Axonal localization of neuritin/CPG15 mRNA is limited by competition for HuD binding (original) (raw)
ABSTRACT
HuD protein (also known as ELAVL4) has been shown to stabilize mRNAs with AU-rich elements (ARE) in their 3′ untranslated regions (UTRs), including Gap43, which has been linked to axon growth. HuD also binds to neuritin (Nrn1) mRNA, whose 3′UTR contains ARE sequences. Although the Nrn1 3′UTR has been shown to mediate its axonal localization in embryonic hippocampal neurons, it is not active in adult dorsal root ganglion (DRG) neurons. Here, we asked why the 3′UTR is not sufficient to mediate the axonal localization of Nrn1 mRNA in DRG neurons. HuD overexpression increases the ability of the Nrn1 3′UTR to mediate axonal localizing in DRG neurons. HuD binds directly to the Nrn1 ARE with about a two-fold higher affinity than to the Gap43 ARE. Although the Nrn1 ARE can displace the Gap43 ARE from HuD binding, HuD binds to the full 3′UTR of Gap43 with higher affinity, such that higher levels of Nrn1 are needed to displace the Gap43 3′UTR. The Nrn1 3′UTR can mediate a higher level of axonal localization when endogenous Gap43 is depleted from DRG neurons. Taken together, our data indicate that endogenous Nrn1 and Gap43 mRNAs compete for binding to HuD for their axonal localization and activity of the Nrn1 3′UTR.
KEY WORDS: RNA-binding protein, Axon, mRNA localization, mRNA transport, Nrn1, HuD
Summary: The stoichiometry of competing target mRNAs (Nrn1 and Gap43), RNA-binding protein levels (HuD), and affinity of mRNA–RNA-binding protein interactions contribute to the efficiency of axonal mRNA localization elements.
INTRODUCTION
Axonally synthesized proteins contribute to axon growth and regeneration (Perry and Fainzilber, 2014). Recent observations point to roles in neurodegeneration as well as neuropathic pain for axonally synthesized proteins (Baleriola et al., 2014; Melemedjian et al., 2010), so the known functions of these axonally synthesized proteins are expanding. Axons isolated to purity from cultured neurons have been used to profile the axonal transcriptome, showing that a surprisingly complex population of hundreds of different mRNAs in axons of the dorsal root ganglion (DRG), hippocampal, retinal (i.e. retinal ganglion cells) and motor neurons (Korsak et al., 2017; Minis et al., 2014; Saal et al., 2014; Taylor et al., 2009; Willis et al., 2007; Zivraj et al., 2010). Several mRNAs have been detected in axons in vivo, and there is recent evidence for axonal mRNA localization in adult brain, spinal cord, and optic nerve axons (Baleriola et al., 2014; Kalinski et al., 2015; Shigeoka et al., 2016; Walker et al., 2012; Zivraj et al., 2010). Despite increasing knowledge of which mRNAs localize into axons, the conditions under which some mRNAs localize, and even distinct functions for proteins generated from some axonal mRNAs (Perry and Fainzilber, 2014), we have relatively little understanding of how axonal mRNA localization is regulated. mRNAs are transported in complex with proteins (Gomes et al., 2014), and a few RNA-binding proteins (RBPs) needed for subcellular localization of mRNAs have been identified (Bi et al., 2007; Cosker et al., 2016; Dombert et al., 2014; Perry et al., 2016; Preitner et al., 2016). We previously showed that mRNAs compete for binding to zip code binding protein 1 (ZBP1; also known as IGF2BP1 and IMP1) in DRG neurons. ZBP1 is an RBP that is expressed at low levels in adult DRGs and is needed for the axonal localization of Gap43 and β-actin mRNAs in adult sensory neurons (Donnelly et al., 2011; Yoo et al., 2013). However, this observation was based on overexpressing the competing 3′ untranslated regions (UTR) elements of Gap43 and β-actin mRNAs, and it was not clear whether endogenous mRNAs also compete with one another for axonal localization.
The AU-rich element (ARE) in the Gap43 3′UTR is a well defined binding site for the ELAV-like protein HuD (ELAVL4), and this interaction with HuD stabilizes the Gap43 mRNA (Bolognani and Perrone-Bizzozero, 2008). HuD interacts with many different mRNAs through binding to AREs (Bolognani et al., 2010). HuD localizes to axons in cultured sensory neurons and in vivo, and HuD has also been implicated in mRNA localization (Smith et al., 2004; Yoo et al., 2013). The Gap43 ARE is necessary and sufficient for the axonal localization of the mRNA in cultured DRG neurons (Yoo et al., 2013). Akten et al. (2011) detected neuritin mRNA [Nrn1; also called cortical plasticity gene 15 (Cpg15)] in RNA immunoprecipitation (RIP) assays from embryonic cortical neuronal extracts using antibodies to HuD and SMN proteins (Akten et al., 2011). Interestingly, the Nrn1 3′UTR drives axonal localization in cultures of embryonic hippocampal and cortical neurons, while its 5′UTR provides the major axonal localizing activity (herein, axonal localizing activity refers to the ability of the mRNA sequence to mediate its own localization to axons) for adult DRG neurons (Akten et al., 2011; Merianda et al., 2013). Given the interaction of HuD with Nrn1 mRNA in CNS neurons (Akten et al., 2011) and axonal localization of HuD in adult DRG neurons (Yoo et al., 2013), we were puzzled as to why the Nrn1 3′UTR has little axon localizing activity in adult DRG neurons. Here, we show that HuD levels are limiting in adult DRG neurons, resulting in a competition between Nrn1 and Gap43 mRNAs for interaction with HuD protein. HuD binds directly to the Nrn1 3′UTR ARE and stabilizes Nrn1 mRNA similar to its effect on Gap43 mRNA. Under normal culture conditions, Gap43 mRNA is expressed at a several-fold excess compared to Nrn1, and the Nrn1 3′UTR can localize reporter mRNA into the DRG axons when endogenous Gap43 mRNA is depleted or HuD is overexpressed. This work shows that endogenous mRNAs can compete for RBP interaction and localization into axons. Our data further emphasize that axonal levels of mRNAs that use shared protein(s) for their localization, are defined by transcriptional activity of the their encoding genes as well as their affinity for and expression levels of their RBP(s).
RESULTS
Availability of HuD limits axonal localization through the 3′UTR of Nrn1
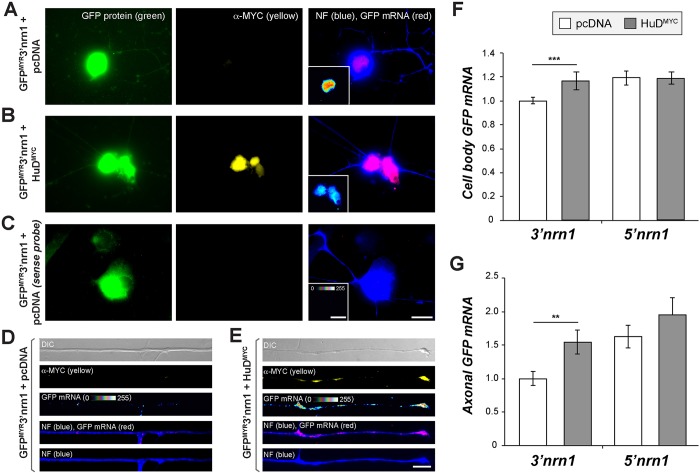
The 3′UTR of the Nrn1 mRNA can localize a heterologous mRNA into axons of cultured hippocampal neurons but has low activity in adult DRG neurons compared to the Nrn1 5′UTR, which is sufficient for axonal localization (Merianda et al., 2013). The RBPs needed for axonal localization of the Nrn1 5′UTR are not known, but work from the Sahin laboratory has indicated that HuD and Smn proteins bind to endogenous Nrn1 mRNA in embryonic CNS neurons where its 3′UTR has higher localizing activity than its 5′UTR (Akten et al., 2011; Merianda et al., 2013). The Gap43 3′UTR has localizing activity in DRG neurons, and the ARE region of Gap43 that HuD binds to is necessary and sufficient for axonal localization of the mRNA (Yoo et al., 2013). Thus, we asked whether HuD levels might limit the axonal localizing activity of the Nrn1 3′UTR in the DRG neurons. For this, we overexpressed HuD in adult DRG neurons by transfection with a Myc-tagged HuD construct (HuDMYC) (Yoo et al., 2013) plus co-transfection with GFPMYR that includes the 5′ or 3′UTR of rat Nrn1 (GFPMYR5′nrn1 and GFPMYR3′nrn1, respectively) and compared these cells to cells transfected with a vector control. The GFPMYR5′nrn1 construct included the 3′UTR of γ-actin mRNA, and the GFPMYR3′nrn1 construct included the 5′UTR of rat calcium/calmodulin kinase IIα mRNA (Camk2a) as previously reported (Merianda et al., 2013). Immunofluorescence showed that HuDMYC protein localized to cell bodies, including the nuclei (Fig. 1B), and axons of the DRG neurons (Fig. 1E). Fluorescence in situ hybridization (FISH) signals for axonal GFPMYR3′nrn1 mRNA were significantly increased in the HuDMYC versus vector control transfected cultures (Fig. 1E,G). However, no change in the axonal GFPMYR5′nrn1 mRNA level was seen upon overexpression of HuD (Fig. 1G). There was a small but significant increase in levels of GFPMYR3′nrn1 mRNA in the cell bodies of the HuD overexpressing DRG neurons (Fig. 1F), which could reflect the known function of HuD protein in mRNA stabilization (also see Fig. 4A). Nonetheless, the increased axonal localization seen with the Nrn1 3′UTR on HuD overexpression suggests that the availability of endogenous HuD limits the axonal localizing activity of the Nrn1 3′UTR in DRG neurons.
Fig. 1.
Increasing HuD levels allows the 3′UTR of Nrn1 to localize into DRG axons. (A–E) Representative images for GFP mRNA and neurofilament protein (NF) in cell body (A–C) and distal axons (D,E) are shown for GFPMYR3′nrn1-expressing DRG neuron co-transfected with pcDNA6-Myc-His (pcDNA) control vector (A,C,D) versus HuDMYC (B,E) as indicated. FISH images using a sense GFP probe are shown in C. For A–C, GFP mRNA is shown in red and NF protein in blue. GFP mRNA is shown as a spectral intensity in insets in A–C (right column) and the RNA-only panels in D and E. Please note that high levels of HuDMYC and GFPMYR mRNA in the cell bodies preclude resolving axonal signals in A. Scale bars: 20 µm for cell body panels, 10 µm for axon panels. (F,G) Quantification of cell body (F) and axonal (G) GFP mRNA levels in cultures transfected as in A–E. mRNA signals are shown relative to the control vector (n is at least 30 neurons in ≥3 separate transfections). **_P_≤0.01; ***_P_≤0.005 (one-way ANOVA with Tukey HSD post-hoc analyses).
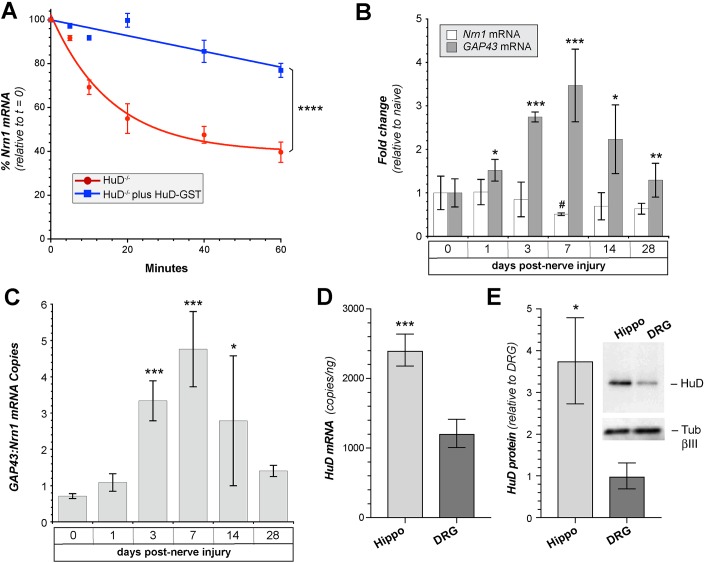
Fig. 4.
HuD binding stabilizes Nrn1 mRNA. (A) In vitro degradation assay using P100 lysates from HuD−/− mouse brain versus HuD−/− mouse brain supplemented with 250 ng HuD-GST as indicated. 500 ng total RNA isolated from wild-type mouse brain was used as the substrate for the degradation assays. The data points show the percentage of remaining Nrn1 mRNA measured by RT-qPCR over the indicated intervals (_n_=3; _F_[1,24]=153.9). ****P<0.0001 (two-way repeated measures ANOVA). (B) Fold change in the relative amount of Nrn1 and Gap43 mRNAs [compared to uninjured (naïve)] in L4/5 DRGs at indicated intervals after mid-thigh sciatic nerve crush (mean±s.d.; _n_=3 per time point). *_P_≤0.05; **_P_≤0.01; ***_P_≤0.001 for indicated Gap43 value versus 0 d time point. #_P_≤0.05 for indicated Nrn1 value versus 0 d time point (Student's _t_-test). (C) Absolute quantification of the ratio of Gap43 to Nrn1 copy numbers taken from ddPCR data in B. *_P_≤0.05; ***_P_≤0.005 (Student's _t_-test). (D) RTddPCR quantification of the amount (mean±s.d.) of HuD mRNA in cultures of 3 DIV adult DRG versus 5 DIV E18 hippocampal neuron (Hippo) cultures (_n_=3; ***=_P_≤0.005 by Student's _t_-test). (E) Relative HuD protein levels in 3 DIV adult DRGs versus 5 DIV E18 hippocampal neuron cultures from western blots (mean±s.d.; _n_=3). *P<0.05 (Student's _t_-test). The inset panel shows a representative western blot.
HuD binds to the Nrn1 mRNA 3′UTR
Nrn1 mRNA has been detected in HuD immunoprecipitates from lysates of embryonic cortical neurons in RNA immunoprecipitation (RIP) assays (Akten et al., 2011), but these authors did not report whether there is any direct interaction of Nrn1 with HuD. To approach this issue, we asked whether endogenous Nrn1 co-precipitates with HuD, using reverse transcriptase (RT)-coupled droplet digital PCR (ddPCR) to detect absolute mRNA copy number. For this, we overexpressed FLAG-tagged HuD (HuDFLAG) followed by immunoprecipitation with anti-FLAG antibodies. Both endogenous GAP43 and Nrn1 mRNAs were detected in these HuDFLAG immunoprecipitates by RTddPCR (Fig. 2A). The observation that Nrn1 mRNA precipitated with a much higher fold enrichment than Gap43 raises the possibility that Nrn1 has a higher binding affinity for HuD than does Gap43.
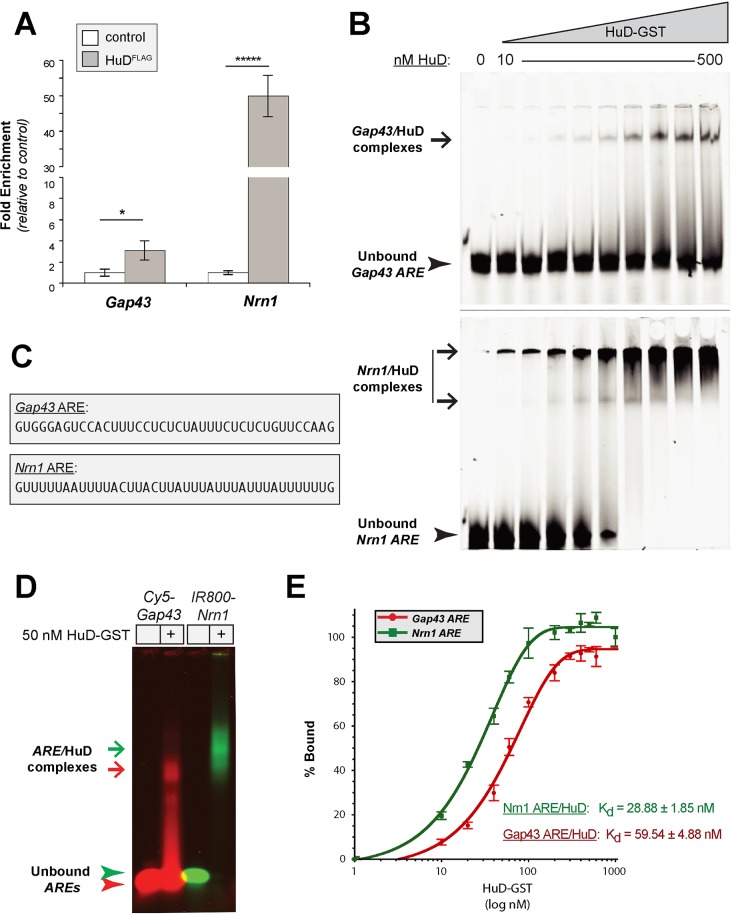
Fig. 2.
Nrn1 mRNA binds directly to HuD. (A) RTddPCR analyses for Gap43 and Nrn1 mRNAs precipitating with FLAG-tagged HuD are shown as the mean± s.d. fold enrichment relative to cells with FLAG control vector (_n_=3). *_P_≤0.05; *****_P_≤0.0001 (Student's _t_-test). (B) Representative polyacrylamide gels from REMSA analyses using 10 nM Cy5.5-tagged Gap43 (top) or Nrn1 (bottom) AREs binding to increasing concentrations of HuD-GST (10, 20, 40, 60, 100, 200, 300, 400 and 500 nM). ARE–HuD-GST complexes (Gap43/HuD; Nrn1/HuD) are indicated by arrows and unbound AREs are indicated by arrowheads. (C) Sequences of Gap43 and Nrn1 AREs used for these studies. (D) Representative agarose gel from REMSA analyses using 10 nM Cy5.5-tagged Gap43 or IR800-tagged Nrn1 AREs plus HuD-GST. The Nrn1 ARE–HuD-GST complex (green arrow) migrates more slowly than the Gap43 ARE–HuD-GST complex (red arrow). Arrowheads indicate the unbound AREs. (E) FPA assays for A488-tagged Gap43 (red) and Nrn1 AREs (green) binding to increasing concentrations of HuD-GST. Average binding values are shown as mean±s.e.m. (_n_≥6). Indicated _K_d values were calculated from the curve-fitting formulae as outlined in the Materials and Methods, with error spanning the maximum and minimum _K_d values calculated using the curve-fitting errors.
To directly test the affinity of the HuD interactions with Gap43 and Nrn1, we generated synthetic RNAs corresponding to the 3′UTR AREs of Nrn1 (nt 937–976, NCBI accession #NM_053346) and Gap43 (nt 1148–1187, NCBI accession #NM_008083) and recombinant GST-tagged HuD protein (HuD-GST) (Fig. 2C). We reasoned this would allow us to determine both whether HuD binds directly to Nrn1 and the relative binding affinities of the HuD interactions with the Nrn1 and Gap43 AREs. We initially used an RNA mobility shift assay (REMSA) with fluorescently tagged Gap43 or Nrn1 AREs plus increasing concentrations of HuD-GST. In a non-denaturing polyacrylamide gel electrophoresis, the mobility of both Gap43 and Nrn1 AREs was reduced upon increasing HuD–GST concentration (Fig. 2B). When using molar equivalents of Nrn1 and Gap43 AREs, free Nrn1 ARE was depleted at a lower HuD-GST concentration than was the free Gap43 ARE (Fig. 2B, arrowheads). This depletion of the free RNA oligonucleotides corresponded to the appearance of higher molecular mass bands that likely represent HuD-GST–RNA complexes, but much of the presumed Nrn1 ARE–HuD-GST complex remained in the wells of these polyacrylamide gels (Fig. 2B, arrows). Despite increasing electrophoresis durations, lowering polyacrylamide concentrations (from 10% to 4%), or adding additional detergents and heparin to the binding reactions, we were not able to completely resolve the Nrn1 ARE–HuD-GST complex (data not shown), suggesting that multimeric HuD–Nrn1 complexes form. Moving to agarose gel electrophoresis allowed us to resolve the Nrn1 ARE–HuD-GST complex indicating that the slower migrating REMSA signals represent RNA–HuD complexes rather than insoluble precipitates of the fluorescent RNA oligonucleotides forming in the presence of increasing protein concentration (Fig. 2D). However, resolution of the reactions was decreased in the agarose gels, with smearing between the Gap43 ARE–HuD-GST and unbound Gap43 ARE oligonucleotides. The reason(s) for the differences in resolution between Gap43 ARE–HuD-GST and Nrn1 ARE–HuD-GST under these electrophoresis conditions is not clear.
As a second measure of direct RNA–protein binding, we used a fluorescence polarization absorption (FPA) assay. In this assay, the random motion of the small fluorescently tagged RNA oligonucleotide is decreased by binding to the larger protein, and this can be measured by an increased emission of polarized light upon excitation of the fluorophore attached to the oligonucleotide (Kuehnert et al., 2015). Addition of increasing amounts of HuD-GST consistently increased the polarized fluorescent emission from stable molar concentrations of Gap43 and Nrn1 AREs (Fig. 2E). Again, the Nrn1 ARE showed higher binding affinity for HuD-GST than did the Gap43 ARE. Calculating the binding affinity of these interactions showed _K_d values of 28.88±1.85 nM (mean±s.e.m.) for Nrn1 ARE–HuD-GST and 59.54±4.88 nM for Gap43 ARE–HuD-GST. The combination of the REMSA, FPA and RIP analyses indicates that Nrn1 ARE not only binds directly to HuD, but also does so with a higher affinity than does the Gap43 ARE.
Nrn1 and Gap43 AREs compete for binding to HuD
Given the apparent higher affinity of the Nrn1 ARE compared to the Gap43 ARE for binding to HuD, we tested whether these two AREs compete for binding to HuD. We initially tested for competitive binding using FPA with stable concentrations of recombinant HuD-GST and fluorescently tagged Gap43 or Nrn1 AREs plus increasing concentrations of non-tagged (‘cold’) Gap43 or Nrn1 AREs. In this assay, competition for binding or displacement of the fluorescent ARE from HuD-GST is represented as a decrease in FPA signals as the cold ARE increases in concentration. Cold Gap43 ARE displaced fluorescent Gap43 ARE from the HuD-GST–RNA complex (Fig. 3A), and cold Nrn1 ARE similarly displaced fluorescent Nrn1 ARE from the HuD-GST–RNA complex (Fig. 3B). Cold Nrn1 ARE displaced fluorescent Gap43 ARE from HuD-GST binding at lower concentrations than required for displacement by cold Gap43 ARE (Fig. 3A). However, the cold Gap43 ARE had little effect on the fluorescent Nrn1 ARE–HuD complex, with no significant decrease in polarized emissions from the Nrn1 ARE–HuD binding despite adding up to an 1000-fold excess cold Gap43 ARE (Fig. 3B). This points to the avidity of the interaction of the Nrn1 ARE–HuD complex. Cold Nrn1 ARE was able to displace its own counterpart but required an ∼200-fold excess, suggesting that the Nrn1 ARE–HuD complex is quite stable.
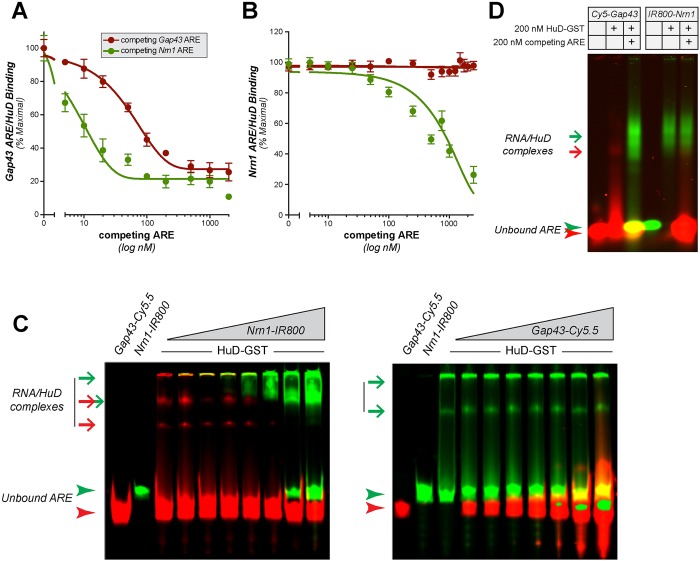
Fig. 3.
Nrn1 ARE competes with Gap43 ARE for binding to HuD_._ (A,B) Displacement FPA assays are shown for 10 nM A488-Gap43 ARE (A) or A488-Nrn1 ARE (B) and 50 nM HuD-GST plus increasing concentrations of unlabeled Gap43 ARE (red) or Nrn1 ARE (green). Values are graphed as the mean±s.e.m. as a percentage of maximal binding (_n_≥4). A fourth-order polynomial was used for curve fitting in A, and a third-order polynomial was used for curve fitting in B. (C) Representative polyacrylamide gels for REMSA analyses using 10 nM Cy5.5-tagged Gap43 ARE (left gel) or IR800-tagged Nrn1 ARE (right gel) and 50 nM HuD-GST plus increasing concentrations of IR800-tagged Nrn1 ARE or Cy5.5-tagged Gap43 ARE (10–200 nM), respectively. RNA–protein complexes are indicated by arrows and unbound AREs are indicated by arrowheads (red for Gap43 ARE and ARE–protein complexes; green for Nrn1 ARE and ARE–protein complexes). (D) Representative agarose gel from REMSA analyses used to resolve RNA–protein complexes. Here, 10 nM Cy5.5-tagged Gap43 or IR800-tagged Nrn1 AREs were bound to 200 nM HuD-GST and then competing ARE was added. RNA–protein complexes and unbound AREs are indicated by arrows and arrowheads, respectively, as in C.
We moved to REMSA to be certain that the altered signals seen in this competition FPA reflect displacement of binding rather than possible inhibition of binding or blockage of polarized fluorescent emissions due to increasing molar RNA oligonucleotide concentrations in these reactions. To address these possibilities, we used Cy5.5-Nrn1 and IR800-Gap43 AREs so that both complexes could be visualized in the same gel. With this approach, Cy5-Gap43 ARE was displaced from HuD-GST binding with increasing concentration of IR800-Nrn1 ARE, with increased IR800-Nrn1 ARE–HuD-GST complex seen commensurate with decrease in Cy5.5-Gap43 ARE–HuD-GST signals (Fig. 3C, left panel). In contrast, increasing concentration of Cy5.5-Gap43 ARE was not able to displace IR800-Nrn1 ARE from its complex with HuD-GST (Fig. 3C, right panel). Again, the IR800-Nrn1 ARE–HuD complex was not completely resolved in these polyacrylamide gels; however, agarose gels showed a clear appearance of a IR800-Nrn1 ARE–HuD-GST complex after displacement of Cy5.5-Gap43 ARE from HuD-GST, but there was no displacement of IR800-Nrn1 ARE from HuD–GST binding with excess Cy5.5-Gap43 (Fig. 3D). This was not a feature of the IR800 conjugation, as an IR800-Gap43 ARE similarly did not displace the Cy5.5-Nrn1 ARE when it was bound to HuD-GST (data not shown). Varying the duration of these reactions or the sequence of binding and competition in an attempt to distinguish competitive binding from displacement did not change the outcome of the results. In each case, a higher proportion of the Nrn1 ARE than the Gap43 ARE bound to HuD, and the Nrn1 ARE displaced the Gap43 ARE from HuD-GST binding, but not the converse (data not shown). These data indicate that the Nrn1 ARE can directly compete with the Gap43 ARE for binding to HuD.
HuD interaction stabilizes Nrn1 mRNA
Since HuD is known to stabilize Gap43 mRNA (Beckel-Mitchener et al., 2002), we asked whether the interaction of HuD also stabilizes Nrn1 mRNA. Thus, we asked whether the degradation rates of endogenous Nrn1 mRNA were affected by the presence of HuD. For this, we used an in vitro RNA decay assay with total RNA isolated from brains of embryonic day (E)18 wild-type mice spiked with S100 protein extracts prepared from brains of HuD−/− mice (Bird et al., 2013) with or without recombinant HuD-GST protein. As determined by RT-coupled quantitative PCR (RT-qPCR), there was a rapid decline in the Nrn1 in the P100 extracts from the HuD−/− mice, and spiking these reactions with recombinant HuD-GST protein resulted in significant stabilization of Nrn1 mRNA (Fig. 4A; _t_1/2=33±5 min for HuD−/− and 171±25 min for HuD−/− plus HuD-GST). This increase in Nrn1 stability in presence of HuD plus the increased axonal localizing activity of the Nrn1 3′UTR seen with HuD overexpression points to a functional relevance for the direct Nrn1 ARE-HuD interactions seen by FPA and REMSA.
Axotomy changes the stoichiometry of Gap43 to Nrn1 mRNA
We have previously shown that Nrn1 localizes into axons of adult DRG neurons through motifs in its 5′UTR (Merianda et al., 2013); the data in Fig. 1 indicate this 5′UTR-dependent axonal localization of Nrn1 mRNA is independent of HuD and that the Nrn1 5′UTR does not contain an ARE (data not shown). We have also shown that sciatic nerve crush injury increases axonal transport of Nrn1 mRNA, with a concomitant decrease in lumbar (L)4/5 DRG levels of Nrn1 mRNA (Merianda et al., 2013). Gap43 transcription is known to increase during periods of active axonal growth, including in DRG neurons (Skene et al., 1986; Yoo et al., 2013). The DRGs used here are effectively axotomized at the time of culture, and show robust growth of axons over the duration of the culture period. This raises the possibility that the ratio of Nrn1 to Gap43 mRNA molecules could be altered by axotomy. Although the Gap43 ARE did not seem to displace the Nrn1 ARE from HuD-GST in the studies above, we questioned whether an increase in Gap43 mRNA relative to Nrn1 mRNA in DRGs might occur after axotomy. Thus, we used RTddPCR to quantify the absolute copy number for Nrn1 and Gap43 mRNAs in the DRGs. Consistent with previous studies, we saw a significant increase in Gap43 mRNA levels in L4/5 DRGs at 3 days after sciatic nerve crush injury compared with that seen in naïve DRG neurons, with peak levels reached at 7 days post axotomy (Fig. 4B). Nrn1 mRNA levels in DRGs showed a significant decrease at 7 days post axotomy (Fig. 4B). The ratio of Gap43 to Nrn1 copies was less than one in naïve DRGs, with Gap43 becoming from 3–5-fold in excess of Nrn1 at 3–7 days post axotomy (Fig. 4C). This in vivo post-injury time point (3 days) corresponds to the culture durations used in Fig. 1. Thus, transcriptional induction of Gap43, but not Nrn1, following axotomy could contribute to the binding of Gap43 to HuD and subcellular localization of Gap43 in the axotomized DRG neurons.
For embryonic CNS neurons, the Nrn1 3′UTR is functional for axonal localization (Akten et al., 2011; Merianda et al., 2013). Thus, we compared the expression of HuD levels in cultured adult DRG neurons versus embryonic hippocampal cultures. As determined by RTddPCR, where we can definitively distinguish HuD from other Hu isoforms, cultures of embryonic hippocampal neurons contained ∼2.5-fold more HuD mRNA than adult DRG neurons (Fig. 4D). Immunoblotting revealed that hippocampal neurons showed ∼4-fold more HuD protein than adult DRG neurons (Fig. 4E). This difference in HuD levels between CNS and PNS neurons likely contributes to the differential activity of the Nrn1 3′UTR for axonal localization in the CNS versus PNS neurons.
Additional sequences in the Gap43 3′UTR increase affinity for HuD binding
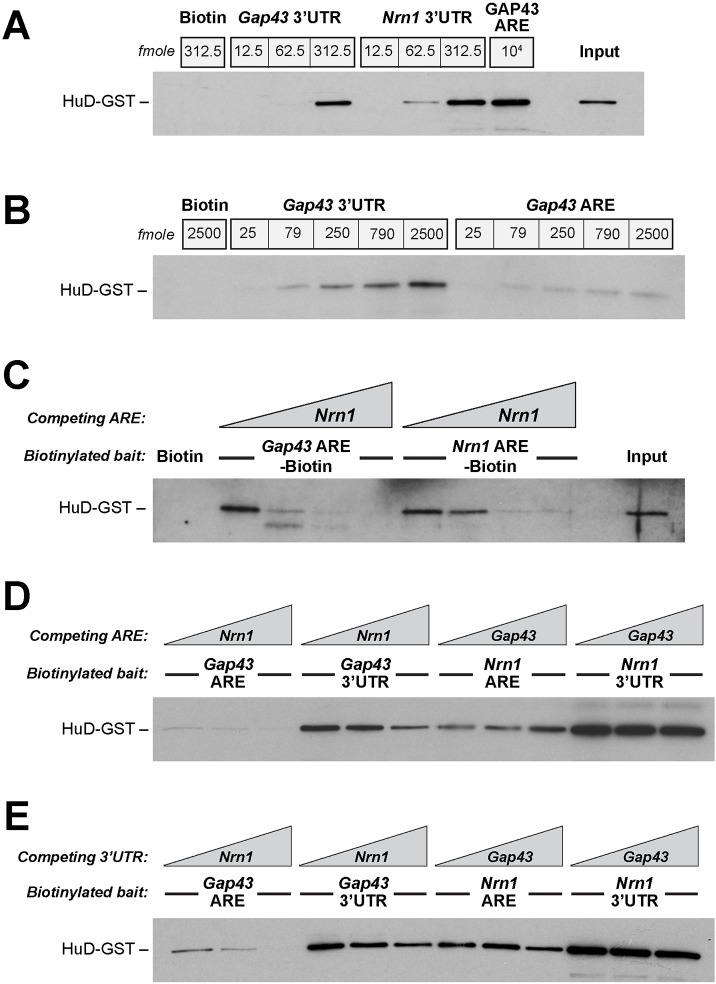
Based on stoichiometry alone, it seems unlikely that a 3–5-fold excess of Gap43 over Nrn1 could fully explain why the Nrn1 3′UTR has little localizing activity in the adult DRG neurons. We considered the possibility that other RBPs might complex with HuD resulting in a higher affinity binding to the Gap43 ARE. Neither ZBP1 (also known as IGF2BP1), which is needed for axonal localization of Gap43 mRNA (Donnelly et al., 2011; Yoo et al., 2013), or IMP2 (also known as IGF2BP2), which has recently been implicated in axonal mRNA transport and has homology to ZBP1 (Preitner et al., 2016), protected the dissociation of the Gap43 ARE–HuD-GST complexes mediated by Nrn1 ARE in FPA assays (data not shown). Thus, we asked whether additional sequences within the Gap43 3′UTR might increase the binding affinity of HuD for the mRNA above that seen with the Gap43 ARE. For this, we used in vitro transcribed biotinylated 3′UTRs of rat Gap43 and Nrn1 for in vitro binding assays with purified HuD-GST protein followed by streptavidin pulldown; the amount of HuD-GST bound was then assessed by immunoblotting. Fig. 5A shows that both Gap43 and Nrn1 3′UTRs bind to HuD-GST in vitro, with increasing concentration of the UTRs pulling down more HuD-GST. At equal molar concentrations, the Gap43 3′UTR showed a much higher binding affinity for HuD-GST than did the Gap43 ARE (Fig. 5B).
Fig. 5.
The Gap43 3′UTR shows higher and more stable binding to HuD than does the Gap43 ARE_._ (A) Representative binding assay for in vitro transcribed biotinylated Gap43 and Nrn1 3′UTRs for HuD-GST. As with the AREs, Nrn1 shows a higher binding at equivalent molar inputs than does Gap43. However, the Gap43 3′UTR appears to have higher HuD binding than the Gap43 ARE alone. 250 fmoles HuD-GST was used in these reactions. (B) Direct comparison of HuD-GST binding for equimolar quantities of biotinylated Gap43 3′UTR and Gap43 ARE. The 3′UTR shows a much higher binding affinity for HuD-GST. 250 fmoles HuD-GST was used in these reactions. (C) Increasing concentrations of cold Nrn1 ARE (0, 0.25, 1.25, 6.25 pmoles) were added to binding reactions with 250 fmoles of biotinylated Gap43 or Nrn1 AREs and 250 fmoles HuD-GST. Immunoblotting for GST shows that cold Nrn1 ARE displaces HuD-GST from interaction with Gap43 ARE and Nrn1 ARE, with Nrn1 ARE again showing higher binding affinity than Gap43 ARE. (D) Increasing concentrations of Nrn1 ARE (0, 0.5 and 2.5 pmoles) also displaces biotinylated Gap43 3′UTR from HuD-GST binding (250 pmoles each), but displacement is less efficient than seen for the biotinylated Gap43 ARE (250 pmoles). As seen in the FPAs and REMSAs in Fig. 3, the Gap43 ARE does not displace either Nrn1 ARE or 3′UTR from HuD-GST binding (concentrations same as above). Refer to Fig. S1 for extended exposure of the three left-most lanes. (E) Competition assays were performed with cold in vitro transcribed Nrn1 and Gap43 3′UTRs at 0, 0.5, and 2.5 pmoles. This shows that the Nrn1 3′UTR displaces both biotinylated Gap43 ARE and 3′UTR from HuD-GST binding (both at 250 fmoles). The cold Gap43 3′UTR resulted in a modest decrease in the amount of Nrn1 ARE–HuD-GST and Nrn1 3′UTR–HuD-GST complexes at highest concentration used. Results are representative of _n_≥3 for all panels.
We then performed competition experiments with increasing concentrations of Nrn1 and Gap43. As in the FPA and REMSA assays, cold Nrn1 ARE displaced both labeled Gap43 ARE and Nrn1 ARE from HuD-GST binding (Fig. 5C). The Gap43 3′UTR–HuD-GST interaction was disrupted by cold Nrn1 ARE, but this required much higher concentrations of cold Nrn1 ARE than did disruption of the Gap43 ARE–HuD-GST complex (Fig. 5D; Fig. S1). Cold Gap43 ARE had essentially no effect on the Nrn1 3′UTR–HuD-GST complex, similar to what was seen in FPA and REMSA for the Nrn1 ARE/HuD-GST interaction (Fig. 5D). Finally, we asked whether cold Gap43 and Nrn1 3′UTRs could disrupt the ARE–HuD-GST or 3′UTR–HuD-GST complexes. Similar to the competition reactions using the cold Nrn1 ARE, cold Nrn1 3′UTR disrupted both the Gap43 ARE–HuD-GST and 3′UTR–HuD-GST complexes, with 3′UTR–HuD-GST being more resistant to displacement than ARE–HuD-GST (Fig. 5E). Interestingly, cold Gap43 3′UTR behaved in a similar manner to the cold Gap43 ARE, with no apparent effect on Nrn1 ARE or Nrn1 3′UTR interaction with HuD, at least up to the concentrations used here. These data indicate that although the ARE is sufficient and necessary for the interaction of HuD with Gap43, additional components of its 3′UTR impact upon the affinity for and stability of binding to HuD.
Endogenous Gap43 mRNA competes with the Nrn1 3′UTR for axonal localization in DRG neurons
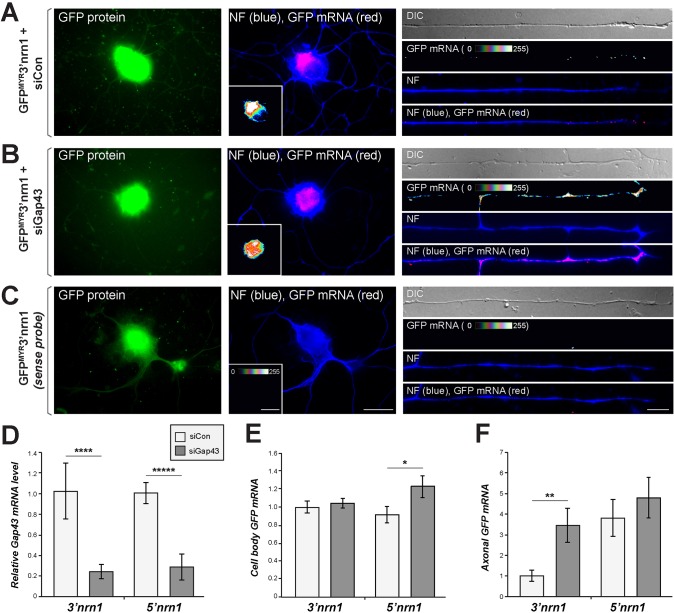
The data above point to a competition between Nrn1 and Gap43 AREs for binding to HuD and subsequent axonal localization in adult DRG neurons. To directly test the possibility that higher Gap43 expression levels restrict activity of the Nrn1 3′UTR in the DRG neurons, we used siRNAs to deplete Gap43 mRNA (si_Gap43_) from DRG cultures and then compared axonal localization of GFPMYR reporters carrying the 5′ or 3′UTR of rat Nrn1. Non-coding siRNA (siCon) was used as a control. There was a clear depletion of Gap43 mRNA, as determined by RTddPCR, with si_Gap43_ in both the _GFPMYR3′nrn1_- and _GFPMYR5′nrn1_-transfected DRGs (Fig. 6D). Quantitative FISH analyses for GFP mRNA showed that GFPMYR3′nrn1 mRNA had a significantly higher axonal localization when endogenous Gap43 was depleted from the DRG neurons (Fig. 6A–C,F) without any significant change in cell body GFPMYR3′nrn1 mRNA levels (Fig. 6E). Depletion of Gap43 mRNA had no significant effect on the axonal levels of GFPMYR5′nrn1 (Fig. 6F), despite the fact that there was a modest increase in the cell body levels of GFPMYR5′nrn1 in the _Gap43_-depleted cultures (Fig. 6E). siRNAs targeting a different region of Gap43 generated similar results, with increased localization of GFPMYR3′nrn1 upon depletion of Gap43 from these neurons (data not shown). These results show that the 3′UTR of Nrn1 can drive axonal mRNA localization in adult DRG neurons when Gap43 mRNA is depleted, indicating that the endogenous Gap43 mRNA prevents the localizing activity of the Nrn1 3′UTR by competing for limiting quantities of HuD in adult sensory neurons. Consistent with this, the amount of endogenous Gap43 co-immunoprecipitating with HuD in DRG neurons was decreased upon expression of GFPMYR3′nrn1 but not upon expression of GFPMYR3′nrn1ΔARE, where the ARE sequence was deleted (Fig. S2).
Fig. 6.
Depletion of Gap43 mRNA activates the 3′UTR of Nrn1 for axonal localization in DRG neurons. (A–C) Representative images for GFP mRNA in the cell body (left and middle columns) and distal axons (right column) for a _GFPMYR3′nrn1_-expressing DRG neuron transfected with non-targeting siRNA (siCon; A) or Gap43_-targeting siRNA (si_Gap43; B) as indicated. FISH images using sense GFP probe are shown in C. The GFP mRNA is shown as a spectral intensity in axonal segments and the inset panels for cell body (the intensity spectrum is shown on the lower right). Scale bars: 20 µm for cell body panels, 10 µm for axon panels. (D) RT-qPCR quantification of Gap43 mRNA levels in GFPMYR3′nrn1- and GFPMYR5′nrn1-transfected DRGs. Gap43 mRNA levels are normalized to that of 18S rRNA and signals are shown relative to siCon+GFPMYR3′nrn1 as mean±s.e.m. (_n_=3). ****_P_≤0.0001 (one-way ANOVA with Tukey HSD post-hoc analyses). (E,F) Quantification of cell body (E) and axonal (F) GFP mRNA levels in cultures transfected as in A–C. mRNA signals are shown relative to the siCon for GFPMYR3′nrn1. There is a significant increase in axonal localization mediated through the 3′UTR of Nrn1 when Gap43 mRNA is depleted. Cell body levels of GFPMYR3′nrn1 mRNA are not affected by Gap43 depletion; curiously, the cell body GFPMYR5′nrn1 mRNA levels increase with depletion of Gap43 (n is at least 30 neurons in ≥3 separate transfections). *_P_≤0.05; **_P_≤0.01; ****_P_≤0.001; *****_P_≤0.0001 for the indicated data sets (one-way ANOVA with Tukey HSD post-hoc analyses).
DISCUSSION
With recognition that intra-axonal protein synthesis contributes to axon growth and pathfinding, there have been concerted efforts to determine which proteins are synthesized in axons. These profiling studies have motivated new approaches to address the potential functions of locally synthesized proteins and how mRNA transport into and translation within axons are regulated. Several lines of evidence indicate that both these mechanisms are tightly regulated. RBPs that recognize regulatory motifs in mRNAs can modulate transport and translation of mRNAs in axons (Gomes et al., 2014). Here, we focused on HuD, a neuronal RBP, that has been implicated in both stabilization and transport of target mRNAs through binding to AREs in those mRNAs (Bird et al., 2013; Mobarak et al., 2000; Yoo et al., 2013). HuD has also been shown to regulate translation through interaction with polyadenylated tails of mRNAs (Fukao et al., 2009), but it is not clear whether this mechanism is relevant for localized mRNAs. Our data indicate that the AREs in the 3′UTRs of Nrn1 and Gap43 compete for binding to HuD and axonal localization. Increasing HuD levels or depleting Gap43 mRNA allows the 3′UTR of Nrn1 to drive axonal localization in the DRGs neurons (Figs 1 and 6). The limited expression of HuD in DRG neurons versus embryonic CNS neurons (Fig. 4D,E) likely contributes to lack of activity of Nrn1 3′UTR for localization in the adult sensory neurons. Gap43 was previously shown to compete with β-actin mRNA for binding to ZBP1 that is needed for axonal localization, but this work was based on overexpressing the localizing 3′UTR of β-actin and Gap43 mRNAs (Donnelly et al., 2011; Yoo et al., 2013). The work on HuD here importantly advances this mechanism of mRNAs competing for RBP interactions to show for the first time that an endogenous mRNA can limit axonal localizing activity of another mRNA through competitive binding for a shared RBP. For evidence of this mRNA–mRNA competition, we find that HuD can bind directly to the Nrn1 3′UTR, HuD expression is lower in neurons where the Nrn1 3′UTR is not active for axonal localization, Gap43 mRNA is expressed in a 3–5-fold molar excess to Nrn1 mRNA in neurons where Nrn1 3′UTR is not active for axonal localization, and depletion of endogenous Gap43 mRNAs from these neurons with low HuD and high Gap43 makes the Nrn1 3′UTR active for axonal localization.
The 5′UTR of Nrn1 mRNA is sufficient for axonal localization in the cultured DRG neurons used here (Merianda et al., 2013). The 5′UTR of Nrn1 does not contain an ARE and, to the best of our knowledge, HuD does not bind to the 5′UTR of Nrn1. Transport of Nrn1 mRNA into axons is increased after sciatic nerve crush injury, with the mRNA shifting from being cell body predominant in naïve neurons to being axon predominant in injury-conditioned neurons (Merianda et al., 2013). This shift of Nrn1 mRNA into axons through its 5′UTR explains the fall in the DRG and cell body levels of Nrn1 mRNA by 3 days after sciatic nerve crush injury (Fig. 4B,C). The depletion of Nrn1 from the DRG soma by 5′UTR-driven Nrn1 mRNA axonal localization may provide Gap43 mRNA with a competitive advantage for HuD binding by promoting a further excess of Gap43 relative to Nrn1 mRNA in the axotomized DRG neurons.
Axon length increases in both cultured DRG neurons and developing chick spinal cord axons when Gap43 mRNA is overexpressed with a heterologous axonal-targeting motif (Donnelly et al., 2013). In addition, overexpressing the Nrn1 coding sequence with the 5′UTR, but not the 3′UTR, in cultured DRG neurons results in a significant increase in axon length (Merianda et al., 2013). Thus, both Gap43 and Nrn1 protein products are growth-promoting when they are locally generated in axons. It is tempting to speculate that the 5′UTR-dependent transport of Nrn1 in the DRG neurons evolved to overcome competition between Nrn1 and Gap43 mRNAs in the PNS neurons used here. Such a mechanism could contribute to the high growth capacity of PNS neurons. Nrn1 and Gap43 AREs only showed a 2-fold difference in HuD-binding affinities, so it is surprising that an excess of Gap43 ARE was not able to displace bound Nrn1 ARE from HuD, while an excess of Nrn1 ARE readily displaced bound Gap43 ARE from HuD in vitro (Fig. 3). Even though the full 3′UTR of Gap43 showed a higher binding affinity for HuD in vitro, this 3′UTR could only mediate minimal displacement of the _Nrn1_–HuD-GST complex. On the other hand, Nrn1 seemed to be quite effective at displacing Gap43 from HuD binding.
Our data suggest that the transcriptional activity of the rat Gap43 gene helps determine whether the Nrn1 3′UTR is active for axonal localization or not. The increased expression of HuD in embryonic CNS neurons compared to adult DRGs argues that RBP levels also must be considered in this competition mechanism. Bird et al. (2013) showed that the stoichiometry of HuD to the KH-type splicing regulatory protein (KHSRP) impacts on the stability of Gap43. Interaction with KHSRP destabilizes ARE-containing target mRNAs, and overexpression of KHSRP depletes neurons of Gap43 and decreases neurite growth (Bird et al., 2013). As we start to consider such competitive interactions of mRNAs with RBPs, the binding affinity that we tested here is a critical parameter that has not been well explored to date. In addition to the _K_d of Nrn1 ARE–HuD binding, the interaction of Nrn1 with HuD appears to be particularly stable based on the quantity of excess unlabeled Nrn1 ARE that was needed to disrupt the Nrn1 ARE-Alexa-Fluor-488–HuD-GST complex (Fig. 3). Since Nrn1 is transported into DRG axons through its 5′UTR, it is intriguing to speculate that Nrn1 could displace Gap43 from the _Gap43_–HuD complex in distal axons. Such displacement of Gap43 from HuD binding by Nrn1 could effectively target Gap43 for degradation through interactions with KHSRP protein. Future studies will be needed to determine whether the full-length Nrn1 mRNA, including the presumed RBPs bound to its 5′UTR that are needed for axonal localization in DRGs, can displace axonal Gap43 from its interaction with HuD and whether this occurs locally in distal axons.
HuD has three RNA recognition motifs (RRMs). HuD stabilizes ARE-containing mRNAs through interactions with RRM1 and RRM2. Considering that HuD is linked to transport of Gap43 through ARE interaction (Yoo et al., 2013), it is likely that the role of HuD in axonal mRNA transport is conferred through RRM1 and RRM2. The RRM3 of HuD has been linked to translational regulation through interaction with poly-adenylate tails of mature mRNAs (Fukao et al., 2009). As determined with RIP analyses, HuD can directly interact with over 800 mRNAs in the mouse forebrain (Bolognani et al., 2010). While some of those mRNAs clearly bind through non-ARE-related mechanisms, those binding through AREs could be subjected to the competitive binding mechanisms uncovered here. Although the in vitro binding assays may not fully reflect the in vivo situation, where additional RBPs and RBP-interacting proteins are present, the approach that we employed can provide new insight into how mRNAs can compete for limiting quantities of RBPs. Based on this and other work, we suggest that fully understanding the dynamics of RBP–mRNA interactions will require incorporating knowledge on mRNA levels and the affinities of these mRNAs for RBP interaction, as well as how other proteins affect these interactions. Interestingly, work from the Raab-Graham laboratory on dendritic Kv1.1 (Kcna1) mRNAs with HuD versus miR-129 (Sosanya et al., 2013) suggests that the mRNA competition mechanisms that we report here should also be considered in relation to modulating the dendritic transcriptome and likely extend to include non-coding RNAs. Furthermore, the ability of competing mRNAs to displace HuD-bound mRNAs may also occur at a subcellular level within distal axons and dendrites, setting up a displaced mRNA for more rapid decay or other fates for localized mRNAs.
MATERIALS AND METHODS
Animal procedures
The Institutional Animal Care and Use Committees of the University of South Carolina or University of New Mexico approved all animal procedures. Sprague Dawley rats (male; 150–225 g) were used for preparation of DRG cultures and survival surgery. Sciatic nerve crush injuries were performed at the mid-thigh as previously described (Twiss et al., 2000), using isoflurane for anesthesia. Hippocampal cultures were prepared from embryonic day 18 rat Sprague Dawley rat pups as previously described (Vuppalanchi et al., 2010).
DNA constructs
Myristoylated GFP (GFPMYR) with the 5′UTR of rat Nrn1 plus the 3′UTR of rat γ-actin, and the 5′UTR of rat Camk2a plus the 3′UTR of rat Nrn1 have been described previously (Merianda et al., 2013). The Myc-tagged HuD (HuDMYC) has also been described (Anderson et al., 2000). pcDNA6-Myc-His (Invitrogen) was used as the control transfection vector for HuDMYC experiments. For Flag-tagged HuD (HuDFLAG), the coding sequence of rat HuD was PCR-amplified and inserted into pFLAG-CMV2 plasmid using EcoRI/SalI restriction enzymes; the pFLAG-CMV2 plasmid served as a control for HuDFLAG.
Cell culture and transfections
DRG neurons were prepared as described previously (Twiss et al., 2000). Hippocampal cultures were prepared as described previously (Gomes et al., 2011). For expression of reporter constructs, dissociated DRGs were transfected immediately after dissociation by using a Basic Neuron SCN Nucleofector kit (Lonza). Transfected DRGs and hippocampal neurons were analyzed at 3 and 5 days in vitro, respectively.
The DRG/neuroblastoma hybrid cell line F11 was maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 2 mM L-glutamine. For transfection, cells were plated on collagen-coated 60 mm plates (0.2 mg/ml, Sigma). After 1 day, cells were transfected using Lipofectamine 2000 (Invitrogen). For this, 2 µg plasmid DNA was diluted into 0.1 ml OPTI-MEM medium (Invitrogen) and mixed with 5 µl of transfection reagent in 100 µl of OPTI-MEM. After 20 min, F11 cells were exposed to the DNA solution. After 4 h, the medium was changed to DMEM supplemented with 0.5% FBS, 2 mM L-glutamine and 10 µM forskolin (Sigma) for differentiation of the F11 cells. Cultures were lysed 72 h after transfection.
RNA isolation and analysis
RNA from cultured cells and L4-5 DRGs was isolated with the RNAqueous total kit (Ambion). RNA was isolated from embryonic mouse brain by using phenol-chloroform extraction followed by ethanol precipitation. RNA concentration was measured by performing fluorimetry using Ribogreen reagent (Invitrogen). Total RNA was reverse transcribed with the SensiFAST cDNA synthesis kit (Bioline) for expression analyses and Superscript II (Invitrogen) for in vitro decay analyses. For RIP analyses, iScript (BioRad) was used for HuDFLAG precipitations and SensiFAST was used for endogenous HuD precipitations. ddPCR was used for absolute quantification of RNA copy number and for the RIP studies. Taqman probes and primer sets for Nrn1 and Gap43 have been described previously (Merianda et al., 2013; Yoo et al., 2013). GAPDH RNA primer sets/probes were used for normalization in siRNA analyses and HMGB1 primers/probes were used for normalization in DRG expression analyses (Merianda et al., 2015). Real-time quantitative PCR (qPCR) was used for testing siRNA depletions and in the RNA survival assays, with SYBR Green (BioRad) detection methods.
Fluorescence in situ hybridization and immunofluorescence
FISH and immunofluorescence (IF) was performed as described previously (Merianda et al., 2013) with minor modifications. Cultures were fixed in 2% paraformaldehyde for 20 min. Digoxigenin (Dig)-labeled cRNA antisense probes (Roche) were used to detect GFP mRNA as described previously (Vuppalanchi et al., 2010). A sense cRNA probe was used for control. Primary antibodies for IF were as follows: chicken anti-neurofilament (NF) H (1:1500; Millipore) and NFM (1:500; Aves), sheep anti-Dig (1:100; Roche) and rabbit anti-GFP (1:500; Abcam) (refer to Table S1 for details on all antibodies in this work). Secondary antibodies were as follows: Cy3-conjugated anti-chicken-IgG, Cy5-conjugated anti-sheep-IgG and FITC-conjugated anti-rabbit-IgG (1:200; Jackson ImmunoResearch). All samples were mounted in Prolong Gold with DAPI (Invitrogen) and analyzed by epifluorescence microscopy using a Leica DM6000B microscope with a 63× oil immersion objective (NA 1.4) and Hamamatsu ER-Flash CCD camera. Signals were quantified from exposure-matched images by using ImageJ (National Institutes of Health). All representative image sets shown are exposure matched and have undergone identical post processing.
Immunoblotting
Adult DRG [3 days in vitro (DIV), ∼60,000 neurons] and E18 hippocampal (5 DIV, ∼100,000 neurons) cultures were lysed in Laemmli sample buffer. Cleared lysates were normalized for protein content by performing a bicinchoninic acid (BCA) assay (BioRad). Lysates were normalized for protein content, fractionated on 8% SDS/PAGE gels and transferred onto a PVDF membrane (GE Healthcare). After blocking in 5% non-fat dried milk powder diluted in Tris-buffered saline with 1% Tween 20 (TBST), membranes were probed overnight at 4°C with rabbit anti-HuD (1:2000; Abcam) or anti-βIII tubulin (1:400; Abcam) antibodies diluted in blocking buffer. Blots were washed in TBST and then incubated with horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (1:2000; Cell Signaling) diluted in blocking buffer for 1 h. Blots were washed in TBST and signals immune complexes were detected by Immobilon Western Chemiluminescent HRP Substrate (GE Healthcare).
For detection of HuD-GST bound to AREs and 3′UTRs, bound GST-HuD protein was resolved on an 8% SDS/PAGE gel, transferred onto nitrocellulose membranes (BioRad), then detected by immunoblotting as above. HRP-conjugated anti-GST rabbit antibody (1:10,000; GE Healthcare) was used. HRP signals were detected by using the ECL Prime reagent (GE Healthcare) according to the manufacturer's instructions.
RNA immunoprecipitation
F11 cells were transfected with either pFLAG vector or pFLAG-HuD plasmid. After 72 h, cells were lysed in 10 mM HEPES pH 7.4, 100 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.5% NP-40, 1 mM Na3VO4, 1 mM NaF, 10 mM PMSF, 1× protease inhibitor cocktail (Roche) and 10 U/ml RNAsin (Promega) prepared under RNase-free conditions with DEPC-treated H2O (RIP buffer). After centrifugation at 15,200 g for 20 min at 4°C, supernatant was further pre-cleared by 1 h incubation with protein-G–Dynabeads (Invitrogen) at 4°C. An aliquot of the lysate was used to isolate input RNA. After quantification of protein concentration in the remaining lysates, normalized lysates were incubated with the FLAG-antibody-conjugated magnetic beads (Sigma) for 4 h with rotation at 4°C. Beads were washed with RIP buffer six times, and bound RNAs were extracted with lysis buffer from the RNeasy Micro kit (Qiagen). Precipitated RNA copy number was normalized to the input quantity for each target, and then relative fold enrichment was calculated in comparison to that in vector control.
For precipitation of endogenous HuD, cultured DRGs were lysed 36 h post transfection in 20 mM HEPES pH 7.4, 10 mM KCl, 1 mM MgCl2, 20% glycerol and 0.2% Triton X-100 (cytoplasm extraction buffer) supplemented with protease inhibitor cocktail (Roche), PhosSTOP (Roche) and 40 units/ml RNasin Plus (Promega). Lysates were cleared by centrifugation at 15,200 g at 4°C for 20 min and normalized for protein input. An aliquot of the lysate was used to isolate input RNA. Supernatants were further pre-cleared with a mixture of Protein A and G Dynabeads (Life Technologies) for 1 h at 4°C on a rotator. Supernatant was collected and 4 µg of E-1 mouse anti HuD (Santa Cruz Biotech) or anti-mouse IgG (Sigma) overnight at 4°C on a rotator. RNA was isolated as above. Reverse transcription and ddPCR were performed as above. Primers used were: GFP sense, 5′-CAACAGCCACAACGTCTATATCAT-3′ and antisense, 5′-GGTGCTCAGGTAGTGGTTGTC-3′; and GAP43 sense, 5′-CAGGAAAGATCCCAAGTCCA-3′ and antisense, 5′-GAGGAAAGTGGACTCCCACA-3′. Precipitated RNA copy number was normalized to the level in the input RNA for each target, and then relative fold enrichment was calculated in comparison to that for cells transfected with GFP vector control.
RNA decay assay
Decay assays were essentially performed as described previously (Bird et al., 2013), with some modifications. Briefly, S100 extracts (30 µg) from the cortices of HuD−/− adult mice were incubated with RNA from E18 wild-type mouse brain (300 ng) with or without GST-HuD (75 ng) in decay buffer [100 mM potassium acetate, 2 mM magnesium acetate, 10 mM creatine phosphate, 1 µg creatine phosphokinase, 1 mM ATP, 0.4 mM GTP, 0.1 mM spermine, 500 ng poly(A) oligonucleotide, 10 mM Tris-HCl pH 7.6, 2 mM DTT and 100 U/ml RNase-OUT (ThermoFisher)]. Decay reactions were carried out at 37°C for 0–60 min. At each time point, 25 µl of the reaction was removed and placed in 400 µl 400 mM NaCl, 25 mM Tris-HCl pH 7.6 and 0.1% SDS (stop buffer). RNA was isolated as above, and RT-qPCR was performed for Nrn1 using the following primers: forward, 5′-AAAGCGAGAGGGAAAAGGAG-3′ and reverse, 5′-TTCGCTTTTCTGGAGGAGAA-3′. GAPDH was used as the reference gene using the primers forward, 5′-TGTGATGGGTGTGAACCACGAGAA-3′ and reverse, 5′-GAGCCCTTCCACAATGCCAAAGTT-3′.
Recombinant protein expression and purification
GST-tagged HuD expression constructs were transformed in BL21 competent cells (Invitrogen). Pre-cultures were grown overnight and diluted (1:50) with 100 ml LB and were then incubated in a shaking incubator at 320 rpm at 37°C for 2 h. Cultures were incubated with 0.1 mM IPTG at 30°C for another 5 h. Cells were pelleted at 4000 g for 20 min, and then resuspended in GST lysis buffer [1× phosphate buffered saline (PBS) supplemented with 1 mM DTT and EDTA-free protease inhibitor cocktail (Roche)]. The cell suspension was lysed on ice with six 10 s pulses in a Sonic Dismembrator (Model F60, Fisher Scientific). The cell suspension was centrifuged at 15,000 g for 20 min at 4°C. Supernatant was applied to GSTrap FF columns (GE Healthcare) according to manufacturer's protocol. Bound proteins were eluted in 50 mM Tris-HCl pH 8.0 and 10 mM reduced glutathione (Sigma). This elution buffer was then exchanged for 20 mM Tris-HCl pH 7.4, 1.5 mM MgCl2, 150 mM NaCl, 0.5 mM EDTA and 0.05% NP-40 (FP buffer) using a Centriprep Centrifugal Filter Device (Millipore). Protein isolates were analyzed for purity by Coomassie-staining SDS/PAGE gels, and with a BCA assay, for concentration. Recombinant ZBP1 was purchased from Origene and IMP2 was purchased from Novoprotein.
RNA electrophoretic mobility shift assay
RNA oligonucleotides corresponding to the rat Gap43 ARE and Nrn1 AREs were synthesized by Integrated DNA Technologies (Coralville, IA) [NM_017195; 40 nt (1211–1250), 5′-GUGGGAGUCCACUUUCCUCUCUAUUUCUCUCUGUUCCAAG-3′ and NM_053346; 39 nt (938–976) 5′ GUUUUUAAUUUUACUUACUUAUUUAUUUAUUUAUUUUUUG-3′, respectively]. For labeling, the vendor synthesized the oligonucleotides conjugated to 5′ IR800CW (IR800), Cy5.5 dye or Alexa Fluor 488 (A488) dyes. Purified GST-HuD protein was incubated with denatured Gap43 ARE and NRN1 ARE oligonucleotide, and binding was allowed for 10 min at room temperature. In competition binding assays, unlabeled RNA oligonucleotides were added in increasing amounts and incubated for another 10 min at room temperature before loading onto 10% native gel (pre-run for 30 min at 140 V). Protein–RNA complexes were resolved at 160 V for 55 min in the dark in a Mini-PROTEAN Tetra Cell apparatus (BioRad). Resolving REMSA reactions on agarose gels was performed as described previously with minor modifications (Das and Reed, 1999). Binding reactions consisted of Cy5.5-_Gap43-_ARE or IR800-_Nrn1_-ARE plus 200 nM HuD-GST. After a 10 min incubation, 200 nM of competing Cy5.5-_Gap43-_ARE or IR800-_Nrn1_-ARE was added to the reaction for an additional 10 min. Reactions were then resolved on a 1.5% agarose gel run for 3.5 h at constant 70 V. Both polyacrylamide and agarose gels were imaged with an Odyssey CLx infrared imaging instrument (Licor). Gel images were processed with Image Studio software (Licor).
Fluorescent polarization assay
Polarization measurements for fluorescence polarization assay (FPA) were conducted with a Spectramax i3 microplate reader (Molecular Devices). A488-labeled oligonucleotides were used for FPA. The excitation wavelength was 485 nm and emission was detected at 525 nm. For each independent experiment, 5 nM of A488-labeled RNA oligonucleotide was used. The final volume of each reaction was brought to 15 μl with 40 mM Tris-HCl pH 7.5, 300 mM NaCl, 1 mM EDTA, 0.1% NP40 and 0.1 mg/ml heparin (FP buffer) supplemented with 50 mg/ml γ-globulins (Sigma) in small volume 384-well plates (Greiner). A scrambled A488-labeled probe (5′-GCAUGGAAGGUUGUGCUGACGUGGGAUCACUUACUCCCAU-3′) was used to test for non-specific binding. Binding data were collected using Softmax Pro 6.4 software (Molecular Devices), and then analyzed using Prism 6 (GraphPad) software. No signals were detected with the scrambled probes (data not shown). Binding affinities (_K_d) were calculated from the formula for exponential rise curve fitting in Kaleidagraph from the curve formula _y_=m1+m2×(1−e−m3*x) solving for y as 50% bound RNA oligonucleotide signal (m1 = _y_min value, m2 = _y_max value, and m3 = x value at mid-point of y).
Binding assays for Nrn1 and Gap43 3′UTRs
A biotinylated Gap43 and Nrn1 ARE RNA oligonucleotide used for assay was purchased from Trilink and cold oligonucleotide was from Integrated DNA Technologies. In vitro transcribed Gap43 and Nrn1 3′UTRs were generated using templates of GFPMYR-3′nrn1 and GFPMYR-3′gap43 plasmids (Merianda et al., 2013; Yoo et al., 2013) by PCR amplification using sense and antisense primers to the 5′ and 3′ ends of the respective UTRs with a T7 promoter added to the sense primer. PCR products were gel purified and then in vitro transcribed with T7 RNA polymerase (Roche). For biotinylated UTRs, biotinyl rNTP labeling mix was used (Roche). Transcribed RNAs were incubated with DNAse I to remove template DNA and then purified by G-50 Sephadex spin columns (Roche). RNA was quantified by fluorimetry using Quant-iT Ribogreen (Invitrogen), and molar concentrations determined from the molecular masses of the full-length transcription product.
Biotinylated UTRs and AREs were incubated with GST-HuD protein in 0.3 ml of assay buffer (10 mM Tris-HCl pH 7.4, 75 mM NaCl, 0.75 mM MgCl2, 0.25 mM EDTA and 0.025% NP-40) supplemented with protease-inhibitor cocktail (Roche) and RNase inhibitor (Promega) for 2 h. For the competition assays, increasing amounts of cold UTR or ARE were included during the incubation period. Streptavidin-Dynabeads M280 (Invitrogen) were used to precipitate biotinylated RNA–protein complexes over 2 h at 4°C. After precipitation, Dynabeads were washed six times with assay buffer followed by elution with Laemmli sample buffer and processed for immunoblotting as noted above.
Image and statistical analysis
Fluorescent intensities from exposure-matched FISH images were quantified with ImageJ_._ Kaleidagraph and Prism 6 were used for statistical analyses. Curve fitting was performed using Prism 6_._ One-way ANOVA with Tukey honest significant difference (HSD) post-hoc analysis was used to compare between data points, and a Student's _t_-test was used to compare between paired data sets. Repeated-measures two-way ANOVAs were used for analyses of Nrn1 mRNA decay rates and to compare means of independent groups. _P_≤0.05 was considered as statistically significant.
Supplementary Material
Supplementary information
Acknowledgements
The authors thank Mike Fainzilber and colleagues in his laboratory for discussions of these data as the project progressed.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.J.L., A.S.G., P.K.S., R.R., T.H., N.P.B., J.L.T.; Methodology: S.J.L., A.S.G., P.P., R.R., R.J.T., T.H., A.N.K.; Formal analysis: C.G., S.J.L., A.S.G., T.S., J.L.T.; Investigation: C.G., S.J.L., A.S.G., T.S., P.K.S., P.P., E.T., R.J.T., A.N.K.; Resources: R.R., S.Y., T.H., N.P.B.; Data curation: E.T., J.L.T.; Writing - original draft: C.G., A.N.K., J.L.T.; Writing - review & editing: C.G., S.J.L., A.S.G., T.S., P.K.S., E.T., S.Y., T.H., A.N.K., N.P.B., J.L.T.; Supervision: T.H., N.P.B., J.L.T.; Project administration: J.L.T.; Funding acquisition: N.P.B., J.L.T.
Funding
This work was supported by grant awards from National Institutes of Health (R01-NS089633 to J.L.T. and N.P.B.; R01-NS041596 to J.L.T.; R01-CA172567 to T.H.; R21-NS099959 to S.Y.), the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (to J.L.T.), and ASPIRE grant program from the University of South Carolina (to S.J.L.). J.L.T. is the endowed Chair in Childhood Neurotherapeutics with the South Carolina SmartState Program. Deposited in PMC for release after 12 months.
References
- Akten B., Kye M. J., Hao le T., Wertz M. H., Singh S., Nie D., Huang J., Merianda T. T., Twiss J. L., Beattie C. E. et al. (2011). Interaction of survival of motor neuron (SMN) and HuD proteins with mRNA cpg15 rescues motor neuron axonal deficits. _Proc. Natl. Acad. Sci. USA_108, 10337-10342. 10.1073/pnas.1104928108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K. D., Morin M. A., Beckel-Mitchener A., Mobarak C. D., Neve R. L., Furneaux H. M., Burry R. and Perrone-Bizzozero N. I. (2000). Overexpression of HuD, but not of its truncated form HuD I+II, promotes GAP-43 gene expression and neurite outgrowth in PC12 cells in the absence of nerve growth factor. _J. Neurochem._75, 1103-1114. 10.1046/j.1471-4159.2000.0751103.x [DOI] [PubMed] [Google Scholar]
- Baleriola J., Walker C. A., Jean Y. Y., Crary J. F., Troy C. M., Nagy P. L. and Hengst U. (2014). Axonally synthesized ATF4 transmits a neurodegenerative signal across brain regions. _Cell_158, 1159-1172. 10.1016/j.cell.2014.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckel-Mitchener A. C., Miera A., Keller R. and Perrone-Bizzozero N. I. (2002). Poly(A) tail length-dependent stabilization of GAP-43 mRNA by the RNA-binding protein HuD. _J. Biol. Chem._277, 27996-28002. 10.1074/jbc.M201982200 [DOI] [PubMed] [Google Scholar]
- Bi J., Tsai N. P., Lu H. Y., Loh H. H. and Wei L. N. (2007). Copb1-facilitated axonal transport and translation of kappa opioid-receptor mRNA. _Proc. Natl. Acad. Sci. USA_104, 13810-13815. 10.1073/pnas.0703805104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird C. W., Gardiner A. S., Bolognani F., Tanner D. C., Chen C.-Y., Lin W.-J., Yoo S., Twiss J. L. and Perrone-Bizzozero N. (2013). KSRP modulation of GAP-43 mRNA stability restricts axonal outgrowth in embryonic hippocampal neurons. _PLoS ONE_8, e79255 10.1371/journal.pone.0079255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognani F. and Perrone-Bizzozero N. I. (2008). RNA-protein interactions and control of mRNA stability in neurons. _J. Neurosci. Res._86, 481-489. 10.1002/jnr.21473 [DOI] [PubMed] [Google Scholar]
- Bolognani F., Contente-Cuomo T. and Perrone-Bizzozero N. I. (2010). Novel recognition motifs and biological functions of the RNA-binding protein HuD revealed by genome-wide identification of its targets. _Nucleic Acids Res._38, 117-130. 10.1093/nar/gkp863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosker K. E., Fenstermacher S. J., Pazyra-Murphy M. F., Elliott H. L. and Segal R. A. (2016). The RNA-binding protein SFPQ orchestrates an RNA regulon to promote axon viability. _Nat. Neurosci._19, 690-696. 10.1038/nn.4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R. and Reed R. (1999). Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. _RNA_5, 1504-1508. 10.1017/S1355838299991501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombert B., Sivadasan R., Simon C. M., Jablonka S. and Sendtner M. (2014). Presynaptic localization of Smn and hnRNP R in axon terminals of embryonic and postnatal mouse motoneurons. _PLoS ONE_9, e110846 10.1371/journal.pone.0110846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly C. J., Willis D. E., Xu M., Tep C., Jiang C., Yoo S., Schanen N. C., Kirn-Safran C. B., van Minnen J., English A. et al. (2011). Limited availability of ZBP1 restricts axonal mRNA localization and nerve regeneration capacity. _EMBO J._30, 4665-4677. 10.1038/emboj.2011.347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly C. J., Park M., Spillane M., Yoo S., Pacheco A., Gomes C., Vuppalanchi D., McDonald M., Kim H. K., Merianda T. T. et al. (2013). Axonally synthesized beta-actin and GAP-43 proteins support distinct modes of axonal growth. _J. Neurosci._33, 3311-3322. 10.1523/JNEUROSCI.1722-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao A., Sasano Y., Imataka H., Inoue K., Sakamoto H., Sonenberg N., Thoma C. and Fujiwara T. (2009). The ELAV protein HuD stimulates cap-dependent translation in a Poly(A)- and eIF4A-dependent manner. _Mol. Cell_36, 1007-1017. 10.1016/j.molcel.2009.11.013 [DOI] [PubMed] [Google Scholar]
- Gomes C., Smith S. C., Youssef M. N., Zheng J.-J., Hagg T. and Hetman M. (2011). RNA polymerase 1-driven transcription as a mediator of BDNF-induced neurite outgrowth. _J. Biol. Chem._286, 4357-4363. 10.1074/jbc.M110.170134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes C., Merianda T. T., Lee S. J., Yoo S. and Twiss J. L. (2014). Molecular determinants of the axonal mRNA transcriptome. _Dev. Neurobiol._74, 218-232. 10.1002/dneu.22123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinski A. L., Sachdeva R., Gomes C., Lee S. J., Shah Z., Houle J. D. and Twiss J. L. (2015). mRNAs and protein synthetic machinery localize into regenerating spinal cord axons when they are provided a substrate that supports growth. _J. Neurosci._35, 10357-10370. 10.1523/JNEUROSCI.1249-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsak L. I. T., Shepard K. A. and Akins M. R. (2017). Cell type-dependent axonal localization of translational regulators and mRNA in mouse peripheral olfactory neurons. _J. Comp. Neurol._525, 2202-2215. 10.1002/cne.24199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehnert J., Sommer G., Zierk A. W., Fedarovich A., Brock A., Fedarovich D. and Heise T. (2015). Novel RNA chaperone domain of RNA-binding protein La is regulated by AKT phosphorylation. _Nucleic Acids Res._43, 581-594. 10.1093/nar/gku1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melemedjian O. K., Asiedu M. N., Tillu D. V., Peebles K. A., Yan J., Ertz N., Dussor G. O. and Price T. J. (2010). IL-6- and NGF-induced rapid control of protein synthesis and nociceptive plasticity via convergent signaling to the eIF4F complex. _J. Neurosci._30, 15113-15123. 10.1523/JNEUROSCI.3947-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merianda T. T., Gomes C., Yoo S., Vuppalanchi D. and Twiss J. L. (2013). Axonal localization of neuritin/CPG15 mRNA in neuronal populations through distinct 5′ and 3′ UTR elements. _J. Neurosci._33, 13735-13742. 10.1523/JNEUROSCI.0962-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merianda T. T., Coleman J., Kim H. H., Kumar Sahoo P., Gomes C., Brito-Vargas P., Rauvala H., Blesch A., Yoo S. and Twiss J. L. (2015). Axonal amphoterin mRNA is regulated by translational control and enhances axon outgrowth. _J. Neurosci._35, 5693-5706. 10.1523/JNEUROSCI.3397-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minis A., Dahary D., Manor O., Leshkowitz D., Pilpel Y. and Yaron A. (2014). Subcellular transcriptomics-dissection of the mRNA composition in the axonal compartment of sensory neurons. _Dev. Neurobiol._74, 365-381. 10.1002/dneu.22140 [DOI] [PubMed] [Google Scholar]
- Mobarak C. D., Anderson K. D., Morin M., Beckel-Mitchener A., Rogers S. L., Furneaux H., King P. and Perrone-Bizzozero N. I. (2000). The RNA-binding protein HuD is required for GAP-43 mRNA stability, GAP-43 gene expression, and PKC-dependent neurite outgrowth in PC12 cells. _Mol. Biol. Cell_11, 3191-3203. 10.1091/mbc.11.9.3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. B. and Fainzilber M. (2014). Local translation in neuronal processes--in vivo tests of a “heretical hypothesis”. _Dev. Neurobiol._74, 210-217. 10.1002/dneu.22115 [DOI] [PubMed] [Google Scholar]
- Perry R. B.-T., Rishal I., Doron-Mandel E., Kalinski A. L., Medzihradszky K. F., Terenzio M., Alber S., Koley S., Lin A., Rozenbaum M. et al. (2016). Nucleolin-mediated RNA localization regulates neuron growth and cycling cell size. _Cell Rep._16, 1664-1676. 10.1016/j.celrep.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N., Quan J., Li X., Nielsen F. C. and Flanagan J. G. (2016). IMP2 axonal localization, RNA interactome, and function in the development of axon trajectories. _Development_143, 2753-2759. 10.1242/dev.128348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saal L., Briese M., Kneitz S., Glinka M. and Sendtner M. (2014). Subcellular transcriptome alterations in a cell culture model of spinal muscular atrophy point to widespread defects in axonal growth and presynaptic differentiation. _RNA_20, 1789-1802. 10.1261/rna.047373.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka T., Jung H., Jung J., Turner-Bridger B., Ohk J., Lin J. Q., Amieux P. S. and Holt C. E. (2016). Dynamic axonal translation in developing and mature visual circuits. _Cell_166, 181-192. 10.1016/j.cell.2016.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene J. H., Jacobson R. D., Snipes G. J., McGuire C. B., Norden J. J. and Freeman J. A. (1986). A protein induced during nerve growth (GAP-43) is a major component of growth-cone membranes. _Science_233, 783-786. 10.1126/science.3738509 [DOI] [PubMed] [Google Scholar]
- Smith C. L., Afroz R., Bassell G. J., Furneaux H. M., Perrone-Bizzozero N. I. and Burry R. W. (2004). GAP-43 mRNA in growth cones is associated with HuD and ribosomes. _J. Neurobiol._61, 222-235. 10.1002/neu.20038 [DOI] [PubMed] [Google Scholar]
- Sosanya N. M., Huang P. P. C., Cacheaux L. P., Chen C. J., Nguyen K., Perrone-Bizzozero N. I. and Raab-Graham K. F. (2013). Degradation of high affinity HuD targets releases Kv1.1 mRNA from miR-129 repression by mTORC1. _J. Cell Biol._202, 53-69. 10.1083/jcb.201212089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A. M., Berchtold N. C., Perreau V. M., Tu C. H., Li Jeon N. and Cotman C. W. (2009). Axonal mRNA in uninjured and regenerating cortical mammalian axons. _J. Neurosci._29, 4697-4707. 10.1523/JNEUROSCI.6130-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twiss J. L., Smith D. S., Chang B. and Shooter E. M. (2000). Translational control of ribosomal protein L4 is required for rapid neurite extension. _Neurobiol. Dis._7, 416-428. 10.1006/nbdi.2000.0293 [DOI] [PubMed] [Google Scholar]
- Vuppalanchi D., Coleman J., Yoo S., Merianda T. T., Yadhati A. G., Hossain J., Blesch A., Willis D. E. and Twiss J. L. (2010). Conserved 3′-untranslated region sequences direct subcellular localization of chaperone protein mRNAs in neurons. _J. Biol. Chem._285, 18025-18038. 10.1074/jbc.M109.061333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B. A., Hengst U., Kim H. J., Jeon N. L., Schmidt E. F., Heintz N., Milner T. A. and Jaffrey S. R. (2012). Reprogramming axonal behavior by axon-specific viral transduction. _Gene Ther._19, 947-955. 10.1038/gt.2011.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis D. E., van Niekerk E. A., Sasaki Y., Mesngon M., Merianda T. T., Williams G. G., Kendall M., Smith D. S., Bassell G. J. and Twiss J. L. (2007). Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. _J. Cell Biol._178, 965-980. 10.1083/jcb.200703209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S., Kim H. H., Kim P., Donnelly C. J., Kalinski A. L., Vuppalanchi D., Park M., Lee S. J., Merianda T. T., Perrone-Bizzozero N. I. et al. (2013). A HuD-ZBP1 ribonucleoprotein complex localizes GAP-43 mRNA into axons through its 3′ untranslated region AU-rich regulatory element. _J. Neurochem._126, 792-804. 10.1111/jnc.12266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivraj K. H., Tung Y. C. L., Piper M., Gumy L., Fawcett J. W., Yeo G. S. H. and Holt C. E. (2010). Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. _J. Neurosci._30, 15464-15478. 10.1523/JNEUROSCI.1800-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information