Structural basis for the functional role of the Shu complex in homologous recombination (original) (raw)
Abstract
The Shu complex, a conserved regulator consisting of Csm2, Psy3, Shu1 and Shu2 in budding yeast, plays an important role in the assembly of the Rad51–ssDNA filament in homologous recombination. However, the molecular basis for the assembly of the Shu complex and its functional role in DNA repair is still elusive. Here, we report the crystal structure of the yeast Shu complex, revealing that Csm2, Psy3, Shu1 and Shu2 interact with each other in sequence to form a V-shape overall structure. Shu1 adopts a structure resembling the ATPase core domain of Rad51 and represents a new Rad51 paralog. Shu2 assumes a novel structural fold consisting of a conserved zinc-finger containing SWIM domain and a small insertion domain. The functional roles of the key residues are validated using mutagenesis and in vitro pull-down and in vivo yeast growth studies. Structural analysis together with available biological data identifies two potential DNA-binding sites, one of which might be responsible for binding the ssDNA region of the 3′-overhang DNA and the other for the dsDNA region. Collectively, these findings reveal the molecular basis for the assembly of the Shu complex and shed new insight on its functional role in homologous recombination.
INTRODUCTION
Homologous recombination (HR) is a highly conserved error-free DNA repair mechanism from yeasts to humans, which is responsible for repair of the most harmful DNA lesions, such as DNA double-strand breaks (DSBs) and inter-strand crosslinks (ICLs), and thus plays a critical role in maintenance of genome integrity (1,2). In addition, HR also plays an important role in maintenance of telomeres, proper segregation of homologous chromosomes in meiosis, and resolving of stalled and collapsed replication forks. Hence, HR is a tightly regulated process and defects in HR can lead to genomic instability. In humans, HR defects can cause many genetic diseases and cancers, such as Fanconi anaemia, Bloom syndrome, and Werner syndrome (3,4). A critical step in HR is the formation of a Rad51-single-stranded DNA (ssDNA) nucleoprotein filament (also called presynaptic filament), which is essential for the homology search and strand invasion steps leading to the formation of a displacement loop (D-loop) (2,5,6). The loading of Rad51 onto the replication protein A (RPA) complex-coated ssDNA is facilitated by a class of proteins called Rad51 mediators, including Rad52 and the Rad51 paralogs Rad55-Rad57 in Saccharomyces cerevisiae, and RAD52 and BRCA2 in humans (7–9).
Saccharomyces cerevisiae primarily uses HR to repair DSBs and thus is the most common model organism for studying the mechanism of DSB repair by HR (7). In the budding yeast, four genes PSY3, CSM2, SHU1, and SHU2 are identified in a genetic screen as suppressors of the slow growth of _top3_Δ or _sgs1_Δ strains under hydroxyurea (HU) or methyl methanesulfonate (MMS) stress (10). Mutants of these genes impair the genome stability in both mitosis and meiosis, and increase the sensitivity of yeast strains to DNA damage reagents, such as MMS, cisplatin, and camptothecin (10–15). In addition, deletion of any individual gene confers similar sensitivity as deletion of the four genes (10). Yeast-two-hybrid (Y2H), immunoprecipitation and recombination expression analyses showed that Psy3, Csm2, Shu1, and Shu2 could form a heterotetrameric complex both in vitro and in vivo, which is referred as the Shu complex (also called the PCSS complex) (16–18). It has been demonstrated that the Shu complex can regulate HR at several types of DNA DSBs (including replication-associated and meiotic DSBs) and specifically functions as a Rad51 mediator which, in cooperation with Rad52 and Rad55–Rad57, promotes the Rad51 loading on the RPA-coated ssDNA and thus the formation of Rad51 presynaptic filament (15,18–21). Genetic and bioinformatic studies have shown that Shu2 orthologs are conserved across eukaryotes from archaea to humans, which define the Shu2/SWS1 protein family; and additionally, orthologs of other components of the Shu complex have also been identified in some species including Schizosaccharomyces pombe, Caenorhabditis elegans, and humans (22–26). These data indicate that the Shu complex might be a conserved regulator in the formation of Rad51–ssDNA filament in eukaryotes.
In the Shu complex, the structure of the Csm2–Psy3 heterodimer has been solved (16,17,27). Although Csm2 and Psy3 share a very low sequence identity (17%), they have a similar structural architecture resembling the ATPase core domain of Rad51 and particularly the Csm2-Pys3 heterodimer resembles a Rad51 homodimer, and therefore both Csm2 and Psy3 are deemed as Rad51 paralogs. In addition, the biochemical studies have shown that Csm2–Psy3 is responsible for the DNA-binding activity of the Shu complex: Csm2–Psy3 binds preferentially to a forked DNA and to a lesser extent, 3′-overhang DNA; whereas Shu1–Shu2 cannot bind to DNA (16,17,20,27). Shu1 is a relatively small protein in the Shu complex and is suggested to be a Rad51 paralog without a clearly defined functional domain, but no ortholog has been identified in other species (26). Shu2 is conserved in other eukaryotes and all members of the Shu2/SWS1 protein family contain a unique Zn finger-like SWIM domain comprising a characteristic sequence motif of CxCx_n_CxH, which is predicted to have DNA-binding and protein-protein interaction functions (28). It is noteworthy that mutations in human SWS1 have been associated with unexplained colorectal adenomatous polyposis, and particularly a homozygous mutation in the SWIM domain of SWS1 results in a frame shift leading to disruption of the zinc coordination of the SWIM domain (29). This observation also underscores the importance of the Shu complex in HR regulation and implies potential biomedical significance. So far, the structures and functions of Shu1 and Shu2 and the molecular basis for the assembly of the Shu complex are still unknown. In addition, it remains elusive how the Shu complex facilitates the formation of the presynaptic filament during HR.
Here, we report the crystal structure of the yeast Shu complex and show that Csm2, Psy3, Shu1, and Shu2 interact with each other in sequence to form a V-shape overall structure. Shu1 assumes a structure resembling the ATPase core domain of Rad51 and thus represents a new Rad51 paralog. As the first structure of the Shu2/SWS1 family members, Shu2 assumes a novel protein fold consisting of a conserved SWIM domain and a small insertion domain. The protein-protein interfaces involve predominantly hydrophobic interactions supplemented with a few hydrophilic interactions. The key residues at the protein-protein interfaces are validated by both in vivo and in vitro functional assays. The structural and functional data reveal the molecular basis for the assembly of the Shu complex, and shed new light on its functional role in the formation of the Rad51–ssDNA filament in HR.
MATERIALS AND METHODS
Cloning, expression and purification of proteins
Full-length PSY3 (residues 1–242), CSM2 (residues 1–213), SHU1 (residues 1–150) and SHU2 (residues 1–223) genes were amplified from S. cerevisiae genomic DNA. For structural study, the PSY3 and CSM2 genes were inserted into the pRSFDuet plasmid (Novagen) and the SHU1 and SHU2 genes were inserted into the pETDuet plasmid (Novagen) with a His6 tag attached to the N-terminus of Shu2. Mutants were generated using the QuikChange® Site-Directed Mutagenesis kit (Strategene) and confirmed by DNA sequencing.
The two reconstructed expression plasmids were co-transformed into Escherichia coli BL21 (DE3) Codon-Plus strain (Novagen). The transformed cells were grown in LB medium at 37°C containing 0.05 mg/ml ampicillin and kanamycin until OD600 reached 0.8 and then induced with 0.2 mM IPTG at 16°C for 24 hr. The cells were harvested by centrifugation and lysed by sonication in a lysis buffer (30 mM Tris–HCl, pH 8.0, 300 mM NaCl and 1 mM PMSF). The target proteins were purified by affinity chromatography using a Ni-NTA column (Qiagen) and then gel filtration chromatography using a Superdex 200 16/600 column (GE Healthcare), and stored in a buffer containing 30 mM Tris–HCl (pH 8.0) and 300 mM NaCl. The purified proteins were of high purity (>95%) and homogeneity as determined by Tricine-SDS-PAGE.
Single-stranded oligonucleotides corresponding to the template and complementary strands were synthesized by Sangon Biotech (Shanghai). The dsDNAs were prepared by annealing of the template and complementary strands from 95°C to 22°C over a period of 6 h in the same storage buffer as for the proteins.
Crystallization, data collection, structure determination and refinement
Prior to crystallization, the dsDNA was incubated with the Shu complex at a molar ratio of 1.2:1 at 4°C overnight. The final concentration of the complex for crystallization was about 20 mg/ml. Crystallization was performed using the hanging drop vapor diffusion method. Crystals were grown at 16°C from drops containing equal volumes (1 μl) of the complex solution and the reservoir solution (0.1 M Bis–Tris, pH 6.5, 0.2 M potassium sodium tartrate, and 10% PEG 10 000). For diffraction data collection, the crystals were cryo-protected using the reservoir solution supplemented with 25% glycerol and then flash-cooled into liquid nitrogen. Diffraction data were collected at 100 K at BL19U1 of National Facility for Protein Science in Shanghai, China, and were processed, integrated and scaled together with HKL3000 (30).
The structure of the Shu complex was solved using a combination of the molecular replacement (MR) and single-wavelength anomalous dispersion (SAD) phasing methods as implemented in Phenix (31). Initial phases were determined by the MR method using the structure of the Csm2–Psy3 heterodimer (PDB code 3VU9) (16) as the search model. In the MR-phased electron density map, the structure of Csm2–Psy3 was well defined, and additionally there was evident and continuous electron density beside Psy3. Considering that Shu2 has a Zn-containing SWIM domain, the SAD was performed using Zn as the anomalous atom and the preliminary MR solution as the partial model, yielding a figure-of-merit (FOM) of 0.411. The resultant electron density map revealed a large portion of the secondary structure elements of Shu1 and Shu2. Iterative cycles of manual model building and refinement gradually developed more electron density that allowed us to build the full model of the Shu1–Shu2 heterodimer. Structure refinement was carried out using Phenix and Refmac5 (32,33) and model building using Coot (34). Structural analyses were carried out using programs in the CCP4 suite (33) and the PISA server (35). The figures were generated using Pymol (http://www.pymol.org). Statistics of the structure refinement and the final structure model are summarized in Table 1.
Table 1. Summary of diffraction data and structure refinement statistics.
| Diffraction data | |
|---|---|
| Wavelength (Å) | 0.9785 |
| Space group | _P_43212 |
| Cell parameters | |
| a, b, c (Å) | 174.2, 174.2, 102.9 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 50.0-3.3 (3.4–3.3)a |
| Observed reflections | 158 237 |
| Unique reflections (I/σ(I) > 0) | 24 373 |
| Average redundancy | 6.5 (6.3) |
| Average I/σ(I) | 20.3 (3.0) |
| Completeness (%) | 99.9 (100.0) |
| R merge (%)b | 8.9 (69.2) |
| Refinement and structure model | |
| No. of reflections (_F_o > 0σ(_F_o)) | 23,864 |
| Working set | 22,642 |
| Free R set | 1222 |
| _R_-factor/free _R_-factor (%)c | 20.8/25.7 |
| No. of atoms | |
| Protein | 6027 |
| Zn | 1 |
| Average B factor (Å2) | |
| Protein | 38.5 |
| Zn | 21.5 |
| RMS deviations | |
| Bond lengths (Å) | 0.011 |
| Bond angles (°) | 1.2 |
| Ramachandran plot (%) | |
| Favored | 96.9 |
| Allowed | 3.1 |
Yeast two-hybrid assays
Yeast-two-hybrid (Y2H) assay plasmid pBTM116 was used to express a fusion of LexA-binding domain, and plasmid pACT2 to express a fusion of GAL4-activation domain (Clontech). The gene fragments with or without indicated mutations were inserted into pBTM116 or pACT2. The Y2H constructs were co-transformed into yeast L40 strain. Cells were cultured on selective SD-Trp-Leu plates. Individual cells were grown at 30°C to early log phase (OD600 of 0.2), and then 5 μl cell culture was spotted onto medium (SD-Leu-Trp) to select for the plasmids or onto medium (SD-Leu-Trp-His, 5 mM 3AT) to select for the expression of reporter gene HIS3 as an indicator of protein-protein interaction. Plates were incubated for 2–4 days at 30°C and then photographed. All experiments were done in triplicate.
Semi-quantitative growth assays
The pRS316-PSY3, pRS316-CSM2, pRS316-SHU1 and pRS316-SHU2 plasmids were constructed by cloning the gene fragments containing the coding regions with or without the indicated mutations and their upstream 1000-bp and downstream 500-bp regions into the BamHI–XhoI sites of pRS316. The plasmids were confirmed by DNA sequencing. The wild-type BY4742 (_MAT_α his3Δ1 leu2Δ0 lys2Δ0 ura3Δ) strain and the _psy3_Δ, _csm2_Δ, _shu1_Δ, and _shu2_Δ BY4742 strains were used. Phenotypic analysis was performed using serial dilutions of the yeast cells grown at 30°C to early mid-log phase (OD600 of 1.0–1.5). Serial dilutions were carried out by spotting the cells and diluting 5-fold for each spot onto the indicated plates. Plates were grown at 30°C for 3–4 days and then photographed. All experiments were done in triplicate.
Affinity pull-down assays
The full-length PSY3 and CSM2 genes were inserted into pETDuet (Novagen) with a His6 tag attached to the N-terminus of Psy3, and the full-length SHU2 and SHU1 were inserted into pETDuet with a His6 tag attached to the N-terminus of Shu2. For later removal of the His6-tag, a human rhinovirus (HRV) 3C protease cleavage site was inserted between the His6 tag and the gene sequence. Mutations and deletions were performed based on these two plasmids. His6-tagged Psy3-Csm2, Psy3Δ(1–7)-Csm2, Psy3Δ(1–15)-Csm2, and His6-tagged Shu2-Shu1, Shu2(V51A)-Shu1, Shu2(V51L)-Shu1, and Shu2(V51D)-Shu1 were expressed and purified as described above. The purified His6-tagged wild-type and mutant Shu2–Shu1 proteins were digested by HRV 3C protease to remove the His6 tag. The purified His6-tagged wild-type and mutant Psy3–Csm2 proteins were covalently conjugated with Ni-NTA agarose beads (Qiagen), and the control beads were prepared using the same method in the absence of protein. For pull-down assay, 20 μg His6–Psy3–Csm2 conjugated beads or control beads were incubated with 100 μg Shu2-Shu1 at 4°C for 2 h, and then washed three times (10 min each) with 1 ml buffer (30 mM Tris–HCl, pH 8.0, 300 mM NaCl, and 0.1% NP-40). The bound proteins were analyzed by Tricine-SDS-PAGE and visualized by Coomassie blue staining.
RESULTS
Crystal structure of the Shu complex
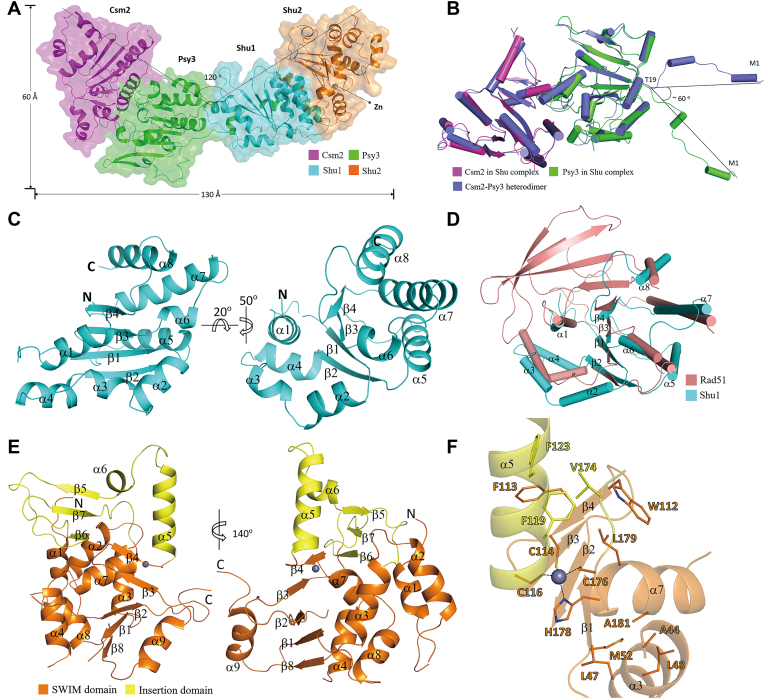
To carry out the structural study of the Shu complex, we co-expressed full-length Psy3, Csm2, Shu1 and Shu2 proteins in E. coli with a His6 tag attached on the N-terminus of Shu2, and purified the Shu complex using a combination of affinity chromatography and gel filtration chromatography. The purified Shu complex is of high purity and homogeneity and exists as a monomer in solution as revealed by gel filtration, SDS-PAGE and SEC-MALS analyses (Supplementary Figure S1). Crystallization experiments of the Shu complex alone did not yield any crystals. Considering that the Shu complex has a reasonably high binding affinity (_K_D = 4.66 μM) for double-stranded DNA (dsDNA) (27), we performed co-crystallization experiments of the Shu complex with several dsDNA of different lengths and successfully obtained crystals of the Shu complex in the presence of 16-bp, 18-bp, or 20-bp dsDNA. Crystals of the Shu complex grown in the presence of 20-bp dsDNA diffracted X-rays better than other crystals and thus were used for diffraction data collection and structure determination. As the Shu1–Shu2 heterodimer could be expressed and purified separately, we also tried to crystallize Shu1–Shu2 alone; however, various attempts failed to produce any crystals. The structure of the Shu complex was solved using a combination of the MR and SAD methods and was refined to 3.3 Å resolution (Table 1). There is one Shu complex in the asymmetric unit, in which most residues of the four proteins are well defined except for some surface exposed loops or residues (residues 99–106, 178–195 and 213 of Csm2, residues 63–66, 145–155, 194–205, 228–231 and 240–242 of Psy3, residues 18–20 and 102 of Shu1, and residues 1–3, 19–20, 60–69 and 83–85 of Shu2) (Supplementary Figure S2A and B). Intriguingly, no electron density was observed for the dsDNA, indicating that the dsDNA does not bind tightly to the Shu complex but its presence might facilitate the crystal packing of the complex.
In the structure of the Shu complex, Csm2, Psy3, Shu1 and Shu2 interact with each other sequentially, and the Csm2–Psy3 and Shu1–Shu2 subcomplexes make an angle of about 120° to form a V-shape overall structure (Figure 1A). Structural comparison shows that Csm2–Psy3 in the Shu complex assumes almost identical structure as the previously reported Csm2–Psy3 alone (16,17,27) with a root-mean-square deviation (RMSD) of 0.4 Å for 345 Cα atoms. However, the N-terminal region (∼19 residues) of Psy3, which forms two short α-helices (α1 and α2), exhibits substantially different conformations with a rotation of ∼60° around Thr19 (Figure 1B). In the Shu complex, the N-terminal region of Psy3 is stretched outside the bulk of Psy3 and interacts directly with Shu1 (see discussion later); in the Csm2–Psy3 structure, this region is also extended outside the bulk of Psy3 but interacts with a symmetry-related Csm2 (Supplementary Figure S2C). Like in the Csm2–Psy3 structure, the L2 loops of Csm2 (residues 175–203) and Psy3 (residues 187–211), which are suggested to be involved in DNA binding, are partially disordered in the Shu complex.
Figure 1.
Crystal structure of the Shu complex. (A) Overall structure of the Shu complex. The Shu complex is shown with a ribbon model in a transparent envelope surface. Csm2, Psy3, Shu1 and Shu2 are colored in magenta, green, cyan, and orange, respectively. The Zn2+ ion is shown with a gray ball. (B) Superposition of the Csm2–Psy3 heterodimer in the Shu complex and in the Csm2–Psy3 structure (colored in slate, PDB code: 3VU9). (C) Overall structure of Shu1 in two different views. Shu1 is shown with a ribbon model with the secondary structure elements labeled. (D) Superposition of Shu1 and the ATPase core domain (residues 191–354) of Rad51 (colored in salmon, PDB code: 1SZP). The secondary structure elements of Shu1 are labeled. (E) Overall structure of Shu2 in two different views. Shu2 is shown with a ribbon model with the SWIM domain and the insertion domain colored in orange and yellow, respectively. The secondary structure elements of Shu2 are labeled. (F) Structure of the Zn-binding site in the SWIM domain of Shu2. The Zn2+ ion is shown with a gray ball and the side chains of key residues are shown in stick models. Interactions between the Zn2+ ion and coordination residues are indicated with dash lines.
The structure of Shu1 consists of a parallel four-stranded β-sheet (β1–β4) flanked by four α-helices on each side (α1–α4 on one side and α5–α8 on the other) (Figure 1C and Supplementary Figure S3A). Previously, Shu1 was suggested to be a Rad51 paralog (26). Structural comparison shows that Shu1 shares a high structural similarity with the ATPase core domain (residues 191-354) of Rad51 with a RMSD of 2.7 Å for 119 Cα atoms (Figure 1D). Nevertheless, the central β-sheet of the Rad51 ATPase domain contains 9 β-strands whereas that of Shu1 contains only 4 β-strands corresponding to β2–β5 of Rad51 (Supplementary Figure S3A and B). These results indicate that Shu1 is indeed a Rad51 paralog with some variations.
Shu2 is a founding member of the Shu2/SWS1 protein family, which contains a Zn finger-like SWIM domain (5,24). Interestingly, Shu2 contains an extremely long insertion in the conserved CxCxnCxH motif (number n = 59), which is much longer than that of a typical SWIM domain (usually n = 6–16) (28) (Supplementary Figure S4A). The structure of Shu2 consists of two relatively separated domains: the SWIM domain forms a large domain comprising a five-stranded β-sheet (β1–β4 and β8) flanked by six α-helices (α1–α4, α7 and α8) on one side and one α-helix (α9) on the other; the insertion region of the conserved motif (residues 117–175) forms a small insertion domain comprising an antiparallel three-stranded β-sheet (β5–β7) and two α-helices (α5-α6) (Figure 1E and Supplementary Figure S3C). The Zn-binding site is located at the interface of the two domains with the Zn2+ coordinated by four invariable residues, Cys114 and Cys116 from the β4–α5 loop, Cys176 from the β7–α7 loop, and His178 from α7 (Figure 1F). Intriguingly, there are two hydrophobic cores flanking the Zn-binding site: one is centered at Phe119 surrounded by Trp112, Phe113, Phe123, Val174 and Leu179; and the other is centered at Ala181 surrounded by Ala44, Leu47, Leu48 and Met52 (Figure 1F). Sequence comparison shows that Phe119 and Ala181 are strictly conserved in all members of the Shu2/SWS1 family identified so far (Supplementary Figure S4A). Our functional studies indicate that both residues play important roles in the stabilization of the Zn-binding site and in the function of Shu2 (see discussion later). As Shu2 is the first structure of the Shu2/SWS1 family, we performed structural similarity search with other known protein structures using the Dali server (36). The search did not reveal significant structural similarities for Shu2 as a whole, or the SWIM domain and the insertion domain separately, indicating that the Shu2 structure represents a new structural fold containing a novel Zn-finger domain.
Interactions at the protein–protein interfaces
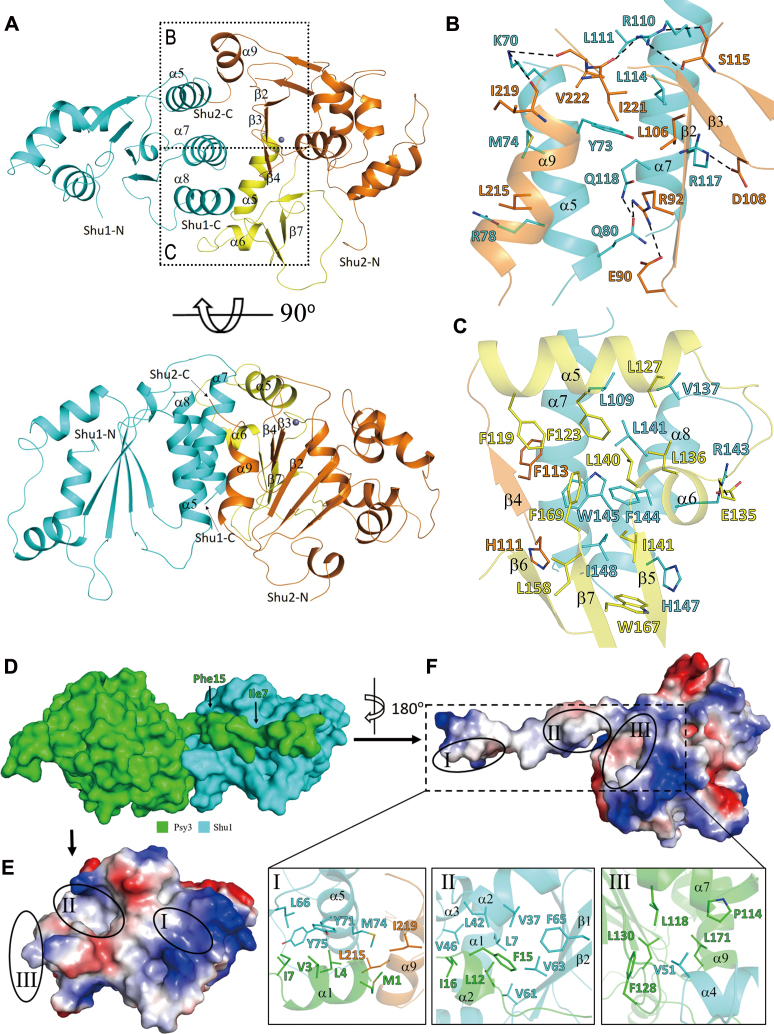
In the Shu complex, the interface of the Csm2–Psy3 heterodimer is almost identical to that in the Csm2–Psy3 structure, which is mediated largely by hydrophobic interactions supplemented with a few hydrogen-bonding interactions (16,17,27). Shu1 and Shu2 form another heterodimer, and the interface is mediated by numerous residues from Shu1 and Shu2 which buries a total solvent-accessible surface area of 3034 Å2 (Figure 2 and Supplementary Figure S5). The interface involving the SWIM domain of Shu2 comprises both hydrophobic and hydrophilic interactions (Figure 2B and Supplementary Figure S5A and C). The hydrophobic interactions are formed between Tyr73, Met74, Arg78, Leu111, Leu114, Trp145 and Ile148 of Shu1 and Leu106, His111, Phe113, Leu215, Ile221 and Val222 of Shu2. Additionally, the side chain of Lys70 of Shu1 makes hydrogen-bonding interactions with the main chains of Ile219 and Val222 of Shu2; the side chain of Gln80 of Shu1 forms a hydrogen bond with the side chain of Arg92 of Shu2; the side chain of Arg110 of Shu1 makes hydrogen-bonding interactions with the main chain and side chain of Ser115 and the main chain of Ile221 of Shu2; and the side chain of Arg117 of Shu1 forms a salt bridge with the side chain of Asp108 of Shu2 (Figure 2B and Supplementary Figure S5A and C). The interface involving the insertion domain of Shu2 comprises predominantly hydrophobic interactions between Leu109, Val137, Leu141, Phe144, His147 and Ile148 of Shu1 and Phe123, Leu127, Leu136, Leu140, Ile141, Leu158, Trp167 and Phe169 of Shu2. In this region, only the side chain of Arg143 of Shu1 forms a salt bridge with the side chain of Glu135 of Shu2 (Figure 2C and Supplementary Figure S5B and C).
Figure 2.
Interactions at the protein-protein interfaces. (A) Overall structure of the Shu1–Shu2 heterodimer in two different views. Shu1 is colored in cyan, and the SWIM domain and insertion domain of Shu2 are colored in orange and yellow, respectively. The Zn2+ ion is shown with a gray ball. (B) Interface between Shu1 and the SWIM domain of Shu2. (C) Interface between Shu1 and the insertion domain of Shu2. The residues involved in the interactions are shown in ball-and-stick models. The hydrophilic interactions are indicated with dashed lines. For clarity, only Ser115, Ile219, Ile221 and Val222 of Shu2 are shown with full models and the other residues are shown with side chains. (D) Surface presentation of the Psy3–Shu1 heterodimer. Psy3 and Shu1 are colored in green and cyan, respectively. (E and F) Electrostatic surfaces of Shu1 and Psy3 showing the interaction interface. The three interaction regions are circled with solid lines. The interaction interfaces of Shu1 and Psy3 exhibit both geometrical and electrostatic complementarities. Detailed interactions in the three regions are shown in the zoom-in windows. The side chains of residues involved in the interactions are shown in stick models.
The interface between the Csm2–Psy3 and Shu1–Shu2 heterodimers is primarily mediated by residues of Psy3 and Shu1, which buries a total solvent-accessible surface area of 2900 Å2 (Figure 2D–F and Supplementary Figure S6). Specifically, the N-terminal region of Psy3 lies across the surface of Shu1 and makes predominantly hydrophobic interactions with residues from several structure elements of Shu1 and additionally a very small portion of Shu2. The interface could be divided into three regions (Figure 2E and F). In region I, Met1, Val3, Leu4 and Ile7 of Psy3 make hydrophobic contacts with Leu66, Tyr71, Met74 and Tyr75 of Shu1, and Leu215 and Ile219 of Shu2. In region II, Leu12, Phe15 and Ile16 of Psy3 are buried into a hydrophobic pocket formed by Leu7, Val37, Leu42, Val46, Val61, Val63, and Phe65 of Shu1. In region III, Val51 of Shu1 is inserted into a hydrophobic cavity formed by Pro114, Leu118, Phe128, Leu130 and Leu171 of Psy3.
Validation of the functional roles of key residues in the assembly of the Shu complex
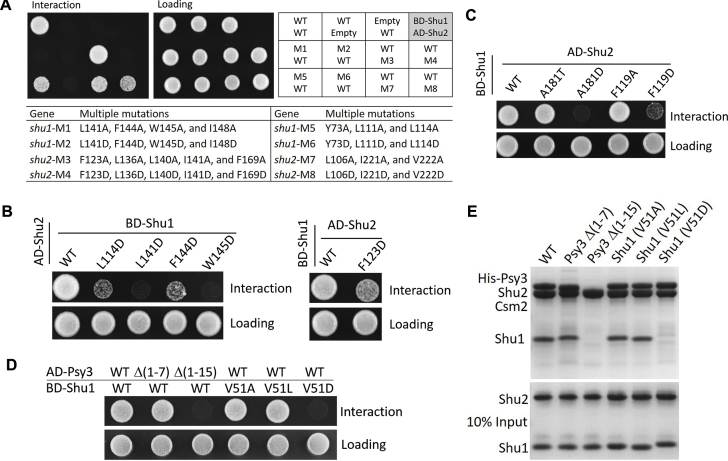
To investigate whether the protein interactions between members of the Shu complex are important for the assembly of the complex, we performed mutagenesis and Y2H or in vitro pull-down assays. The functional roles of key residues at the Csm2–Psy3 interface have been previously validated by in vitro pull-down assays (27) and thus we would not repeat here. Firstly, we analyzed the functional roles of the key residues at the Shu1–Shu2 interface. As Shu1 and Shu2 could not be expressed and purified separately, we performed Y2H instead of pull-down assays to analyze the effects of mutations on the Shu1–Shu2 interaction. The Shu1–Shu2 interface involves numerous residues from both Shu1 and Shu2, and we designed four sets of residues and mutated them to either Ala or Asp (Figure 2B and C). At the interface involving the insertion domain of Shu2, the first set consists of Leu141, Phe144, Trp145 and Ile148 of Shu1 (designated as M1 and M2 for mutations to Ala and Asp, respectively), and the second set consists of Phe123, Leu136, Leu140, Ile141, and Phe169 of Shu2 (M3 and M4). At the interface involving the SWIM domain of Shu2, the third set consists of Tyr73, Leu111 and Leu114 of Shu1 (M5 and M6), and the fourth set consists of Leu106, Ile221 and Val222 of Shu2 (M7 and M8). Our Y2H results show that M1, M2, M4 and M6 completely disrupt the Shu1–Shu2 interaction; M5, M7, and M8 significantly impair the interaction; and M3 has no notable effect on the interaction (Figure 3A and Supplementary Figure S7B). To analyze whether the multiple mutations would affect expression of the proteins, we constructed a series of co-expression plasmids of Shu1 and Shu2 containing the eight sets of mutations and expressed them using the same method as the WT co-expression plasmid in E. coli. The results show that these mutations have no significant impact on the expression of Shu1 and Shu2, but impair the Shu1–Shu2 interaction to differed degrees as analyzed by purification of the Shu1–Shu2 complex using His6-Shu2 to pull-down Shu1 (Supplementary Figure S8A and B). Among the four Shu1 mutants, M1, M2, and M6 have no interaction with Shu2 and M5 of Shu1 has weak interaction with Shu2 (Supplementary Figure S8A). Among the four Shu2 mutants, M4 has no interaction with Shu1 and M3, M7 and M8 have weak interaction with Shu1 (Supplementary Figure S8B). These results are largely consistent with the Y2H assay results except for the M3 mutation of Shu2, which weakens moderately the interaction with Shu1 in the purification experiment but has no significant effect on the interaction with Shu1 in the Y2H assay. To further identify the key residues at the Shu1–Shu2 interface, we performed single mutations of the above residues to Ala or Asp, and the Y2H results show that mutations L141D and W145D of Shu1 completely disrupt the Shu1–Shu2 interaction; mutations L114D and F144D of Shu1 and F123D of Shu2 severely impair the interaction; and the other mutations have no significant effects on the interaction (Figure 3B and Supplementary Figure S7C). Structural analysis shows that Leu114, Leu141, Phe144, and Trp145 of Shu1, and Phe123 of Shu2 are located at the core of the hydrophobic interactions, and their changes to Asp apparently destabilize the Shu1–Shu2 interaction (Supplementary Figure S9). Furthermore, sequence alignment shows that the four key residues of Shu1 are conserved in other fungal species, of which Leu114 and Trp145 are strictly conserved and Leu141 and Phe144 are highly conserved (Supplementary Figure S4B). Additionally, among all members of the Shu2/SWS1 family identified so far, Phe123 of Shu2 is also highly conserved with two variations to Ala or Val (Supplementary Figure S4A).
Figure 3.
Validation of the functional roles of key residues in the assembly of the Shu complex. (A) Y2H interactions between Shu1 and Shu2 containing without or with multiple mutations at the interface. The lower panel lists the detailed mutations of the M1-M8 mutants. (B) Y2H interactions between Shu1 and Shu2 containing without or with point mutations at the interface. (C) Y2H interactions between Shu1 and Shu2 containing without or with mutations of Phe119 and Ala181 surrounding the Zn-binding site of Shu2. (D) Y2H interactions between the Csm2–Psy3 and Shu1–Shu2 heterodimers containing without or with mutants of Psy3 or Shu1. (E) Pull-down assays between the His6-tagged Csm2–Psy3 heterodimer (WT or mutants of Psy3) and the Shu1–Shu2 heterodimer (WT or mutants of Shu1). Tricine-SDS-PAGE gels were analyzed by Coomassie blue staining.
Secondly, we analyzed the functional roles of the key residues at the Zn-binding site of Shu2 in the Shu1–Shu2 interaction. Consistent with the previous Y2H assays (24), our Y2H assays show that mutations C114S, C116S, C176S, and H178A of Shu2 disrupt the Shu1–Shu2 interaction (Supplementary Figure S7D). Although the Zn2+ and the coordination residues have no direct interactions with Shu1, disruption of the Zn-binding would affect the stability of the nearby structural elements (including α5, β3 and β4) which are involved in the interactions with Shu1 (Figure 2A). Structural analysis shows that there are two hydrophobic cores flanking the Zn-binding site centered at the conserved Phe119 and Ala181, respectively. The previous Y2H assays showed that mutations F119A and A181T have no significant effects on the Shu1–Shu2 interaction (24). We mutated Phe119 into Ala and Asp and Ala181 into Thr and Asp, respectively. Our Y2H results show that mutations F119A and A181T indeed have no significant effects on the Shu1–Shu2 interaction; however, mutation A181D completely disrupts and mutation F119D severely impairs the Shu1–Shu2 interaction (Figure 3C). It is possible that these mutations destabilize the hydrophobic cores and thus affect the Shu1–Shu2 interaction.
Thirdly, we analyzed the functional roles of the key residues at the interface of the Csm2–Psy3 and Shu1–Shu2 heterodimers on the formation of the Shu complex using both Y2H and pull-down assays. Based on the Psy3–Shu1 interaction, we prepared two Psy3 truncates [Δ(1–7) and Δ(1–15)] and three Shu1 mutants (V51A, V51L, and V51D). Our results show that the truncations and mutations have no effects on the expression and stability of the two subcomplexes (Supplementary Figure S8C and D). Truncation Δ(1–15) of Psy3 and mutation V51D of Shu1 completely disrupt the Psy3–Shu1 interaction in the Y2H assays and the assembly of the Shu complex in the pull-down assays; however, truncation Δ(1–7) of Psy3 and mutations V51A and V51L of Shu1 have no significant effects on the interaction and the assembly (Figure 3D and E). In addition, the sequence alignment shows that Val51 of Shu1 is highly conserved in other fungal species with two variations to Leu or Ile (Supplementary Figure S4B). The Y2H assay results suggest that residues in the region 8–15 of Psy3 are important for the Psy3–Shu1 interaction. Our structural analysis shows that in the region 8–15 of Psy3, residues Leu12 and Phe15 are buried in a hydrophobic pocket of Shu1 and have tight interactions with Shu1 (Figure 2F and Supplementary Figure S10A). The adjacent residue Ile16 of Psy3 is located on the edge of the hydrophobic cavity of Shu1 but has few contacts with residues of Shu1 (Figure 2F and Supplementary Figure S10A). To analyze the functional role of these residues in the Psy3–Shu1 interaction, we performed mutagenesis and Y2H assays. The results show that mutations L12D and F15D completely disrupt the interaction and mutation I16D has no significant impact on the interaction (Supplementary Figure S10B). These results together indicate that the interactions at region I of the Psy3–Shu1 interface are not essential, and the interactions at regions II and III are critical for the assembly of the Shu complex and particularly Val51 of Shu1 plays an important role in the assembly.
In vivo functional analyses of the Shu complex
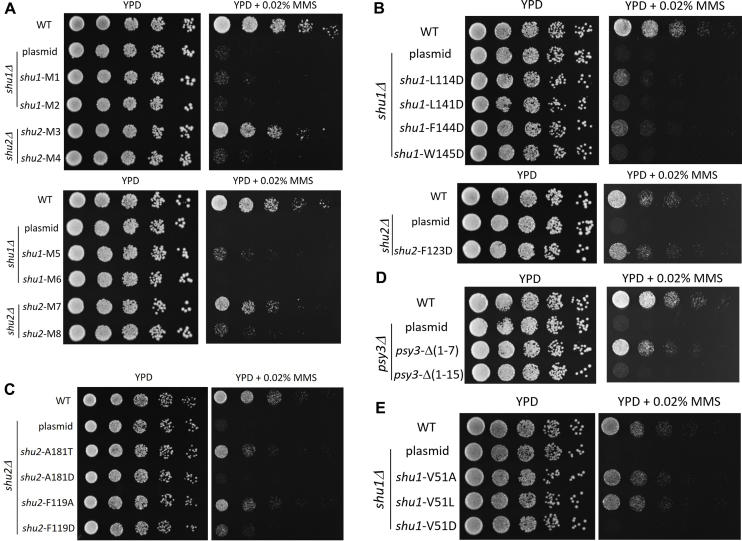
To investigate the functional role of the Shu complex in vivo, we performed mutagenesis and yeast MMS sensitivity assays to examine whether mutations of the key residues important for the assembly of the Shu complex affect its function in the repair of MMS-induced DNA damage. Previous genetic data demonstrate that all four components of the Shu complex are essential for its function in DNA repair (10,15). Indeed, our yeast growth experiments show that depletion of any component of the Shu complex abolishes its function in DNA repair (Supplementary Figure S11A–C). All mutations or truncations examined in the Y2H assays are also analyzed in the yeast MMS sensitivity assays. The results show that at the Shu1–Shu2 interface, M1, M2, M4, and M6 completely abolish, M5 and M8 severely impair, and M3 and M7 slightly affect the function of the Shu complex (Figure 4A). The single mutations of Shu1 and Shu2 that significantly impair the Shu1–Shu2 interaction either completely abolish (L141D and W145D), or severely impair (L114D and F144D), or moderately compromise (F123D) the function of the Shu complex (Figure 4B). At the Zn-binding site of Shu2, mutations C114S, C116S, C176S and H178A completely abolish the function of the Shu complex (Supplementary Figure S11D). In addition, mutation A181D completely abolishes and mutation F119D severely impairs the function of the Shu complex, but mutations A181T and F119A also moderately compromise the function of the complex (Figure 4C). At the Psy3–Shu1 interface, truncation Δ(1–15) and mutations L12D and F15D of Psy3 and mutation V51D of Shu1 completely abolish the function of the complex, and truncation Δ(1–7) and mutation I16D of Psy3 and mutations V51A and V51L of Shu1 only slightly impair the function of the complex (Figure 4D, E and Supplementary Figure S10C). All of these results are very consistent with our Y2H results. Collectively, our structural and functional data indicate that the Shu1–Shu2 interaction, the Zn-binding site of the SWIM domain of Shu2, and the Csm2–Psy3 and Shu1–Shu2 interface are critical for the assembly of the Shu complex and important for its function in the DNA repair.
Figure 4.
In vivo functional analyses of the Shu complex. (A and B) MMS sensitivity assays for multiple or single mutations of the residues involved in the Shu1–Shu2 interaction. (C) MMS sensitivity assays for mutations of Phe119 and Ala181 of Shu2. (D and E) MMS sensitivity assays for mutants of Psy3 and Shu1.
DISCUSSION
The yeast Shu complex consisting of Csm2, Psy3, Shu1 and Shu2, is a conserved regulator of homologous recombination in DNA repair. The previous biological studies show that the Shu complex, in cooperation together with the Rad55–Rad57 complex, can promote the formation of the Rad51–ssDNA filament in the HR process (15,18–21). In this work, we solved the crystal structure of the Shu complex and performed the structure-based functional studies. Our structural and functional data together reveal the molecular basis for the assembly of the Shu complex. In addition, our data together with the previous biological data shed light on the functional role of the Shu complex in the formation of the Rad51 presynaptic filament during HR.
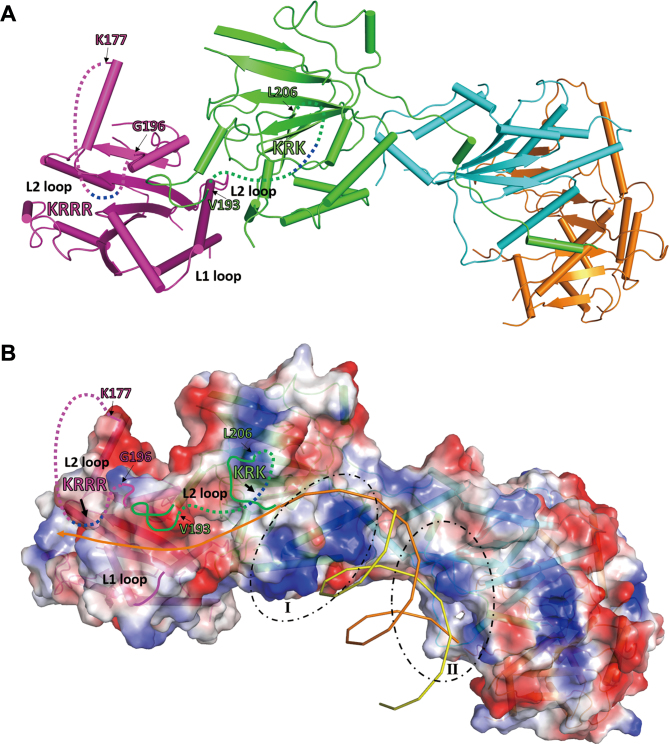
The previous biochemical data show that the Csm2–Psy3 subcomplex is able to bind preferentially to forked DNA and 3′-overhang DNA, both of which are DNA structures recognized and repaired by HR; whereas the Shu1–Shu2 subcomplex cannot bind to DNA (16,17,20,27). These results led to the suggestion that Csm2–Psy3 is responsible for the DNA-binding activity of the Shu complex, and might bind to the 5′-end of ssDNA and stabilize the Rad51 filaments (16). Intriguingly, the previous biochemical data also show that Psy3 alone lacks DNA-binding activity, but the Psy3–Shu1–Shu2 complex exhibits high affinity for both ssDNA and dsDNA (27). The previous structural and mutagenesis data show that both Csm2 and Psy3 contain a surface exposed, positively charged L2 loop; and mutations of the L2 loop of Csm2 completely abolish the DNA-binding ability of Csm2–Psy3, whereas those of Psy3 have no effect on the binding affinity of Csm2–Psy3 for dsDNA, but decrease the affinity of Csm2–Psy3 for ssDNA and abolish the binding ability of Psy3–Shu1–Shu2 for both ssDNA and dsDNA (27). These results suggest that the Shu complex might contain two DNA-binding sites: one site involves the Csm2 L2 loop, and the other is formed by the Psy3 L2 loop and the Shu1–Shu2 heterodimer (27). Like in the structure of the Csm2–Psy3 subcomplex, the L2 loops of both Csm2 and Psy3 are largely disordered, probably due to the absence of DNA (Figure 5A). As the basic residues on the L2 loops (189-KRRR-192 of Cms2 and 199-KRK-201 of Psy3) are located in the disordered regions, there are no obvious positively charged regions on the surface of the Csm2–Psy3 heterodimer (Figure 5B). On the other hand, there are two positively charged regions around the Csm2–Psy3 and Shu1–Shu2 interface: region I is located on the surface of Psy3 and consists of basic residues Lys156, Lys159, Lys162, Arg165 and Lys166; and region II is located on a surface cleft at the Psy3–Shu1 interface and consists of basic residues Lys5 and Arg8 of Psy3 and Arg58, Arg60 and Arg78 of Shu1 (Figure 5B and Supplementary Figure S12). In the Shu complex, the cluster of basic residues on the C-terminal region of the Csm2 L2 loop appears to be positioned in the close vicinity of the N-terminal region of the L2 loop of Psy3 and the L1 loop of Csm2, and these three structure elements together form a shallow positively charged surface groove (Figure 5A and B). We propose that this surface groove is likely to form a major DNA-binding site (site I). In addition, the cluster of basic residues on the Psy3 L2 loop is positioned in the middle of the disordered region. It is possible that the disordered Psy3 L2 loop might be oriented towards the positively charged regions (I and II) at the Psy3 and Shu1–Shu2 interface, and these three structure elements together form another DNA-binding site (site II). Site I is a shallow surface groove which is structurally similar to the ssDNA-binding site in the Rad51 presynaptic complex (37), and site II consists of a flat surface and a large cleft. As the Shu complex prefers to bind forked and 3′-overhang DNAs both of which contain ssDNA and dsDNA regions, it is possible that site I may be responsible for binding the ssDNA region of the substrate and site II may be involved in binding of the dsDNA region of the substrate, and the two binding sites work cooperatively to ensure a tight binding of the substrate (Figure 5B). Further structural and biochemical studies are needed to elucidate the exact molecular mechanism for the recognition and binding of DNA substrates by the Shu complex.
Figure 5.
Possible DNA binding model for the Shu complex. (A) A ribbon diagram of the Shu complex. The proteins are colored the same as in Figure 1. The disordered L2 loops of Csm2 and Psy3 are indicated by dashed lines with the two clusters of positively charged residues colored in blue. (B) An electrostatic surface of the Shu complex in the same orientation as in (A). The two positively charged regions at the Psy3–Shu1 interface are circled by dashed lines. A 3′-overhang DNA is modeled to bind to the Shu complex. The ssDNA region of the DNA substrate binds to the potential binding site I formed by the L2 loop and L1 loop of Csm2 and the N-terminal region of the L2 loop of Psy3, and the dsDNA region of the DNA substrate binds to the potential binding site II formed by the C-terminal region of the Psy3 L2 loop and the two positively charged regions at the Psy3–Shu1 interface.
The previous functional data demonstrate that the Shu complex stimulates Rad51 loading onto the RPA-coated ssDNA in cooperation with Rad52 and Rad55–Rad57 (21). The Y2H and pull-down assays show that the Shu complex interacts directly with Rad55–Rad57 via the Csm2–Rad55 interaction (19–21). On the other hand, Rad55-Rad57 can also bind directly to Rad51 via Rad55, and the interactions of Rad55–Rad57 with Rad51 and Csm2–Psy3 are non-competitive (21,38,39). These results suggest that Rad55 might contain two binding interfaces for Rad51 and/or its paralog to extend the filament smoothly. Based on these results, we can propose a working model for the functional role of the Shu complex in promoting the formation of the presynaptic filament. During the HR process, the ssDNA generated by DNA end resection is firstly coated and protected by the RPA proteins. Secondly, the Shu complex recognizes and binds to the 5′-end of the ssDNA. Thirdly, the Rad55–Rad57 complex is recruited to the damage site via the Csm2–Rad55 interaction and then loaded on the ssDNA. Subsequently, the RPAs are gradually replaced by Rad51 with the help of Rad52 and then the Rad51–ssDNA filament extends in the 5′-3′ direction.
AVAILABILITY
The crystal structure of the Shu complex has been deposited with the Protein Data Bank under accession code 5XYN.
Supplementary Material
Supplementary Data
ACKNOWLEDGEMENTS
We thank the staff members at BL19U1 of the National Facility for Protein Science in Shanghai, China for providing technical support in diffraction data collection, and the other members of our group for valuable discussion. We are very grateful to Prof. Jinqiu Zhou at Shanghai Institute of Biochemical and Cell biology for providing the pRS316 plasmid and the wild-type, _psy3_Δ, _csm2_Δ, _shu1_Δ, and _shu2_Δ yeast BY4742 strains, and Prof. Ming Lei for providing the pBTM116 and pACT2 plasmids and the yeast L40 strain.
Author Contributions: S.Z. participated in the cloning, protein purification, and crystallization, performed the structure determination and structural analysis, and the Y2H and MMS assays, and drafted the manuscript. L.W. performed the mutagenesis and pull-down experiments and participated in the Y2H assays. Y.T. performed the cloning, protein purification, and crystallization. T.B. and R.L. participated in the mutagenesis, Y2H assays and MMS assays. T.Z. participated in the structure determination. J.C. participated in the experimental design and data analyses. J.D. conceived the study, participated in the experimental design and data analyses, and wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
National Natural Science Foundation of China [31521061]; Strategic Priority Research Program of the Chinese Academy of Sciences [XDB08010302]; Ministry of Science and Technology of China [2013CB910404]. Funding for open access charge: National Natural Science Foundation of China [31521061]; Strategic Priority Research Program of the Chinese Academy of Sciences [XDB08010302]; Ministry of Science and Technology of China [2013CB910404].
Conflict of interest statement. None declared.
REFERENCES
- 1.Krejci L., Altmannova V., Spirek M., Zhao X.. Homologous recombination and its regulation. Nucleic Acids Res. 2012; 40:5795–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.San Filippo J., Sung P., Klein H.. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008; 77:229–257. [DOI] [PubMed] [Google Scholar]
- 3.Ceccaldi R., Sarangi P., D’Andrea A.D.. The Fanconi anaemia pathway: new players and new functions. Nat. Rev. Mol. Cell Biol. 2016; 17:337–349. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein K.A., Gangloff S., Rothstein R.. The RecQ DNA helicases in DNA repair. Annu. Rev. Genet. 2010; 44:393–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Godin S.K., Sullivan M.R., Bernstein K.A.. Novel insights into RAD51 activity and regulation during homologous recombination and DNA replication. Biochem. Cell Biol. 2016; 94:407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kowalczykowski S.C. An Overview of the Molecular Mechanisms of Recombinational DNA Repair. Cold Spring Harb. Perspect. Biol. 2015; 7:a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Symington L.S., Rothstein R., Lisby M.. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics. 2014; 198:795–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karpenshif Y., Bernstein K.A.. From yeast to mammals: recent advances in genetic control of homologous recombination. DNA Repair (Amst.). 2012; 11:781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zelensky A., Kanaar R., Wyman C.. Mediators of homologous DNA pairing. Cold Spring Harb. Perspect. Biol. 2014; 6:a016451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shor E., Weinstein J., Rothstein R.. A genetic screen for top3 suppressors in Saccharomyces cerevisiae identifies SHU1, SHU2, PSY3 and CSM2: four genes involved in error-free DNA repair. Genetics. 2005; 169:1275–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang M.E., Rio A.G., Nicolas A., Kolodner R.D.. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:11529–11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rabitsch K.P., Toth A., Galova M., Schleiffer A., Schaffner G., Aigner E., Rupp C., Penkner A.M., Moreno-Borchart A.C., Primig M. et al. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr. Biol. 2001; 11:1001–1009. [DOI] [PubMed] [Google Scholar]
- 13.Wu H.I., Brown J.A., Dorie M.J., Lazzeroni L., Brown J.M.. Genome-wide identification of genes conferring resistance to the anticancer agents cisplatin, oxaliplatin, and mitomycin C. Cancer Res. 2004; 64:3940–3948. [DOI] [PubMed] [Google Scholar]
- 14.Smith S., Hwang J.Y., Banerjee S., Majeed A., Gupta A., Myung K.. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:9039–9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Godin S.K., Zhang Z., Herken B.W., Westmoreland J.W., Lee A.G., Mihalevic M.J., Yu Z., Sobol R.W., Resnick M.A., Bernstein K.A.. The Shu complex promotes error-free tolerance of alkylation-induced base excision repair products. Nucleic Acids Res. 2016; 44:8199–8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasanuma H., Tawaramoto M.S., Lao J.P., Hosaka H., Sanda E., Suzuki M., Yamashita E., Hunter N., Shinohara M., Nakagawa A. et al. A new protein complex promoting the assembly of Rad51 filaments. Nat. Commun. 2013; 4:1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.She Z., Gao Z.Q., Liu Y., Wang W.J., Liu G.F., Shtykova E.V., Xu J.H., Dong Y.H.. Structural and SAXS analysis of the budding yeast SHU-complex proteins. FEBS Lett. 2012; 586:2306–2312. [DOI] [PubMed] [Google Scholar]
- 18.Ball L.G., Zhang K., Cobb J.A., Boone C., Xiao W.. The yeast Shu complex couples error-free post-replication repair to homologous recombination. Mol. Microbiol. 2009; 73:89–102. [DOI] [PubMed] [Google Scholar]
- 19.Xu X., Ball L., Chen W., Tian X., Lambrecht A., Hanna M., Xiao W.. The yeast Shu complex utilizes homologous recombination machinery for error-free lesion bypass via physical interaction with a Rad51 paralogue. PLoS One. 2013; 8:e81371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godin S., Wier A., Kabbinavar F., Bratton-Palmer D.S., Ghodke H., Van Houten B., VanDemark A.P., Bernstein K.A.. The Shu complex interacts with Rad51 through the Rad51 paralogues Rad55-Rad57 to mediate error-free recombination. Nucleic Acids Res. 2013; 41:4525–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaines W.A., Godin S.K., Kabbinavar F.F., Rao T., VanDemark A.P., Sung P., Bernstein K.A.. Promotion of presynaptic filament assembly by the ensemble of S. cerevisiae Rad51 paralogues with Rad52. Nat. Commun. 2015; 6:7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor M.R., Spirek M., Chaurasiya K.R., Ward J.D., Carzaniga R., Yu X., Egelman E.H., Collinson L.M., Rueda D., Krejci L. et al. Rad51 paralogs remodel pre-synaptic Rad51 filaments to stimulate homologous recombination. Cell. 2015; 162:271–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McClendon T.B., Sullivan M.R., Bernstein K.A., Yanowitz J.L.. Promotion of homologous recombination by SWS-1 in complex with RAD-51 paralogs in Caenorhabditis elegans. Genetics. 2016; 203:133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Godin S.K., Meslin C., Kabbinavar F., Bratton-Palmer D.S., Hornack C., Mihalevic M.J., Yoshida K., Sullivan M., Clark N.L., Bernstein K.A.. Evolutionary and functional analysis of the invariant SWIM domain in the conserved Shu2/SWS1 protein family from Saccharomyces cerevisiae to Homo sapiens. Genetics. 2015; 199:1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu T., Wan L., Wu Y., Chen J., Huang J.. hSWS1.SWSAP1 is an evolutionarily conserved complex required for efficient homologous recombination repair. J. Biol. Chem. 2011; 286:41758–41766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin V., Chahwan C., Gao H., Blais V., Wohlschlegel J., Yates J.R. 3rd, McGowan C.H., Russell P.. Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. EMBO J. 2006; 25:2564–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao Y., Li X., Liu Y., Ruan J., Qi S., Niu L., Teng M.. Structural analysis of Shu proteins reveals a DNA binding role essential for resisting damage. J. Biol. Chem. 2012; 287:20231–20239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makarova K.S., Aravind L., Koonin E.V.. SWIM, a novel Zn-chelating domain present in bacteria, archaea and eukaryotes. Trends Biochem. Sci. 2002; 27:384–386. [DOI] [PubMed] [Google Scholar]
- 29.Spier I., Kerick M., Drichel D., Horpaopan S., Altmuller J., Laner A., Holzapfel S., Peters S., Adam R., Zhao B. et al. Exome sequencing identifies potential novel candidate genes in patients with unexplained colorectal adenomatous polyposis. Fam. Cancer. 2016; 15:281–288. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z., Minor W.. Carter CW, Sweet RM. Macromolecular Crystallography: Part A. 1997; 276:NY: Academic Press; 307–326. [Google Scholar]
- 31.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murshudov G.N., Vagin A.A., Dodson E.J.. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997; 53:240–255. [DOI] [PubMed] [Google Scholar]
- 33.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 35.Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 36.Holm L., Rosenstrom P.. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010; 38:W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu J., Zhao L., Xu Y., Zhao W., Sung P., Wang H.W.. Cryo-EM structures of human RAD51 recombinase filaments during catalysis of DNA-strand exchange. Nat. Struct. Mol. Biol. 2017; 24:40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson R.D., Symington L.S.. Functional differences and interactions among the putative RecA homologs Rad51, Rad55, and Rad57. Mol. Cell. Biol. 1995; 15:4843–4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997; 11:1111–1121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data