Chloropicophyceae, a new class of picophytoplanktonic prasinophytes (original) (raw)
Abstract
Prasinophytes are a paraphyletic group of nine lineages of green microalgae that are currently classified either at the class or order level or as clades without formal taxonomic description. Prasinophyte clade VII comprises picoplanktonic algae that are important components of marine phytoplankton communities, particularly in moderately oligotrophic waters. Despite first being cultured in the 1960s, this clade has yet to be formally described. Previous phylogenetic analyses using the 18S rRNA gene divided prasinophyte clade VII into three lineages, termed A, B and C, the latter formed by a single species, Picocystis salinarum, that to date has only been found in saline lakes. Strains from lineages A and B cannot be distinguished by light microscopy and have very similar photosynthetic pigment profiles corresponding to the prasino-2A pigment group. We obtained phenotypic and genetic data on a large set of prasinophyte clade VII culture strains that allowed us to clarify the taxonomy of this important marine group. We describe two novel classes, the Picocystophyceae and the Chloropicophyceae, the latter containing two novel genera, Chloropicon and Chloroparvula, and eight new species of marine picoplanktonic green algae.
Subject terms: Biodiversity, Marine biology
Introduction
Prasinophytes are a paraphyletic group of nine lineages of green microalgae that are currently classified either at the class or order level or family or as clades without formal taxonomic description1. The taxonomy of prasinophytes has proved particularly challenging in part due to the small size and simple morphology of many of its members2,3. A good example of this is prasinophyte clade VII4 that are coccoid cells ranging in size from 2 to 3 µm with few specific morphological features.
Phylogenetic analysis of the 18S rRNA gene divided prasinophyte clade VII into three lineages, termed A, B and C4, the latter formed only by Picocystis salinarum, a picoplanktonic species described from saline lakes5–7. Strains from lineages A and B cannot be distinguished by light microscopy and have very similar photosynthetic pigment profiles corresponding to the prasino-2A pigment group8. P. salinarum cells tend towards an easily observed tri-lobed shape under conditions of nutrient depletion and possess monadoxanthin and diatoxanthin as major carotenoids5. In contrast to results from phylogenetic analyses using the 18S rRNA gene, analyses using partial plastid 16S rRNA gene sequences9, complete nuclear and plastid encoded rRNA operons10,11 and chloroplast genomes12 suggest that P. salinarum forms a lineage that is separate from prasinophyte clade VII A and B. In the absence of morphological differentiation, molecular data obtained from culture strains and environmental samples have allowed the delimitation of at least 10 different phylogenetic clades, termed A1 to A7 and B1 to B39, within prasinophyte clade VII.
From an ecological point of view, prasinophyte clade VII appears to be a major group of picoplanktonic green algae in marine waters9,13–17. In moderately oligotrophic areas it is often the main Chlorophyta group, replacing Mamiellophyceae which tends to dominate in coastal waters9,14. Clades B1 and A4 typically dominate in oceanic waters and different sub-clades seem to occupy distinct niches, although the precise habitat of each clade is still unclear9.
Prasinophyte clade VII remains without formal taxonomic description despite the fact that members of this clade have been maintained in culture since 196518. In recent years the principle of combining morphological and molecular data to delineate species has increasingly been adopted in microalgal taxonomy. Intra- and inter-specific genetic variation in molecular markers are used to describe individuals and determine DNA-based species19. In addition to sequence divergence methods, analysis of the secondary structure of the Internal Transcribed Spacer 2 (ITS2) has been used for delimiting biological species. The ITS2 is part of the eukaryotic nuclear ribosomal operon, located between the 5.8S and 28S rRNA genes (ITS1 is located between the 18S and 5.8S rRNA genes). The primary sequence and length of ITS2 vary extensively among different taxa, however its secondary structure, when transcribed into RNA, retains features that are important for its biological function and thought to be universal among eukaryotes20–23. To generate new rRNA molecules, the entire operon is transcribed as a single rRNA precursor and the new 18S and 28S rRNA molecules are obtained after a complex excision process of both ITS regions primarily guided by their transcripts secondary structure24,25. The use of secondary structure of ITS2 in microalgal taxonomy increased after Coleman et al.21 and Muller et al.26 suggested a link between the presence of compensatory base changes (CBCs) and species boundaries. The ITS2 secondary structure includes four helices. A double-sided base change of a nucleotide pair in a given helix retaining the secondary structure is considered a CBC, while a single-side change is called hemi-CBC (hCBC).
In this paper, we analyze a wide set of phenotypic and genetic characters of members of prasinophyte clade VII, including ultrastructure, cell size, DNA content, pigment profiles, multigene phylogeny and ITS2 secondary structure. The data obtained lead us to describe two novel classes, the Picocystophyceae and the Chloropicophyceae, which contain two novel genera, Chloropicon and Chloroparvula, and eight species that are new to science.
Material and Methods
Cultured strains
The prasinophyte clade VII strains used in this study are listed in Table 1. These strains were selected from the Roscoff Culture Collection (RCC, http://www.roscoff-culture-collection.org) and Microbial Culture Collection at NIES (National Institute for Environmental Studies, http://mcc.nies.go.jp). Strains were grown at 22 °C in L1 seawater medium27 under an average light intensity of 100 µmoles photons.m−2.s−1 and a 12:12 h LD (Light:Dark) regime. NIES strains were grown in ESM seawater medium28.
Table 1.
Strains of prasinophytes clade VII used in this study.
Pigments Analysis
Approximately 50 ml of cultures listed in Table 1 (except NIES-3669 which required 200 ml) were collected in late exponential or early stationary phase by filtration onto glass fiber GF/F filters (Whatman, Maidstone, UK) without applying vacuum. Prior to sample collection, cell concentration was determined by flow cytometry using a Becton Dickinson Accuri C6. Total time for filtration did not exceed 10 minutes and total volume filtered was recorded. Filters were removed as soon as they became clogged, protected from light at all processing stages and immediately frozen in liquid nitrogen and stored at −80 °C. Frozen filters were extracted with 3 mL of 90% acetone in screw cap glass tubes with polytetra-fluoroethylene (PTFE) lined caps, placed in an ice-water bath. After 15 minutes, filters were homogenized using a clean stainless steel spatula for filter grinding. Tubes were placed in an ultrasonic bath with water and ice for 5 minutes. The slurries were then centrifuged for 5 minutes at 3940 g and supernatants filtered through 13 mm diameter polypropylene syringe filters (MS PTFE, 0.22 µm pore size) to remove cell and filter debris. Before injection, 0.4 ml of Milli-Q water was added to 1 ml of each sample extract to avoid peak distortion. Pigments extracted from clade VII strains were analyzed using the method of Zapata et al.29 as modified by Garrido et al.30 to improve the separation of loroxanthin and neoxanthin (Table 2).
Table 2.
Concentration of Chl a per cell and ratios (mol.mol−1) of pigment to Chl a concentration of 22 strains of prasinophytes clade VII grown under an average of 100 µmoles photons.m−2.s−1 New data are presented along with data from a previous study8.
| Strain | Species | sub - clade | Light | fg Chl a/cell | Chl b | Chlide a | Chlide b | Loro | Neo | Viola | Asta | Anth | Zea | Lut | ββ - car | βε - car | Allo | Diato | Monado |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RCC719* | C. mariensis | A1 | 140 | 14.89 | 0.683 | 0 | 0 | 0.035 | 0.086 | 0.617 | 0.264 | 0 | 0.478 | 0.193 | 0.149 | 0.107 | 0 | 0 | 0 |
| RCC998* | A1 | 140 | 26.07 | 0.783 | 0 | 0 | 0.014 | 0.117 | 0.877 | 0.157 | 0.058 | 0.168 | 0.285 | 0.131 | 0.095 | 0 | 0 | 0 | |
| RCC15 * | C. primus | A2 | 140 | 20.34 | 0.901 | 0.066 | 0.018 | 0.058 | 0.003 | 0.327 | 0.165 | 0.043 | 0.103 | 0.376 | 0.093 | 0.154 | 0 | 0 | 0 |
| RCC287* | C. sieburthii | A3 | 140 | 4.99 | 0.986 | 0.106 | 0 | 0.024 | 0.167 | 0.572 | 0.210 | 0.024 | 0.043 | 0.363 | 0.079 | 0.051 | 0 | 0 | 0 |
| RCC857* | A3 | 140 | 4.10 | 1.000 | 0 | 0 | 0.026 | 0.122 | 0.534 | 0.187 | 0.024 | 0.094 | 0.291 | 0.064 | 0.056 | 0 | 0 | 0 | |
| RCC1124* | C. roscoffensis | A4 | 140 | 8.61 | 0.960 | 0.099 | 0 | 0 | 0.116 | 0.525 | 0.148 | 0.020 | 0.086 | 0.399 | 0.082 | 0.095 | 0 | 0 | 0 |
| RCC1871* | A4 | 100 | 3.30 | 1.196 | 0 | 0 | 0 | 0.119 | 0.460 | 0.064 | 0.015 | 0.091 | 0.444 | 0.025 | 0.078 | 0 | 0 | 0 | |
| RCC917 | A4 | 100 | 18.49 | 1.081 | 0.025 | 0.006 | 0 | 0.132 | 0.603 | 0.110 | 0.006 | 0.046 | 0.388 | 0.046 | 0.078 | 0 | 0 | 0 | |
| RCC726 | A4 | 100 | 7.73 | 0.956 | 0.020 | 0 | 0 | 0.118 | 0.537 | 0.151 | 0.010 | 0.120 | 0.369 | 0.067 | 0.088 | 0 | 0 | 0 | |
| NIES-2755 | A4 | 100 | 10.01 | 1.125 | 0.011 | 0 | 0 | 0.133 | 0.608 | 0.110 | 0.005 | 0.036 | 0.359 | 0.044 | 0.075 | 0 | 0 | 0 | |
| RCC4429 | A4 | 100 | 10.62 | 1.044 | 0.027 | 0 | 0 | 0.118 | 0.697 | 0.115 | 0.010 | 0.038 | 0.373 | 0.052 | 0.094 | 0 | 0 | 0 | |
| NIES-3667 | A4 | 100 | 30.17 | 1.108 | 0.024 | 0.013 | 0 | 0.129 | 0.570 | 0.152 | 0.006 | 0.044 | 0.354 | 0.067 | 0.070 | 0 | 0 | 0 | |
| RCC856* | C. laureae | A5 | 140 | 23.37 | 0.860 | 0 | 0 | 0.113 | 0.079 | 0.291 | 0.570 | 0.042 | 0.310 | 0.461 | 0.136 | 0.075 | 0 | 0 | 0 |
| RCC3374* | C. maureeniae | A7 | 140 | 4.14 | 0.726 | 0.229 | 0 | 0.012 | 0.082 | 0.272 | 0.778 | 0.041 | 0.118 | 0.332 | 0.146 | 0.059 | 0 | 0 | 0 |
| RCC996* | A | 140 | 51.57 | 0.931 | 0.074 | 0 | 0.043 | 0.095 | 0.119 | 0.191 | 0.039 | 0.245 | 0.36 | 0.074 | 0.09 | 0 | 0 | 0 | |
| RCC3376 * | A | 140 | 4.08 | 0.813 | 0.105 | 0 | 0.008 | 0.112 | 0.572 | 0.324 | 0.054 | 0.129 | 0.048 | 0.194 | 0.085 | 0 | 0 | 0 | |
| NIES-3669 | C. pacifica | B1 | 100 | 6.34 | 0.907 | 0 | 0 | 0 | 0.115 | 0.252 | 0 | 0.153 | 0.298 | 0.766 | 0 | 0.107 | 0 | 0 | 0 |
| RCC696 | B2 | 100 | 26.30 | 1.030 | 0.061 | 0.042 | 0 | 0.125 | 0.569 | 0.095 | 0.009 | 0.050 | 0.437 | 0.081 | 0.058 | 0 | 0 | 0 | |
| NIES-2756* | B2 | 140 | 4.37 | 0.882 | 0.394 | 0.236 | 0.031 | 0.137 | 0.504 | 0.302 | 0.051 | 0.154 | 0.706 | 0.240 | 0.061 | 0 | 0 | 0 | |
| RCC4572 | B3 | 100 | 27.19 | 1.032 | 0.130 | 0.100 | 0 | 0.136 | 0.534 | 0.108 | 0.006 | 0.019 | 0.454 | 0.088 | 0.055 | 0 | 0 | 0 | |
| NIES-2758* | C. japonica | B | 140 | 14.58 | 0.624 | 0 | 0 | 0.026 | 0.078 | 0.613 | 0.045 | 0.024 | 0.074 | 0.302 | 0.330 | 0.028 | 0 | 0 | 0 |
| RCC3402* | P. salinarum | C | 100 | 60.40 | 0.283 | 0 | 0 | 0 | 0.039 | 0.035 | 0 | 0.003 | 0.018 | 0.071 | 0.129 | 0.015 | 0.047 | 0.107 | 0.119 |
Light microscopy
Two milliliters of cultures in exponential or early stationary phase were harvested by centrifugation (2000 g, 5 minutes) and observed with light microscopy under an Olympus BX 51 microscope equipped with differential interference contrast (DIC), phase contrast and blue fluorescence filters. Microphotographs were obtained with a SPOT RT-slider digital camera (Diagnostics Instruments, Sterling Heights, MI). For cell size, about 100 randomly chosen cells were measured using the Fiji open - source platform31.
Transmission electron microscopy
For thin sections, cells were fixed in 2% glutaraldehyde (final concentration) in growth medium for 1 h at room temperature and centrifuged (4000 rpm, 30 min) to form a pellet that was rinsed three times in growth medium (5 min each) and then three times in 0.1 M sodium cacodylate (5 min each). The cells were post-fixed in a mixture of 1% osmium tetroxide and 1% potassium ferricyanide in 0.1 M sodium cacodylate (final concentrations) for 2 hours at 4 °C and subsequently rinsed three times (10 min each) in 0.1 M cacodylate and twice in MilliQ water (5 min each). The cells were stained for 1 h in 1% aqueous uranyl acetate. Samples were dehydrated in an aqueous ethanol series (10 min in 30%, 50%, 70%, 90%, 96%, and four times in 100%, 5 min each) and rinsed twice with propylene oxide (5 min each). Samples were then left overnight in a 1:1 mixture of propylene oxide and Epon’s resin (EMBed-812 based on EPON-812). The next morning the cells were transferred to Epon and three changes (1 h each) were made before they were polymerized at 60 °C overnight. Ultrathin sections of embedded samples were made with a Leica Ultracut UCT microtome (Wetzlar, Germany), using a diamond knife. Sections were mounted on copper grids coated with Formvar film and some of the samples were stained with uranyl acetate (saturated solution in 50% ethanol) and lead citrate (saturated solution in 0.1 M NaOH). All chemicals were obtained from Sigma-Aldrich (St. Louise, USA). Sections were viewed with a Philips CM-100 TEM (Hillsboro, Oregon, USA) at the Electron Microscopy Unit of the Department of Molecular Biosciences, University of Oslo.
Scanning electron microscopy
Cells were fixed in 2% glutaraldehyde for 1 h and 5–10 mL of fixed cell suspensions gravity filtered onto Nuclepore filters (13 mm diameter, 2 µm pore size, volumes used depended on cell density and filter clogging). Filters were rinsed in growth medium (10 min) and subsequently in 0.1 M cacodylate (10 min). Cells were post-fixed in 1% osmium tetroxide in 0.1 M cacodylate (final concentrations). Three subsequent rinses in 0.1 M cacodylate were performed (5 min each) and the cells were dehydrated in an aqueous ethanol series (10 min in 70%, 90%, 96% and three changes in 100%, 10 min each). The filter-holders were transferred in 100% ethanol to a Critical Point Dryer (Baltec CPD 030, Balzers, Liechtenstein) and the dried filters were mounted on stubs on carbon tabs. An additional protocol was followed for some of the samples; a drop of culture was placed on a poly-L-lysin coated coverslip and fixed in the vapor of 2% osmium tetroxide and left to sink overnight in a moist chamber before they were rinsed, dehydrated and critical point dried as above. All chemicals were obtained from Sigma-Aldrich (St. Louise, USA). The coverslips were mounted on stubs, sputter coated with platinum and viewed in a Hitachi S-4800 (Pleasanton, California, USA) field-emission scanning electron microscope at the Electron Microscopy Unit of the Department of Molecular Biosciences, University of Oslo and at Microbial Culture Collection at NIES (National Institute for Environmental Studies, http://mcc.nies.go.jp), Tokyo.
Genome size
The genome size of strains was estimated by flow cytometry. Cultures were harvested before onset of the light phase during exponential growth (we observed that the seventh day after trasnfer provided more consistent results). Nuclei were released by injection of 5–10 µL of culture into 250 µl of Nuclei Isolation Buffer (NIB), previously described in Marie et al.32, diluted to 50% concentration with distilled water. The mix of cultures and NIB was incubated at 98 °C for five minutes. Micromonas commoda (RCC299) was added as an internal standard (genome size = 21 Mbp). The nucleic acid specific stain SYBR Green I (Molecular Probes) was added at a final dilution of 1:5000 of the commercial solution. Samples were incubated for 15 minutes before analysis on a FACS Canto II flow cytometer (Becton Dickinson) equipped with a 488 nm excitation and the standard filter setup. The procedure was repeated twice for each strain and measurements were taken in triplicate.
Genomic DNA extraction, PCR amplification and Cloning
Cells were harvested in exponential growth phase and concentrated by centrifugation. Total nucleic acids were extracted using the Nucleospin Plant II kit (Macherey-Nagel, Düren, DE) following the manufacturer’s instructions. The nearly full length nuclear 18S rRNA gene33, the nuclear region containing the Internal Transcribed Spacers (ITS) 1 and 2, as well as the 5.8S rRNA gene34 and partial plastid 16S rRNA gene35–37 were obtained by PCR amplification using universal primers (Supplementary Table 1).
PCR products for 18S and plastid 16S rRNA were purified with the QIAquick PCR purification kit (QIAGEN, Hilden, Germany) and directly sequenced either at the Roscoff Biological Station Genomer platform as described below or sent to the Macrogen Company (Korea). ITS gene amplicons were cloned into PCR4-TOPO vectors (Invitrogen, Carlsbad, CA, USA) and transformed into Escherichia coli competent cells following the manufacturer’s instructions before sequencing. An average of ten clone inserts per strain were then amplified using M13 vector primers and purified using Exosap (USB products, Santa Clara, CA, USA). The sequences were determined using Big Dye Terminator V3.1 (Applied Biosystems) and T3 forward and T7 reverse vector primers. DNA was sequenced using an ABI prism 3100 sequencer (Applied Biosystems). Sequences have been deposited to GenBank under the following accession numbers: MF077471 - MF077519.
ITS2 secondary structures
Forty-two ITS2 sequences (second internal transcribed spacer, separating the 5.8S and 28S rRNA genes) were obtained from the strains listed in Table 1. The ITS2 boundaries (5.8 and 28S rRNA flanking regions) were annotated using Hidden Markov Models (HMMs) and a Viridiplantae database38 as implemented in the ITS2 database annotation tool with the default parameters (http://its2.bioapps.biozentrum.uni-wuerzburg.de/)39. The partial B9 helix formed by the hybridization of 5.8 and 28S rRNA ITS2 flanking regions was checked for structural motifs known to be required for the precise removal of ITS2 during ribosomal RNA processing25. RNA secondary structure predictions were performed using the Mfold web interface40 under the default options with the folding temperature fixed at 37 °C, resulting in multiple alternative folding patterns per sequence. The preliminary structure for each sequence was chosen based on the presence of previously defined ITS2 hallmarks defined by Coleman21,22,41,42 and similarities among the other structures found within and between the clades. This occasionally coincided with the minimum free energy configuration. Exported secondary structures in Vienna format and the respective nucleotide sequences were aligned and visualized using 4SALE version 1.743,44, and manually edited through extensive comparative analysis of each position (nucleotide) in sequences from the same clade, between clades and finally between lineages of prasinophyte clade VII. The hallmarks proposed by Caisová45 were used to unambiguously set the helices. The resulting consensus intramolecular folding pattern (secondary structure) for Choropicophyceae was drawn using CorelDRAW × 7. The proposed ITS2 folding pattern included: nucleotides conserved at 70% and 60% in lineages A and B, clades and branches, 100% conserved nucleotides within lineages A and B and each separate lineage. Regions without length and base pair conservation, for example the apical part of helices I and II as well as the lateral helix IIIa, were also represented. Putative CBC type changes were identified by pairwise comparison of the sequences in the conserved regions of the helices I, II and III within each clade and between clades. All changes, including hCBCs and non-CBC (e.g. N – N ↔ N × N) in all helices and positions analyzed and the positions of each nucleotide pair in the alignment are provided in Supplementary Table 5. The final alignment with the secondary structures in Vienna format is available as Supplementary Material 1.
Phylogenetic analyses (ITS2 and concatenated 18S/Plastid 16S rRNA)
Nuclear 18S rRNA and partial plastid 16S RNA sequences obtained from the strains listed in Table 1 as well GenBank sequences belonging to members of the core chlorophytes and streptophytes were concatenated using Geneious 10.0.546. Streptophytes sequences were used as outgroup. Accession numbers are provided on the phylogenetic trees. The concatenated sequences were aligned with MAFFT using the E-INS-i algorithm47. For ITS2, only the sequences from Chloropicophyceae strains were aligned with MAFFT using the G-INS-i algorithm47. For each sequence within the alignment, the preliminary secondary structure annotated in dot-bracket format was associated, generating a Vienna file which was imported to 4SALE43,44. The final alignment was edited on the basis of conserved secondary structures. Phylogenetic reconstructions were performed with two different methods: maximum likelihood (ML) and Bayesian analyses. The substitution models TN93 + G + I and K2 + G were selected for concatenated 18S rRNA/plastid 16S rRNA and ITS2 sequence datasets respectively, based on Akaike information criterion (AIC) and the Bayesian information criterion (BIC) options implemented in MEGA 6.0648. ML analysis was performed in PhyML 3.049 with SPR (Subtree Pruning and Regrafting) tree topology search operations and the approximate likelihood ratio test with the Shimodaira-Hasegawa-like procedure. Markov chain Monte Carlo iterations were conducted for 1,000,000 generations sampling every 100 generations with burning length 100,000 using MrBayes 3.2.250. MAFFT and MrBayes programs were run within Geneious 10.0.546. Clade nodes were considered as well supported when SH-like support values and Bayesian posterior probabilities were higher than 0.7 and equal to 1.0, respectively.
Intra- and inter-clade sequence distances (_p_-distance) were calculated with combined nuclear and plastid datasets as well as for ITS2. The analysis was conducted using MEGA v. 648 and all positions containing gaps and missing data were removed. All alignments are available as Supplementary Material 2.
Multigene phylogeny
Forty five transcriptomes (Supplementary Table 2) from the Moore Foundation Marine Microbiology Transcriptome Sequencing Program (MMETSP)51 were selected to determine the phylogenetic placement of Chloropicophyceae based on a multigene alignment. Reads were downloaded from the MMETSP archive (http://data.imicrobe.us/project/view/104) to the ABiMS platform in Roscoff (http://abims.sb-roscoff.fr). The quality of the reads was checked by FastQC v.0.52 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were separated into ribosomal and non-ribosomal sequences with RiboPicker v.0.4.352 using as a reference the Small Subunit RNA database (SSR database) from the SILVA project (release 119)53. Ribosomal and non-ribosomal sequences were assembled separately using the de novo reconstruction method of Trinity release r2014071754 using default parameters. Contig abundance was estimated based on Fragments Per Kilobase Of Exon Per Million Fragments Mapped55 using RSEM v.1.1.1756. We only retained non-ribosomal contigs for which FPKM ≥ 2000 and percent of isoform ≥ 1.
Non-ribosomal contigs were analyzed by the Core Eukaryotic Genes Mapping Approach - CEGMA v.257 using the Iplant Collaborative platform58. Using KOGs (Cluster of eukaryotic genes), CEGMA identifies a set of 458 core proteins that are highly conserved and present in a large number of taxa59. We selected 127 genes which were present in all transcriptomes (Supplementary Table 3) and included the Arabidopsis thaliana genome as a reference. Nucleic sequences were translated to amino acids and concatenated. The set of sequences was aligned with MAFFT yielding an alignment with 30,548 amino acid positions. The alignment was trimmed with Gblocks60 using the default parameters resulting in an alignment with 22,073 positions without gaps (Supplementary Material 3). The most appropriate model of protein evolution was determined with ProtTest v.3.261 using the Akaike Information Criterion to be LG + I + G + F. Two different methods were used for phylogenetic inferences: Maximum Likelihood using PhyML 3.049 and Bayesian using MrBayes v.3.2.250,62. 100 bootstrap replicates were set for ML and 500,000 generations for Bayesian analysis. Convergence was checked with Tracer (http://tree.bio.ed.ac.uk/software/tracer/) and all effective sample sizes (ESS) were in excess of 350.
Results and Discussion
Analysis of morphology and genome size of members of prasinophyte clade VII reveals few discriminating characters between clades
Large genetic divergences, such as those observed between clades and lineages of prasinophyte clade VII9, are commonly associated with morphological variations. A large set of culture strains of members of prasinophyte clade VII (Table 1) were grown under identical culture conditions and morphological and genome size analyses were performed to determine whether patterns characteristic of lineages or clades exist.
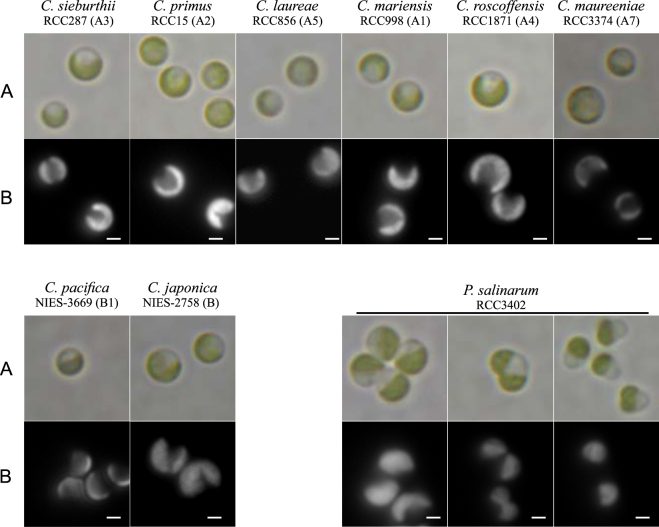
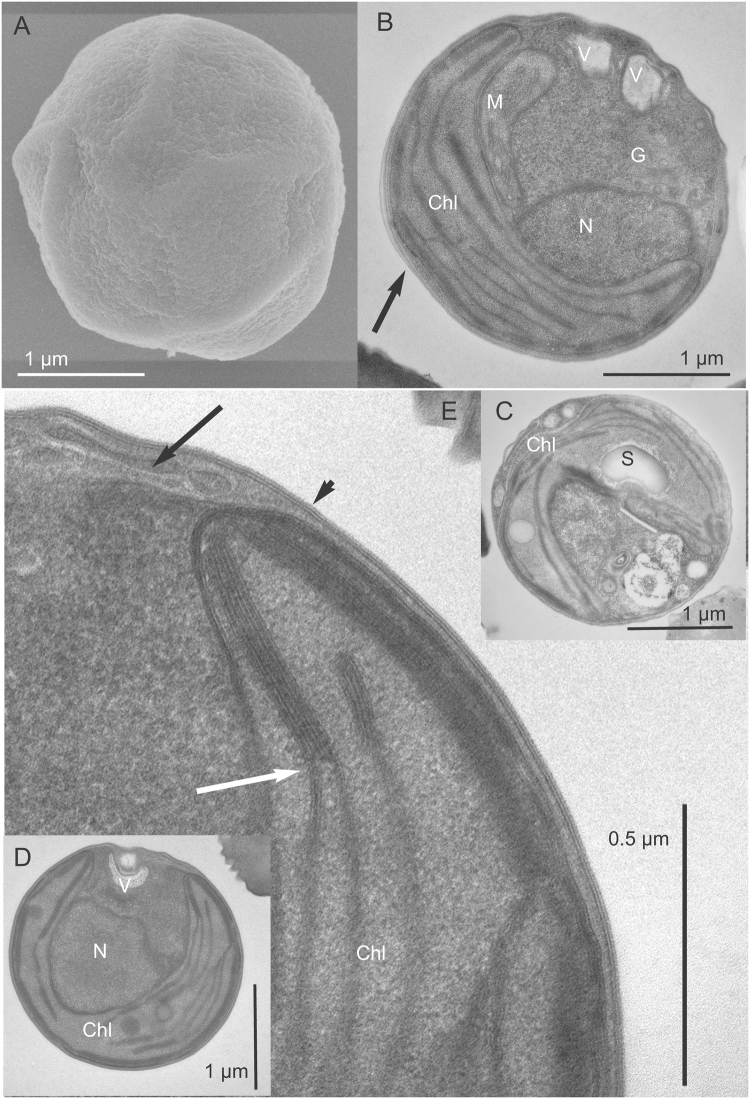
In cultures, cells belonging to lineages A and B usually occur as solitary coccoid green cells, about 1.5–3 µm in size (Fig. 1 and Supplementary Figure 1A). At high cell densities, cells seem to secrete a substance that enables them to stick together and form loose colonies or aggregates (data not shown). Picocystis salinarum (lineage C) cells have a very different morphology. They do occur as coccoid green cells but two additional morphologies are often observed in culture: ovoid and tri-lobed (Fig. 1). As described by Lewin et al.5, these two distinct morphologies are often observed in old, nutrient-depleted cultures and their average cell size is about 2.5 µm (Supplementary Figure 1A). Transmission electron microcopy images clearly confirmed the observations of Lewin et al.5, and illustrated with more detail the bilobed chloroplast and the nucleus and single mitochondrion that occupies the third lobe of the cell in tri-lobed cells (Supplementary Figure 1).
Figure 1.
Light and fluorescence micrographs of Chloropicon sieburthii (A3), C. primus (A2), C. laureae (A5), C. mariensis (A1), C. roscoffensis (A4), C. maureeniae (A7), Chloroparvula japonica (NIES-2758), C. pacifica (B1, NIES-3669) and Picocystis salinarum (RCC3402). (A) Bright field micrographs showing cell outline, shape of chloroplasts and their color. (B) Black and white micrographs showing chlorophyll auto fluorescence of live cells. Scale bar: 1 µm.
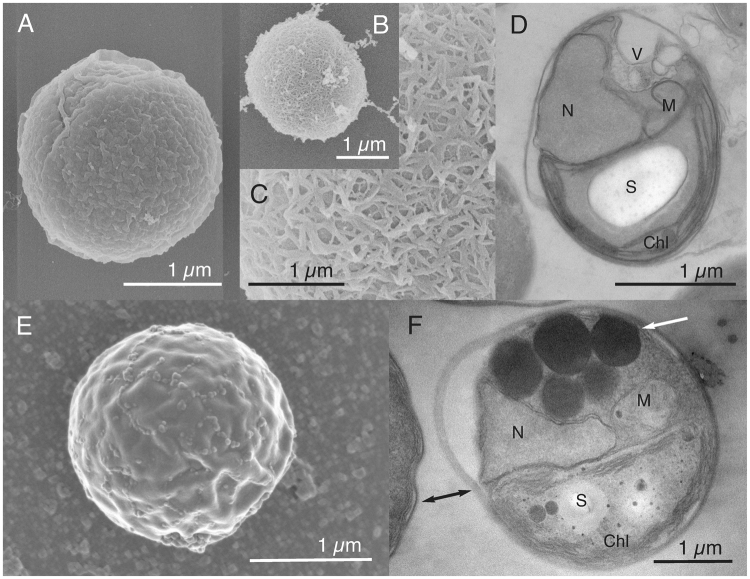
Among the strains from lineages A and B, small differences in cell size were observed between cells from the same strain or from different clades (Supplementary Figure 1A), but these differences were not consistent enough to use size as an appropriate criterion to delineate clades. Transmission and scanning electron microcopy images were also obtained for 5 strains belonging to different clades within lineage A (A1, A2, A3, A4, A5 and A7) (Figs 2 and 3) and 2 strains from lineage B (NIES-2758 and NIES-3669 from clade B1) (Fig. 4). Cells from lineages A and B normally contain one chloroplast that is often crescent shaped and harbors a starch grain (Figs 2 and 4). In dividing cells, two chloroplasts are observed (not shown). Thylakoids are commonly arranged singly or in stacks of three (Fig. 2E). The mitochondrion is located between the nucleus and chloroplast (Figs 2B and 4D and F). The Golgi apparatus seems inconspicuous and is observed only in a few sections (Fig. 2B). The cell wall is delicate and composed of layers (Fig. 2B and E). Opposite the chloroplast, vacuoles containing particles showing Brownian movement can be observed under light microscopy (Figs 2B and D and 4D). The only notable difference between the cells from lineages A and B is the fibrous cell wall (Fig. 4C), the bigger size of the starch grain (Fig. 4D) and the presence of impregnate granules in the cytoplasm (Fig. 4F), however these features were not always present.
Figure 2.
Chloropicon sieburthii (RCC287, A3), type species. TEM-graphs of thin sections and SEM-graphs. (A) Single whole cell with smooth, slightly irregular surface. (B) Section showing chloroplast (Chl), mitochondrion, nucleus (N), vacuoles (V), an inconspicuous Golgi apparatus and cell wall (arrow). (C) Starch grain (S) in chloroplast. (D) Cell with chloroplast showing the organization of lamella. (E) Enlarged part of cell showing chloroplast with lamella consisting of 1 to 4 thylakoids (white arrow) and tri-layered cell wall. The inner layer may contain inclusions (arrow).
Figure 3.
TEM-graphs of thin sections and SEM-graphs. (A,B) Chloropicon primus (RCC15, A2), (C,D) C. laureae (RCC856, A5). (E,F) C. mariensis (RCC998, A1), (G,H) C. roscoffensis (RCC1871, A4), (I,J) C. maureeniae (RCC3374, A7).
Figure 4.
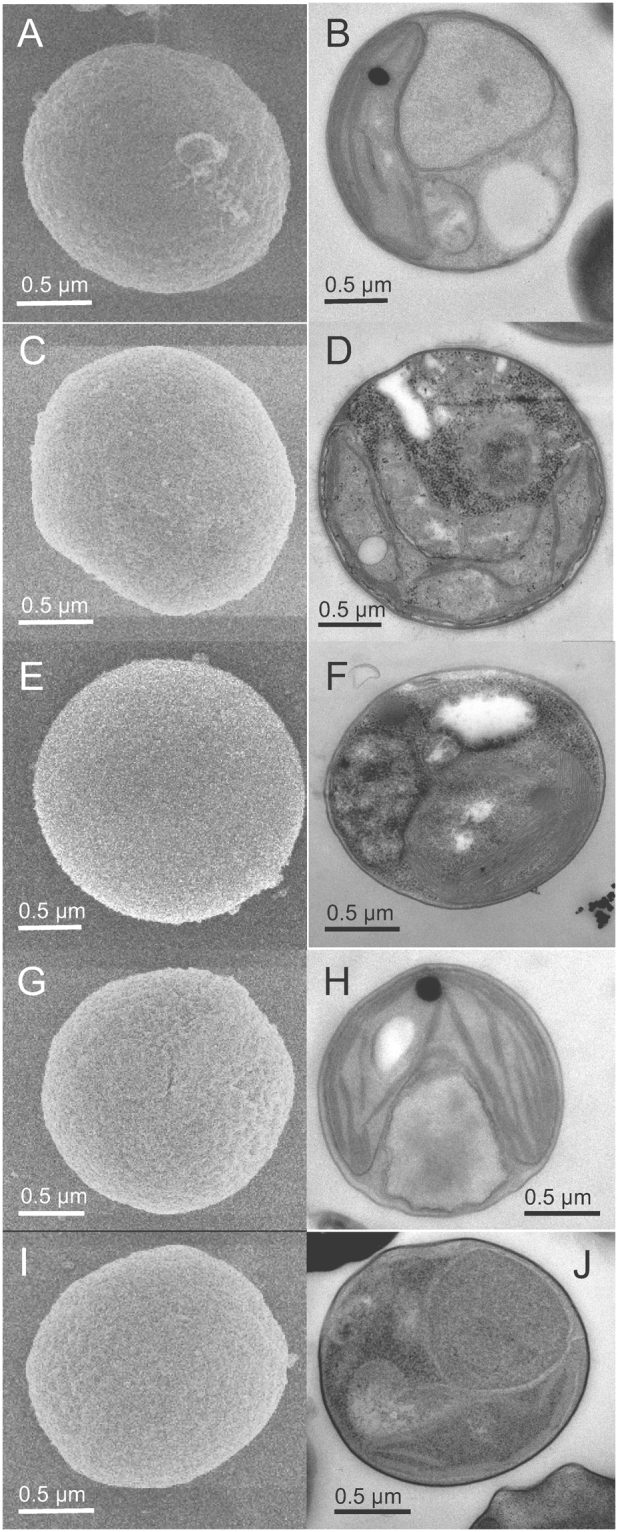
TEM-graphs of thin sections and SEM-graphs. (A–D) Chloroparvula japonica (NIES-2758). (E,F) C. pacifica (type species, NIES-3669, B1).
Genome size estimated by flow cytometry ranged from 20 to almost 70 Mbp (Supplementary Figure 1B), which is higher than for picoplanktonic oceanic green algae such as Bathycoccus, Micromonas and Ostreococcus for which genome size is around 20 Mbp63–65. Karyotype analysis has not been performed for the different strains of clade VII thus the total number of chromosomes remains unknown. There are no clear differences between lineages A and B or among clades. Estimated genome size also varied between strains from the same genetic clade. These differences were particularly marked among the strains belonging to clades A4 and B2 (Supplementary Figure 1B). Strains from clade A4 formed a group with “low genome size” (RCC722, RCC726 and NIES-2755) and a group with “high genome size” (RCC917, RCC1124, RCC1871, RCC3376, RCC4429 and RCC4430) (Supplementary Figure 1B). Two strains from the “low genome size” group (RCC722 and NIES-2755) also had the smallest average cell size of all strains analyzed (Supplementary Figure 1A). The lower estimates of genome size could be the result of incomplete isolation of cell nuclei due to different composition of the cell wall in these isolates or a different level of DNA condensation. Alternatively, it has been shown for Ostreococcus that the size of at least two chromosomes can vary between individuals from same clade (D), which ultimately influences the DNA content of a given cell66. However, the global pattern of genome size was species-specific66. Another possibility would be that some strains have undergone diploidy as previously observed in macroalgae67 although this does not seem to have been observed in microalgae. Despite these differences in genome size, concatenated 18S and 16S rRNA sequence divergence within clade A4 was very low (0.1%, one substitution in 1579 analyzed positions), and ITS2 sequences were identical for all strains (see below). In contrast, B2 showed the highest intra-clade sequence divergence for both the combined dataset and ITS2 sequences (see below) which may correspond with the differences in estimated genome size.
Pigment composition differs among the different lineages and clades of prasinophyte clade VII
Pigment signature has traditionally been applied as a taxonomic proxy of algal diversity in oceanography68. Pigment composition can be closely connected to environmental adaption69. The pigment composition of prasinophyte clade VII strains belonging to lineages A and B is typical of green algae (Chlorophyta). They all contain the following set of carotenoids: neoxanthin, violaxanthin, antheraxanthin, zeaxanthin, lutein and β,ε – carotene8,70. The pigment composition of lineage C, Picocystis salinarum, is unusual in that it contains alloxanthin and monadoxanthin (typical of cryptophytes71) and diatoxanthin (typical of heterokonts68) in addition to chlorophyll a and b and the basic set of carotenoids commonly found in green algae5,8. Alloxanthin and diatoxanthin have also been reported in _Coccomyxa_–like algae72, a chlorophyceaen pathogen of the mussel Mytilus galloprovincialis.
In the present study, we provide pigment data for 7 additional strains from lineages A and B, meaning that pigment signatures are available for 21 strains (Table 2). Violaxanthin and lutein were the two most abundant carotenoids for most of the strains from lineages A and B, astaxanthin coming third when it is present. Astaxanthin has previously been shown to increase with light intensity in prasinophyte clade VII (from 2- to 4-fold depending on the strain), suggesting a photoprotective role for this carotenoid8. Of the 21 strains, only NIES-3669 belonging to clade B1 did not possess astaxanthin and β,β – carotene as accessory carotenoids. In contrast, this strain had an antheraxanthin to Chl a ratio 10 times higher than that of the other strains (Table 2). Antheraxanthin is part of the photoprotective epoxidation and de-epoxidation cycle VAZ (violaxanthin-antheraxanthin-zeaxanthin) and its content can be variable depending on light conditions73.
Loroxanthin was also detected among some strains from lineages A and B (Table 2) and has previously been suggested to have a major light harvesting role since it increases at low light intensity8. Loroxanthin was absent in clade B1 (NIES-3669), B3 (RCC4572), in one strain from clade B2 (RCC696) and clade A4 (Table 2). Several strains from clade A4 were isolated from northern or southern temperate latitudes (~49° N and ~33° S) or from tropical regions (Table 1). The two clade A4 strains previously analyzed (RCC1124 and RCC1871), isolated from temperate North Atlantic Ocean waters, lacked loroxanthin. In the present study five other clade A4 strains from a wider range of latitudes were analyzed. The data confirm that all strains of clade A4, whatever the latitude from which they were isolated, lacked loroxanthin (Table 2). Clearly the absence of loroxanthin is a phenotypic characteristic of clade A4, but it is also absent from most strains from lineage B, and therefore cannot be used as a biomarker to distinguish A4 from all other members of prasinophyte clade VII.
Phylogenomic analysis of chloroplast sequences have suggested that prasinophyte clade VII lineage A is a sister group of the core Chlorophyta12,74,75. The core Chlorophyta comprise a well-supported clade containing the classes Chlorophyceae, Ulvophyceae, Trebouxiophyceae, Chlorodendrophyceae and Pedinophyceae. The presence of astaxanthin and loroxanthin carotenoids in clade VII lineage A and B strains, as in core Chlorophyta, while it is absent in other prasinophytes is indicative of another common feature between this group of prasinophytes and core Chlorophyta.
Nuclear and plastid SSU rRNA as well as ITS2 phylogeny support clade separation
Our previously published phylogenetic analyses based on nuclear and plastid SSU rRNA gene sequences demonstrated that lineage C did not contain any sub-division. In contrast, lineages A and B could be further divided intoclades: A1 to A7 and B1 to B39. Each clade was defined based on the presence of at least two environmental or strain sequences obtained from different locations (and/or samples at a given location) with strong phylogenetic support for at least one of the gene markers used. Clade B3 was composed only by environmental sequences9. Strain RCC4572, recently isolated from the Atlantic Ocean76, had signatures for both nuclear and plastid SSU rRNA genes previously identified as clade B3. This means that all clades known from the environment9 have now been brought into culture.
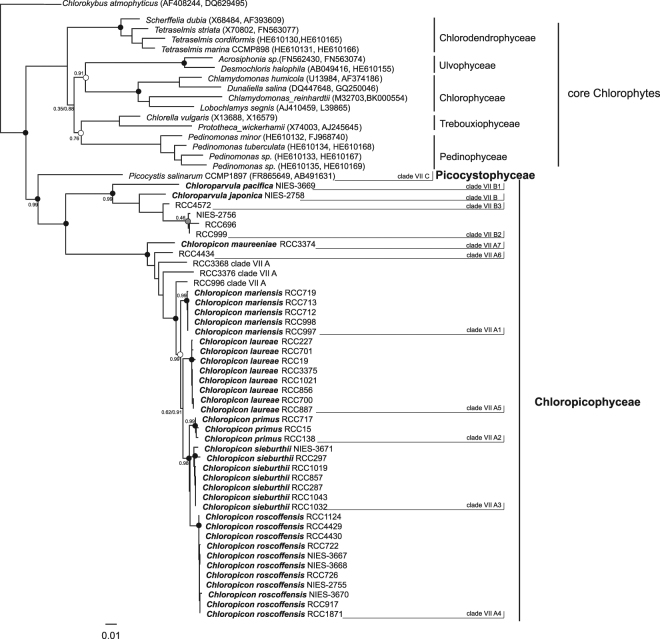
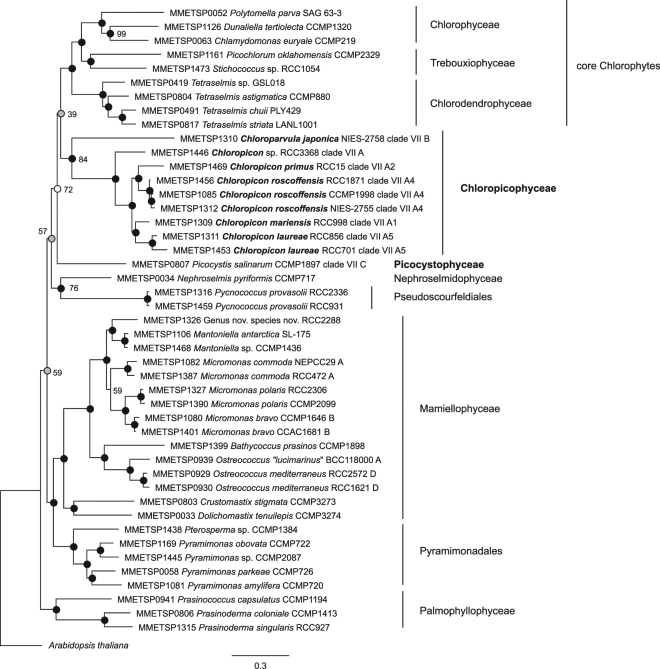
A phylogenetic analysis combining partial plastid SSU rRNA gene sequences with a congruent data set of nuclear 18S rDNA sequences (Fig. 5) recovered the major diverging clades within prasinophyte clade VII lineages A and B with high support values for maximum likelihood (ML) and Bayesian analyses. The only exception was clade B2 which had no support from ML (0.46) analysis (Fig. 5). Phylogenetic analysis based on ITS2 (internal transcribed spacer 2) sequences from 41 unique strains from lineages A and B also confirmed the major divergent clades described above (Supplementary Figure 3). However, both analyses (combined SSU rRNA and ITS2) failed to resolve the relationships between the different clades (low bootstrap and variable tree topologies).
Figure 5.
Maximum-likelihood tree inferred from concatenated plastid and nuclear sequences of prasinophytes clade VII. Sequences belonging to members of the core Chlorophytes and Streptophytes were included in addition to the sequences obtained from the cultures. Streptophytes were used as an outgroup. Solid dots correspond to significant support (>0.7) for ML analysis and full support (1.0) by Bayesian analysis. When ML support is below 1.0 the percentage is indicated next to the symbol. Grey dots correspond to non-significant ML support (<0.7) and full support from Bayesian analysis. Empty dot corresponds to ML support but no support from Bayesian analysis.
The uncorrected sequence distance (_p_-distance) within clades calculated with the combined 18S nuclear and 16S plastid datasets was below 0.04% (for clade B2), while interclade divergence was as high as 13.5% (between A5 and B2, Supplementary Table 4). The distance observed between lineages A and B (12.6%) is similar to the divergence found for example between Chlorodendrophyceae and lineage A. The interclade sequence distance varied from 1.0% (i.e. A2 vs. A3) to 4.3% (i.e. A2 vs. A7) for A and 2.2% (i.e. B2 vs. B3) to 7.7% (i.e. B1 to B3) for B (Supplementary Table 4).
Interclade distances using ITS2 sequences were higher with a maximum value of 42% between A2 and B1. Within the clades, the distance varied from 0% in A4 to 5.6% in B2. The high sequence _p_-distance found in B2 suggests that this clade may represent different species (see below). However, these values should be taken with caution given the inherent difficulty in aligning ITS regions (Supplementary Table 4).
The genetic divergence found between clades within prasinophyte clade VII suggests that they may each represent different species. In another group of prasinophytes, Micromonas, similar genetic divergence values have been observed for the highly conserved 18S rRNA gene among different clades recently erected to species status77. For the picoplanktonic Ostreococcus, the highest sequence distance, also based on 18S rRNA analyses, was 1.8% between two clades that are now considered to represent different species66.
ITS2 structure confirms clade separation
In order to examine in more depth the level of inter- and intra-clade diversity of prasinophyte clade VII we analyzed the secondary structure of ITS2 for 41 strains listed in Table 1 (Fig. 6). Unfortunately, since data are only available for one strain of Picocystis (clade C), it is not possible to determine the ITS2 folding pattern for this lineage.
Figure 6.
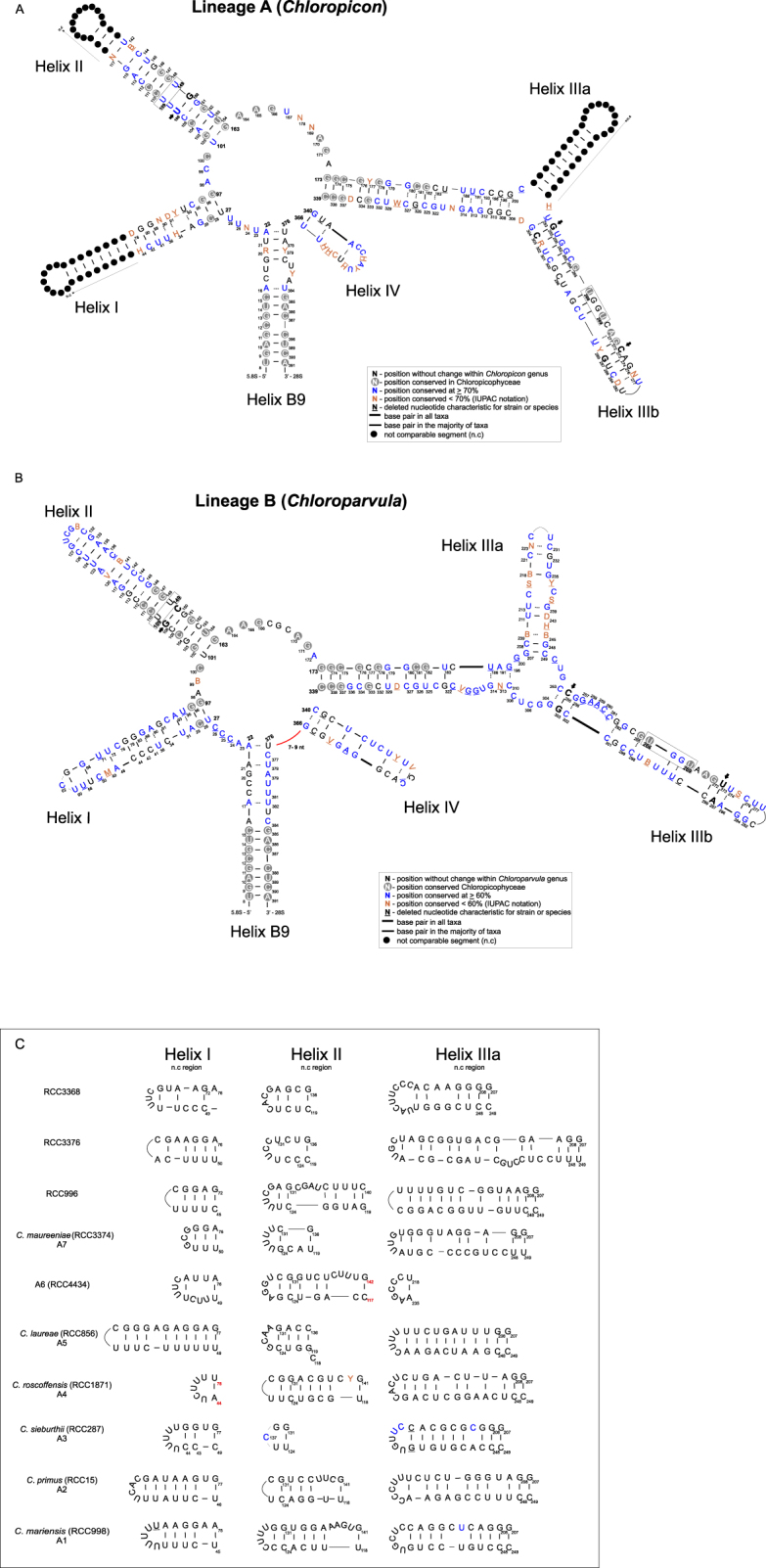
Consensus secondary structure model of the ITS2 molecule of Chloropicophyceae with the two genera, A) Chloropicon (lineage A) and B) Chloroparvula (lineage B). The four major helices are labeled as Helix I – Helix IV and the interaction region of 5.8S and 28S rRNA as B9. Nucleotide letters shown in blue in both ITS2 diagrams refer to those present in 70% (A) and 60% (B) of the clades and branches analyzed. Any position with less than the majority rule applied are shown as IUPAC ochre symbols. Invariable positions within each lineage are drawn in black and circled in grey when common to both A and B lineages. Arrows and nucleotides in bold indicate the major three CBCs between the two lineages. Positions with deletions are underlined. Regions without length and base pair conservation are shown as black dots. These regions, corresponding to the apical part of helices I and II as well as the lateral helix IIIa are drawn for each clade/branch in the panels on the right side.
The ITS2 secondary structure of lineages A and B contained the four-helix domains known in many eukaryotic taxa in addition to helix B9 (Fig. 6). Helix B9, a region of the 5.8S and 28S rRNA interaction, shows the highly conserved eight base pair stem required for the precise excision of the ITS225. Helices II and III harbor the universal hallmarks proposed by Mai and Coleman20 and Schultz et al.78: the pyrimidine-pyrimidine (Y-Y) mismatch in helix II and YRRY (pyrimidine – purine – pyrimidine) motif on the 5′ side of Helix III (boxes, Fig. 6). The Y-Y mismatch was U × U for all sequences analyzed, with the exception of clade A6 (C × U) and the solitary branch RCC3368 (U × C). The YRRY motif of helix III was represented by the sequence UGGU in all strains analyzed, except for NIES-2756 (clade B2) where the guanidine is replaced by adenine (UAAU). The spacers between helices I and II and between II and III displayed the fixed number of nucleotides proposed by Caisová et al.45 (Fig. 6). Spacers between helices B9 and I, III and IV and IV and B9 were less conserved when compared to the Chlorophyta consensus structure and within lineage B. Remarkably, the secondary structure of lineage B exhibited an insertion of 7–9 nucleotides between helices III and B9 which was completely absent in lineage A and NIES-2758 presented a unique deletion of 3 nucleotides within the spacer between helices B9 (Fig. 6B). Among the conserved spacers (between helices I – II and II – III), not only the length was conserved but also the nucleotides occupying the alignment positions 98 (A), 100 (C), 164 to 166 (AAG) and 170 to 172 (AGA) were conserved with respect to the ITS2 Chlorophyta consensus secondary structure (for details see Fig. 3 of Caisová et al.45) (Fig. 6). The only exceptions were clades A6 (RCC4434) with one uracil at position 98 and B1 (NIES-3669), also with a replacement of A by uracil at position 172. Since these clades were represented by a single strain, we cannot confirm whether these changes are characteristic of these clades.
The first two base pairs of helices I, II and III are another important hallmark for the unambiguous identification of these helices. They were conserved within lineages A and B (Fig. 6) and in agreement with the consensus ITS2 structure of Chlorophyta (for details see Fig. 3 of Caisová et al.45). However, the three-nucleotide motif (AGG) on the 5′ side of the base of Helix IV, also proposed by Caisová et al.45, was not detected in our structures (Fig. 6).
Two approaches have been proposed for CBC identification: 1) phenetic, whereby in base pair sequence comparison all CBCs between two sequences are considered, without direct reference to their evolutionary origin22,26, and 2) phylogenetic, which considers the status of a given base pair in the ancestor of two sister taxon a priori the determination of the CBC45,79,80. Unfortunately, the phylogenetic approach could not be used in our study given the conflicting branching pattern among phylogenies (Fig. 5, Supplementary Figure 3). Table 3 details the number of CBCs between two branches or clades and the nucleotide pair identification number where CBCs were found (numbers in brackets).
Table 3.
CBCs in the conserved regions of the helices I, II and III within each clade and between the clades. The numbers in bold represent the number of CBCs found between two branches or clades. The numbers in brackets represent the CBC identification number (see Supplementary Table 5 for the list of all CBC and for their position on the ITS2 alignment).
| Chloropicon mariensis (A1) | Chloropicon primus (A2) | Chloropicon sieburthii (A3) | Chloropicon roscoffensis (A4) | Chloropicon laureae (A5) | RCC4434 (A6) | Chloropicon maureeniae (A7) | RCC996 (A) | RCC3368 (A) | RCC3376 (A) | RCC2337 and RCC999 (B2) | RCC696 (B2) | Chloroparvula japonica (NIES-2758) | Chloroparvula pacifica (B1) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chloropicon mariensis (A1) | 0 | |||||||||||||
| Chloropicon primus (A2) | 1 [13] | 0 | ||||||||||||
| Chloropicon sieburthii (A3) | 4 [9, 13, 50, 57] | 2 [13, 20] | 0 | |||||||||||
| Chloropicon roscoffensis(A4) | 2 [9, 13] | 0 | 4 [13, 20, 51, 57] | 0 | ||||||||||
| Chloropicon laureae (A5) | [20, 36, 50] | 4 [9, 13, 29, 36] | 5 [9, 13, 20, 36, 57] | 3 [9, 13, 20] | 0 | |||||||||
| RCC4434 (A6) | 4 [18, 19, 20, 57] | 5 [9, 13, 18, 19, 20] | 4 [9, 18, 19, 50] | 6 [9, 13, 18, 19, 20, 57] | 5 [13, 18, 20, 50, 57] | 0 | ||||||||
| Chloropicon maureeniae (A7) | 5 [9, 10, 14, 15, 57] | 5 [10, 13, 14, 15, 48] | 8 [10, 14, 15, 20, 50, 51, 57, 58] | 5 [10, 13, 14, 15, 57] | 8 [9, 10, 13, 14, 15, 20, 50, 57] | 9 [9, 10, 14, 15, 18, 19, 20, 57, 58] | 0 | |||||||
| RCC996 (A) | 0 | 2 [10, 13] | 4 [10, 13, 20, 57] | 1 [13] | 4 [10, 20, 36, 50] | 6 [10, 13, 18, 19, 20, 57] | 5 [10, 13, 14, 15, 57] | 0 | ||||||
| RCC3368 (A) | 5 [9, 10, 13, 30, 57] | 3 [9, 10, 36] | 8 [9, 10, 13, 20, 30, 36, 50, 57] | 4 [9, 10, 30, 57] | 4 [9, 10, 30, 57] | 7 [9, 10, 13, 18, 19, 20, 57] | 5 [9, 13, 14, 15, 58] | 5 [10, 13, 30, 36, 57] | 0 | |||||
| RCC3376 (A) | 3 [19, 34, 35] | 4 [9, 13, 19, 35] | 6 [9, 19, 20, 35, 51, 57] | 4 [9, 13, 19, 35] | 4 [19, 20, 35, 36] | 4 [18, 20, 50, 57] | 6 [9, 14, 19, 35, 50, 57] | 4 [13, 19, 35, 50, 51] | 7 [9, 13, 19, 35, 36, 50, 57] | 0 | ||||
| RCC2337 and RCC999 (B2) | 14 [8, 9, 14, 16, 18, 35, 40, 41, 49, 54, 55, 56, 57, 58] | 16 [7, 8, 13, 14, 16, 18, 35, 37, 40, 41, 48, 50, 54, 55, 56, 58] | 15 [8, 14, 16, 18, 20, 35, 37, 40, 41, 49, 50, 54, 55, 56, 58] | 17 [7, 8, 13, 14, 16, 18, 35, 40, 41, 49, 50, 51, 54, 55, 56, 57, 58] | 10 [8, 9, 11, 13, 14, 54, 55, 56, 57, 58] | 14 [7, 9, 14, 16, 18, 19, 20, 37, 49, 50, 54, 55, 56, 58] | 16 [7, 8, 10, 15, 16, 18, 35, 40, 41, 49, 50, 51, 54, 55, 56, 58] | 18 [7, 8, 10, 13, 14, 16, 18, 33, 35, 40, 41, 49, 50] | 18 [7, 8, 9, 10, 13, 14, 16, 18, 30, 35, 40, 41, 49, 50, 54, 55, 56, 58] | 16 [7, 8, 14, 16, 18, 19, 40, 41, 49, 50, 51, 54, 55, 56, 57, 58] | 0 | |||
| RCC696 (B2) | 12 [8, 9, 14, 16, 18, 40, 41, 49, 54, 55, 56, 57] | 13 [7, 8, 13, 14, 16, 18, 37, 40, 41, 49, 54, 55, 56] | 12 [8,14, 16, 18, 20, 37, 40, 41, 49, 54, 55, 56] | 15 [7, 8, 13, 14, 16, 18, 37, 40, 41, 49, 51, 54, 55, 56, 57] | 15 [7, 8, 9, 11, 13, 14, 16, 18, 20, 41, 49, 54, 55, 56, 57] | 10 [9, 14, 16, 18, 19, 20, 49, 54, 55, 56] | 11 [7, 8, 10, 15, 16, 18, 40, 51, 54, 55, 56] | 11 [7, 8, 10, 13, 14, 16, 18, 41, 54, 55, 57] | 17 [4, 7, 8, 9, 10, 13, 14, 16, 18, 30, 37, 40, 41, 49, 54, 55, 56] | 15 [7, 8, 14, 16, 18, 19, 37, 40, 41, 49, 51, 54, 55, 56, 57] | 7 [21, 41, 42, 43, 44, 45, 47] | 0 | ||
| Chloroparvula japonica (NIES-2758) | 11 [8, 9, 14, 16, 18, 20, 40, 41, 49, 50, 54] | 12 [8, 13, 14, 18, 40, 41, 48, 49, 50, 54, 58] | 12 [8, 14, 16, 20, 40, 41, 49, 50, 54, 57, 58] | 12 [8, 13, 14, 16, 18, 40, 41, 49, 50, 51, 54, 58] | 16 [8, 9, 13, 14, 16, 18, 20, 27, 29, 40, 41, 48, 49, 50, 54, 58] | 11 [9, 14, 16, 18, 19, 20, 49, 50, 54, 57, 58] | 12 [8, 15, 16, 18, 40, 41, 49, 50, 51, 54, 57, 58] | 10 [4, 8, 13,14, 16, 18, 49, 50, 54, 58] | 15 [8, 9, 13, 14, 16, 18, 30, 40, 41, 48, 49, 50, 54, 57, 58] | 13 [8, 9, 14, 16, 18, 19, 40, 41, 49, 50, 51, 54, 58] | 8 [20, 21, 22, 23, 42, 43, 56, 57] | 10 [20, 21, 22, 23, 24, 41, 43, 45, 56, 57] | 0 | |
| Chloroparvula pacifica (B1) | 12 [9, 14, 16, 19, 20, 29, 30, 40, 41, 49, 50, 54] | 9 [13, 14, 16, 19, 29, 40, 41, 49, 54] | 11 [14, 16, 19, 20, 29, 30, 40, 41, 49, 54, 57] | 9 [13, 14, 10, 19, 29, 30, 49, 54, 58] | 11 [9, 13, 14, 16, 19, 20, 29, 30, 40, 49, 54] | 11 [9, 14, 16, 19, 20, 29, 30, 49, 50, 54, 57] | 10 [15, 16, 19, 29, 30, 49, 50, 54, 57, 58] | 13 [8, 13, 14, 16, 18, 49, 50, 54, 58, 40, 41, 49, 50, 54] | 12 [9, 13, 14, 16, 19, 29, 30, 40, 49, 50, 54, 57] | 9 [9, 14, 16, 29, 30, 40, 41, 49, 54] | 15 [8, 20, 21, 22, 23, 24, 25, 29, 30, 37, 40, 45, 50, 56, 57, 58] | 15 [8, 19, 20, 21, 22, 23, 24, 25, 29, 30, 37, 40, 41, 56, 57] | 10 [8, 19, 21, 24, 29, 30, 40, 45, 50, 58] | 0 |
Caisová et al., using a phylogenetic approach, showed that in two classes of green algae, Ulvales79 and Chlorophyceae45, CBCs on Helix II and III were often correlated with divergences at supra-specific taxonomic levels, for example genus. A significant number of CBCs, mainly localized in helices II and III, were observed between clades belonging to different lineages (Table 3). Three CBCs (bp position 16, 49 and 54, Supplementary Table 5) in helices II and III distinguished lineage A from B (Fig. 6).
Within clades, only clade B2 had CBCs (and hCBCs) in helices II and III, between strain RCC696 on the one hand and the other two, RCC999 and NIES-2756, on the other (Table 3). In fact, sequence divergence within this clade was highest among the clades for both sequence datasets used (Supplementary Table 4) and the pigment composition of RCC696 was slightly different from that of NIES-2756 since it lacked loroxanthin (Table 2). Within the other clades of lineages A and B, the ITS2 sequences were nearly identical and neither CBCs nor hCBCs were detected (Table 2, Supplementary Table 5).
In general, several CBCs were detected between clades or solitary branches. There were two exceptions for which CBCs were not observed: between RCC996 and A1 and between A4 and A2 (Table 3), although hCBCs (7 and 6 respectively) were found in helices I, II and III (Supplementary Table 5). All clades and branches, including these ones, can be differentiated by molecular signatures present in both plastid and nuclear SSU rRNA genes, a fact that was further confirmed here. The branch formed by RCC996 has not been classified within a specific clade9 given the absence of similar complete 18S rRNA gene sequence from other cultures or environmental samples. In addition, A1 possesses a 500 bp long 18S rRNA intron around position 1,000 which is not detected in any other clade. While few sequences corresponding to that of RCC996 have been found in metabarcoding datasets9, clade A1 was relatively more important at the DCM (deep chlorophyll maximum) of some Pacific Tara Ocean stations fitting the fact that all A1 strains have been isolated from deep euphotic waters9. Another example of distinct molecular signatures congruent with ecological specificities was provided by contrasting clades A4 and A2. Clade A4 is the second most abundant clade after B1 and is mainly found in coastal waters (OSD dataset), probably indicating a habitat preference, while the abundance and distribution of A2 is very sporadic9. Strains belonging to A4 differed from others by the absence of loroxanthin (Table 2). Thus, despite the absence of CBCs between clade A1 and RCC996 or between A2 and A4, each probably represents organisms with distinct biological and ecological properties.
Nuclear multigene phylogeny confirms the monophyly of lineages A and B
A multigene phylogenetic analysis was performed using the transcriptome sequence database obtained in the frame of the MMETSP Marine Microbiology Initiative51. Forty-five transcriptomes were selected including all those available for prasinophyte clade VII as well as from related Chlorophyta lineages (Supplementary Table 2). The Core Eukaryotic Genes Mapping Approach (CEGMA) defines a set of 458 core nuclear genes for which HMM profiles are available. Of these, 127 genes (Supplementary Table 3) were found in all of the selected transcriptomes and these were used to establish a multigene phylogeny based on a concatenated amino acid alignment of 22073 positions. In this analysis, the transcriptomes from 9 strains belonging to lineages A and B formed a moderately supported clade, independent of lineage C, confirming their monophyly (Fig. 7). This clade was a sister clade to core Chlorophyta, although with low ML support in contrast to the strong support reported by Lemieux et al.12 for a phylogenomic analysis based on chloroplast sequences. The position of lineage C (Picocystis salinarum) varied depending on the method used. In the ML analysis, Picocystis formed a branch of its own, weakly related to prasinophyte clade VII and core chlorophytes (Fig. 7). In previous chloroplast genome phylogenies Picocystis branched with Pseudoscourfeldiales75 or formed an independent branch12.
Figure 7.
Maximum Likelihood (ML) phylogenetic tree based on a concatenated alignment of 22,073 amino acids corresponding to 127 nuclear core genes extracted from transcriptomes obtained for 45 Chlorophyta strains obtained in the framework of the Marine Microbiology Initiative (MMETSP)51. Solid dots correspond to significant support (>70%) for ML analysis and full support (100% probability) by Bayesian analysis. When ML support is below 100%, the percentage is indicated next to the symbol. Grey dots correspond to non-significant ML support (<70%) and full support from Bayesian analysis. Empty dot corresponds to ML support but no support from Bayesian analysis.
Within lineage A, the major clades for which transcriptomes were available (A1, A2, A4 and A5) and solitary branches defined by strains NIES-2758 and RCC3368 (CCMP2111) were recovered in the multigene analysis (Fig. 7). The latter strains cannot be ascribed to any of the clades previously defined9 because no other culture or environmental sequences are similar to them. In accordance with 16S/18S and ITS phylogenies (Fig. 5, Supplementary Figure 3), clades A2 and A4 were closely related in the multigene analysis (Fig. 7). Clades A1 and A5 formed a cluster with 100% support, but this association was not observed in 18S/16S and ITS2 rRNA analyses (Fig. 5, Supplementary Figure 3). In the multigene analysis all other Chlorophyta groups (Chlorodendrophyceae, Chlorophyceae and Trebouxiophyceae which belong to the core Chlorophyta), as well as Mamiellophyceae, Pseudoscourfeldiales, Nephroselmidophyceae and Palmophyllophyceae were monophyletic with 100% bootstrap support in both methods.
Prasinophyte clade VII comprises 2 new classes containing three genera and 8 species
A reassessment of the taxonomy of prasinophytes has been needed for some time. The idea of raising prasinophyte lineages to class status was proposed by Nakayama et al.81, based upon the paraphyletic nature of prasinophyte lineages, their genetic dissimilarity based on rRNA sequences and the recognition of well supported classes in the core Chlorophyta. The differentiation of clades proposed by Guillou et al.4 was a first step, followed by the erection of novel classes replacing some of these clades: Mamiellophyceae for clade II10, Chlorodendrophyceae for clade IV82, and Palmophyllophyceae for clade VI75. The phenotypic and genetic data that we obtained on a large set of culture strains in the present study allows clarification of the taxonomy of clade VII which is ecologically important in oceanic waters9. All of the analyses performed on strains from lineages A and B converge to establish that these two lineages share many phenotypic and genetic traits, including similar morphology (Figs 1–4), similar pigment composition (Table 2), and monophyly in all phylogenetic analyses (Figs 5 and 7 and Supplementary Figure 3). P. salinarum was originally grouped with prasinophyte clade VII based on 18S rRNA phylogenetic analyses, forming lineage C restricted to this species4. The degree of sequence similarity between the nuclear 18S RNA gene sequence of P. salinarum and those of other prasinophyte clade VII (around 88%) is comparable to that between lineages A and B9. The concatenated nuclear 18S/plastid 16S rRNA phylogenetic tree (Fig. 5) gave the same result. However, in phylogenetic analysis using only plastid 16S rRNA gene sequences, lineage C formed an independent lineage from prasinophyte clades VII A and B9. Moreover, phylogenetic analyses using the complete nuclear11, plastid encoded rRNA operons10,11 and chloroplast genomes12 already suggested that P. salinarum forms a separate lineage from prasinophyte clade VII A and B. In all of these analyses, only the data from RCC15 (CCMP1205, clade A2) and RCC3402 (CCMP1897, P. salinarum) were used. Multigene phylogeny (Fig. 7), morphology (Supplementary Figure 2) and pigment composition, in particular the presence of red lineage carotenoids5,8 (Table 2) provide compelling evidence that prasinophyte clade VII and P. salinarum should be considered independent lineages and that these represent distinct classes of prasinophytes. Therefore, we have raised prasinophyte clade VII lineages A and B together to class status as the Chloropicophyceae and we also create the class Picocystophyceae to accommodate the genus Picocystis described by Lewin5.
Few morphological characters distinguish Chloropicophyceae from other picoplanktonic prasinophytes. Among described green algae, there are four genera containing naked coccoid non-motile cells: Prasinoderma, Picochlorum, Pycnococcus and Ostreococcus. The pyrenoid is easily observed in Pycnococcus83 and Prasinoderma84, whereas it is absent in Chloropicophyceae (Figs 2–4). Sexual reproduction or auto-sporulation have been proposed for Pycnococcus83,85 and Picochlorum86, but have never been observed in Chloropicophyceae cells. Pigments composition is perhaps the most distinctive character between Chloropicophyceae and these four genera. Pycnococcus, Prasinoderma and Ostreococcus belong to pigment group prasino-3A and 3B68, all containing prasinoxanthin70, which is absent in Chloropicophyceae (Table 1) and Picochlorum86. The carotenoids astaxanthin and loroxanthin are found in cells of Chloropicophyceae, while they are absent in Picochlorum.
Within the Chloropicophyceae we establish two new genera: Chloropicon and Chloroparvula, corresponding to lineages A and B, respectively. Certain morphological features distinguish lineage B (Chloroparvula) cells from those of lineage A (Chloropicon): presence of a fibrous cell wall (Fig. 4C), the larger size of the starch grain (Fig. 4D) and the presence of impregnate granules in the cytoplasm (Fig. 4F). Despite overall morphological similarity, lineages A and B formed independent monophyletic lineages in our multigene phylogeny, with Chloropicon receiving 100% support with both methods used (Fig. 5). Unfortunately only one transcriptome was available for the genus Chloroparvula (previously clade VIIB), so it was not possible to assess multigene phylogeny support for the erection of this genus. The average genetic distance observed between lineages A and B (around 12%) in our concatenated dataset is similar to the divergence between well-established classes of the core Chlorophyta (Supplementary Table 4), justifying separation of these two lineages (at least) at the genus level.
Müller et al.26 and Caisová et al.45,79 showed that the absence of CBCs in ITS2 secondary structures is not an indicator that two organisms belong to the same species. This is particularly true for Ulvales and Chlorophyceae for which a lack of correlation between CBCs in ITS2 at the species level was reported45,79. However, the presence of at least one CBC is a good indicator that two organisms represent distinct species (93.1% confidence for plants and fungi26). For picoeukaryotes that are indistinct morphologically66,77, form complex species80,87,88 or are even uncultured89, this distinguishing character may be particularly useful. ITS2 secondary structure analyzed together with molecular signatures of nuclear and plastid SSU rRNA genes support the hypothesis that Chloropicophyceae clades and branches represent distinct species, despite the absence of clear morphological differences. In addition to knowledge on their ecological distribution, these results lead us to erect to species status 7 clades (A1, A2, A3, A4, A5, A7 and B1) and one solitary branch (NIES-2758) for which we have ultrastructural information. The other clades (A6, B2 and B3) were not erected to species level due to the absence of EM images necessary to establish holotypes. The new species of Chloropicophyceae are: Chloropicon mariensis (A1), Chloropicon primus (A2), Chloropicon sieburthii (A3), Chloropicon roscoffensis (A4), Chloropicon laureae (A5), Chloropicon maureeniae (A7), Chloroparvula japonica (NIES-2758) and Chloroparvula pacifica (B1).
The formal taxonomic description of prasinophyte clade VII as the new class Chloropicophyceae will facilitate interpretation of large-scale metabarcoding and/or metagenomics analyses that aim at investigating the ecological patterns and the role in the marine environment of this enigmatic group of oceanic picoplanktonic green algae.
Taxonomy section
Chlorophyta Reichenbach 1834
Chloropicophyceae Lopes dos Santos and Eikrem classis nov.
Diagnosis: Coccoid green cells, with a diameter of 1.5–4 µm, found in marine waters. One nucleus, one mitochondrion, one chloroplast surrounded by two membranes, containing starch grain. Chloroplast with chlorophylls a and b. Pyrenoid absent. Flagella absent. Coccoid cells with layered cell wall. Sexual reproduction unknown.
Chloropicales Lopes dos Santos and Eikrem ord. nov.
Diagnosis: With characters of class. Additional characters; accessory pigments are neoxanthin, violaxanthin, antheraxanthin, zeaxanthin, lutein, loroxanthin, astaxanthin, β,β- carotene, β,ε- carotene.
Chloropicaceae Lopes dos Santos and Eikrem fam. nov.
Diagnosis: With characters of order. Additional characters; cell wall thin and delicate.
Chloropicon Lopes dos Santos and Eikrem gen. nov.
Diagnosis: With characters of order. Coccoid cells measure 2–4 µm. One green chloroplast, often crescent shaped with starch grain. Thylakoids occur singly and in stacks of three. Central nucleus, mitochondrion located between nucleus and chloroplast. Vacuoles (1–2) present at cell periphery may contain particles. Surface of cell wall smooth.
Etymology: Named for its green color and small size.
Type species: Chloropicon sieburthii
Chloropicon sieburthii Lopes dos Santos and Eikrem sp. nov.
Diagnosis: With characters of the genus. Additional characters; combined nucleotide sequences of the nuclear 18S rRNA (AY425302), rRNA ITS (MF077490) and plastid 16S rRNA (AY702147) are species specific.
Holotype: Cells embedded in resin block deposited at the Natural History Museum, University of Oslo, accession number O-A-10001. Figure 2B–E show cells from the resin block. Authentic culture deposited in the Roscoff Culture Collection as RCC287.
Type locality: Strain RCC287 was isolated from water sampled in the Equatorial Pacific Ocean (0°, 179°49′ W) at 120 m depth.
Etymology: Named in honour of John McN. Sieburth, who published the first electron microscopy images of natural populations of marine picoeukaryotes.
Chloropicon primus Lopes dos Santos and Eikrem sp. nov.
Diagnosis: With characters of genus. Additional characters; combined nucleotide sequences of nuclear 18S rRNA (U40921), rRNA ITS (HE610139) and plastid 16S rRNA (AY702121, FN563080) are species specific.
Holotype: Cells embedded in resin block and thin-sections deposited at the Natural History Museum, University of Oslo, accession number O-A-10002. Figure 4B shows cell from the thin sections. Culture deposited in the Roscoff Culture Collection as RCC15.
Type locality: RCC15 was isolated in 1965 from a sample collected during the Trident cruise 26 in the Gulf Stream, North East Atlantic.
Etymology: The first species of the genus to be isolated into culture and have its 18S rRNA gene sequence published.
Chloropicon roscoffensis Lopes dos Santos and Eikrem sp. nov.
Diagnosis: With characters of genus. Additional characters; loroxanthin absent, combined nucleotide sequences of nuclear 18S rRNA (KF899840), rRNA ITS (MF077510) and plastid 16S rRNA (LN735295) are species specific.
Holotype: Cells embedded in resin block deposited at the Natural History Museum, University of Oslo accession number O-A-10003. Figure 4H shows a cell from the resin block. Culture deposited in the Roscoff Culture Collection as RCC1871.
Type locality: RCC1871 was isolated from the English Channel off Roscoff (48° 45′ N, 3° 57′ W).
Etymology: From the type locality.
Chloropicon mariensis Eikrem, Lopes dos Santos sp. nov.
Diagnosis: With characters of genus. Additional characters; combined nucleotide sequences of nuclear 18S rRNA (KF422632), rRNA ITS (MF077504) and plastid 16S rRNA (LN735516) are species specific.
Holotype: Cells embedded in resin block deposited at the Natural History Museum, University of Oslo, accession number O-A-10004. Figure 4F shows cell from the resin block. Culture deposited in the Roscoff Culture Collection as RCC998.
Type locality: RCC998 was isolated from water sampled at 100 m depth in the South Pacific Ocean (9° 04′ S, 136° 59′ W).
Etymology: Named in recognition of Dominique Marie who isolated the culture and his efforts in picoplankton research.
Chloropicon laureae Lopes dos Santos and Eikrem sp. nov.
Diagnosis: With characters of genus. Additional characters; combined nucleotide sequences of nuclear 18S rRNA (KF422631), rRNA ITS (MF077480) and plastid 16S rRNA (LN735470) are species specific.
Holotype: Cells embedded in resin block deposited at the Natural History Museum, University of Oslo, accession number O-A-10005. Figure 4D shows cell from the resin block. Culture deposited in the Roscoff Culture Collection as RCC856.
Type locality: RCC856 was isolated from water sampled at 10 m depth in the South Pacific Ocean off Marquesas Islands (8° 20′ S, 141° 15′ W).
Etymology: Named after Laure Guillou who first distinguished the prasinophyte clades, including clade VII.
Chloropicon maureeniae Lopes dos Santos and Eikrem sp. nov.
Diagnosis: With characters of genus. Additional characters; combined nucleotide sequences of nuclear 18S rRNA (KU843595), rRNA ITS (MF077515) and, plastid 16S rRNA (KU843568) are species specific.
Holotype: Cells embedded in resin block deposited at the Natural History Museum, University of Oslo, accession number O-A-10006. Figure 4J shows cell from the resin block. Culture deposited in the Roscoff Culture Collection as RCC3374.
Type locality: RCC3374 (CCMP2152) was isolated from the North Pacific Ocean off Hawaii (22° 45′ N, 158° 00′ W).
Etymology: Named in recognition of Maureen Keller who developed K medium that has facilitated the isolation of oceanic species into culture.
Chloroparvula Lopes dos Santos, Noël and Eikrem gen. nov.
Diagnosis: With characters of the family. Additional characters; cell wall thick and smooth or sometimes with fibrils, string like ornamentation.
Etymology: Named for its green color and small size.
Type species: Chloroparvula pacifica
Chloroparvula pacifica Lopes dos Santos, Noël and Eikrem sp. nov.
Diagnosis: With characters of genus with loroxanthin absent. Combined nucleotide sequences of nuclear 18S rRNA (KU843574), rRNA ITS (MF077486) and plastid 16S rRNA (KU843560) are species specific.
Holotype: Cells embedded in resin block deposited at the Natural History Museum, University of Oslo, accession number O-A-10007. Figure 3D shows cells from the resin block. Original culture deposited in NIES Microbial Culture Collection as NIES-3669; sub-culture deposited in Roscoff Culture Collection as RCC4656.
Type locality: NIES-3669 (RCC4656) was isolated from a surface water sample collected from the North Pacific Ocean off Japan (42°16′ N, 145°07′ E).
Etymology: Named for its abundance in the Pacific Ocean.
Chloroparvula japonica Lopes dos Santos, Noël and Eikrem sp. nov.
Diagnosis: With characters of genus. Additional characters; combined nucleotide sequences of nuclear 18S rRNA (KF422628), rRNA ITS (MF077482) and plastid 16S rRNA (LN735350) are species specific.
Holotype: Cells embedded in resin block deposited at the Natural History Museum, University of Oslo, accession number O-A-10008. Figure 3F shows cell from the resin block. Original culture deposited in NIES Microbial Culture Collection as NIES-2758; sub-culture deposited in Roscoff Culture Collection as RCC2339.
Type locality: NIES-2758 (RCC2339) was isolated in surface from the North Pacific Ocean off the coast of Japan (33° 46′ N, 129° 41′ E).
Etymology: Named for the origin of the authentic culture off the coast of Japan.
Picocystophyceae Eikrem and Lopes dos Santos classis nov.
Diagnosis: Green coccoid cells with chlorophylls a and b. Layered cell wall containing polyarabinose, mannose, galactose and glucose. Chloroplast surrounded by two membranes and containing starch grain.
Picocystales Eikrem and Lopes dos Santos order nov.
Diagnosis: With characters of class. Additional characters; accessory pigments are alloxanthin, diatoxanthin, monadoxanthin, chlorophyll b, neoxanthin, lutein and β,β- carotene.
Picocystaceae Eikrem and Lopes dos Santos fam. nov.
Diagnosis: With characters of order. Additional characters; coccoid cells contain green chloroplasts with starch grain.
_Picocysti_s R. A Lewin. Characters of type species Picocystis salinarum.
Picocystis salinarum R.A. Lewin emend. Eikrem and Lopes dos Santos.
Diagnosis: Cells measuring 2-3 µm with 1-2 chloroplasts, a mitochondrion and dictyosome. Combined nucleotide sequences of nuclear 18S rRNA (FR865649), rRNA ITS (HE610138, MF077484) and plastid 16S rRNA (AB491631) are species specific.Paratype: Cells embedded in resin block deposited at the Natural History Museum, University of Oslo, accession number O-A-10009. Supplement figure 2A shows cell from embedding. Original Culture CCMP1897 deposited in Roscoff Culture Collection as RCC3402.Type locality: Pacific ocean (37°47′N, 122°21′W).
Availability of materials and data
All material including data, figures and tables are available from: 10.6084/m9.figshare.5027375.
Electronic supplementary material
Acknowledgements
We are thankful to Dr. Ian Probert, Jahn Throndsen and Øjvind Moestrup for a critical reading of the manuscript and providing invaluable help for the taxonomic treatment. We thank Dr. Matthias Wolf for advice on the use of 4SALE software, Dr. Nathalie Simon for helpful discussions on ITS2 secondary structures and the Roscoff ABIMS platform for access to bioinformatics resources. Financial support for this work was provided by the European Union program MaCuMBA (FP7-KBBE-2012–6–311975) and FONDECYT regular PiSCOSouth (1171802). This work is a contribution of Microalgas Nocivas (IEO) and Unidad Asociada al IIM (CSIC).
Author Contributions
Lopes dos Santos, A. Vaulot, D. and Eikrem, W. conceived the study. Gourvil, P. and Noël, M.-H. isolated and cultivated some of the strains used. Lopes dos Santos, A. and Gourvil P. obtained the sequences. Pollina, T., Corre, E. and Vaulot, D. analyzed the MMETSP dataset and performed multigene phylogenetic analyses. Lopes dos Santos, A. and Marie, D. carried out the genome size analyses. Lopes dos Santos, Pollina, T., Noël, M.-H. and Eikrem, W. obtained the morphological data. Rodríguez, F. and Garrido, J.L. performed the pigment analyzes. Lopes dos Santos, A., Eikrem, W. and Vaulot, D. wrote the manuscript. All authors discussed data, read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
4/17/2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-12412-5.
References
- 1.Tragin ML, dos Santos A, Christen R, Vaulot D. Diversity and ecology of green microalgae in marine systems: an overview based on 18S rRNA gene sequences. Perspect. Phycol. 2016;3:141–154. [Google Scholar]
- 2.Leliaert F, et al. Phylogeny and molecular evolution of the green algae. CRC. Crit. Rev. Plant Sci. 2012;31:1–46. [Google Scholar]
- 3.Leliaert F, Verbruggen H, Zechman FW. Into the deep: New discoveries at the base of the green plant phylogeny. BioEssays. 2011;33:683–692. doi: 10.1002/bies.201100035. [DOI] [PubMed] [Google Scholar]
- 4.Guillou L, et al. Diversity of picoplanktonic prasinophytes assessed by direct nuclear SSU rDNA sequencing of environmental samples and novel isolates retrieved from oceanic and coastal marine ecosystems. Protist. 2004;155:193–214. doi: 10.1078/143446104774199592. [DOI] [PubMed] [Google Scholar]
- 5.Lewin RA, Krienltz L, Oerickei RG, Takeda H, Hepperle D. Picocystis salinarum gen. et sp. nov. (Chlorophyta) - a new picoplanktonic green alga. Phycologia. 2000;39:560–565. [Google Scholar]
- 6.Roesler CS, et al. Distribution, production, and ecophysiology of Picocystis strain ML in Mono Lake, California. Limnol. Oceanogr. 2002;47:440–452. [Google Scholar]
- 7.Krienitz L, Bock C, Kotut K, Luo W. Picocystis salinarum (Chlorophyta) in saline lakes and hot springs of East Africa. Phycologia. 2012;51:22–32. [Google Scholar]
- 8.Lopes dos Santos A, Gourvil P, Rodriguez-Hernandez F, Garrido JL, Vaulot D. Photosynthetic pigments of oceanic Chlorophyta belonging to prasinophytes clade VII. J. Phycol. 2016;52:148–155. doi: 10.1111/jpy.12376. [DOI] [PubMed] [Google Scholar]
- 9.Lopes dos Santos A, et al. Diversity and oceanic distribution of prasinophytes clade VII, the dominant group of green algae in oceanic waters. ISME J. 2017;11:512–528. doi: 10.1038/ismej.2016.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marin B, Melkonian M. Molecular phylogeny and classification of the Mamiellophyceae class. nov. (Chlorophyta) based on sequence comparisons of the nuclear- and plastid-encoded rRNA Operons. Protist. 2010;161:304–336. doi: 10.1016/j.protis.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Marin B. Nested in the Chlorellales or independent class? Phylogeny and classification of the Pedinophyceae (Viridiplantae) revealed by molecular phylogenetic analyses of complete nuclear and plastid-encoded rRNA operons. Protist. 2012;163:778–805. doi: 10.1016/j.protis.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Lemieux C, Otis C, Turmel M. Six newly sequenced chloroplast genomes from prasinophyte green algae provide insights into the relationships among prasinophyte lineages and the diversity of streamlined genome architecture in picoplanktonic species. BMC Genomics. 2014;15:857. doi: 10.1186/1471-2164-15-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moon-van der Staay SY, De Wachter R, Vaulot D. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature. 2001;409:607–610. doi: 10.1038/35054541. [DOI] [PubMed] [Google Scholar]
- 14.Shi, X. L., Marie, D., Jardillier, L., Scanlan, D. J. & Vaulot, D. Groups without cultured representatives dominate eukaryotic picophytoplankton in the oligotrophic South East Pacific Ocean. PLoS One4 (2009). [DOI] [PMC free article] [PubMed]
- 15.Viprey M, Guillou L, Ferréol M, Vaulot D. Wide genetic diversity of picoplanktonic green algae (Chloroplastida) in the Mediterranean Sea uncovered by a phylum-biased PCR approach. Environ. Microbiol. 2008;10:1804–1822. doi: 10.1111/j.1462-2920.2008.01602.x. [DOI] [PubMed] [Google Scholar]
- 16.Wu W, Huang B, Liao Y, Sun P. Picoeukaryotic diversity and distribution in the subtropical-tropical South China Sea. FEMS Microbiol. Ecol. 2014;89:563–579. doi: 10.1111/1574-6941.12357. [DOI] [PubMed] [Google Scholar]
- 17.Romari K, Vaulot D. Composition and temporal variability of picoeukaryote communities at a coastal site of the English Channel from 18S rDNA sequences. Limnol. Oceanogr. 2004;49:784–798. [Google Scholar]
- 18.Potter D, Lajeunesse TC, Saunders GW, Andersen RA. Convergent evolution masks extensive biodiversity among marine coccoid picoplankton. Biodivers. Conserv. 1997;6:99–107. [Google Scholar]
- 19.Leliaert F, et al. DNA-based species delimitation in algae DNA-based species delimitation in algae. Eur. J. Phycol. 2014;49:179–196. [Google Scholar]
- 20.Mai JC, Coleman AW. The internal transcribed spacer 2 exhibits a common secondary structure in green algae and flowering plants. J. Mol. Evol. 1997;44:258–271. doi: 10.1007/pl00006143. [DOI] [PubMed] [Google Scholar]
- 21.Coleman AW. The significance of a coincidence between evolutionary landmarks found in mating affinity and a Dna sequence. Protist. 2000;151:1–9. doi: 10.1078/1434-4610-00002. [DOI] [PubMed] [Google Scholar]
- 22.Coleman AW. Is there a molecular key to the level of ‘biological species’ in eukaryotes? A DNA guide. Mol. Phylogenet. Evol. 2009;50:197–203. doi: 10.1016/j.ympev.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 23.Joseph N, Krauskopf E, Vera MI, Michot B. Ribosomal internal transcribed spacer 2 (ITS2) exhibits a common core of secondary structure in vertebrates and yeast. Nucleic Acids Res. 1999;27:4533–4540. doi: 10.1093/nar/27.23.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pöll, G. et al. rRNA maturation in yeast cells depleted of large ribosomal subunit proteins. PLoS One4 (2009). [DOI] [PMC free article] [PubMed]
- 25.Schillewaert S, Wacheul L, Lhomme F, Lafontaine DLJ. The Evolutionarily Conserved Protein LAS1 Is Required for Pre-rRNA Processing at Both Ends of ITS2. Mol. Cell. Biol. 2012;32:430–444. doi: 10.1128/MCB.06019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller T, Philippi N, Dandekar T, Schultz J, Wolf M. Distinguishing species. RNA. 2007;13:1469–1472. doi: 10.1261/rna.617107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guillard RRL, Hargraves PE. Stichochrysis immobilis is a diatom, not a chrysophyte. Phycologia. 1993;32:234–236. [Google Scholar]
- 28.Okaichi, T., Nishio, S. & Imatomi, Y. In Toxic phytoplankton - occurrence, mode of action and toxins (ed. Japanese Fisheries Society, T.)23–34 (1982).
- 29.Zapata M, Rodriguez F, Garrido JL. Separation of chlorophylls and carotenoids from marine phytoplankton: a new HPLC method using a reversed phase C-8 column and pyridine-containing mobile phases. Mar. Ecol. - Prog. Ser. 2000;195:29–45. [Google Scholar]
- 30.Garrido JL, Rodríguez F, Zapata M. Occurrence of loroxanthin, loroxanthin decenoate, and loroxanthin dodecenoate in Tetraselmis species (Prasinophyceae, Chlorophyta) J. Phycol. 2009;45:366–374. doi: 10.1111/j.1529-8817.2009.00660.x. [DOI] [PubMed] [Google Scholar]
- 31.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marie, D., Simon, N., Guillou, L., Partensky, F. & Vaulot, D. DNA/RNA analysis of phytoplankton by flow cytometry. Curr. Protoc. Cytom. Chapter 11, Unit11.12 (2001). [DOI] [PubMed]
- 33.Lepère C, et al. Whole Genome Amplification (WGA) of marine photosynthetic eukaryote populations. FEMS Microbiol. Ecol. 2011;76:516–523. doi: 10.1111/j.1574-6941.2011.01072.x. [DOI] [PubMed] [Google Scholar]
- 34.White, T. J., Bruns, T., Lee, S. & Taylor, J. In PCR protocols: a guide to methods and applications. 315–322 (Academic Press, Orlando, Florida, 1990).
- 35.West NJ, et al. Closely related Prochlorococcus genotypes show remarkably different depth distributions in two oceanic regions as revealed by in situ hybridization using 16S rRNA-targeted oligonucleotides. Microbiology. 2001;147:1731–1744. doi: 10.1099/00221287-147-7-1731. [DOI] [PubMed] [Google Scholar]
- 36.Fuller NJ, et al. Analysis of photosynthetic picoeukaryote diversity at open ocean sites in the Arabian Sea using a PCR biased towards marine algal plastids. Aquat. Microb. Ecol. 2006;43:79–93. [Google Scholar]
- 37.Fuller NJ, et al. Molecular analysis of photosynthetic picoeukaryote community structure along an Arabian Sea transect. Limnol. Oceanogr. 2006;51:2502–2514. [Google Scholar]
- 38.Keller A, et al. 5.8S–28S rRNA interaction and HMM-based ITS2 annotation. Gene. 2009;430:50–57. doi: 10.1016/j.gene.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 39.Ankenbrand MJ, Keller A, Wolf M, Schultz J, Föster F. ITS2 database V: Twice as much. Mol. Biol. Evol. 2015;32:3030–3032. doi: 10.1093/molbev/msv174. [DOI] [PubMed] [Google Scholar]
- 40.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coleman AW. ITS2 is a double-edged tool for eukaryote evolutionary comparisons. Trends Genet. 2003;19:370–375. doi: 10.1016/S0168-9525(03)00118-5. [DOI] [PubMed] [Google Scholar]
- 42.Coleman AW. Pan-eukaryote ITS2 homologies revealed by RNA secondary structure. Nucleic Acids Res. 2007;35:3322–3329. doi: 10.1093/nar/gkm233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seibel PN, Müller T, Dandekar T, Schultz J, Wolf M. 4SALE–a tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinformatics. 2006;7:498. doi: 10.1186/1471-2105-7-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seibel P, Müller T, Dandekar T, Wolf M. Synchronous visual analysis and editing of RNA sequence and secondary structure alignments using 4SALE. BMC Res. Notes. 2008;1:91. doi: 10.1186/1756-0500-1-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caisová L, Marin B, Melkonian M. A consensus secondary structure of ITS2 in the Chlorophyta identified by phylogenetic reconstruction. Protist. 2013;164:482–496. doi: 10.1016/j.protis.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 46.Kearse M, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guindon S, Dufayard J-F, Lefort V, Anisimova M. New algorithms and methods to estimate maximum- likelihoods phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 50.Ronquist F, Huelsenbeck JP. MrBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 51.Keeling PJ, et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014;12:e1001889. doi: 10.1371/journal.pbio.1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmieder R, Lim YW, Edwards R. Identification and removal of ribosomal RNA sequences from metatranscriptomes. Bioinformatics. 2012;28:433–435. doi: 10.1093/bioinformatics/btr669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pruesse E, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 58.Goff SA, et al. The iPlant collaborative: cyberinfrastructure for plant biology. Plant Genet. Genomics. 2011;2:34. doi: 10.3389/fpls.2011.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tatusov RL, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 61.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 63.Derelle E, et al. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. USA. 2006;103:11647–11652. doi: 10.1073/pnas.0604795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Worden AZ, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- 65.Moreau H, et al. Gene functionalities and genome structure in Bathycoccus prasinos reflect cellular specializations at the base of the green lineage. Genome Biol. 2012;13:R74. doi: 10.1186/gb-2012-13-8-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Subirana L, et al. Morphology, genome plasticity, and phylogeny in the genus Ostreococcus reveal a cryptic species, O. mediterraneus sp. nov. (Mamiellales, Mamiellophyceae) Protist. 2013;164:643–659. doi: 10.1016/j.protis.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 67.Wang B, Zarka A, Trebst A, Boussiba S. Astaxanthin accumulation in Haematococcus pluvialis (Chlorophyceae) as an active photoprotective process under high irradiance. J. Phycol. 2003;39:1116–1124. [Google Scholar]
- 68.Jeffrey, S. W., Wright, S. W. & Zapata, M. In Phytoplankton Pigments: Characterization, Chemotaxonomy and Applications in Oceanography (ed. Roy S, Egeland ES, Johnsen G, L. C.) 3–77 (Cambridge University Press, Cambridge., 2011).
- 69.Rodríguez F, et al. Ecotype diversity in the marine picoeukaryote Ostreococcus (Chlorophyta, Prasinophyceae) Environ. Microbiol. 2005;7:853–859. doi: 10.1111/j.1462-2920.2005.00758.x. [DOI] [PubMed] [Google Scholar]
- 70.Latasa M, Scharek R, Le Gall F, Guillou L. Pigment suites and taxonomic groups in Prasinophyceae. J. Phycol. 2004;40:1149–1155. [Google Scholar]
- 71.Takaichi S. Carotenoids in Algae: Distributions, biosyntheses and functions of carotenoids in algae. Mar. Drugs. 2011;9:1101–1118. doi: 10.3390/md9061101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Crespo C, Rodríguez H, Segade P, Iglesias R, García-Estévez JM. Coccomyxa sp. (Chlorophyta: Chlorococcales), a new pathogen in mussels (Mytilus galloprovincialis) of Vigo estuary (Galicia, NW Spain) J. Invertebr. Pathol. 2009;102:214–219. doi: 10.1016/j.jip.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 73.Jahns P, Latowski D, Strzalka K. Mechanism and regulation of the violaxanthin cycle: The role of antenna proteins and membrane lipids. Biochim. Biophys. Acta - Bioenerg. 2009;1787:3–14. doi: 10.1016/j.bbabio.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 74.Lemieux C, Otis C, Turmel M. Chloroplast phylogenomic analysis resolves deep-level relationships within the green algal class Trebouxiophyceae. BMC Evol. Biol. 2014;14:211. doi: 10.1186/s12862-014-0211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leliaert F, et al. Chloroplast phylogenomic analyses reveal the deepest-branching lineage of the Chlorophyta, Palmophyllophyceae class. nov. Sci. Rep. 2016;6:25367. doi: 10.1038/srep25367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marie D, Le Gall F, Edern R, Gourvil P, Vaulot D. Improvement of phytoplankton culture isolation using single cell sorting by flow cytometry. J. Phycol. 2017;53:271–282. doi: 10.1111/jpy.12495. [DOI] [PubMed] [Google Scholar]
- 77.Simon, N. et al. Revision of the genus Micromonas (Manton et Parke) (Chlorophyta, Mamiellophyceae), of the type species M. pusilla (Butcher) Manton & Parke and of the species M. commoda (van Baren, Bacry and Worden) and description of two new species based on the genetic and phenotypic characterization of cultured isolates. Protist in press (2017). [DOI] [PubMed]
- 78.Schultz J, Maisel S, Gerlach D, Müller T, Wolf M. A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA. 2005;11:361–364. doi: 10.1261/rna.7204505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Caisová L, Marin B, Melkonian M. A close-up view on ITS2 evolution and speciation - a case study in the Ulvophyceae (Chlorophyta, Viridiplantae) BMC Evol. Biol. 2011;11:262. doi: 10.1186/1471-2148-11-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kaczmarska I, Mather L, Aluddington I, Muise F, Ehrman JM. Cryptic diversity in a cosmopolitan diatom known as Asterionellopsis glacialis (Fragilariaceae): Implications for ecology, biogeography, and taxonomy. Am. J. Bot. 2014;101:267–286. doi: 10.3732/ajb.1300306. [DOI] [PubMed] [Google Scholar]
- 81.Nakayama T, et al. The basal position of scaly green flagellates among the green algae (Chlorophyta) is revealed by analyses of nuclear-encoded SSU rRNA sequences. Protist. 1998;149:367–80. doi: 10.1016/S1434-4610(98)70043-4. [DOI] [PubMed] [Google Scholar]
- 82.Massjuk NP. Chlorodendrophyceae class. nov. (Chlorophyta, Viridiplantae) in the Ukrainian flora: I. The volume, phylogenetic relations and taxonomical status. Ukr. Bot. J. 2006;63:601–6014. [Google Scholar]
- 83.Guillard RRL, Keller MD, O’Kelly CJ, Floyd GL. Pycnococcus provasolii gen. et sp. nov., a coccoid prasinoxanthin containing phytoplankter from the western North Atlantic and Gulf of Mexico. J. Phycol. 1991;27:39–47. [Google Scholar]
- 84.Jouenne F, Eikrem W, Le Gall F, Johnsen G, Vaulot D. Prasinoderma singularis sp. nov., a solitary coccoid prasinophyte from the South East Pacific Ocean. Protist. 2011;162:70–84. doi: 10.1016/j.protis.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 85.Fawley MW, Qin M, Yun Y. The relationship between Pseudoscourfieldia marina and Pycnococcus provasolii (Prasinophyceae, Chlorophyta): Evidence from 18S rDNA sequence data. J. Phycol. 1999;35:838–843. [Google Scholar]
- 86.Henley WJ, et al. Phylogenetic analysis of the ‘Nannochloris-like’ algae and diagnoses of Picochlorum oklahomensis gen. et sp nov (Trebouxiophyceae, Chlorophyta) Phycologia. 2004;43:641–652. [Google Scholar]
- 87.Balzano S, et al. Morphological and genetic diversity of Beaufort Sea diatoms with high contributions from the Chaetoceros neogracilis species complex. J. Phycol. 2017;53:161–187. doi: 10.1111/jpy.12489. [DOI] [PubMed] [Google Scholar]
- 88.Percopo I, et al. Pseudo-nitzschia arctica sp. nov., a new cold-water cryptic Pseudo-nitzschia species within the P. pseudodelicatissima complex. J. Phycol. 2016;52:184–199. doi: 10.1111/jpy.12395. [DOI] [PubMed] [Google Scholar]
- 89.Rodríguez-Martínez R, Rocap G, Logares R, Romac S, Massana R. Low evolutionary diversification in a widespread and abundant uncultured protist (MAST-4) Mol. Biol. Evol. 2012;29:1393–1406. doi: 10.1093/molbev/msr303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.