Molecular organization of the Ndc80 complex, an essential kinetochore component (original) (raw)
Abstract
The four-protein Ndc80 complex, an essential kinetochore component conserved from yeast to humans, plays an indispensable role in proper chromosome alignment and segregation during mitosis. In higher eukaryotes, the homologous complex probably resides in the middle domain of the trilaminar kinetochore, linking centromeric heterochromatin with microtubule-associated structures. We have prepared recombinant Ndc80 complex by pairwise coexpression of its components (Ndc80p and Nuf2p; Spc24p and Spc25p) and shown that they form independently stable subcomplexes. Rotary shadowing electron microscopy, combined with limited proteolysis and antibody labeling, demonstrates that the heterotetrameric Ndc80 complex is an ≈570-Å-long rod, with globular regions at either end. The shaft contains α-helical coiled-coil segments from each of the two subcomplexes, linked end-to-end. When integrated with published observations derived from inactivating the components of Ndc80, the molecular organization we deduce suggests that the Spc24p/Spc25p end of the rod faces the centromere and the Ndc80p/Nuf2p end faces a spindle microtubule.
Keywords: coiled coil, electron microscopy, mitosis
Kinetochores are multiprotein complexes that assemble on centromeric DNA and mediate attachment of chromosomes to microtubules. These attachments are stable during phases of microtubule polymerization and depolymerization, and they generate forces involved in chromosome movement (1). Kinetochores also regulate cell cycle progression at the metaphase-to-anaphase transition (2). In higher eukaryotes, cytological studies and electron microscopy (EM) of kinetochores reveal a trilaminar structure, with an electron-dense inner domain containing centromeric heterochromatin, a less dense middle domain, and a diffuse outer domain containing microtubule binding activities (3). Very little is known about the three-dimensional structures of the proteins within these kinetochore layers.
Kinetochores of the budding yeast, Saccharomyces cerevisiae, are simpler than those of most other organisms. Although regional centromeres span megabases of DNA in human cells, and 100 kb or more in fission yeast, short 125-bp point centromeres suffice for accurate chromosome segregation in S. cerevisiae (4–6). Kinetochores in higher eukaryotes bind 30 or more microtubules, those of budding yeast, a single microtubule (7–9). S. cerevisiae kinetochores are, nonetheless, large protein assemblies containing 60 or more different subunits organized into at least 14 multiprotein complexes (10). Moreover, sequence analysis of kinetochore proteins from various species reveals that many S. cerevisiae kinetochore proteins have been conserved from yeast to humans (P. Meraldi, A. D. McAinsh, E. Rheinbay, and P.K.S., unpublished results).
The four-protein “Ndc80 complex” contains Ndc80p, Nuf2p, Spc24p, and Spc25p. Close homologs of these proteins are found in all eukaryotes (11–15), and human Hec1 can substitute functionally for yeast Ndc80p (13). Loss-of-function mutations in any of the four essential Ndc80 complex subunits cause yeast chromosomes to detach from spindle microtubules and inactivate the spindle checkpoint, resulting in extremely high rates of chromosome loss (11, 16). Inactivation of Ndc80 complex proteins in Schizosaccharomyces pombe, Caenorhabditis elegans, Xenopus laveis, and humans gives rise to similar chromosome missegregation and checkpoint phenotypes as in budding yeast, suggesting substantial conservation of function (12, 16–20). However, the Ndc80p complex is not thought to contact microtubules or DNA directly, nor to have a specific role in the checkpoint. Rather, the complex seems to link microtubule-binding and Mad-Bub checkpoint proteins to proteins that contact centromeric (CEN) DNA, in particular CBF3, on which Ndc80 depends for CEN association (21, 22).
We describe in this article the reconstitution of the S. cerevisiae Ndc80 complex from recombinant proteins and an analysis of its structure using biochemical methods and EM. We find the Ndc80 complex to be a long rod with globular “heads” at either end of an α-helical coiled-coil shaft. Ndc80p and Nuf2p contribute to one head and to the intervening shaft; Spc24p and Spc25p complete the shaft and contribute to the other head. This molecular organization helps to explain a number of the functional properties of the Ndc80 complex in yeast and humans.
Materials and Methods
Plasmids and Cloning. Baculoviral expression vectors encoding Ndc80, Nuf2, Spc24, and Spc25 were provided by C. Espelin (Massachusetts Institute of Technology, Cambridge, MA). The full-length, His-6-tagged, and truncated versions of Ndc80, Nuf2, Spc24, and Spc25 were cloned individually between the _Nde_I and _Bam_HI sites of pET3aTr (23). Spc24 and Spc25 were subcloned into pST39 (23) to generate a single dicistron for coexpression of the two full-length proteins. Ndc80p (381–691) and Nuf2p (222–451) were subcloned into pST39 to generate a single dicistron for coexpression of the Ndc80/Nuf2 coiled coil. Full-length Ndc80 and Nuf2 were cloned into pETDuet-1 (Novagen) to generate a coexpression vector.
Protein Expression and Purification. For insect cell expression, Hi5 cells were coinfected with four baculoviruses, each encoding one of the four Ndc80 complex components, and harvested 48 h after infection. For Escherichia coli expression, BL21-AI cells (Invitrogen Life Technologies) transformed with the expression plasmid were grown at 37°C, and induced for 4 h with 0.2% arabinose at OD600 of 0.6. All of the protein complexes and subcomplexes contained a subunit with a His-6 tag. For purification of the complexes, cells were lysed in 20 mM 1,3-bis[tris(hydroxymethyl)methylamino]propane (Bistris propane) (pH 7.6), 250 mM NaCl, 4 mM 2-mercaptoethanol, 1 mM PMSF. The lysate was passed through Ni-nitrilotriacetic acid (NTA) agarose (Qiagen, Valencia, CA), and the beads were washed in the same buffer with 10 mM imidazole (and no PMSF). The complexes were eluted with a 20- to 250-mM imidazole gradient. Fractions containing the desired protein complex were pooled, concentrated, and further purified by size exclusion chromatography on Superdex S200 10/30. (Amersham Pharmacia Biosciences).
EM. Protein samples were rotary shadowed at 5 μg/ml in 112 mM ammonium bicarbonate, 45% glycerol (pH 7.5), following published procedures (24). For antibody decoration experiments, complexes bearing a His-6 tag were mixed first with anti-His-6 monoclonal antibody (Novagen). Samples were imaged at ×50,000 magnification on a JOEL-1200EX microscope operated at 60 kV. EM images were digitized on an Epson Perfection 3170 scanner. Lengths of Ndc80 molecules were measured by using the program imagej (NIH).
Limited Proteolysis. Proteolysis with trypsin/chymotrypsin was carried out on ice at protein concentrations between 1–3 mg/ml, and a substrate:protease ratio of 100:1 (wt/wt). Digestion was terminated by adding PMSF to 0.1 mM. For Edman N-terminal sequencing, cleaved protein samples were mixed with 2× SDS/PAGE loading buffer, boiled for 2 min, and loaded onto a polyacrylamide gel (Bio-Rad). After electrophoresis, protein fragments were transferred to a 0.2 μm PVDFSQ membrane (Millipore), before being submitted for Edman sequencing at the Biopolymers Core Facility of Tufts University. For liquid chromatography (LC)-MS, the cleaved protein fragments were injected into a Superdex S200 10/30 size exclusion column, and eluted peaks were collected and lyophilized before being subjected to LC-MS at the University of California, Berkeley.
Results
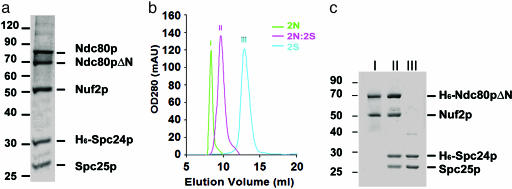
Recombinant Ndc80 Complex. We coexpressed in insect cells, using baculoviral expression vectors, the four proteins of the S. cerevisiae Ndc80 complex: Ndc80p (691 residues), Nuf2p (451 residues), Spc24p (213 residues), and Spc25p (221 residues). Purification of recombinant complexes was facilitated by fusing a His-6 tag to one of the subunits. When recombinant Ndc80 complex, purified by metal-chelate and size exclusion chromatography, was analyzed by SDS/PAGE, five bands were observed (Fig. 1_a_). Four corresponded to the molecular weights expected of full-length Ndc80p, Nuf2p, Spc24p, or Spc25p. The fifth band, with an apparent molecular mass of 70 kDa, contained an N-terminally truncated version of Ndc80p; Edman sequencing revealed an N terminus at residue 106. Comparison of Ndc80p sequences from 29 organisms reveals that the first 100 residues of the S. cerevisiae protein are poorly conserved among fungi and largely absent from higher eukaryotes (P. Meraldi, A. D. McAinsh, E. Rheinbay, and P.K.S., unpublished results, and data not shown). Because the human homolog of Ndc80 can substitute for yeast Ndc80, it is likely that the first 106 residues are not essential for function. We therefore chose to work with a recombinant Ndc80p variant comprising only residues 106–691 (Ndc80pΔN); residue numbers quoted below still follow the native yeast sequence.
Fig. 1.
Analysis of recombinant Ndc80 complex. (a) Purified recombinant Ndc80 complex separated by 10% SDS/PAGE, showing four components of the Ndc80 complex with His-6-tagged Spc24, and a band corresponding to proteolytically truncated Ndc80 (labeled Ndc80pΔN). His-6-Spc24p migrates more slowly than Spc25p, despite its lower molecular weight, consistent with the properties of native protein extracted from yeast (11). (b) Gel filtration chromatograms. Trace I, Ndc80p/Nufp (2N) subcomplex; trace III, Spc24p/Spc25p (2S) subcomplexes; trace II, tetramer reconstituted from the two subcomplexes (2N:2S). (c) SDS/PAGE of samples from each gel filtration peak in b. The designation of the peak appears above the corresponding lane; identities of the bands are on the right.
The four-protein Ndc80 complex can be reconstituted from two subcomplexes: Ndc80pΔN/Nuf2p (the 2N subcomplex) and Spc24p/Spc25p (the 2S subcomplex) (Fig. 1_b_). Each subcomplex can be formed by coexpressing the two respective subunits in either insect cells or E. coli. We purified each subcomplex, with one of two components fused to a His-6 tag, by metal-chelating, followed by gel filtration chromatography. The 2N complex on its own forms aggregates (Fig. 1_b_, trace I), but, when combined with 2S (Fig. 1_b_, trace III), all four proteins migrate as a single peak (Fig. 1_b_, trace II). Unlike the 2N subcomplex, the four-protein assembly is stable. On Coomassie blue-stained gels, bands corresponding to the four components of the 2N plus 2S recombinant complex had equal intensity, suggesting equimolar proportions and consistent with results obtained with complexes purified from yeast (11). Recombinant Ndc80 complex has an estimated molecular mass of 175 kDa from analytical ultracentrifugation (data not shown), in good agreement with the mass determined for Ndc80p complex purified from yeast (22). We conclude that recombinant Ndc80 complex has the same subunit composition as the native complex, and that it is a heterotetramer with a stoichiometry of 1:1:1:1.
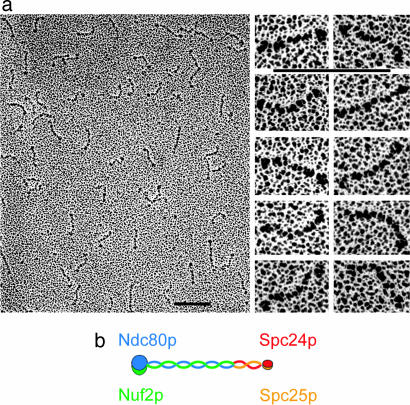
Rod-Like Structure of the Ndc80 Complex. All four proteins in the Ndc80 complex contain heptad-repeat regions, of the kind found in parallel, two-chain, α-helical coiled-coils [paircoil (25)]. Rotary shadowing EM reveals a rod-like assembly (Fig. 2_a_). Although the complex seems to have adopted a variety of conformations when deposited onto the mica surface, the rods are generally quite uniform, with no apparent interruption of a homogeneous structure. The ends present small, globular elements, and often one end has a larger head than the other. Using tropomyosin of known length as a reference (data not shown), we measured the Ndc80 complex to be 620 ± 40 Å long (n = 103), including the globular heads, without correcting for the platinum-carbon particle size. A corrected estimate, allowing ≈25 Å for platinum-carbon accumulation at each end, is 570 Å. The equivalent Stokes radius of such a rod is ≈80 Å, consistent with the hydrodynamic properties of the Ndc80 complex extracted from yeast (22), and with the elution volume of the recombinant complex (Fig. 1_b_).
Fig. 2.
Rotary shadowing EM of Ndc80. (a Left) Overview of a field. (Right) Selected images of the rod-like complexes. (Scale bars, 100 nm.) (b) Diagram showing overall molecular organization of the Ndc80 complex as deduced from the micrographs and from the lengths and positions of predicted coiled-coil segments in the various polypeptide chains. Blue, Ndc80p; green, Nuf2p; red, Spc24p; orange, Spc25p, orange.
The lengths of the predicted coiled-coil regions in Ndc80p and Nuf2p are both ≈300–400 Å. Those of Spc24p and Spc25p are ≈150 Å. Thus, the experimentally measured length of the complex visualized in the micrograph correlates well with the calculated combined lengths of 2N and 2S. Heptad repeat sequences in Ndc80p and Nuf2p are at the C termini, whereas those of Spc24p and Spc25p are at the N termini. We thus propose that the two subcomplexes join together in a “C-to-N” fashion, as illustrated in Fig. 2_b_.
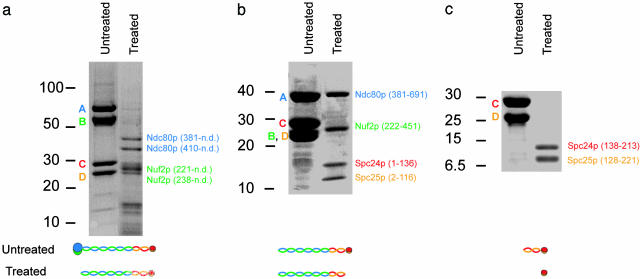
Coiled-Coil Core of the Ndc80 Complex. To determine experimentally the locations of coiled-coil domains in each protein molecule, we used limited proteolysis by a combination of trypsin and chymotrypsin to map proteolytically resistant segments of the complex. We treated the intact recombinant complex with proteases at 100:1 (wt/wt) ratio for 5 h on ice, separated the proteolytic fragments by SDS/PAGE (Fig. 3_a_), and observed five major bands. Four of the bands were identified by Edman sequencing. The two largest contain Ndc80p fragments cleaved at K380-M381 (≈40 kDa) and at K409-G410 (≈35 kDa), respectively; the two smallest, Nuf2 fragments cleaved at K220-Q221 (≈27 kDa) and L237-E238 (≈25 kDa). We deduce from the mobility of these four fragments that each extends from the cleavage site to a position at or near the C terminus of the corresponding protein. The protein bands at ≈15 kDa yielded ambiguous results with Edman sequencing and remained unidentified, but are probably fragments of Spc24p/Spc25p (see below). The cleavage sites at residues K380 of Ndc80p, K220 of Nuf2p are close to the N termini of the predicted heptad-repeat regions in these proteins and seem to mark the start of the coiled-coil domains. The cleavage at K409 of Ndc80p and L237 of Nuf2p may reflect a local instability in the colied coil.
Fig. 3.
Limited proteolysis of the Ndc80 complex. Each panel shows paired lanes from SDS/PAGE of untreated and protease-treated samples. Diagrams showing the presumed molecular organization of the fragments before dissociation in SDS are beneath the corresponding pair of lanes. (a) Proteolysis of intact Ndc80 complex. Bands designated A, B, C, and D are Ndc80p (blue), Nuf2p (green), Spc24p (red), and Spc25p (orange), respectively. Peptides identified by Edman sequencing are labeled on the right. The C termini of the fragments were not determined (n.d.), but in all cases probably corresponded to the C terminus of the parent molecule. (b) Proteolysis of 2NCC:2S. Bands in the untreated lane are Ndc80p (381–691) (A, blue), Nuf2p (222–451) (B, green), Spc24p (C, red), and Spc25p (D, orange), respectively. The protease-treated sample was purified by gel filtration before SDS/PAGE analysis (see Materials and Methods). The bands identified by LC-MS are labeled. (c) Proteolysis of the Spc24p/Spc25p (2S) subcomplex. Protein fragments in the right-hand lane were identified as in b.
To determine whether the C-terminal coiled-coil segments of Ndc80p and Nuf2p dimerize on their own and whether that dimer can form a complex with full-length 2S, we expressed Ndc80p (381–691) and Nuf2p (222–451) in E. coli. The fragments copurified as a heterodimer (2NCC), which formed a heterotetramer with the 2S subcomplex (data not shown). Reassembled 2NCC:2S heterotetramer was treated with 1% (wt/wt) protease for 1 h on ice, and resulting proteolytic fragments were separated by gel filtration chromatography. Two major protein peaks eluted from the column. The first contained four peptides, which were analyzed by LC-MS and Edman sequencing. They were Ndc80p (381–691), Nuf2p (222–451), Spc24p (1–136), and Spc25p (2–116), fragments that match very well with each protein's predicted coiled-coil segment. The four protease-resistant fragments apparently form a heterotetramer in the absence of the head domains. The second proteolytic peak contained two fragments of ≈10 and 13 kDa (estimated from SDS/PAGE mobilities), which were not analyzed by MS. They presumably correspond to the head of 2S (see below and Fig. 3_c_)
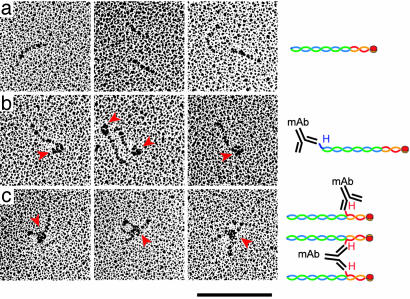
An End-to-End Connection Between Two Coiled Coils. Results of limited proteolysis are consistent with our inference from EM that coiled-coils from the two subcomplexes make up the shaft of the elongated complex and join N-C (Fig. 2_b_). To test this picture further, we bound anti-His-6 monoclonal antibodies to recombinant 2NCC:2S complexes containing N-terminally tagged His-6-Ndc80p (381–691) or His-6-Spc24p and visualized the antibody–antigen complexes in the EM after contrasting by rotary shadowing (Fig. 4). Antibody labeling of the Ndc80p coiled-coil N terminus added a large triangular element, characteristic of an IgG, to one end of the complex. In contrast, antibody labeling of the Spc24p N terminus added a triangular element to the middle of the complex and occasionally generated X-shaped structures. The latter arise when the centers of two rods are linked together by a single antibody. These images confirm our placement of the the Ndc80p N terminus at one end of the rod and the Spc24p N terminus in the middle.
Fig. 4.
Rotary shadowing EM of antibody labeled 2NCC:2S complex. (a) Untagged 2NCC:2S with no added antibody. His-6-tagged complexes are indistinguishable from untagged complexes (data not shown). (b) 2NCC:2S containing N-terminally tagged His-6-Ndc80p (381–691) (H2NCC:2S) was labeled with anti-His-6 mAb. (c) 2NCC:2S containing Spc24p with an N-terminal His-6-tag was labeled with anti-His-6 mAb. Red arrowheads point to the triangular outlines of IgG molecules. Diagrams to the Right illustrate our interpretation of the image. H, the position of the His-6-tag in the same color as the tagged protein. mAb, anti-His-6 monoclonal antibody, not to scale. Blue, Ndc80p (381–691); green, Nuf2p (222–451); red, Spc24p; orange, Spc25p. (Scale bar, 100 nm.)
The protease-resistant fragments of 2N contain 311 residues from Ndc80p and 230 from Nuf2p. Assuming that the heptad repeats are roughly aligned at their N termini and that the ≈80-residue C-terminal extension of Ndc80p is protected by association with 2S, we estimate a coiled-coil length of 345 Å. A similar calculation for 2S yields a coiled-coil length of 170 Å. The measured length of the complex, corrected for platinum-carbon accumulations, is 570 Å. Taking 30 Å as the diameter for the globular regions at each end, we estimate 510 Å for the central rod, in good agreement with the total coiled-coil length of 515 Å. We conclude that the two coiled coils abut directly, with minimal overlap.
We obtained some information about the end-to-end joint by subjecting the 2S subcomplex to limited digestion with trypsin and chymotrypsin. When we separated the products by gel filtration chromatography, we detected one major peak, which we analyzed by LC-MS. Two protein fragments from Spc24p and Spc25p survived the proteolysis and gel filtration (Fig. 3_c_): Spc24p 138–213, and Spc25p 128–221. These correspond to the C-terminal globular domains. Thus, the coiled coil of Spc24p and Spc25p is resistant to digestion as part of a 2NCC:2S complex, whereas it is protease sensitive in 2S alone. Evidently, the Spc24p/Spc25p coiled coil is protected by its association with Ndc80p and Nuf2p. One likely source of this protection is the 80-residue C-terminal “overhang” from Ndc80, which could reinforce the 2S coiled-coil, for example, by creating a three-strand rather than two-strand structure.
Discussion
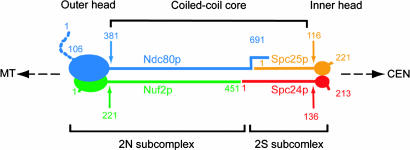
The data reported here demonstrate that the Ndc80 complex comprises two discrete subcomplexes: Ndc80pΔN/Nuf2p (2N) and Spc24p/Scp25p (2S), each containing a coiled-coil (Fig. 5). The two subcomplexes are joined end-to-end through their coiled-coils to form a heterotetrameric complex ≈570 Å long, with a 510-Å central shaft capped at either end by globular head domains. The joint connecting the coiled-coils at the middle of the Ndc80 complex is unusual. The coiled-coils seem to abut directly. Electron micrographs show no evidence for a globular element at the connection, nor does the rod seem to bend or distort preferentially at a specific position along its length. The C terminus of Ndc80p probably extends ≈80 residues beyond the C terminus of Nuf2p, and the coiled-coil region of Spc24p, ≈20 residues beyond the end of Spc25p. These termini could provide overhanging sequences involved in formation of the 2N:2S complex, and the two coiled-coils would then have a stiff, extended overlap, rather than a hinge-like connection.
Fig. 5.
Schematic model of the Ndc80 complex. MT, microtubule; CEN, centromere. Each subunit of the Ndc80 complex is represented by an oval (the globular domain) and a stick (the coiled-coil region). The dashed line indicates the N-terminal segment of Ndc80p that was deleted in all of our constructs. Numbers label the approximate first and last residues of each protein segment. The coiled-coils of 2N and 2S form the coiled-coil core. The globular domains of 2N form the outer head (toward the microtubule); the globular domains of the 2S, the inner head (toward the centromere).
Studies in yeast using temperature-sensitive mutants and in human cells using RNA interference (RNAi) show that inactivation of either Ndc80 or Nuf2 causes the other protein to be lost from kinetochores and apparently degraded (20, 26, 27). Spc24 and Spc25 remain attached to kinetochores in the absence of Nuf2 and Ndc80 (20). Our model explains these findings if we postulate that the 2S head is oriented toward the centromere and the 2N head toward the microtubule and that the 2S complex is stable in vivo in the absence of 2N, as it is in vitro.
The structure of the Ndc80 complex is consistent with its postulated role as a linker between microtubule-binding proteins and DNA-binding proteins (22). The stiff linkage between 2N and 2S subcomplexes will allow it to maintain tension between spindle and chromosome. At the spindle end, Ndc80 seems to interact with the DASH/Dam1 complex (28, 29), which in turn may assemble into a ring encircling a microtubule (30, 31). Ndc80 rods could extend from such a ring and link it to chromatin-proximal elements. Sequence and functional comparisons indicate that key structural features of the Ndc80 complex have been conserved through evolution. Thus, in higher eukaryotes, the homologous complex should also link microtubule-bound and chromatin-bound structures. Images from immuno-gold staining with a monoclonal antibody to Hec1, the human homologue of Ndc80, place its epitope within the outer plate of the trilaminar kinetochore (32). The full complex is of suitable length to span the middle domain and to project the 2S end toward the inner plate. One or more of the other coiled-coil-containing, kinetochore-associated proteins could provide additional struts.
Acknowledgments
We thank David S. King (Howard Hughes Medical Institute, University of California, Berkeley) for LC-MS analysis of the protein fragments, and Chris Espelin, Viji Draviam, Kelly Arnette, Kim Simons, J. Miranda, and Nick Larsen for advice and discussions. This work was supported by National Institutes of Health Grant GM51464 (to P.K.S.). S.C.H. is an investigator of the Howard Hughes Medical Institute.
Abbreviations: 2N, Ndc80pΔN/Nuf2p; 2S, Spc24p/Spc25p; LC, liquid chromatography.
References
- 1.Koshland, D. E., Mitchison, T. J. & Kirschner, M. W. (1988) Nature 331**,** 499–504. [DOI] [PubMed] [Google Scholar]
- 2.Cleveland, D. W., Mao, Y. & Sullivan, K. F. (2003) Cell 112**,** 407–421. [DOI] [PubMed] [Google Scholar]
- 3.Roos, U. P. (1973) Chromosoma 41**,** 195–220. [DOI] [PubMed] [Google Scholar]
- 4.Chikashige, Y., Kinoshita, N., Nakaseko, Y., Matsumoto, T., Murakami, S., Niwa, O. & Yanagida, M. (1989) Cell 57**,** 739–751. [DOI] [PubMed] [Google Scholar]
- 5.Clarke, L. & Carbon, J. (1980) Proc. Natl. Acad. Sci. USA 77**,** 2173–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cottarel, G., Shero, J. H., Hieter, P. & Hegemann, J. H. (1989) Mol. Cell. Biol. 9**,** 3342–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peterson, J. B. & Ris, H. (1976) J. Cell Sci. 22**,** 219–242. [DOI] [PubMed] [Google Scholar]
- 8.McDonald, K. L., O'Toole, E. T., Mastronarde, D. N. & McIntosh, J. R. (1992) J. Cell Biol. 118**,** 369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winey, M., Mamay, C. L., O'Toole, E. T., Mastronarde, D. N., Giddings, T. H., Jr., McDonald, K. L. & McIntosh, J. R. (1995) J. Cell Biol. 129**,** 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McAinsh, A. D., Tytell, J. D. & Sorger, P. K. (2003) Annu. Rev. Cell Dev. Biol. 19**,** 519–539. [DOI] [PubMed] [Google Scholar]
- 11.Wigge, P. A. & Kilmartin, J. V. (2001) J. Cell Biol. 152**,** 349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howe, M., McDonald, K. L., Albertson, D. G. & Meyer, B. J. (2001) J. Cell Biol. 153**,** 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng, L., Chen, Y. & Lee, W. H. (1999) Mol. Cell. Biol. 19**,** 5417–5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bharadwaj, R., Qi, W. & Yu, H. (2004) J. Biol. Chem. 279**,** 13076–13085. [DOI] [PubMed] [Google Scholar]
- 15.McCleland, M. L., Kallio, M. J., Barrett-Wilt, G. A., Kestner, C. A., Shabanowitz, J., Hunt, D. F., Gorbsky, G. J. & Stukenberg, P. T. (2004) Curr. Biol. 14**,** 131–137. [DOI] [PubMed] [Google Scholar]
- 16.Janke, C., Ortiz, J., Lechner, J., Shevchenko, A., Magiera, M. M., Schramm, C. & Schiebel, E. (2001) EMBO J. 20**,** 777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hori, T., Haraguchi, T., Hiraoka, Y., Kimura, H. & Fukagawa, T. (2003) J. Cell Sci. 116**,** 3347–3362. [DOI] [PubMed] [Google Scholar]
- 18.Martin-Lluesma, S., Stucke, V. M. & Nigg, E. A. (2002) Science 297**,** 2267–2270. [DOI] [PubMed] [Google Scholar]
- 19.McCleland, M. L., Gardner, R. D., Kallio, M. J., Daum, J. R., Gorbsky, G. J., Burke, D. J. & Stukenberg, P. T. (2003) Genes Dev. 17**,** 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillett, E. S., Espelin, C. W. & Sorger, P. K. (2004) J. Cell Biol. 164**,** 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, X., Rines, D. R., Espelin, C. W. & Sorger, P. K. (2001) Cell 106**,** 195–206. [DOI] [PubMed] [Google Scholar]
- 22.De Wulf, P., McAinsh, A. D. & Sorger, P. K. (2003) Genes Dev. 17**,** 2902–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan, S. (2001) Protein Expression Purif. 21**,** 224–234. [DOI] [PubMed] [Google Scholar]
- 24.Fowler, W. E. & Erickson, H. P. (1979) J. Mol. Biol. 134**,** 241–249. [DOI] [PubMed] [Google Scholar]
- 25.Berger, B., Wilson, D. B., Wolf, E., Tonchev, T., Milla, M. & Kim, P. S. (1995) Proc. Natl. Acad. Sci. USA 92**,** 8259–8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeLuca, J. G., Howell, B. J., Canman, J. C., Hickey, J. M., Fang, G. & Salmon, E. D. (2003) Curr. Biol. 13**,** 2103–2109. [DOI] [PubMed] [Google Scholar]
- 27.Meraldi, P., Draviam, V. M. & Sorger, P. K. (2004) Dev. Cell 7**,** 45–60. [DOI] [PubMed] [Google Scholar]
- 28.Cheeseman, I. M., Anderson, S., Jwa, M., Green, E. M., Kang, J., Yates, J. R., 3rd, Chan, C. S., Drubin, D. G. & Barnes, G. (2002) Cell 111**,** 163–172. [DOI] [PubMed] [Google Scholar]
- 29.Shang, C., Hazbun, T. R., Cheeseman, I. M., Aranda, J., Fields, S., Drubin, D. G. & Barnes, G. (2003) Mol. Biol. Cell 14**,** 3342–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miranda, J., De Wulf, P., Sorger, P. K. & Harrison, S. C. (2005) Nat. Struct. Mol. Biol. 12**,** 138–143. [DOI] [PubMed] [Google Scholar]
- 31.Westermann, S., Avila-Sakar, A., Wang, H. W., Niederstrasser, H., Wong, J., Drubin, D. G., Nogales, E. & Barnes, G. (2005) Mol. Cell 17**,** 277–290. [DOI] [PubMed] [Google Scholar]
- 32.Deluca, J. G., Dong, Y., Hergert, P., Strauss, J., Hickey, J. M., Salmon, E. D. & McEwen, B. F. (2005) Mol. Biol. Cell 16**,** 519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]