Molecular phylogenetic analyses of the mitochondrial ADP-ATP carriers: The Plantae/Fungi/Metazoa trichotomy revisited (original) (raw)
Abstract
We investigated the basal phylogeny of eukaryotes through analyses of sequences from the ADP–ATP mitochondrial carrier, a transmembrane protein that is stable in function across eukaryote kingdoms. The ADP–ATP data strongly suggest the grouping of Plantae and Fungi to the exclusion of Metazoa. We implemented several procedures to avoid pervasive analytical artifacts such as erroneous alignment, random rooting, long branch attraction, and misidentification of noisy characters. The quest of an eukaryote tree that would be largely consistent across multiple loci might be essentially illusory because of differential lineage sorting, horizontal gene transfer, and the chimeric nature of early eukaryotes. Better understanding of these evolutionary parameters, requiring separate phylogenetic analyses of multiple independent loci, is fundamental for resolution of the modes of emergence and evolution of the major eukaryote lineages.
Keywords: eukaryote crown, AAC transporters, multigene family, phylogeny
Phylogenetic relationship among Plantae, Metazoa, and Fungi is generally considered solved, as an exclusive animal–fungal monophyletic group is widely accepted. Indeed, in an attempt to compare and combine molecular data, several authors have phylogenetically analyzed amino acid sequences from multiple proteins and concluded that the Metazoa/Fungi sister relationship is the best supported hypothesis (e.g., ref. 1 and refs. therein). However, several of these analyses reveal striking conflicts among gene trees (e.g., ref. 2). Furthermore, several of these analyses are problematic, as (i) the statistical supports under reasonable models of evolution (i.e., adapted to the investigation of very deep divergences) were usually weak or absent, and (ii) a priori assumptions used for rooting the tree have not been tested extensively (as discussed in ref. 3). The latter point is of paramount importance, as defining the phylogenetic relationships among Fungi, Metazoa, and Plantae boils down to rooting the eukaryote tree. Unfortunately, the large divergences between eukaryotes and unambiguous outgroups (i.e., prokaryotes) favor “random rooting” (4, 5), a phenomenon related to the very classical and well-described “long branch attraction” artifact (4, 6–9). Keeling and Doolittle (10) attempted to avoid the use of prokaryote sequences (hence, potentially, random rooting) by analyzing paralogous genes (α, β, and γ tubulins). Indeed, different genes that originated through duplication events of an ancestral sequence actually root each other at the nodes corresponding to the duplication events. Unfortunately, the three inferred gene subtrees yielded inconsistent results. The recent analysis of Baldauf et al. (1) is of particular interest, because it incorporates multiple protein sequences from representatives of all major eukaryote groups and confirms the existence of conflict among gene trees, although the authors concluded that, overall, there was support for a [Fungi + Metazoa] clade.
Conflict Among Gene Trees.
It is, in fact, quite unreasonable to expect that phylogenetic analyses of multiple genes will provide consistent results for a coherent and easily defined topology of the eukaryote phylogenetic tree, because many specific parameters will cause analytical and resolution problems. First, the deepest eukaryote nodes are very old, constraining phylogeneticists to use slowly evolving genes to avoid saturation of substitutions, i.e., erosion of the phylogenetic signal (11, 12). Furthermore, the eukaryote crown likely consists of a major explosive radiation (e.g., ref. 3) of multiple lineages (of which Plantae, Metazoa, and Fungi represent only a small portion), which defines a so-called “Felsenstein's zone,” i.e., a succession of very short internal branches followed by very long edges. The presence of a Felsenstein's zone is indicative of an extremely difficult phylogenetic endeavor, as the short internal branches are unlikely to bear many informative changes that, in turn, are likely to be erased on the very long terminal edges.
Second, the rapid succession of nodes in a phylogeny is prone to yield multiple conflicting true gene trees. Indeed, although a nonrecombining piece of DNA will have a strictly nonreticulated bifurcating history, that true gene tree can conflict with the phylogeny of another (unlinked) nonrecombining piece of DNA (Fig. 1). This process of “stochastic lineage sorting” (13–16) is caused by the fact that each allelic lineage has a non-null probability to go extinct and can cause multiple gene trees to conflict in their branching patterns, although each of them indicates the correct historical relationships with respect to the corresponding gene (e.g., refs. 17–19).
Figure 1.
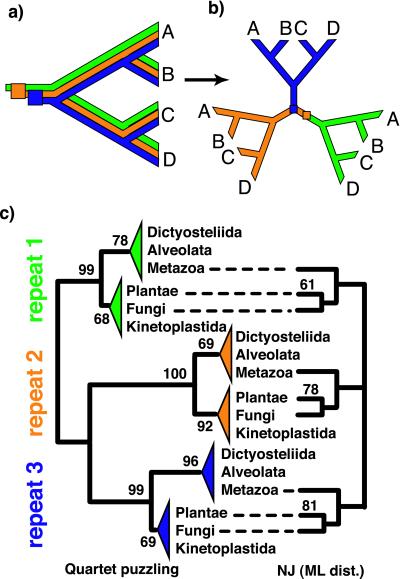
Schematic representation of phenomena that can obscure phylogeny inference from multiple loci (colored lines) that originated from duplication events (colored squares). (a) Green, the copy of that locus became extinct in the lineage of species B; orange, extinction within the lineage of species A was followed by lateral transfer of an homologous copy from the lineage of species B; blue, polymorphism was maintained through two bifurcation events and followed by stochastic extinction of allelic lineages such that sequences from that locus define a true tree (with respect to the phylogeny of this gene copy), in which A and C form a clade to the exclusion of B; black, none of the phenomena described above applied to that copy whose phylogeny is consistent with the species phylogeny (dotted outline). (b) True phylogeny that can be inferred from a complete analysis of the four loci present in the three extant species A, B, and C.
Third, eukaryotes are chimeric organisms. Indeed, whereas mitochondria and chloroplasts of eukaryotes are the result of primary endosymbiosis of prokaryotic cells, it is likely that different major eukaryote lineages have independently experienced secondary and tertiary endosymbiosis, i.e., eukaryotes incorporated other eukaryotes (20, 21). These multiple endosymbiotic events were followed by both drastic reduction in plastid genome complexity and transfer of genes from organelles to the nucleus. This well-accepted pattern of endosymbiosis might just be the tip of a lateral gene transfer (LGT) iceberg. As recently reviewed and articulated by Doolittle (22, 23), there is a growing recognition of the evolutionary importance of LGT in prokaryotes. Molecular data available at the time the present paper was written are probably insufficient for proper investigation of the same issue in eukaryotes. As the full genomes of representatives of several major eukaryote lineages have recently been, or will soon be, made publicly available (24), one can hope that these sequencing efforts will allow estimation of the extent and significance of LGT in early eukaryote evolution and whether it blurs phylogenetic relationships among the genomes of modern Plantae, Metazoa, Fungi, and Protista.
In short, the quest for a eukaryote tree that would be largely consistent across multiple loci might be essentially illusory, because (i) differential lineage sorting and horizontal gene transfer might have been extensive enough to yield a large proportion of conflict among true gene trees (Fig. 1), and (ii) the old age and “explosive” nature of early eukaryote evolution make it potentially difficult to establish with confidence the topology of each individual true gene tree.
ADP–ATP Carrier (AAC) Transporters and the Phylogeny of Eukaryotes.
The membrane transport proteins are phenetically classified into more than 150 different families of genes (25). The mitochondrial carrier family (MCF) forms a compact group of more than 200 sequenced members. All of the known MCF members are at work in eukaryotic organelles, even though they are nuclearly encoded. Most MCF proteins exchange substrates through the mitochondrial (mt) membrane (25, 26) and participate in the massive traffic of metabolites associated with respiration. MCF proteins are around 300 residues in length and share a common structure consisting of six transmembrane α-helical spanners that define three domains of transmembrane–loop–transmembrane form. The three domains are bridged in a row and are highly significantly similar, indicating they originated through duplication events (25, 27).
We report here extensive phylogenetic analyses of all available protein sequences from one member of the MCF: the AAC. This protein is stable and essential in function across eukaryote kingdoms. First, to avoid artifactual results because of ambiguities in alignment—homology assessment is typically locally ambiguous even among reasonably similar sequences, especially in sections of the alignment where positions are potentially informative for deep nodes—we used an approach consisting of removing the amino acid positions that are most unstable to variations of the heuristic alignment parameters (28, 29). Second, we used two rooting procedures that are independent of each other and do not require the use of noneukaryote taxa. We rooted the AAC tree by using outgroup sequences from the carnitine–acyl–carnitine carriers, the tissue differentiation protein, the mt phosphate carrier, the mt RNA splicing protein, the oxoglutarate–malate carrier, and the uncoupling protein. In our analyses, the likelihood of random rooting is lower than in the analyses of tubulin sequences, because we analyzed members of a gene family that could be more recent because they are shared by the mitochondriate eukaryotes only. Furthermore, as AAC carriers are proteins consisting of three domains repeated in tandem, we introduce an original rooting procedure making use of these duplication events. Because these are internal repeats, the likelihood that recombination event(s) might be the source of different phylogenies across repeats (because LGT or differential sorting) is much lower than for genetically unlinked copies (e.g., different members of a multigene family). Third, we tested the stability of our phylogenetic results to sequential removal of maximum likelihood (ML) γ rate categories of sites, i.e., the successive exclusion of fast evolving, hence potentially noisy amino acid sites (see Materials and Methods). Finally, we discuss the impact of removing unstable aligned positions and γ rate categories of sites in the 18S ribosomal RNA, a gene previously reported as supporting a [Fungi + Metazoa] clade.
Materials and Methods
Data.
The AAC sequences were searched in nonredundant DNA GenBank with the program tblastn (30) with a broad set of AAC query sequences from ref. 25. All hits characterized by a higher similarity than known AAC paralogous genes were interpreted as AAC orthologs. In other words, all AAC sequences available at the time this analysis was initiated were included. The resulting number of AAC sequences was impractically high for phylogenetic analyses, and redundant sequences were removed with the following algorithm: all possible pairwise alignments among ingroup sequences were performed with clustalw (31) with default settings. The corresponding pairwise ML distances were calculated by using protdist from the phylip package (32), with the Dayhoff scoring matrix. When a pairwise alignment yielded a distance < 0.01, one of the two sequences was randomly excluded. All sequences with a high proportion of missing data and a distance < 0.03 from any other sequence were also excluded. Thirty-seven AAC sequences were kept after the full procedure. After performing an initial multiple alignment of these 37 sequences, none had amino acid frequencies significantly different from the frequency distribution of a ML model [χ2 test in the program puzzle (33)], indicating that artifactual inference of relationships caused by differences in nucleotide compositions is unlikely. Candidate outgroup sequences—from each of the 11 other MCF subfamilies—were similarly searched, and a subsample of 34 sequences was chosen to maximize both the number of mitochondrial carrier (MC) subfamilies represented and taxonomic diversity. The outgroup and ingroup were combined, giving an initial alignment of 71 sequences.
Alignment.
First, an alignment stability test was performed with the 37 AAC ingroup sequences. A guide tree for the multiple alignment was produced with clustalw by using default protein settings. By using the same guide tree, the alignment procedure was repeated 35 times with different sets of alignment parameters (gap opening penalties from 6 to 14 by steps of 2, and extension penalties from 0.02 to 0.14 by steps of 0.02). We then calculated the strict consensus among these alignments and excluded positions at which they differed (28). The whole procedure of generating/comparing alignments and producing the consensus was performed by using the program soap (29). Second, for each of various combinations of outgroup taxa, we performed a few alignments by using different gap opening/extension penalties to quickly identify, hence exclude, the outgroup sequences bringing marked instability in the alignments. The remaining outgroup sequences were combined with the 37 ingroup sequences into a full alignment stability test (soap; opening penalties 8–14, and extension penalties 0.04–0.08). Third, a single representative AAC sequence from each kingdom (human, AC004000; Arabidopsis, AL021749; yeast, AL023634) was aligned against itself with dotplot [(Wisconsin Package (34)] to roughly demarcate the positions of the three internal repeat units. For each of the representative sequences, the three repeats were pairwise-aligned with the local alignment program bestfit (Wisconsin Package) for more precisely defining their edges. The three internal repeat fragments were then identified in each of the 37 ingroup sequences by extrapolating, in the full length ingroup alignment, the edges identified in the three representative sequences. All of the internal fragments were then piled up in a multiple alignment after masking by gap signs the sites previously found unstable in the stability analysis involving the full length ingroup sequences. This 3 × 37 sequence alignment was then subjected to an alignment stability test (soap, opening penalties 8–14, and extension penalties 0.04–0.10).
Phylogenetic Analyses.
Phylogenetic analyses were first performed on two different data sets: (i) “MC”, i.e., the alignment of the 37 AAC full sequences rooted by the eleven MC sequences, and (ii) “REPEAT”, i.e., the alignment of the 3 × 37 AAC internal repeat units. In some cases (when computation burden was impractically high), the REPEAT data set was reduced to 3 × 13 sequences (= “REPEAT_reduced”). All data sets were analyzed with ML methods by using the programs protml [molphy-package (35)] and puzzle (33). The stability of nodes was tested by bootstrapping (36) by using the protein distance methods in the phylip package. The quartet-puzzling trees [with JTT substitution model (37)] were also produced, and searches were performed both with uniform substitution rate and with rates following a γ distribution (four rate categories). We also tested the stability of the nodes to exclusion/inclusion of character columns containing gaps. protml searches were performed on the MC and REPEAT_reduced data sets as follows: by using the “quickadd” algorithm (with JTT model and amino acid frequencies estimated from the data), the 3,000 tree topologies with the best approximate likelihoods were saved. Branch lengths were fully optimized for each of these topologies, which were then reordered according to their exact likelihoods. To perform exhaustive searches on the MC data set, we first reduced search space by constraining monophyly of Plantae, of Metazoa, and of Fungi, as well as—for vertebrates, nematodes, insects, Plantae, and Fungi—the nodes well supported in the heuristic ML searches (bold branches in Fig. 2). We then produced a reduced data set (“MC44”; 33 ingroup and 11 outgroup sequences) by removing the four taxa (Drosophila pseudoobscura, Rana rugosa, Gossypium hirsutum, and Lupinus albus), whose deletion did not significantly reduce the taxonomic diversity represented in the data set, although it decreased computation burden to practicable levels. The MC44 unconstrained data sets was subjected to bootstrapping (1,000 replicates) under neighbor-joining (NJ) with ML distances (Dayhoff PAM matrix) by using the phyllip package.
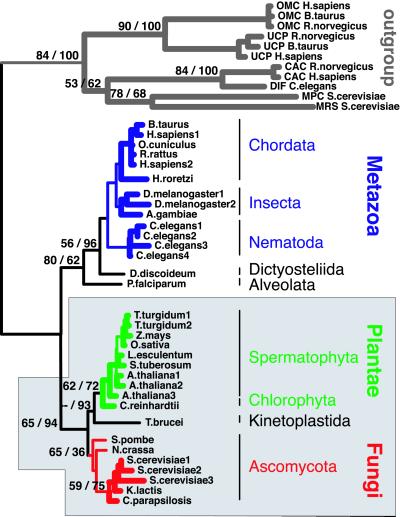
Figure 2.
Best ML phylogeny (−lnL = 8,617.7; exhaustive search) of eukaryotes based on amino acid sequences of the AAC mt carrier. Quartet-puzzling supports (10,000 replicates)/NJ bootstrap values with ML distances (1,000 replicates) are indicated (when >50%) at the nodes. Bold lines indicate subtopologies constrained during the exhaustive ML searches and signal decay analyses. All analyses of the AAC locus support the grouping (shaded box) of Plantae (green) and Fungi (red) in a clade to the exclusion of Metazoa (blue). Outgroup includes members of other mt carriers.
As some taxa clearly define long edges, we repeated all analyses after their exclusion to test whether they would cause artifactual results because of the “long-branch” attraction phenomenon (6).
Signal Decay.
To identify which characters support alternative hypotheses of relationships among Plantae, Fungi, and Metazoa, we assigned each aligned site to one of eight ML γ rate categories as follows: the program puzzle was provided with partially resolved user trees in which the Metazoa/Plantae/Fungi trichotomy was left unresolved (strongly supported monophyletic groups were constrained; see Fig. 2); a ML search was performed with JTT model and substitution rates from a γ distribution with eight categories; and the category contributing most to the likelihood was recorded for each site. After sorting sites according to their substitution rate class, we generated six partitions by sequentially removing categories 8 to 3, i.e., the six fastest-evolving classes of sites. The largest (i.e., original) and smallest data sets contain categories 1–8 and 1–2, respectively. Each partition was subjected to: (i) a heuristic quartet-puzzling ML analysis with JTT model and γ distribution (eight rate categories), and (ii) an exhaustive protml search, as described above. This procedure is an extension of that proposed by ref. 38.
Supplementary Material.
Additional information about reanalyses of an 18S data set as well as MCF data processing/analysis is available in the supplemental text and Figs. 4 and 5, which are published as supplemental data on the PNAS web site, www.pnas.org.
Results and Discussion
Alignment.
The stability analyses [using the program soap (29)] revealed that the gap opening/extension parameters have a very heterogeneous effect along the multiple alignments. It is likely that optimal alignments would require a dynamic assignment of gap penalties, as different portions of a molecule generally differ in their rate of insertion/deletion events. Our approach of removing positions that are unstable across many different multiple alignments is conservative and probably greatly reduces the risk of artifactual results caused by fast-evolving portions (with regard to insertions/deletions and, to a lesser extent, to substitutions) of the molecules investigated. Therefore, it is likely well suited to the analysis of deep divergences (the mean pairwise uncorrected distance among the AAC protein sequences of the ingroup taxa is 30%).
The outgroup sampling, which both minimizes alignment instability and maximizes taxonomic and MC subfamily diversities, includes 11 sequences: one tissue differentiation protein, one mt phosphate carrier, one mtRNA splicing protein, two carnitine–acyl–carnitine carriers, three oxoglutarate–malate carriers, and three uncoupling proteins. Alignment stability analysis detected 159 of the 433 columns (in the ingroup + outgroup reference alignment) as unstable. When considering the predicted tertiary structure of a yeast AAC protein, all unstable sites were located in the loop and coil regions protruding out of the membrane. Still, some highly stable residues are located outside the membrane; hence, the result of the stability test cannot be equated to exclusion of the nontransmembrane portions of the AAC molecule. After removal of the unstable regions and noninformative trailing sites, the final MC alignment includes 250 amino acid residues.
Self-pairwise local alignments of human, Arabidopsis, and fission yeast each identified three internal homologous repeats of 70–90 residues. Corresponding sites in each of the 37 AAC ingroup sequences were extrapolated as homologous. An alignment stability test was then performed on the 3 × 37 sequences (internal repeats). This final “REPEAT” alignment has a length of 70 amino acids.
Phylogenetic Analyses.
MC.
Exhaustive ML analysis of the MC44 data set (33 AAC and 11 outgroup sequences) yields a single best tree (minus the natural logarithm of the likelihood = 8,615.9), in which Plantae and Fungi grouped to the exclusion of Metazoa, a result that contradicts the classical hypothesis of a sister relationship between Fungi and Metazoa. However, 312 suboptimal trees (−lnL = 8,616.2–8,633.4) do not exclude the lnL value of the best tree within their standard error (SE). Although most (78.2%) of these trees exhibit a monophyletic [Plantae + Fungi], neither [Metazoa + Plantae] nor [Metazoa + Fungi] monophyly could be fully rejected, as these groups were found in 6.4 and 8.3% of the suboptimal trees, respectively. When all of the 312 non-“significantly” (1 SE criterion) worse trees are sorted by decreasing likelihood, the best trees supporting monophyly of [Metazoa + Plantae] and [Metazoa + Fungi] are the 119th and 146th trees, respectively. The low statistical power of these ML analyses may be explained by the species Halocynthia roretzi (a Urochordata) that has a particularly long branch within Metazoa. In the best ML tree, Halocynthia groups not with vertebrates but as a sister taxon to nematodes. We performed three additional analyses to test whether, indeed, this long branch caused artifactual results elsewhere in the tree: (i) monophyly of chordates (i.e., urochordates + vertebrates) was constrained; (ii) Halocynthia was removed; and (iii) all long branches (Halocynthia, Plasmodium, Dictyostelium [slime mold], and Trypanosoma) were removed. Constraining the monophyly of chordates reduces the lnL value of the best tree by 1.8 units but still yields a monophyletic [Plantae + Fungi + _Trypanosoma_] (Fig. 2). Sixty-one suboptimal trees (−lnL = 8,618.2–8,630.2) cannot be rejected by the one SE criterion, but this time all of them exhibit a [Plantae + Fungi] clade (often invaded by Trypanosoma), excluding Metazoa. In the best tree (Fig. 2), slime mold (Dictyostelium discoideum) is the closest sister taxon to Metazoa, Plasmodium is a sister taxon to [Metazoa + slime mold], and trypanosome groups next to Plantae before Fungi, even though many other tree topologies in respect to these “deep nodes” cannot be rejected with significance. On the other hand, the branch separating the [Plantae + Fungi + _Trypanosoma_] monophyletic group from the root is highly significant, as it is nearly four times the corresponding SE. When Halocynthia is removed from the data set, all these results remain unchanged. After removing all four taxa defining long branches (leaving 29 ingroup taxa and 11 outgroup taxa), ML analyses yield a single best tree of −lnL 7,618.5 and 11 non-“significantly” worse trees (−lnL = 7,618.5–7,624.1). The eight best of these 11 trees group Plantae and Fungi to the exclusion of Metazoa, whereas the last three trees group Plantae and Metazoa in a clade.
The MC44 data set was also analyzed with the quartet-puzzling method. When applying uniform substitution rates, the monophyly of [Plantae + Fungi + _Trypanosoma_] and the monophyly of [Metazoa + slime mold] are supported by 65 and 56% of quartets, respectively. Excluding sites with gaps marginally affects these values. We identified that this apparent low support for the relevant nodes is because of a very long branch defined by Trypanosoma. Results are very different when analyses are repeated without Trypanosoma (43 sequences, 250 amino acids): the monophyly of [Plantae + Fungi] is increased to 92%. No internal structure of Metazoa is resolved with significant quartet-puzzling values. When a four-rate category γ distribution is implemented, the topology of the tree slightly changes: the monophyly of [Plantae + Fungi] and of [Metazoa + slime mold] is still supported by 80 and 84% of quartets, respectively, but Plasmodium is now left outside as the first offshoot of the ingroup, a result consistent with previous phylogenetic analyses of other molecular markers [e.g., Enolase (39), EF-1α (40)]. Again, removing the gap sites in the alignment yields very similar trees with marginally higher branch support values.
The main results of the ML analyses described above are supported by bootstrapping under distance methods: by using NJ on the MC44 data sets (44 sequences, 250 amino acids), the monophylies of [Plantae + Fungi + _Trypanosoma_] and of [Metazoa + slime mold] are supported by bootstrap values of 94 and 96%, respectively (Fig. 2). None of the internal branches within Metazoa is supported with bootstrap proportions higher than 50%.
REPEAT.
To root the AAC gene tree without the need to use members from other mt carrier subfamilies, we performed ML analyses of the alignment among 3 × 13 internal repeat sequences (i.e., the “REPEAT_reduced” data set), because it should yield a “supertree” with three subtrees (one for each internal repeat) connected to each other by their respective roots (Figs. 3 a and b). Note that LGT and/or differential sorting can yield topological incompatibility among true subtrees only when recombination(s) have occurred among these loci. Hence, the comparison of internal repeats should virtually eliminate this problem: any observed conflict among topologies should reflect tree inference inaccuracy rather than authentic historical differences. Still, inference of true and identical topologies among internal repeats of a given set of sequences could still yield erroneous conclusions (at the organismal phylogeny level) if orthology of the sequences is mistaken for paralogy.
Figure 3.
Schematic representation of the evolution of a locus that experienced duplications without translocation of the duplicated fragments. (a) Given the low probability of recombination among them, it is very likely the three fragments tandemly repeated in the AAC gene will share the same history such that (b) the true phylogeny that can be inferred from a complete analysis of the three internal repeats should exhibit three fully compatible subtrees connected to each other by their respective roots. (c) Quartet-puzzling analysis (left phylogram; values at the nodes indicate puzzling support, 10,000 replicates), NJ analysis with ML distances (right cladogram; values indicate bootstrap support, 1,000 replicates), and protein ML analysis (data not shown) of the three internal repeats all support a monophyletic [Plantae + Fungi].
A heuristic ML analysis of this REPEAT_reduced data set yields a best tree of −lnL = 2,722.0 as well as 148 suboptimal trees (−lnL = 2,722.8–2,743.0), not excluding the lnL value of the best tree within their SE. Strikingly, the best tree, in each of the three subtrees, exhibits the grouping of Plantae and Fungi to the exclusion of Metazoa. The branch separating the [Plantae + Fungi] group from the next deeper node is significantly positive in two of the three cases. However, the resolution of the tree is low (and is further reduced when gapped positions are excluded), and no competing tree topology can be rejected with significance. Unfortunately, given the high number of sequences involved, an exhaustive ML analysis is computationally impractical even on this reduced data set.
An analysis of the full “REPEAT” data set (3 × 37 sequences) was performed under ML quartet puzzling. Changing the substitution rate model (uniform or γ distribution) has a marginal effect on the results. The resulting supertree exhibits three subtrees that have the same general topology (Fig. 3c Left), with each a well supported monophyletic [Plantae + Fungi + _Trypanosoma_] clade. These results are further supported by the bootstrap analyses performed by using distance methods: 55, 61, and 71% of the 1,000 bootstrap replicates support the monophyly of [Plantae + Fungi + _Trypanosoma_] in the subtrees corresponding to the first, second, and third repeats, respectively. If we consider only the grouping of Plantae and Fungi, regardless of whether Trypanosoma belongs to that clade, these numbers increase to 61, 78, and 81% (Fig. 3c Right). Very few internal branches are resolved with quartets puzzling proportions >50%.
Despite the short size of the repeats, the stability of the [Plantae + Fungi] clade (i) across the tree repeats and (ii) to variations of analyses is striking and adds significant credence to this grouping.
Signal Decay.
By performing alignment stability tests, we effectively reduced the risk that the grouping of Plantae and Fungi would be an artifact because of erroneous inference of homology among characters. Moreover, this result was stable to exclusion of “problematic” (because fast-evolving) taxa. However, inaccurate phylogenetic relationships can be inferred (especially when Felsenstein's zones are involved) if a large number of homoplasic (i.e., noisy) characters are included in the analyses. It is important to note that ML approaches will greatly reduce (in comparison with maximum parsimony; see, e.g., ref. 41), but not eliminate, the negative impact of these sites. To investigate whether our ML results could be because of deceptive homoplasic characters, we assigned each site to one of eight categories of rates from a γ distribution. We then reanalyzed the data set after sequentially removing six of the eight classes from the alignment. We realize that the procedure should cause a loss of resolution. This loss, however, is not really problematic, as removing fastest evolving sites should reduce resolution first at tip nodes, whereas we are trying to test the [Plantae + Fungi] clade, a very deep node indeed. A striking result of these signal decay analyses is that sequential exclusion of the six partitions of sites did not collapse the [Plantae + Fungi] clade (see supplemental data on the PNAS web site). Very similar results are obtained under exclusion of Halocynthia and Trypanosoma and under ML puzzling analyses.
Comparison with 18S.
Several loci [e.g., EF-1α (1), enolase (39), and 18S (42)] have been documented as supporting a [Metazoa + Fungi] clade. The 18S data set is of particular interest, because it has been studied extensively (e.g., refs. 3, 42) and constitutes, taxonomically, the best sampled gene for eukaryotes. The ML trees resulting from our reanalysis of the full-length 18S alignment support a previously reported [Metazoa + Fungi] clade. Furthermore, this group (with the exception of one fungus sequence) seems to remain somewhat stable through decay analysis: the clade is observed during iterative exclusion of rate categories 8 to 6 (data not shown), exclusion of category 5 yields [Plantae + Metazoa], and exclusion of further categories collapses Plantae, Metazoa, and Fungi into a polytomy. The situation, however, is radically different when sites detected as unstable to alignment parameters are removed. In the ML tree of this “cleaned” 18S data set, a [Plantae + Fungi + _Trypanosoma_] clade is observed (see supplemental data on the PNAS web site), even though it is invaded by Plasmodium, which clusters with Trypanosoma. It is important to realize that the original alignment (42) was included in the sensitivity analysis as the master sequence to which all other alignments were compared. Therefore, the “cleaned” alignment is not a realignment of the 18S data set but a sample (i.e., the stable positions) of the master alignment, which was itself based on secondary structure of the molecule (42) (http://rrna.uia.ac.be). Decay analysis on the “cleaned” data set yields fluctuating topologies of the best trees.
Conclusion
The AAC locus yields a strong signal possibly relevant for the resolution of the Plantae/Fungi/Metazoa trichotomy. Our analyses suggest that [Metazoa + slime molds] and [Plantae + Fungi + _Trypanosoma_] form two major clades within the eukaryote tree. This result is robust and stable and is obtained through two independent rooting procedures that are less likely to produce artifactual random rooting than previous investigations of other genes. Hence, we think it is relatively premature to conclude that an exclusive animal/fungal monophyletic group, suggested in previous analyses, provides the definitive solution to the phylogenetic relationships among Plantae, Metazoa, and Fungi (without even considering the large number of protist lineages). We certainly do not claim that our analyses (and suggested sister relationship between Plantae and Fungi) conclusively put the question to rest.
First, despite our efforts to avoid them, analytical problems relevant to the AAC data set might have remained undetected during our analyses. For example, although both signal decay and soap analyses reduce the overall noise of data sets, they could also cause potentially informative characters (such as gaps) to be removed. Furthermore, some analyzed data sets (e.g., in ref. 1) incorporated a larger taxonomic sampling (especially for protists) than that used in our AAC analyses. One could advocate that selection of a larger set of ingroup species increases the likelihood of producing accurate phylogenies (but see ref. 43), whereas it could simultaneously decrease the likelihood of obtaining a high support for it. It is likely that our AAC phylogeny will be tested in the future when new AAC sequences become available. Given the difficulties intrinsic to deep phylogeny inference, testing the AAC tree through the use of all available phylogeny inference techniques and model parameters is impractical and unlikely to yield a fully unquestionable result (e.g., ref. 44). Still, the AAC putative historical signal exposed here is sufficiently stable to warrant consideration despite it conflicts with the widely accepted resolution of the Metazoa/Fungi/Plantae trichotomy.
Second, we think the real issue is whether the eukaryote species tree can be resolved through comparison and integration of molecular phylogenetic analyses of multiple loci. Obviously, this question cannot be answered before extensive analyses of a large number of independent loci are performed (with each locus receiving analytical treatments similar to those used here: alignment stability test, decay analysis, etc.). We expect that, if LGT and differential lineage sorting have been extensive enough, the notion of a single eukaryote species tree might be nonsensical. Even if one could resolve with high confidence the true gene trees inferred from a large number of unlinked loci, one might never reach a conclusive high-majority consensus on the cladistic cohesion of certain groups of species, simply because such a consensus might not exist. Nevertheless, even if simple resolution of the eukaryote crown were not achievable—i.e., even in the pessimistic view where the three possible groupings of Metazoa, Plantae, and Fungi would each be supported by one-third of all available unlinked loci—phylogenetic analyses of multiple independent markers would still be of paramount importance. Indeed, we anticipate that similar data-mining approaches, i.e., on the basis of extensive analyses of data that were not initially gathered for evolutionary inference endeavor, will eventually provide an accurate understanding of how the cells and genomes of eukaryotes evolved, merging the gap between character variation at the molecular level and organismal evolution. These issues are more far-reaching than the quest for a single tree of eukaryotes.
Supplementary Material
Supplemental Data
Acknowledgments
We are grateful to Daniel Van Belle, Pekka Pamilo, and three anonymous reviewers for helpful comments on previous versions of this manuscript. Daniel Van Belle also managed the cluster of multiprocessor computers used for our analyses. This work was supported by the Communauté Française de Belgique (ARC 98/03–223), the National Fund for Scientific Research Belgium (FNRS), the Free University of Brussels (ULB), the Van Buuren Fund, the Defay Fund, and the Finnish Cultural Foundation.
Abbreviations
ML
maximum likelihood
SE
standard error
LGT
lateral gene transfer
AAC
ADP–ATP carriers
MC
mitochondrial carrier
MCF
MC family
mt
mitochondrial
NJ
neighbor joining
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Baldauf S, Roger A J, Wenk-Siefert I, Doolittle W F. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- 2.Nikoh N, Hayase N, Iwabe N, Kuma K, Miyata T. Mol Biol Evol. 1994;11:762–768. doi: 10.1093/oxfordjournals.molbev.a040156. [DOI] [PubMed] [Google Scholar]
- 3.Philippe H, Lopez P, Brinkmann H, Budin K, Germot A, Laurent J, Moreira D, Müller M, Le Guyader H. Proc R Soc London Ser B. 2000;267:1213–1221. doi: 10.1098/rspb.2000.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wheeler W C. Cladistics. 1990;6:363–367. doi: 10.1111/j.1096-0031.1990.tb00550.x. [DOI] [PubMed] [Google Scholar]
- 5.Leipe D D, Gunderson J H, Nerad T A, Sogin M L. Mol Biochem Parasit. 1993;59:41–48. doi: 10.1016/0166-6851(93)90005-i. [DOI] [PubMed] [Google Scholar]
- 6.Felsenstein J. Syst Zool. 1978;27:401–410. [Google Scholar]
- 7.Philippe H, Laurent J. Curr Opin Genet Dev. 1998;8:616–623. doi: 10.1016/s0959-437x(98)80028-2. [DOI] [PubMed] [Google Scholar]
- 8.Hirt R, Logsdon J, Healy B, Dorey M, Doolittle W, Embley T. Proc Natl Acad Sci USA. 1999;96:580–585. doi: 10.1073/pnas.96.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stiller J W, Hall B. Mol Biol Evol. 1999;16:1270–1279. doi: 10.1093/oxfordjournals.molbev.a026217. [DOI] [PubMed] [Google Scholar]
- 10.Keeling P, Doolittle W. Mol Biol Evol. 1996;13:1297–1305. doi: 10.1093/oxfordjournals.molbev.a025576. [DOI] [PubMed] [Google Scholar]
- 11.Moreira D, Le Guyader H, Philippe H. Mol Biol Evol. 1999;16:234–245. doi: 10.1093/oxfordjournals.molbev.a026105. [DOI] [PubMed] [Google Scholar]
- 12.Roger A J, Sandblom O, Doolittle W F, Philippe H. Mol Biol Evol. 1999;16:218–233. doi: 10.1093/oxfordjournals.molbev.a026104. [DOI] [PubMed] [Google Scholar]
- 13.Tajima F. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahata N, Nei M. Genetics. 1985;110:325–344. doi: 10.1093/genetics/110.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pamilo P, Nei M. Mol Biol Evol. 1988;5:568–583. doi: 10.1093/oxfordjournals.molbev.a040517. [DOI] [PubMed] [Google Scholar]
- 16.Takahata N. Genetics. 1989;122:957–966. doi: 10.1093/genetics/122.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Avise J C. Molecular Markers, Natural History and Evolution. New York: Chapman & Hall; 1994. [Google Scholar]
- 18.Maddison W. In: Experimental and Molecular Approaches to Plant Biosystematics. Hoch P C, Stephenson A G, editors. St. Louis, MO: Missouri Botanical Garden; 1995. pp. 273–287. [Google Scholar]
- 19.Avise J C, Wollenberg K. Proc Natl Acad Sci USA. 1997;94:7748–7755. doi: 10.1073/pnas.94.15.7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delwiche C. Am Nat. 1999;154:S164–S177. doi: 10.1086/303291. [DOI] [PubMed] [Google Scholar]
- 21.Katz L A. Am Nat. 1999;154:S137–S145. doi: 10.1086/303289. [DOI] [PubMed] [Google Scholar]
- 22.Doolittle W. Trends Genet. 1999;15:M5–M8. [PubMed] [Google Scholar]
- 23.Doolittle W. Curr Opin Struct Biol. 2000;10:355–358. doi: 10.1016/s0959-440x(00)00096-8. [DOI] [PubMed] [Google Scholar]
- 24.Kondrashov A S. Curr Opin Genet Dev. 1999;9:624–629. doi: 10.1016/s0959-437x(99)00027-1. [DOI] [PubMed] [Google Scholar]
- 25.Saier H M. Adv Microb Physiol. 1998;40:81–136. doi: 10.1016/s0065-2911(08)60130-7. [DOI] [PubMed] [Google Scholar]
- 26.Walker J E. Curr Opin Struct Biol. 1992;2:519–526. [Google Scholar]
- 27.Kuan J, Saier M H. Crit Rev Biochem Mol Biol. 1993;28:209–233. doi: 10.3109/10409239309086795. [DOI] [PubMed] [Google Scholar]
- 28.Gatesy J, DeSalle R, Wheeler W. Mol Phylogenet Evol. 1993;2:152–157. doi: 10.1006/mpev.1993.1015. [DOI] [PubMed] [Google Scholar]
- 29.Löytynoja A, Milinkovitch M C. Bioinformatics. 2001;17:573–574. doi: 10.1093/bioinformatics/17.6.573. [DOI] [PubMed] [Google Scholar]
- 30.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 31.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Felsenstein J. phylip (Phylogeny Inference Package) Ver. 3.5c. Univ. of Washington, Seattle, WA: Dept. of Genetics; 1993. [Google Scholar]
- 33.Strimmer K, von Haeseler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 34.gcg (2000) (Genetics Computer Group, Madison, WI).
- 35.Adachi J, Hasegawa M. molphy: Programs for Molecular Phylogenetics, Ver. 2.3. Tokyo: Institute of Statistical Mathematics; 1995. [Google Scholar]
- 36.Felsenstein J. Evolution (Lawrence, KS) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 37.Jones D, Taylor W, Thornton J. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 38.Ruiz-Trillo I, Riutort M, Littlewood D T J, Herniou E A, Baguna J. Science. 1999;283:1919–1923. doi: 10.1126/science.283.5409.1919. [DOI] [PubMed] [Google Scholar]
- 39.Hannaert V, Brinkmann H, Nowitzki U, Lee J A, Albert M-A, Sensen C W, Gaasterland T, Müller M, Michels P, Martin W. Mol Biol Evol. 2000;17:989–1000. doi: 10.1093/oxfordjournals.molbev.a026395. [DOI] [PubMed] [Google Scholar]
- 40.Baldauf S, Palmer J, Doolittle W. Proc Natl Acad Sci USA. 1996;93:7749–7754. doi: 10.1073/pnas.93.15.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huelsenbeck J, Hillis D M. Syst Biol. 1993;42:247–264. [Google Scholar]
- 42.Van de Peer Y, De Wachter R. J Mol Evol. 1997;45:619–630. doi: 10.1007/pl00006266. [DOI] [PubMed] [Google Scholar]
- 43.Kim J. Syst Biol. 1996;45:363–374. [Google Scholar]
- 44.Sanderson M J, Kim J. Syst Biol. 2000;49:817–829. doi: 10.1080/106351500750049860. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data