiotic system.
Keywords: EST, quantitative RT-PCR
Although aphids feed only on a nutritionally poor diet, phloem sap, they show explosive reproductivity, which makes them notorious agricultural pests. Nutritional aspect of this fecundity is based on an intimate symbiotic association with a microorganism. Within the haemocoel, almost all aphid species have dozens of bacteriocytes, specialized cells for harboring intracellular symbiotic bacteria of the genus Buchnera that belong to the γ-subdivision of the Proteobacteria (1). Physiological studies demonstrated that Buchnera provide host aphids with essential amino acids (amino acids that metazoa cannot synthesize; tryptophan, lysine, methionine, phenylalanine, threonine, valine, leucine, isoleucine, arginine, and histidine) (2–6) and riboflavin (vitamin B2) (7) that aphids cannot synthesize and are scarce in the phloem sap diet (8, 9). Since the initial infection >100 million years ago (10), Buchnera have been subjected to strict vertical transmission through host generations, and the mutualism between the host and Buchnera has reached to such an extent that neither can reproduce in the absence of the other (2, 3).
Recent studies on the complete genome sequences of three lineages of Buchnera (those in association with Acyrthosiphon pisum, Schizaphis graminum, and Baizongia pistaceae) provided comprehensive knowledge on potential functions of Buchnera in the symbiotic system (11–13). The sequenced Buchnera genomes were only 0.61–0.65 Mb in size and encoded 510–570 proteins. The gene composition confirmed and extended the previous physiological data that Buchnera are able to provide their hosts with essential amino acids and riboflavin. Whereas massive genome reduction is generally found in endocellular bacteria of both parasitic and mutualistic nature (11–19), the genomes of the endocellular parasites retain only a few, if any, genes for synthesis of these nutrients (14, 15, 18). Thus, the gene repertoire of the Buchnera genomes clearly reflects the mutualistic nature of Buchnera for the host aphids (11).
The genome studies of Buchnera have provided not only genetic insights into how the symbiotic system operates but also have raised a number of questions that can be answered only with the genomic information of the host side. Whereas Buchnera retain genes for the biosynthesis of nutrients that are required by the hosts, they lack many genes that seem to be essential for their own living. For example, most genes involved in biosyntheses of nonessential amino acids (amino acids that metazoa can synthesize; alanine, asparagine, aspartic acid, cysteine, glutamic acid, glutamine, glycine, proline, serine, and tyrosine) and phospholipids are lost, implying that Buchnera can synthesize neither nonessential amino acids nor even their own cell membrane. Most genes encoding transcriptional regulators are also missing, suggesting that Buchnera may scarcely be able to regulate their own metabolic and cellular activities (11). It appears likely that these and other incomplete aspects of Buchnera functions must be compensated for by the activities of the host bacteriocyte.
In an effort to understand the host's role in this symbiotic system, we assessed the mRNA population of the host bacteriocyte of the pea aphid, Acyrthosiphon pisum, by EST analysis and real-time quantitative RT-PCR.
Materials and Methods
Aphids. Strain ISO, a parthenogenetic clone of the pea aphid Acyrthosiphon pisum, was used in this study. Diagnostic PCR has verified that this clone is free of secondary symbionts such as R-type, T-type, U-type, Rickettsia, Wolbachia, Spiroplasma, or Arsenophonus (data not shown). The insects were reared on Vicia faba at 15°C in a long-day regime of 16 h of light and 8 h of dark. Parthenogenetic apterous adults (12–15 days old) were used for the experiments.
RNA Preparation from Bacteriocytes. The aphids were dissected in buffer A (20) on 1% agarose plate. Bacteriocytes freed from the insect body were collected with a micropipette and immediately lysed in TRIzol reagent (GIBCO/BRL). Total RNA was extracted according to the manufacturer's instructions. In total, 60,000–80,000 bacteriocytes collected from ≈2,000 insects were used for the library construction.
cDNA Library Construction. Full-length cDNA library was generated by cap-trapper method (21, 22) with a slight modification. Oligo(dT) primer for the first-strand cDNA synthesis: 5′-GAGAGAGAGAGGATCCTTCTGGAGAGTTTTTTTTTTTTTTTTVN-3′. Double-stranded linkers for the second-strand cDNA synthesis: GN5 linker and N6 linker (molar ratio of N6:GN5 = 1:4) (23). Second-strand cDNA was digested with _Bam_HI and _Xho_I, and ligated to lambda FLC-I vector, which carries two loxP sites (24). After amplification in C600 cells, the phage DNA was converted into plasmid with Cre recombinase.
Sequence of Clones. Sequencing reactions were performed by using ABI big dye terminator v3.1 (Applied Biosystems) along with the M13 forward primer: 5′-TGTAAAACGACGGCCAGT-3′. Reaction products were run on an ABI 3700 capillary sequencer (Applied Biosystems).
Sequence Processing and Clustering. The raw chromatogram files were base-called with phred (25, 26), and the vector sequence was masked with crossmatch. Only sequences that exhibited >100 bases with a phred quality value of >20 were used for the subsequent analyses. Contaminants derived from the vector, Escherichia coli genome, or Buchnera genome were detected by using blastn (27) with high-stringency parameters (E value of <1.0 × 10–20, cost to open a gap = 1, cost to extend a gap = 3, word size = 10). After these quality assessments, the high-quality ESTs were deposited in the DNA Data Bank of Japan database (accession nos. BP535536–BP537955). These sequences were assembled by using the cap3 program (28).
Sequence Analysis and Annotation. The combined set of the consensus sequences was analyzed as a putative “unigene set.” The unigene set was annotated based on the blast similarity searches against protein databases. First, each unigene was compared against the fly protein database [gadf ly version 3.1, Berkeley _Drosophila_ Genome Project, which can be accessed at www.fruitfly.org] by using blastx, and automatically assigned a gene annotation of the top hit (E ≤ 1.0 × 10–10), regarding it as a putative fly homolog. Gene Ontology (GO) classifications of the corresponding fly homologs were transferred from FlyBase (which can be accessed at http://flybase.org, FlyBase Consortium, 2003). Second, similarity search was performed against the nonredundant protein database [www.ncbi.nlm.nih.gov, National Center for Biotechnology Information (NCBI), March 2004]. Unigenes with no apparent fly homolog were annotated in this step if similar proteins were found. In addition, if the blast bitscore of top hit was significantly higher than that of fly homolog, the annotation was replaced. These annotation procedures were automated by the custom Ruby script. Finally, the gene assignments were inspected manually and annotated by using a specifically developed annotation interface. All of the information on our unigenes with their annotations is available upon request.
Real-Time Quantitative RT-PCR. RNA was isolated from whole bodies and bacteriocytes of 12- to 15-day-old parthenogenetic apterous adults by using TRIzol reagent, followed by RNase-free DNase I treatment. Each whole-body sample and bacteriocyte sample derived from one individual and a batch of bacteriocytes that were collected from ≈10 individuals, respectively. First-strand cDNAs were synthesized by using pd(N)6 primer and the First-Strand cDNA synthesis kit (Amersham Biosciences, Piscataway, NJ). External standards were constructed by PCR using whole-body cDNA and gene-specific primer sets that were designed referring to cap3 base confidence scores of contigs generated by the EST analysis (Table 3, which is published as supporting information on the PNAS web site). Quantification was performed with the LightCycler instrument and FastStart DNA Master SYBR green I kit (Roche Diagnostics, Mannheim, Germany). Running parameters were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 55°C for 10 s, and 72°C for 4 s. Signal intensity was measured at the end of each elongation phase unless otherwise stated. When primer dimers were detected in preliminary experiments, signal intensity was measured at an additional step after elongation (Table 3). Results were analyzed by using the lightcycler software, version 3.0 (Roche Diagnostics), and relative expression levels were normalized to mRNA for ribosomal protein (Rp)L7. All quantitative RT-PCRs were performed in triplicate. Figures thus show each value as the mean ± SE of 10 independent experiments of 10 independent samples (n = 10), with each one of them represented by the mean of three separate quantitative RT-PCRs. Statistical analyses were performed by using the Mann–Whitney U test. Levels of transcripts for neither ribosomal protein S3A nor elongation factor 1α showed significant difference between the bacteriocyte and the whole body (P > 0.05), proving appropriateness of the quantification system (data not shown).
Results and Discussion
Construction of the Bacteriocyte cDNA Library. The aphid bacteriocyte contains a large number of Buchnera cells in the cytoplasm. Thus, to construct a cDNA library of the host bacteriocyte, contaminants of the bacterial symbionts must be removed. First, we attempted to synthesize the host cDNAs selectively by using an oligo(dT) primer that targets the 3′ poly(A) tail of eukaryotic mRNAs. However, this conventional method did not work, because the genome of Buchnera is highly AT-rich (11–13), and thus, allowed frequent annealing of the oligo(dT) primer to their RNAs. Within a cDNA library preliminarily constructed by using the conventional method, 75% of clones were of Buchnera origin (data not shown). Therefore, we adopted an alternative method, the cap trapper that can select cDNAs derived from eukaryotic mRNAs with the 5′ cap structure (21, 22). Application of this method reduced the Buchnera contaminants in the cDNA library to an acceptable level (9.9%, see below).
Generation and Assembly of Bacteriocyte ESTs. In total, 2,870 cDNA clones were sequenced from the 5′ end. After removal of low-quality sequences and contaminants derived from the vector and E. coli genome, 2,602 high-quality sequences were obtained. Of these sequences, 257 ESTs matched to the genome of Buchnera aphidicola str. APS (GenBank accession no. NC_002528). After removal of the Buchnera sequences, 2,345 ESTs of 656.6 bp average length were clustered by using the cap3 program. The ESTs were assembled into 246 contigs with 91 ESTs remaining as singlets. Further analyses were performed by using these 337 nonredundant sequences as a putative unigene set.
Similarity Search and Annotation. The unigene set was subjected to blastx similarity searches. Of 337 unigenes, 233 (183 contigs and 50 singlets, 69.1%) and 244 (190 contigs and 54 singlets, 72.4%) showed significant similarities (E ≤1.0 × 10–10) to protein-encoding genes in the fly database (gadf ly, version 3.1) and the NCBI nonredundant database, respectively. These blast hits were used to annotate the unigenes as described in Materials and Methods. The unigenes were ranked by the number of corresponding EST clones (Table 1 and Table 4, which is published as supporting information on the PNAS web site).
Table 1. Highly expressed genes in the bacteriocyte.
| Local ID | No. of ESTs | Protein homolog | Source organism | NCBI accession no. | E value |
|---|---|---|---|---|---|
| R2C00037 | 134 | LSZ i-1 | A. gambiae | AAT51799 | 2.3 × 10-35 |
| R2C00204 | 71 | LSZ i-1 | A. gambiae | AAT51799 | 1.1 × 10-34 |
| R2C00172 | 70 | ANT2 | Drosophila melanogaster | AAB31734 | 6.9 × 10-103 |
| R2C00040 | 55 | GS2 (CG1743) | D. melanogaster | NP_727525 | 2.2 × 10-80 |
| R2C00101 | 55 | α-Tubulin at 84B (CG1913; α Tub84B) | D. melanogaster | NP_476772 | 9.3 × 10-116 |
| R2C00059 | 45 | No hits | |||
| R2C00253 | 34 | Cytosolic malate dehydrogenase | Homo sapiens | NP_005908 | 1.8 × 10-66 |
| R2C00050 | 33 | RpS9 (CG3395) | D. melanogaster | NP_524004 | 1.8 × 10-95 |
| R2C00038 | 27 | CAT2 | H. sapiens | NP_003037 | 1.9 × 10-46 |
| R2C00023 | 26 | HSC70 (CG4264) | D. melanogaster | NP_524356 | 1.1 × 10-92 |
| R2C00089 | 26 | PC | D. melanogaster | Q9V7S5 | 4.5 × 10-28 |
| R2C00113 | 26 | Rp L15 (CG17420) | D. melanogaster | NP_652103 | 1.1 × 10-88 |
| R2C00020 | 25 | OT | Schizosaccharomyces pombe | NP_593169 | 1.7 × 10-22 |
| R2C00100 | 24 | Phosphoenolpyruvate carboxykinase (CG17725; Pepck) | D. melanogaster | NP_523784 | 2.2 × 10-73 |
| R2C00132 | 24 | GCVT | H. sapiens | NP_000472 | 1.4 × 10-41 |
| R2C00244 | 23 | AS | Manduca sexta | Q9U505 | 1.0 × 10-46 |
| R2C00124 | 22 | RpS14a (CG1524) | D melanogaster | NP_524884 | 2.5 × 10-76 |
| R2C00011 | 21 | No hits | |||
| R2C00108 | 21 | RpS3A (CG2168) | D. melanogaster | NP_524618 | 1.6 × 10-83 |
| R2C00022 | 20 | Diacetyl/l-xylulose reductase | Xenopus tropicalis | NP_989100 | 1.6 × 10-30 |
| R2C00106 | 20 | 5′-Nucleotidase precursor | Lutzomyia longipalpis | Q9XZ43 | 2.2 × 10-47 |
Comparative Analysis of Aphid Transcriptomes. To obtain an overview of the aphid bacteriocyte transcriptome (ESTBC), we compared EST population in ESTBC with those of other aphid transcriptomes. As of May 2004, two aphid EST sets were available in the dbEST depository of GenBank; one is from the pea aphid whole-body library (ESTWB) containing 1,071 sequences, and the other is from a whole-body library of the brown citrus aphid, Toxoptera citricida (ESTTC), containing 4,267 sequences (29). Note that all these aphid libraries were constructed without using PCR, which might bias cDNA populations. The data were extracted, analyzed, and annotated by the same method. ESTWB and ESTTC were assembled into 742 unigenes (151 contigs plus 591 singlets) and 2,176 unigenes (465 contigs plus 1,711 singlets), respectively. Sequence comparisons demonstrated that 25.2% (70 contigs plus 15 singlets) and 36.5% (97 contigs plus 26 singlets) of unigenes in ESTBC were similar to those in ESTWB and ESTTC, respectively.
Selective Up-Regulation of Genes Related to Amino Acid Metabolism, Transport, and Defense Response in the Bacteriocyte. The cap3 program assembled 2,345 ESTs of ESTBC into 337 unigenes, indicating 85.6% redundancy. This value is much higher than that of ESTWB (30.7%) and ESTTC (49.0%), suggesting that the bacteriocyte expresses relatively small number of genes at conspicuously high levels. The numbers of EST clones under each of the GO categories were compared among ESTBC, ESTWB, and ESTTC (Table 2; a complete list is available upon request). Among these categories, “amino acid metabolism (GO:0006520),” “transport (GO:0006810),” and “defense response (GO:0006952)” contained significantly higher percentage of ESTs in ESTBC than in the whole-body transcriptomes, ESTWB and ESTTC (P < 0.001, Fisher's exact test with Bonferroni correction for multiple comparisons).
Table 2. GO classification of ESTs in aphid transcriptomes.
| Percent of total fly-hit ESTs | |||
|---|---|---|---|
| GO* | ESTBC | ESTWB | ESTTC |
| GO:0008150: Biological process | 89.76 | 80.61 | 79.83 |
| [i] GO:0007610: behavior | 0.17 | 1.20 | 1.36 |
| [i] GO:0007582: physiological process | 88.20 | 79.01 | 77.16 |
| [i] GO:0008152: metabolism | 62.33 | 68.18 | 63.97 |
| [i] GO:0006520: amino acid metabolism | 9.13 | 4.14 | 2.15 |
| [i] GO:0009987: cellular process | 34.89 | 30.35 | 35.33 |
| [i] GO:0007154: cell communication | 9.63 | 5.88 | 7.21 |
| [i] GO:0030154: cell differentiation | 1.22 | 1.87 | 2.43 |
| [i] GO:0050875: cellular physiological process | 32.44 | 27.94 | 32.43 |
| [i] GO:0008151: cell growth and/or maintenance | 27.55 | 24.47 | 29.15 |
| [i] GO:0006810: transport | 24.48 | 16.58 | 15.30 |
| [i] GO:0006865: amino acid transport | 1.56 | 0 | 0.05 |
| [i] GO:0007275: development | 11.35 | 13.24 | 12.21 |
| [i] GO:0050896: response to stimulus | 18.59 | 7.09 | 11.51 |
| [i] GO:0006952: defense response | 15.86 | 4.55 | 3.46 |
| [i] GO:0042742: defense response to bacteria | 11.41 | 0 | 0.05 |
| [i] GO:0050789: regulation of biological process | 2.84 | 8.82 | 7.11 |
Up-Regulated Genes Involved in Amino Acid Metabolism. ESTBC contained 164 EST clones relevant to amino acid metabolism. Despite the relative abundance of EST clones related to amino acid metabolism in ESTBC (Table 2), these ESTs corresponded to only 13 unigenes, which was less than those in ESTWB (18 unigenes) and ESTTC (25 unigenes). To further verify the abundance of transcripts detected in ESTBC, real-time quantitative RT-PCR was carried out. As representatives, unigenes for glutamine synthetase 2 (GS2; 55 clones) (EC 6.3.1.2), cationic amino acid transporter 2 (CAT2; 27 clones), glycine cleavage system T protein (GCVT; 24 clones) [an aminomethyl transferase (EC 2.1.2.10), which is a part of the glycine cleavage multienzyme complex catalyzing the degradation of glycine], Henna (13 clones)[an enzyme with activities of phenylalanine 4-monooxygenase (EC 1.14.16.1) and tryptophan 5-monooxygenase (EC 1.14.16.4), which are involved in catabolisms of l-phenylalanine and l-tryptophan, respectively], glutaryl-CoA dehydrogenase (GCDH; 6 clones) (EC 1.3.99.7; an enzyme that catalyzes the oxidative decarboxylation of glutaryl-CoA, which is involved in l-tryptophan metabolism and degradative pathways of l-lysine and l-hydroxylysine) and phosphoserine aminotransferase (PSAT; 4 clones) (EC 2.6.1.52; an enzyme that is involved in serine biosynthesis) were selected. Unigenes for GS2, CAT2, and GCVT were among those with the largest number of EST clones in ESTBC (Table 1).
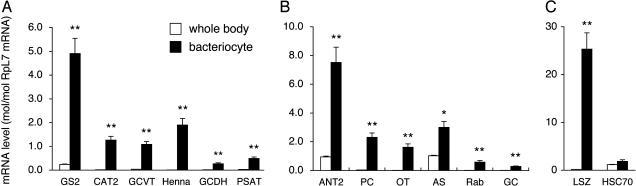
Quantitative RT-PCR confirmed significantly higher expression of these genes in the bacteriocyte. Genes for GS2, CAT2, GCVT, Henna, GCDH, and PSAT were expressed 20.7-, 93.3-, 30.4-, 297-, 23.5- and 18.7-fold higher in the bacteriocyte than in the whole body, respectively (Fig. 1_A_).
Fig. 1.
Quantitative RT-PCR of aphid genes expressed in the bacteriocyte. (A) Genes related to amino acid metabolism: GS2, CAT2, GCVT, Henna, GCDH, and PSAT. (B) Genes related to transport: ANT2, PC, OT, AS, Ras-like Rab GTPase, and GC. (C) Genes related to defense response: LSZ and HSC70. White columns, expression levels in the whole body; black columns, expression levels in the bacteriocyte; bars, SE (n = 10). The expression levels are shown in terms of mRNA copies of target genes per copy of mRNA for RpL7. Asterisks indicate statistically significant differences (Mann–Whitney U test; *, P < 0.05; **, P < 0.01).
Role of the Bacteriocyte in Amino Acid Metabolism of Aphids. The bacteriocyte is the specialized cell for harboring the essential symbiont Buchnera, whose pivotal role is the synthesis of essential amino acids that are scarce in the phloem sap diet (2, 3). The genome of Buchnera is specialized for production of essential amino acids: Genes for synthesis of essential amino acids are retained, whereas genes for synthesis of nonessential amino acids are mostly lost (11). In the bacteriocyte, therefore, it is expected that essential amino acids are supplied by the symbiont whereas nonessential amino acids must be synthesized in excess by the host cell. In agreement with the expectation, we found that genes relevant to utilization of essential amino acids (i.e., CAT2, Henna, and GCDH) and genes relevant to synthesis of nonessential amino acids (i.e., GS2 and PSAT) were highly expressed in the bacteriocyte. A gene involved in catabolism of a nonessential amino acid, glycine, (i.e., GCVT) was also highly expressed in the bacteriocyte. Notably, glycine is among the few nonessential amino acids that Buchnera is able to synthesize (11). These results unveiled an important aspect of the molecular basis of interdependency between the host and symbiont.
Up-Regulated Genes Involved in Transport. The bacteriocyte transcriptome ESTBC contained 440 EST clones related to transport, representing 38 unigenes. Within the category of “transport,” its subcategory “amino acid transport (GO:0006865)” contained significantly higher percentage of ESTs in ESTBC than in the whole-body transcriptomes ESTWB and ESTTC (P < 0.001) (Table 2). This subcategory was represented by 28 clones corresponding to two ESTBC unigenes, a contig R2C00038 and a singlet BCA014030, which were similar to genes encoding cationic amino acid transporters, type 2 (CAT2) and type 1 (CAT1), respectively. R2C00038 (27 clones) was one of the most highly expressed unigenes in ESTBC as already described (Table 1). Moreover, the transport category contained several other unigenes with remarkably large numbers of EST clones: genes for ADP/ATP translocase (ANT2; 70 clones, a translocator that exchanges ADP and ATP across the mitochondrial inner membrane), inorganic phosphate cotransporter (PC; 26 clones, an integral membrane protein that belongs to the sodium/anion cotransporter family), mitochondrial oxaloacetate transport protein (OT; 25 clones, a mitochondrial inner membrane protein that transports oxaloacetate and sulfate), and ATP synthase subunit c (AS; 23 clones, a component of mitochondrial ATP synthase) (Table 1). Other transport-related unigenes were also examined: genes for Ras-like Rab GTPase (10 clones, a GTP-binding protein that regulates vesicle transport) and mitochondrial glutamate carrier (GC; 7 clones, an integral membrane protein involved in the transport of glutamate across the inner mitochondrial membrane).
Quantitative RT-PCR confirmed significantly higher expression of these genes in the bacteriocyte. Genes for ANT2, PC, OT, AS, Ras-like Rab GTPase, and GC were expressed 7.98-, 68.1-, 182-, 2.92-, 40.7-, and 24.5-fold higher in the bacteriocyte than in the whole body, respectively (Fig. 1_B_).
Importance of Transport in the Bacteriocyte. In the aphid symbiotic system, metabolic integration and interdependency between the host and symbiont are so intricate that the partners are regarded as comprising an almost inseparable biological entity (2, 3). Located at the symbiotic interface, the bacteriocyte is expected to be involved in exchange of various metabolites and substrates between the host and the symbiont. In agreement with the expectation, we identified a number of transport-related genes that were strikingly up-regulated in the bacteriocyte.
The aphid–Buchnera mutualism is principally based on the provision of essential amino acids from the symbiont to the host (2, 3). The high expression of the gene encoding CAT2, which is involved in the import of cationic amino acids such as lysine and arginine (essential amino acids) from the environment into the eukaryotic cells (30), is intriguing in this context. In the bacteriocyte, Buchnera cells are encased in a membrane of host origin (31, 32). Transporters of this type located on the host membrane will enable the transport of amino acids synthesized by Buchnera into the cytoplasm of the bacteriocyte.
We identified several genes for mitochondria-related transporters (ANT2, OT, AS, and GC) that were significantly up-regulated in the bacteriocyte. The abundance of their transcripts probably reflects high mitochondrial activity in the bacteriocyte, where active ATP synthesis and energy transfer are required for energy-consuming amino acid metabolisms. Certainly, electron microscopic studies have identified a dense population of mitochondria in aphid bacteriocytes (31, 32). The genome of Buchnera lacks most genes for tricarboxylic acid (TCA) cycle, whereas complete gene sets for glycolysis and respiratory chain are retained (11). Because TCA cycle operates in mitochondria, although speculative, the up-regulated mitochondrial activity and transport in the bacteriocyte might be relevant to cooperative metabolic interactions between Buchnera and the organelle.
One of the up-regulated genes encoded Ras-like Rab GTPase, which regulates vesicular transport of proteins and lipids between compartments in eukaryotic cells (33). Because Buchnera cells are encased in a host membrane, intracellular trafficking mechanism of this type may play important roles in the symbiotic system. The genome of Buchnera lacks key genes for phospholipid biosynthesis, implying that Buchnera is unable to synthesize its own cell membrane (11). Phospholipids of host origin might be delivered to Buchnera cells by using the vesicular transport system.
Most Abundant Transcripts in the Bacteriocyte Encoded Invertebrate-Type Lysozymes. The unigenes corresponding to the most abundant transcripts, R2C00037 (134 clones) and R2C00204 (71 clones), were similar to each other (97.7% nucleotide sequence similarity), representing 8.7% of total ESTs in ESTBC (Table 1). The top blast hit for these unigenes was f ly CG6426 (R2C00037: E = 1.4 × 10–37 and R2C00204: E = 4.1 × 10–37) whose function was unknown, but GO annotation “defense response to bacteria” (GO:0042742) was assigned to the gene at the “inferred from electronic annotation (IEA)” level of evidence. In addition, subordinate hits with significant similarity (E ≤ 1.0 × 10–10) included lysozyme (LSZ) i-1 from Anopheles gambiae (R2C00037: E = 2.3 × 10–35 and R2C00204: E = 1.1 × 10–34), destabilase 2 homolog from Drosophila melanogaster (R2C00037: E = 1.7 × 10–27 and R2C00204: E = 8.7 × 10–27), destabilase 2 from medicinal leech, Hirudo medicinalis (R2C00037: E = 5.1 × 10–11 and R2C00204: E = 7.9 × 10–12) and destabilase I from H. medicinalis (R2C00037: E = 8.7 × 10–11 and R2C00204: E = 1.3 × 10–11).
Lysozymes (EC 3.2.1.17) are the enzymes that destroy bacterial cell walls, and are structurally classified into several types; i.e., chicken, goose, invertebrate, plant, and bacteria types (34). Phylogenetic analysis indicated that lysozymes encoded by R2C00037 and R2C00204 belong to the invertebrate type (data not shown).
We performed quantitative RT-PCR by using the primer set for conserved regions between R2C00037 and R2C00204, whereby their transcripts were quantified collectively (Table 3). The expression level was 156 times higher in the bacteriocyte than in the whole body (Fig. 1_C_). It is also notable that the level of the transcripts was strikingly higher, 25.3-fold, than that of the control gene encoding RpL7. These results confirmed that the lysozyme-encoding genes represent the most highly expressed genes in the aphid bacteriocyte.
It is of great interest as to why the bacteriocyte, whose function is to harbor the symbiotic bacteria, highly expresses the antibacterial genes. This finding might be relevant to lysosomal break-down of Buchnera (31, 32) or elimination of microbial intruders (35). Future studies should focus on the biochemical properties, substrate specificity, and antimicrobial spectrum of the bacteriocyte-specific lysozymes.
Other Up-Regulated Genes. Based on comparative analysis between ESTBC and ESTWB, several other genes seemingly up-regulated in the bacteriocyte were selected and further examined by quantitative RT-PCR (Fig. 1_C_; see also Fig. 2, which is published as supporting information on the PNAS web site). Whereas the gene for a heat shock protein cognate 4 (HSC70; 26 clones) was among the most highly expressed unigenes in the defense response category (see Table 1), quantitative RT-PCR revealed no significant up-regulation of the gene in the bacteriocyte (P > 0.05) (Fig. 1_C_).
Unigenes Encoding Transcription Factors. Unigenes BCA016027 (a singlet), R2C00051 (two clones), R2C00028 (nine clones), and R2C00237 (two clones) encoded putative transcription factors such as Beadex, CG17870, CG1101, and CG5033, respectively (Table 4), none of which showed significant similarity to Distalless, Ultrabithorax/Abdominal-A, or Engrailed that were detected with antibodies in bacteriocytes of several species of aphids (36).
ESTs Similar to Only Prokaryotic Genes. In the bacteriocyte transcriptome, we identified two unigenes that showed significant similarity only to prokaryotic genes, but not to those of Buchnera. R2C00193 (10 clones) and R2C00214 (4 clones) matched to RlpA (rare lipoprotein A) precursor of Yersinia pestis (E = 7.3 × 10–11) and a hypothetical protein of Wolbachia pipientis (E = 7.1 × 10–22), respectively (Table 4). It is notable that ESTTC also contained a transcript similar to R2C00193 (CD450666: E = 1.0 × 10–44). Southern blot analysis confirmed that these transcripts have corresponding loci in the aphid genome (data not shown). Whereas genes with significant similarity to prokaryotic genes have been found in the genomes of a bruchid beetle and a silkworm (37, 38), high level of expression of these genes in the bacteriocyte is intriguing in the context of its obligatory interdependency with the symbiotic bacterium, Buchnera.
Conclusion
In this study, we demonstrated that a number of genes that are related to amino acid metabolism, intra- and intercellular transport, antibacterial activity and other biological processes, are highly expressed in the bacteriocyte. The up-regulation of host genes relevant to amino acid metabolism corroborated our previous finding that _Buchnera_-mediated production of essential amino acids from nonessential amino acids is among the most important processes in the symbiotic system, and, furthermore, profoundly enriched our understanding of the complementary metabolic features that underpin the integrity of the host–symbiont relationship. The up-regulated genes related to transport highlighted an important aspect of the bacteriocyte that mediates exchange of various metabolites and substrates at the host–symbiont interface. The bacteriocyte-specific lysozyme genes provided promising candidate molecules that might be involved in the control and maintenance of the bacterial flora in the host cell. The other up-regulated host genes, although their roles are currently obscure, would provide clues to understanding of previously unrecognized aspects of the host–symbiont interactions. Of course, these hypothetical processes that we suggest for the bacteriocyte must be verified by functional analysis of the up-regulated host genes.
Supplementary Material
Supporting Information
Acknowledgments
We thank Dr. Satoru Kobayashi for supporting computer analysis. This work was supported by the Special Postdoctoral Researchers Program of RIKEN, Japan Society for the Promotion of Science Grant-in-Aid for Young Scientists Grant (B) 14760031, and a Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists.
Author contributions: A.N. designed research; A.N., N.S., and T.S. performed research; Y.H. and P.C. contributed new reagents/analytic tools; A.N. and S.S. analyzed data; and A.N., S.S., H.I., T.K., and T.F. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: GO, Gene Ontology; GS2, glutamine synthetase 2; CAT2, cationic amino acid transporter 2; GCVT, glycine cleavage system T protein; GCDH, glutaryl-CoA dehydrogenase; PSAT, phosphoserine aminotransferase; ANT2, ADP/ATP translocase; PC, inorganic phosphate cotransporter; OT, mitochondrial oxaloacetate transport protein; AS, ATP synthase subunit c; GC, mitochondrial glutamate carrier; Rp n, ribosomal protein n; LSZ, lysozyme; NCBI, National Center for Biotechnology Information.
Data deposition: The sequences reported in this paper have been deposited in DNA Data Bank of Japan database (accession nos. BP535536–BP537955).
References
- 1.Munson, M. A., Baumann, P. & Kinsey, M. G. (1991) Int. J. Syst. Bacteriol. 41**,** 566–568. [Google Scholar]
- 2.Baumann, P., Baumann, L., Lai, C. Y., Rouhbakhsh, D., Moran, N. A. & Clark, M. A. (1995) Annu. Rev. Microbiol. 49**,** 55–94. [DOI] [PubMed] [Google Scholar]
- 3.Douglas, A. E. (1998) Annu. Rev. Entomol. 43**,** 17–37. [DOI] [PubMed] [Google Scholar]
- 4.Sasaki, T. & Ishikawa, H. (1995) J. Insect Physiol. 41**,** 41–46. [Google Scholar]
- 5.Febvay, G., Liadouze, I., Guillaud, J. & Bonnot, G. (1995) Arch. Insect Biochem. 29**,** 45–69. [Google Scholar]
- 6.Nakabachi, A. & Ishikawa, H. (1997) Insect Biochem. Mol. Biol. 27**,** 1057–1062. [DOI] [PubMed] [Google Scholar]
- 7.Nakabachi, A. & Ishikawa, H. (1999) J. Insect Physiol. 45**,** 1–6. [DOI] [PubMed] [Google Scholar]
- 8.Ziegler, H. (1975) in Transport in Plants I, eds. Zimmermann, M. H. & Milburn, J. A. (Springer, New York), Vol. 1, pp. 59–100. [Google Scholar]
- 9.Sandstrom, J. & Moran, N. (1999) Entomol. Exp. Appl. 91**,** 203–210. [Google Scholar]
- 10.Moran, N. A., Munson, M. A., Baumann, P. & Ishikawa, H. (1993) Proc. R. Soc. London Ser. B 253**,** 167–171. [Google Scholar]
- 11.Shigenobu, S., Watanabe, H., Hattori, M., Sakaki, Y. & Ishikawa, H. (2000) Nature 407**,** 81–86. [DOI] [PubMed] [Google Scholar]
- 12.Tamas, I., Klasson, L., Canback, B., Naslund, A. K., Eriksson, A. S., Wernegreen, J. J., Sandstrom, J. P., Moran, N. A. & Andersson, S. G. (2002) Science 296**,** 2376–2379. [DOI] [PubMed] [Google Scholar]
- 13.van Ham, R. C., Kamerbeek, J., Palacios, C., Rausell, C., Abascal, F., Bastolla, U., Fernandez, J. M., Jimenez, L., Postigo, M., Silva, F. J., et al. (2003) Proc. Natl. Acad. Sci. USA 100**,** 581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stephens, R. S., Kalman, S., Lammel, C., Fan, J., Marathe, R., Aravind, L., Mitchell, W., Olinger, L., Tatusov, R. L., Zhao, Q., et al. (1998) Science 282**,** 754–759. [DOI] [PubMed] [Google Scholar]
- 15.Andersson, S. G., Zomorodipour, A., Andersson, J. O., Sicheritz-Ponten, T., Alsmark, U. C., Podowski, R. M., Naslund, A. K., Eriksson, A. S., Winkler, H. H. & Kurland, C. G. (1998) Nature 396**,** 133–140. [DOI] [PubMed] [Google Scholar]
- 16.Akman, L., Yamashita, A., Watanabe, H., Oshima, K., Shiba, T., Hattori, M. & Aksoy, S. (2002) Nat. Genet. 32**,** 402–407. [DOI] [PubMed] [Google Scholar]
- 17.Gil, R., Silva, F. J., Zientz, E., Delmotte, F., Gonzalez-Candelas, F., Latorre, A., Rausell, C., Kamerbeek, J., Gadau, J., Holldobler, B., et al. (2003) Proc. Natl. Acad. Sci. USA 100**,** 9388–9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oshima, K., Kakizawa, S., Nishigawa, H., Jung, H. Y., Wei, W., Suzuki, S., Arashida, R., Nakata, D., Miyata, S., Ugaki, M. & Namba, S. (2004) Nat. Genet. 36**,** 27–29. [DOI] [PubMed] [Google Scholar]
- 19.Wu, M., Sun, L. V., Vamathevan, J., Riegler, M., Deboy, R., Brownlie, J. C., McGraw, E. A., Martin, W., Esser, C., Ahmadinejad, N., et al. (2004) PLoS Biol. 2**,** E69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishikawa, H. (1982) Comp. Biochem. Physiol. B Biochem. Mol. Biol. 72**,** 239–247. [Google Scholar]
- 21.Carninci, P., Kvam, C., Kitamura, A., Ohsumi, T., Okazaki, Y., Itoh, M., Kamiya, M., Shibata, K., Sasaki, N., Izawa, M., et al. (1996) Genomics 37**,** 327–336. [DOI] [PubMed] [Google Scholar]
- 22.Carninci, P. & Hayashizaki, Y. (1999) Methods Enzymol. 303**,** 19–44. [DOI] [PubMed] [Google Scholar]
- 23.Shibata, Y., Carninci, P., Watahiki, A., Shiraki, T., Konno, H., Muramatsu, M. & Hayashizaki, Y. (2001) BioTechniques 30**,** 1250–1254. [DOI] [PubMed] [Google Scholar]
- 24.Carninci, P., Shibata, Y., Hayatsu, N., Itoh, M., Shiraki, T., Hirozane, T., Watahiki, A., Shibata, K., Konno, H., Muramatsu, M. & Hayashizaki, Y. (2001) Genomics 77**,** 79–90. [DOI] [PubMed] [Google Scholar]
- 25.Ewing, B., Hillier, L., Wendl, M. C. & Green, P. (1998) Genome Res. 8**,** 175–185. [DOI] [PubMed] [Google Scholar]
- 26.Ewing, B. & Green, P. (1998) Genome Res. 8**,** 186–194. [PubMed] [Google Scholar]
- 27.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25**,** 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, X. & Madan, A. (1999) Genome Res. 9**,** 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunter, W. B., Dang, P. M., Bausher, M. G., Chaparro, J. X., McKendree, W., Shatters, R. G., Jr., McKenzie, C. L. & Sinisterra, X. H. (2003) J. Insect Sci. 3**,** 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoshide, R., Ikeda, Y., Karashima, S., Matsuura, T., Komaki, S., Kishino, T., Niikawa, N., Endo, F. & Matsuda, I. (1996) Genomics 38**,** 174–178. [DOI] [PubMed] [Google Scholar]
- 31.Hinde, R. (1971) J. Insect Physiol. 17**,** 1791–1800. [DOI] [PubMed] [Google Scholar]
- 32.Griffiths, G. W. & Beck, S. D. (1973) J. Insect Physiol. 19**,** 75–84. [Google Scholar]
- 33.Zerial, M. & McBride, H. (2001) Nat. Rev. Mol. Cell Biol. 2**,** 107–117. [DOI] [PubMed] [Google Scholar]
- 34.Bachali, S., Jager, M., Hassanin, A., Schoentgen, F., Jolles, P., Fiala-Medioni, A. & Deutsch, J. S. (2002) J. Mol. Evol. 54**,** 652–664. [DOI] [PubMed] [Google Scholar]
- 35.Nakabachi, A., Ishikawa, H. & Kudo, T. (2003) J. Invertebr. Pathol. 82**,** 152–161. [DOI] [PubMed] [Google Scholar]
- 36.Braendle, C., Miura, T., Bickel, R., Shingleton, A. W., Kambhampati, S. & Stern, D. L. (2003) PLoS Biol. 1**,** E21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kondo, N., Nikoh, N., Ijichi, N., Shimada, M. & Fukatsu, T. (2002) Proc. Natl. Acad. Sci. USA 99**,** 14280–14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daimon, T., Hamada, K., Mita, K., Okano, K., Suzuki, M. G., Kobayashi, M. & Shimada, T. (2003) Insect Biochem. Mol. Biol. 33**,** 749–759. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information