The stress granule transcriptome reveals principles of mRNA accumulation in stress granules (original) (raw)
. Author manuscript; available in PMC: 2018 Nov 16.
SUMMARY
Stress granules are mRNA-protein assemblies formed from nontranslating mRNAs. Stress granules are important in the stress response and may contribute to some degenerative diseases. Here we describe the stress granule transcriptome of yeast and mammalian cells through RNA-Seq analysis of purified stress granule cores and smFISH validation. While essentially every mRNA, and some ncRNAs, can be targeted to stress granules, the targeting efficiency varies from <1% to >95%. mRNA accumulation in stress granules correlates with longer coding and UTR regions and poor translatability. Quantifying the RNA-Seq analysis by smFISH reveals only 10% of bulk mRNA molecules accumulate in mammalian stress granules, and only 185 genes have more than 50% of their mRNA molecules in stress granules. These results suggest stress granules may not represent a specific biological program of mRNP assembly, but instead form by condensation of nontranslating mRNPs in proportion to their length and lack of association with ribosomes.
INTRODUCTION
Stress granules (SGs) are non-membrane bound assemblies of RNA and protein that form when translation initiation is limited (Protter and Parker, 2016; Buchan and Parker, 2009). SGs also share many protein components with neuronal granules, and mutations that increase SG formation or perturb SG clearance are implicated in degenerative diseases such as ALS and multisystem proteinopathy, where aberrant SG-like assemblies form (Buchan et al., 2013; Dewey et al., 2012; Kim et al., 2013b; Li et al., 2013; Mackenzie et al., 2017; Ramaswami et al., 2013). SGs are thought to assemble via nontranslating mRNAs serving as scaffolds for RNA binding proteins, which interact with each other through a variety of protein-protein interactions (Panas et al., 2016; Protter and Parker, 2016).
By super-resolution microscopy, SGs show denser regions of proteins and mRNAs (based on oligo(dT) FISH), referred to as cores, which can be biochemically purified (Jain et al., 2016). Cores are linked together by less concentrated regions of SG components referred to as a “shell”, although whether the composition of the shell is different from cores has not been addressed (Jain et al., 2016). Purification of SG cores revealed the yeast and human SG core proteome is enriched in translation initiation factors, RNA binding proteins, proteins with predicted prion-like domains (PrLDs), and proteins involved in neurodegenerative diseases (Jain et al., 2016). Intriguingly, SGs cores were also found to be composed of many proteins that were neither known to bind RNA nor predicted to contain PrLDs. Thus, this analysis identified many known classes of SG proteins, while providing evidence for additional SG components.
Little is known about the RNA composition of SGs. SGs are known to contain non-translating mRNA from early reports showing that SGs contain PABP, stain positive for poly(A) mRNA, lack large ribosomal subunits, and are sensitive to drugs that alter the translation (Kedersha et al., 2000, 2002, 1999; McEwen et al., 2005). A few specific mRNAs have been shown to localize to SG (Kedersha and Anderson, 2002; Nelles et al., 2016; Stoecklin et al., 2004; Stöhr et al., 2006), although the full population of mRNAs in SG, and whether SG contain specific non-coding RNAs, or lincRNAs, has not been addressed.
Herein, we adapt our purification of SG cores to determine the transcriptome of both yeast and mammalian SG cores. We find that SGs are composed of over 80% mRNAs, although some lincRNAs and ncRNAs are enriched in SG. SGs contain mRNAs from essentially every expressed gene, with no single mRNA or ncRNA representing more than 1% of the SG RNA molecules. Partitioning of specific mRNAs into SGs is decreased by efficient translation properties and increased by longer coding and UTR regions. By quantifying the composition of the SG transcriptome, we discover only 10% of the bulk mRNA molecules accumulate in SGs, and only 185 genes have more than 50% of their mRNA molecules present in SGs. These results suggest SGs do not represent a specific biological program of mRNP assembly but instead form by condensation of untranslating mRNPs in proportion to their length and lack of association with ribosomes.
RESULTS
Mammalian SG cores are enriched for specific mRNAs
To determine the RNAs in mammalian SG cores, we purified and performed RNA-Seq in triplicate on SG cores isolated from U-2 OS cells after 60’ of arsenite exposure (Figure S1A and Methods), referred to as SGcoreRNA. For each sample, 5% of the lysate was extracted for RNA-Seq from the total RNA population, referred to as TotalRNA. Pairwise correlation coefficients indicate reliable transcriptomes between SGcoreRNA library triplicates and TotalRNA library triplicates (Figure S1B).
Pairwise correlation analysis between SGcoreRNA and TotalRNA libraries demonstrate SG core transcriptomes are different from total RNA transcriptomes **(R < 0.04**, Figure S1B). Supporting this analysis, out of the 15689 transcripts identified (restricted to >1 FPKM in TotalRNA), 1841 transcripts are enriched in SG cores (>2-fold change, p<0.01) and 2539 transcripts are depleted from SG cores (<0.5-fold change, p<0.01) (Figure 1A and Table S1). The remainder of the transcripts partitioned similarly between SG cores and the total RNA.
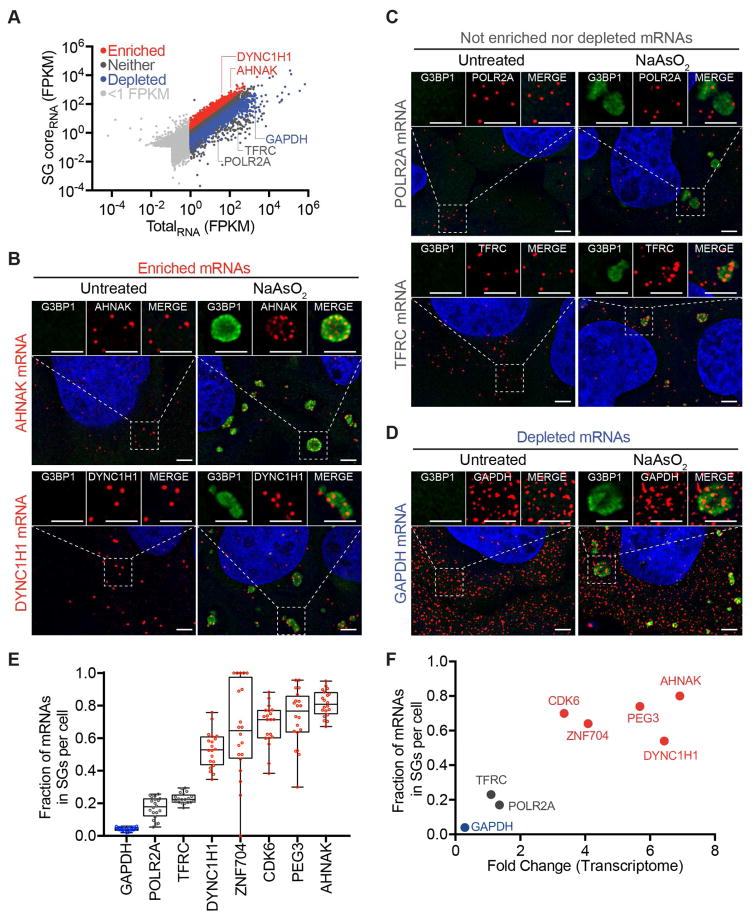
Figure 1. mRNAs differ in degree of recruitment to SGs.
(A) Scatter plot depicting RNA abundance (FPKM) in SG purified RNA vs Total RNA. Red dots indicate RNAs that are significantly enriched (Fold change > 2 and p < .01) in SG purified RNA compared to Total RNA. Blue dots indicate RNAs that are significantly depleted (Fold change <.05 and p >.01) in SG purified RNA compared to TotalRNA. Dark gray dots indicate RNAs that are either not significantly enriched or fail to meet the fold change requirement. Light gray dots indicate mRNAs below <1FPKM. (B) smFISH validation of mRNAs enriched in SGs (AHNAK and DYNC1H1). (C) smFISH validation of RNAs that are neither enriched nor depleted from SGs (POLR2A and TFRC). (D) smFISH validation of a mRNA that is depleted from SGs (GAPDH). Scale bar = 2μm. (E) Quantification of the fraction of each indicated transcript in SGs per cell. 20 cells were counted. (F) Scatterplot depicting fraction of each indicated transcript in Sgs vs. FPKM ratios of SGcoreRNA-Seq: TotalRNA-Seq data (Fold-change). Twenty cells were counted for each experiment.
Validation of RNA-Seq by smFISH
To determine if the SGcoreRNA transcriptome was accurate, we examined several mRNAs that were, based on RNA-Seq, preferentially enriched, neither enriched nor depleted, or preferentially depleted in SG cores by smFISH. We performed smFISH analysis on both stressed and unstressed U-2 OS cells for the following mRNAs: AHNAK (fold change (FC) is calculated as FPKMSGcore/FPKMTotal, FC = 6.92), DYNC1H1 (FC = 6.44), POLR2A (FC = 1.36), TFRC (FC = 1.10), PEG3 (FC=5.69), CDK6 (FC = 3.35), ZNF704 (FC = 4.09), and GAPDH (FC = 0.29) (Figure 1B–D, S1C).
The smFISH analyses revealed that AHNAK, DYNC1H1, PEG3, ZNF704, and CDK6 mRNAs are more enriched in SGs compared to POLR2A, TFRC, and GAPDH mRNAs (Figure 1B–D, S1C), consistent with the RNA-Seq results. By quantifying the number of transcripts in SGs and cells detected by smFISH, we observed that on average, 80% of AHNAK, 74% of PEG3, 70% of CDK6, 64% of ZNF704 and 53% of DYNC1H1 mRNAs are in SGs while 17%, 23% and 4% of POLR2A, TFRC, and GAPDH mRNAs are in SGs respectively (Figure 1E). These proportions parrellel the enrichment of these mRNAs in SG cores by RNA-Seq (Figure 1F). These numbers also reflect the SGcore FPKM reads. For example, DYNC1H1 has roughly twice as many FPKM reads as POLR2A (183.8 and 80, respectively) and roughly twice as many average number transcripts in SG in cells as measured by smFISH (22 and 10, respectively) (Figure 1F). Therefore, the smFISH experiments indicate our transcriptome analysis of SG cores is valid.
The preferential recruitment of specific RNAs to SGs was examined with other SG markers including PABP and Pumilio 2 (PUM2) (Figure S2A, B), indicating we are purifying RNAs enriched in bona fide SGs. Moreover, in ΔΔG3BP1/2 U-2 OS cells, which fail to form SGs under arsenite treatment (Kedersha et al., 2016), we no longer observe the clustering of SG-enriched mRNAs by smFISH (Figure S2B). These results argue we are identifying SG-enriched mRNAs.
Quantitative analysis of the bulk transcriptome and SG transcriptome
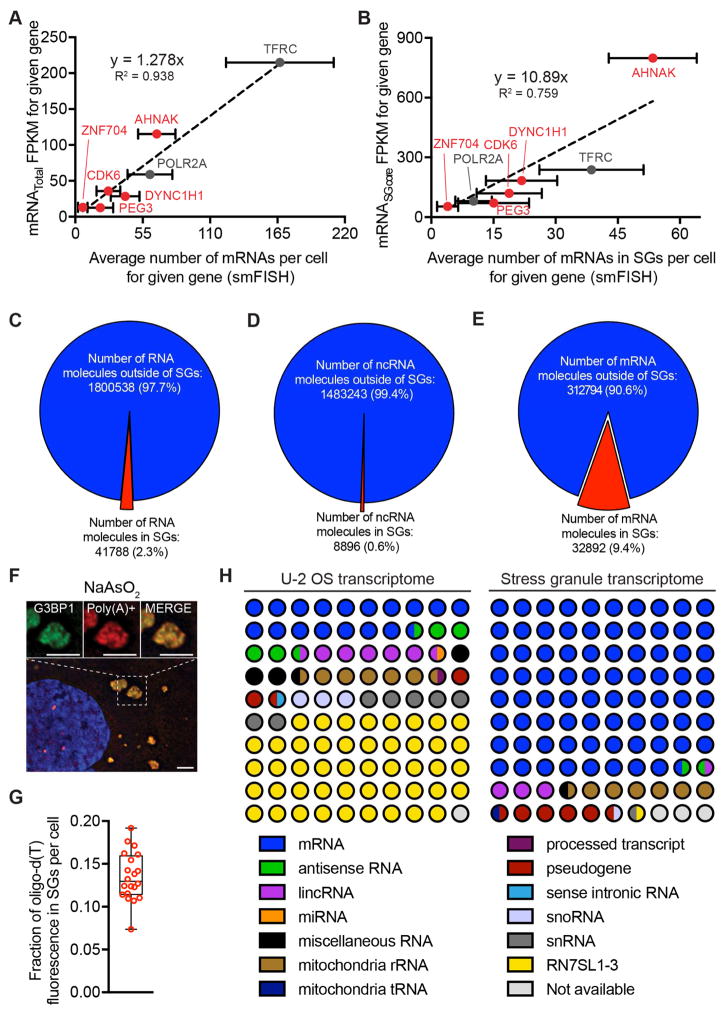
The combination of smFISH, which allowed us to count the number of specific mRNAs in the cell and within SGs, and RNA-Seq data for the total and SG transcriptome allowed us to standardize our RNA-Seq reads to numbers of mRNAs per cell. We observed a linear relationship between individual mRNAs identified per cell by smFISH and FPKM values for each of these mRNAs (Figure 2A). Linear regression analysis allowed us to estimate the relative number of molecules of every mRNA in the cell based on its FPKM value (Table S1). We did not consider genes where the FPKM was less than 1, which allowed us to examine 11195 mRNAs (See STAR methods). From this analysis, we estimate there are an approximate 300,000 total mRNA molecules per cell, with mRNA abundance for different genes spanning from ~3500 to <1 mRNA per cell. These numbers are in the same range as other estimates for mammalian cells (Schwanhaussser et al., 2011)
Figure 2. Quantitative analysis of SG composition.
(A) Scatterplot depicting an average number of mRNAs per cell by smFISH vs. FPKM values from TotalRNA-Seq data. (B) Scatterplot depicting an average number of mRNAs in SGs per cell by smFISH vs. FPKM values from SGcoreRNA-Seq data. Error bars indicate 1 strandard deviation. (C) Pie chart depicting the fraction of total RNA inside and outside SGs. (D) Pie chart depicting the fraction of ncRNAs inside and outside SGs. (E) Pie chart depicting the fraction of mRNAs inside and outside SGs. (F) Oligo(dT) staining of sodium arsenite induced SGs. Scale bar = 2μm. (G) Quantitation of the fraction of oligo(dT) staining within SGs per cell. (H) RNA composition of the U2-OS transcriptome and the SGs transcriptome. RNA designations are from ensemble release 90.
One possibility is that we are purifying only a subpopulation of mRNAs from SGs by isolating G3BP1 cores and this is not an accurate representation of the complete RNA in SGs. To examine this, we looked at the relationship of the number of mRNA molecules in SGs for eight different mRNAs by smFISH and the FPKM values as determined by the SG core purification (Figure 2B). We excluded GAPDH mRNAs from this analysis because it is so abundant, we are unable to count all GAPDH mRNAs by smFISH reliably. We observed a linear relationship between the number of mRNAs within SGs identified by smFISH and the FPKM values from RNA-Seq analysis of SG core-purified RNA with good correlation (Figure 2B, R2 = 0.759). At a minimum, this correlation of RNA-Seq data with smFISH implies that the RNA composition of the shell is fairly similar to the core. More simply, we suggest that RNAs detected in the shell region by oligo(dT) FISH (Jain et al., 2016) are attached to, and co-purify with, cores once cells are lysed (see discussion).
With this analysis, we can provide an approximation of the percent enrichment of every mRNA in SGs (Table S1). It should be noted that these estimates are based on extrapolation from the transcripts we have examined by smFISH, and it remains possible that not all mRNAs will follow the same pattern. Nevertheless, we can estimate several features of the RNA content in SGs in 60’ arsenite stress. We estimate that there are roughly 42,000 RNA molecules in SGs, of which ~78% are mRNAs (Figure 2C, H). Only .6% of ncRNAs localize to SGs, while roughly 10% of mRNA molecules localize to SGs (Figure 2D, E). Consistent with this finding, we see a similar fraction of oligo(dT) staining (~12%) in SGs (Figure 2F, G), suggesting that our transcriptome analysis of SG cores captures most of the RNA content of SGs.
mRNA partitioning to SGs is largely similar in diverse stresses
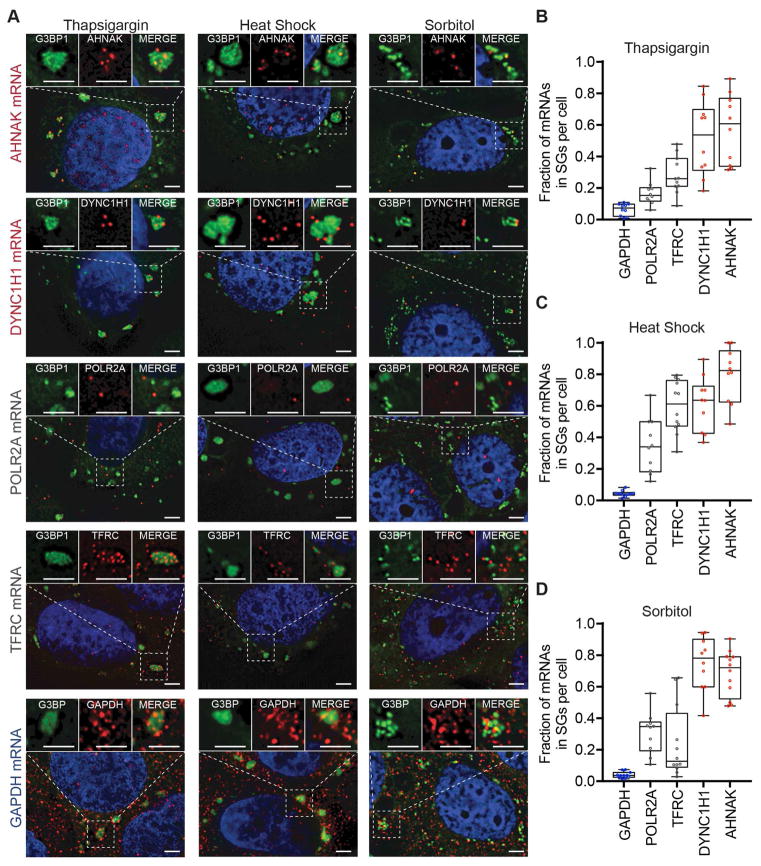
Since the protein components of SGs can vary under different stresses (Kedersha et al., 1999, Stoecklin et al., 2004), we examined whether the RNA composition varies under different stresses. We performed smFISH for several mRNAs (AHNAK, DYNC1H1, POLR2A, TFRC, and GAPDH) and quantified their enrichment in SGs in arsenite, thapsigargin, heat shock, or sorbitol-induced SGs (Kedersha et al., 2016; Kim et al., 2007).
We observed that there are similarities and differences between different stresses (Figure 3A–D). For example, in all stresses, the GAPDH mRNA is depleted from SGs, while the AHNAK and DYNCH1 mRNAs are enriched. In contrast, the TFRC is only enriched in SG during heat shock with an ~twofold increase in the fraction of mRNAs in SGs compared to other stresses, and the POLR2A mRNA is more highly enriched in SG during heat shock or sorbitol stress as compared to arsenite or thapsigargin stress. These results demonstrate that there are differences in the quantitative enrichment of specific mRNAs in SG dependent on the stress condition, which could be due to differences in the specificity of translation repression during different stresses.
Figure 3. mRNA localization is conserved during multiple stresses.
(A) smFISH images acquired for five transcripts (AHNAK, DYNC1H1, TFRC, POLR2A and GAPDH) during three different types of stresses (thapsigargin, heat shock, and sorbitol). (B) Quantitation of each transcript’s enrichment in SGs. Ten or more cells were counted for each experiment.
We also examined the localization of AHNAK and POLR2A mRNAs in U-2 OS ΔΔG3BP1/2 cells under heat shock. U-2 OS ΔΔG3BP1/2 cells can not assemble SGs under arsenite and thapsigargin stresses but can assemble SGs under heat shock and sorbitol stresses (Figure S2B) (Kedersha et al., 2016). We see enrichment of AHNAK mRNAs in heat shock-induced SGs in U-2 OS ΔΔG3BP1/2 but not POLR2A mRNAs (Figure S2B). This finding implies the RNA content of SGs is at least partially conserved in G3BP1/2 independent SGs.
mRNAs enriched in mammalian SGs have less ribosome density
To determine how mRNAs are partitioned into SGs, we examined the properites of SG enriched, depleted, and distributed RNAs. We reasoned that translation efficiency might be a major determinant for localization during stress since (1) decreased translation initiation induces SGs, and (2) trapping ribosomes on transcripts with cycloheximide is sufficient to repress SG formation (reviewed in Panas et al., 2016; Protter and Parker, 2016). This rationale predicts that efficiently translated transcripts should be depleted from SGs, while inefficiently translated transcripts preferentially accumulate in SGs.
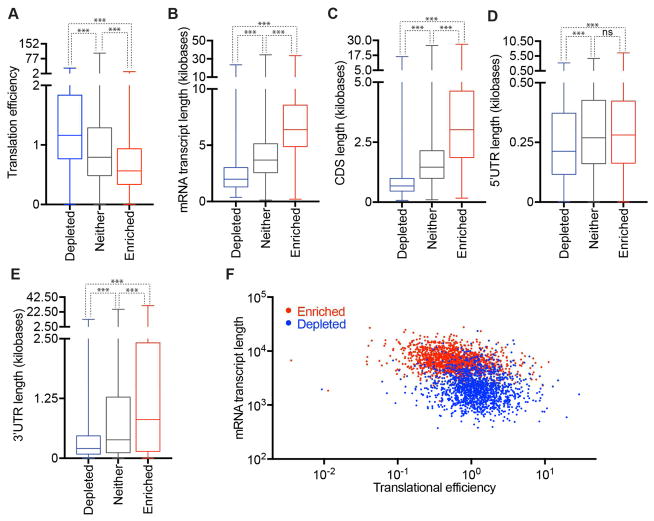
We compared our sequencing results to a previous report that examined translation efficiency (Sidrauski et al., 2015). For statistical purposes, we restrict our analysis for mRNA with FPKM in total RNA greater than 1 (approximately one transcript per cell) (Figure 2A). Out of this restricted 11195 mRNAs list, 14.5% were enriched in SGs (> 2-fold change, p < 0.01) and 15.9% were depleted from SGs (<0.5 fold-change, p <0.01) (Figure S3A, B). We found that transcripts with higher translation efficiency tend to be depleted from SGs, while transcripts with lower translation efficiency tend to localize to SGs (Figure 4A and Methods). This finding demonstrates that one component that influences mRNA localization to SGs is the translation status of specific mRNAs. Since mRNAs that are poorly translated are generally less abundant and stable (Radhakrishnan and Green, 2016), we also observed mRNAs enriched in SGs have a shorter half life, are less abundant, and have less GC content (Figure S3C–E).
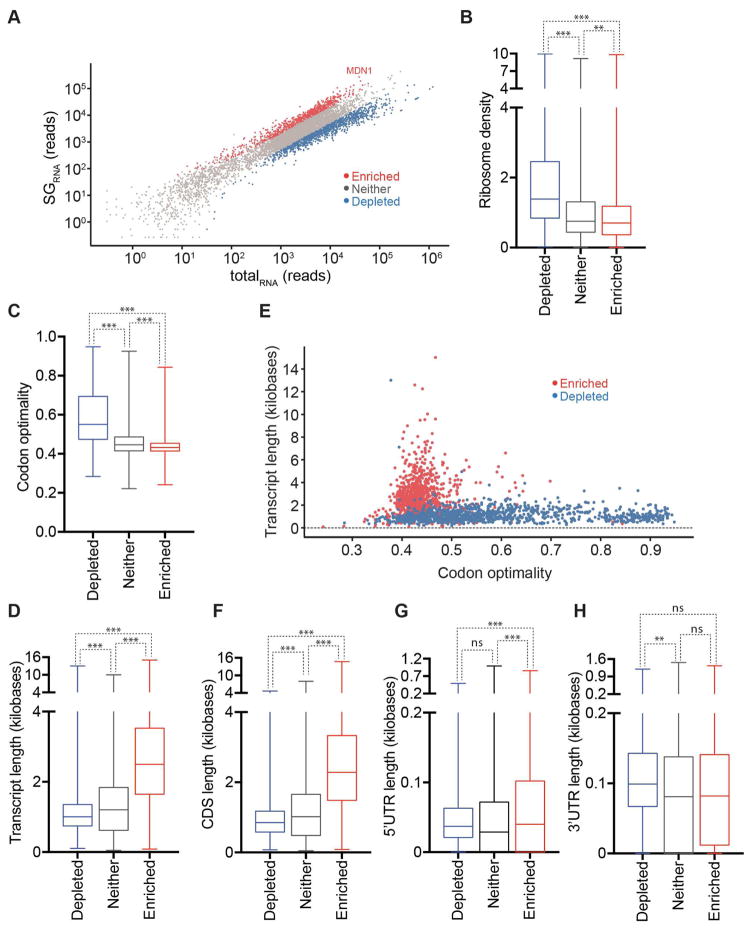
Figure 4. Physical basis of differential mRNA recruitment to mammalian SGs.
(A–E) Boxplots depicting (A) translational efficiency, (B) total transcript length, (C) CDS length, (D) 5’ UTR length, and (E) 3’UTR length for each of the three classes of mRNA localization during stress: SG enriched mRNAs, SG depleted mRNAs, or neither. (F) Scatter plot depicting the mRNA transcript length vs. translational efficiency for SG enriched (red) and SG depleted (blue) mRNAs. n.s. (not significant) = p > .05 and *** = p value ≤0.001 (Student’s t-test).
Mammalian SG-enriched mRNAs are long
Examination of other metrics revealed SG-localized mRNAs were much longer (average length = 7.1 kb) than SG-depleted mRNAs (average length = 2.5 kb) and mRNAs that showed no preference for SGs (average length = 4.2 kb) (Figure 4B). Examination of the different contributions of the 5’ UTR, ORF, or 3’ UTR revealed that the length of the coding region showed the largest difference between enriched and depleted mRNAs in SGs, although the 3’ UTR also had an effect (Figure 4C–E). We only observed slightly longer 5’ UTRs on SG-enriched mRNAs (Figure 4D), which could be because longer 5’ UTRs can decrease translation initiation due to RNA structure.
Given the overall dependence on mRNA length for accumulation in SGs, we also examined how the contributions of the 5’ and 3’ UTRs varied with the length of the coding region. The logic for this analysis was that for mRNAs with short coding regions, the 5’ UTR and 3’ UTR length might compensate for a shorter coding region. Therefore the 5’ and 3’ UTR effects might be more obvious at shorter ORF lengths. Thus, we examined the contributions of the 5’ and 3’ UTR by first binning mRNAs by their ORF length and then determining if there was a difference in the 5’ or 3’ UTR length for mRNAs accumulating in SGs compared to those depleted from SGs. We observed that 3’ length made a significant difference for all coding regions size bins, but with a small effect once the mRNA coding region was over 3000 bases (Figure S3F). A similar analysis for 5’ UTR length revealed the impact of longer 5’ UTR lengths was insignificant (Figure S3G). Taken together, these results indicate the length of the coding region and 3’ UTRs are important metrics for determining SG accumulation.
Mammalian SG-enriched ncRNAs are long
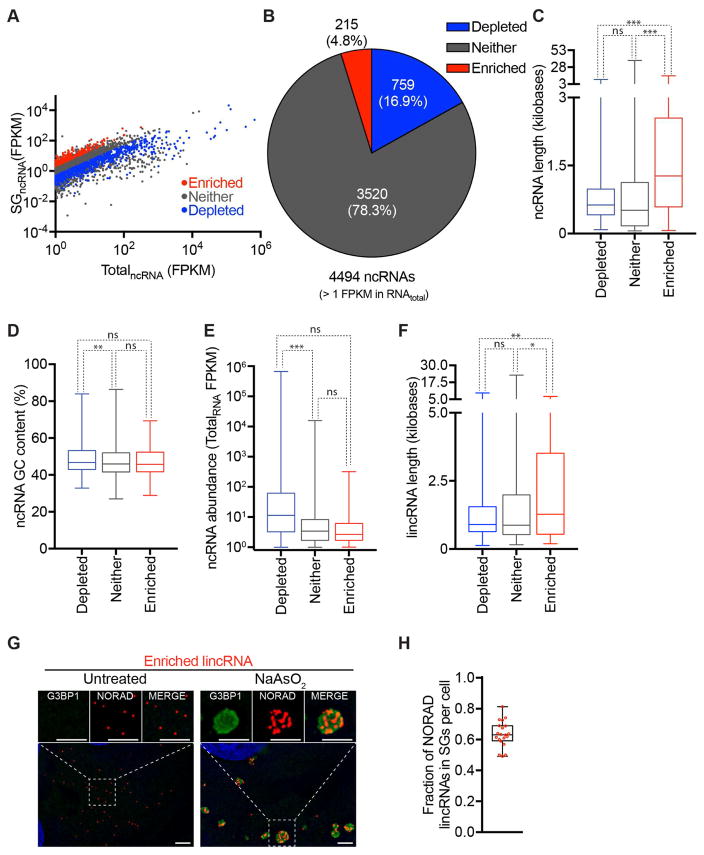
We also examined the ncRNAs that are preferentially recruited to SGs. We restricted our analysis for ncRNA with FPKM in total RNA greater than 1. Out of these restricted 4494 ncRNAs, 4.8% were enriched in SGs (> 2-fold change, p < 0.01) and 16.9% were depleted from SGs (<0.5 fold-change, p <0.01) (Figure 5A–B). Enriched ncRNAs (average length = 1.9 kb) tend to be longer than depleted ncRNA (average length = 0.9 kb) and ncRNA that showed no preference for SGs (average length = 0.9 kb) (Figure 5C). GC content and abundance of ncRNA are not significant predictors of accumulation in SGs (Figure 5D–E).
Figure 5. Physical basis of differential ncRNA recruitment to mammalian SGs.
(A) Scatter plot depicting ncRNA abundance (FPKM) in SG purified RNA vs. Total RNA. Red dots indicate ncRNAs that are significantly enriched (Fold change > 2 and p < .01) in SG purified RNA compared to Total RNA. Blue dots indicate ncRNAs that are significantly depleted (Fold change <.05 and p >.01) in SG purified RNA compared to Total RNA. Gray dots indicate ncRNAs that are either not significantly enriched or fail to meet the fold change requirement. (B) Pie chart depicting the relative contribution of each class of ncRNA (SG-enriched, SG-depleted, or neither) to the total transcriptome SG transcriptome (right). (C–F) Boxplot depicting the (C) transcript length, (D) GC content, (E) abundance of ncRNA, and (F) length of lincRNAs for the three classes of localization: SG enriched mRNAs, SG depleted mRNAs, or neither. n.s., *, **, and *** = p value (Student’s t-test) > 0.05, ≤0.05, ≤0.01, and ≤0.001 respectively. (G) smFISH images of an SG enriched lincRNA, NORAD, during no stress and sodium arsenite induced stress. Scale bar = 2μm. (H) Quantitation of the fraction of NORAD RNAs in SGs. Twenty cells were counted.
Specific examination of lincRNAs revealed SG-enriched lincRNAs also tend to be longer (Figure 5F). We validated a lincRNA, NORAD, is enriched in SGs by smFISH (Figure 5G–H). NORAD is one of the top 10 most enriched lincRNA in SGs (Table S3A) and is thought to function as a Pumulio sponge (Lee et al., 2016; Tichon et al., 2016).
Length and Translation efficiency influence SG targeting of RNAs in yeast
To determine if the principles of mRNA targeting to SGs observed in mammalian cells were conserved, we purified and sequenced the mRNAs within SG cores from Saccharomyces cerevisiae after induction of SGs by sodium azide for 30’ (Buchan et al., 2011). SGRNA and TotalRNA were isolated from three independent biological replicates and sequenced. Reads from biological replicates of SGRNA as well as TotalRNA were highly reproducible (Figure S4A).
Yeast RNA-Seq reads from SGRNA and TotalRNA showed little similarity to each other indicating a subset of cellular RNA is significantly enriched in SGs (Figure 6A, Table S2, Figure S4A). By comparing reads across RNAs in SGRNA and TotalRNA, we found that 916 mRNAs were significantly (p-value <0.01) enriched in SGRNA as compared to TotalRNA, while 1107 mRNAs were significantly enriched in TotalRNA as compared to SGRNA (Figure 6A). Thus, during stress in yeast, we observe the same three classes of mRNAs as in mammalian cells: one subset preferentially localized to SGs, another subset preferentially depleted from SGs, and a third subset partitioning similarly between SGs and the cytosol. smFISH for a SG-enriched mRNA, MDM1, validated that it accumulated in SGs (Figure S4B).
Figure 6. The physical basis of mRNA localization to SGs is conserved from mammals to yeast.
(A) Scatterplot depicting normalized RNA-Seq reads from libraries made from yeast SG purified RNA vs. total RNA. (B and C) Boxplots depicting two metrics of translatability, (B) ribosome density and (C) codon optimality, are correlated with altered localization during stress. (D) Boxplot depicting how transcript length correlates with altered localization. (E) Scatterplot of transcript length vs. fraction optimal codons for SG enriched (red) and SG depleted mRNAs (blue). (F–H) Boxplots depicting how length correlates with localization for (F) the CDS, (G) the 5’UTR, and (H) the 3’ UTR. n.s., **, and *** = p value (Student’s t-test) > 0.05, ≤0.01, and ≤0.001 respectively.
Multiple lines of evidence suggest that poorly translated mRNAs localize to yeast SGs, while efficiently translated mRNAs are depleted. First, we observed that predicted ribosome density and translation initiation rates for SG-depleted mRNAs are significantly higher than the predicted rates for SG-enriched mRNAs (p-value <0.0001, Figure 6B and S4C) (Siwiak and Zielenkiewicz, 2010). Second, we observe codon optimality, which is a metric for translation efficiency (Presnyak et al., 2016), strongly correlates with the distribution of mRNAs. Specifically, SG-enriched mRNAs have an average of 43.9% optimal codons, SG-depleted mRNAs have an average of 59.6% optimal codons (p-value <0.0001, Figure 6C). Consistent with the observation that SG-enriched mRNAs are composed of non-optimal codons, and the observation that non-optimal codons decrease a transcript’s stability (Presnyak et al., 2016), we also found that SG-enriched mRNAs are less stable than SG-depleted mRNAs (Figure S5D).
Similar to mammalian cells, we found that the average SG-enriched mRNA length in yeast (2.7 kb) is significantly longer than SG-depleted mRNAs (1.1kb) (Figure 6D) (Nagalakshmi et al., 2008). This length dependence in yeast is driven primarily by ORF length, and not by 5’ UTR or 3’ UTR length, which is consistent with the overall shorter 3’ UTR length in yeast (Figures 6F–H).
Thus, similar to mammalian cells, the major metrics for mRNA accumulation in yeast SGs are length and poor translation efficiency (Figure 4F, 6E). Indeed, mathematical models made by machine learning and based solely on length and codon optimality (or ribosome density for mammalian mRNAs) are sufficient to accurately predict whether a mRNA will be enriched or depleted from SGs for 75.9% of the yeast mRNAs and 74.1% of mammalian mRNAs (Figure S5, see Methods).
Yeast SGs contain non-coding RNAs
Since we observed that mammalian SGs contain ncRNAs, we wanted to examine if yeast SGs contain ncRNA as well. In yeast, we identified some stable untranslated transcripts (SUTs) (Xu et al., 2009) that accumulate in SGs (Figure S4E). The SUTs enriched in SGs were significantly longer than SG-depleted SUTs (Figure S4G). In addition, cryptic unstable transcripts (CUTs), which are a class of non-coding RNAs that are related to SUTs (Xu et al., 2009), but are much shorter (200–800nts), are rarely enriched in SGs (Figure S4F). Again, SG-enriched CUTs were longer than SG-depleted CUTs (Figure S4H). The inclusion of ncRNAs in SGs demonstrates that prior translation per se is not a requisite for RNA accumulation in SGs and that length plays a role in targeting both coding and noncoding transcripts to SGs.
Membrane association limits SG partitioning of mRNA
Analysis of the SG transcriptome reveals that mRNAs associated with membranes are depleted from SGs, which was suggested by early observations that mRNAs localizing to the ER do not localize to SGs (Unsworth et al., 2010). Specifically, by comparison of the yeast SG-enriched mRNAs to mRNAs associated with the ER through SRP interactions (Chartron et al., 2016), we found that SG-enriched mRNAs are significantly distinct from the subset of mRNAs encoding proteins that localized via SRP (Figure S6A), while SG-depleted mRNAs showed significant overlap with ER-localized mRNAs. Consistent with this observation, we find that human SG-enriched mRNAs are significantly distinct from the subset of mRNAs encoding proteins that are secreted (Figure S6C). We also found that SG-enriched mRNAs in both yeast and mammalian cells were significantly distinct from mRNAs that encode proteins that localize to the mitochondria (Figure S6B, D) (Chartron et al., 2016), while SG-depleted mRNAs showed significant overlap with mRNAs encoding mitochondria-localized proteins (Figure S6B, D).
Taken together, these findings suggest that mRNA localization to other organelles may preclude mRNA from efficiently partitioning into SGs, which could be explained by restricted diffusion when mRNAs are bound to membrane surfaces. It should be noted that we cannot rule out the formal possibility that a subset of SGs form on the ER or mitochondrial surface and such membrane-associated SGs do not purify efficiently in our biochemical preparations.
SG-enriched mRNAs are modestly enriched for SG-resident proteins
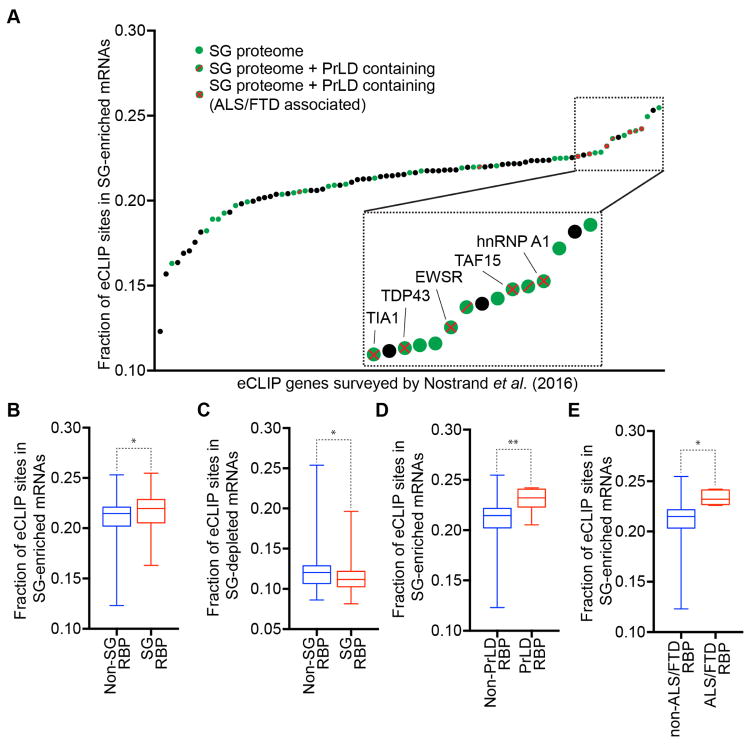
One model for mRNA recruitment to SGs is that SG-enriched mRNAs would bind more SG-targeted RNA binding proteins, and the accumulation of those proteins in SG would then partition specific mRNAs into SGs. A prediction of this model is that SG-enriched RNAs would be enriched for RNA-binding sites of known SG-localized RNPs. To test this prediction, we mined previously published eCLIP datasets on both SG resident (n = 36) and non-SG resident (n =51) RNPs by scoring eCLIP binding sites in SG-enriched RNAs (Figure 7A) (Nostrand et al., (2016)). We observed that RNA binding proteins on average had approximately 22% of their eCLIP sites in mRNAs enriched in SG.
Figure 7. SG-enriched mRNAs are enriched for SG-resident proteins.
(A) Survey of RNA binding proteins from Nostrand et al. (2016)) plotted against the fraction of eCLIP sites found in SG-enriched mRNAs. Green dots are SG-resident RBPs. Black dots are non-SG resident RBPs. Green dots with a red diagonal line are SG-resident RBPs with PrLD domain. Green dots with a red X are SG-resident RBPs with PrLD domain and are associated with ALS/FTD. Box plot illustrating the fraction of eCLIP sites found in (B) SG-enriched mRNAs or (C) SG-depleted mRNAs from SG-resident RBPs or non-SG RBPs. Box plot depicting the fraction of eCLIP sites found in SG-depleted mRNAs from SG-RBPs or non-SG RBPs. Box plot illustrating the fraction of eCLIP sites found in SG-enriched RNAs from (D) SG-RBPs containing PrLD and (E) RBPs associated with ALS/FTD. * and ** = p value (Student’s t-test) ≤0.05, and ≤0.01 respectively.
Surprisingly, we observed SG-localized RNA binding proteins were only modestly enriched on SG-enriched mRNAs as compared to non-SG resident RNA binding proteins (Figure 7B, p-value = 0.0484). Similarly, SG-localized RNA binding proteins were slightly underrepresented in SG-depleted mRNAs as compared to non-SG resident RNA binding proteins (Figure 7C, p-value = 0.0486). Whether this modest enrichment is because these SG-resident RNA binding proteins play a role in targeting mRNAs into SGs, or are identified as SG components because they show a slight preference for mRNAs that accumulate in SGs will require future experiments.
We also observed that RNA binding proteins found in SGs with prion-like domains (PrLD), or with an association with degenerative diseases such as ALS or FTD, showed an even greater bias to bind to mRNAs enriched in SGs (Figure 7A, D–E). The enrichment of PrLD containing proteins involved in disease with SG mRNAs is consistent with a model whereby the prevalent recruitment of these proteins into SGs may both enhance the formation of aberrant pathological aggregates, and contribute to disease progression (Li et al., 2013; Ramaswami et al., 2013).
DISCUSSION
Based on determining the SG transcriptome in yeast and mammalian cells we demonstrate several principles of RNA accumulation in SGs. One key observation is that SGs are primarily accumulations of diverse mRNAs. For example, analyses in mammalian cells showed SG are >78% mRNAs, although some ncRNAs can accumulate within SGs (Figure 2, 5). Similarly, in yeast RNA-Seq genes indicates ~95% of the molecules in SGs are mRNAs. More strikingly, there is no overly abundant RNA within SGs, with the most abundant mRNAs in mammalian SGs, actin, only representing ~0.5% of the SG mRNAs (Figure 2E, Table S1B). Thus, SGs are a true composite of many mRNAs, which is also supported by the fact that essentially every mRNA is present in SGs to some extent. This is not an artifact of contaminating abundant RNA molecules since smFISH for GAPDH, an abundant mRNA depleted from SGs, affirms that 4% of the GAPDH mRNA molecules are present within SGs (Figure 1). The fact that essentially every mRNA can accumulate in SGs suggests that the interactions required for SG accumulation are generic and are not limited to a specific subset of mRNAs.
Two key observations argue that we are effectively identifying the majority of RNAs within SGs. The first observation that we effectively identify the full spectrum of SG RNAs is that smFISH analysis of individual mRNAs, which identifies mRNAs anywhere within SGs, and the RNA-Seq analysis, which by definition is only sequencing RNAs present in the core region, show a strong linear correlation (Figure 2B). The second key observation is that based on the quantification of the RNA-Seq of SG cores, we calculate that ~9.4% of the total mRNAs in the cell accumulate in SGs, which is similar to the 13% of total oligo(dT) staining that resides in SGs under the same conditions (Figure 2F, G). Thus, while it is possible we are selectively losing a subset of RNAs loosely associated with SGs, these two observations suggest we are efficiently identifying the vast majority of mRNAs associated with SGs. One simple explanation is that the staining of both proteins and RNAs in the shell region of SGs (Jain et al., 2016) is due to mRNAs of sufficient length that they have portions residing within the core region, and thus co-purify with cores after cell lysate. However, two caveats should be noted. First, since we are purifying SG cores using G3BP1 as an affinity ligand, it is possible we are missing some mRNAs in SG, particularly if they accumulate in SG cores that are lacking in G3BP. Second, we may be underestimating the fraction of some RNAs in SGs if specific mRNAs in the shell region region are lost during the purification process.
Two observations led to the striking conclusion that only 10–12% of the total mRNA molecules in U-2 OS cells accumulate in SG. First, 12–13% of the poly(A)+ RNA signal accumulates in SG based on oligo(dT) FISH (Figure 2F, G). Second, by quantifying the distribution of the mRNA population, we calculated that approximately 9–10% of the mRNA molecules in the cell assemble into SG (Figure 2E). Moreover, based on our standardization of the RNA-Seq data by smFISH analysis (Figure 2A, B), we estimate that there are only 185 genes in which greater than 50% of the mRNA molecules are present in SGs. Thus, only a minority of the bulk mRNA population enters SGs and only for <2% of the analyzed genes are the majority of the transcripts within SGs.
The diversity and overall minor amount of mRNA within SGs force us to reconsider their nature and possible function. First, the small amount of mRNA within SGs suggests that these structures will not have large global effects on bulk mRNA. This is consistent with observations that cell lines lacking G3BP, which fail to form SG, still efficiently repress global translation of mRNAs (Kedersha et al., 2016), and cells with defective SG formation do not show altered mRNA degradation (Bley et al., 2015). Thus, if SGs directly affect the function of mRNAs within them, SG formation will only have a substantial impact on the few mRNAs with a high percentage of their total molecules present in SG. It remains possible that sequestering ~10–12% of the mRNAs in the cell has other impacts on cell physiology, such as affecting signaling pathways (Kedersha et al., 2013).
The transcriptome of SGs argues that every SG will be different in composition. Since total SG volume in U-2 OS cells is 10 μm3, and each core is ~0.0066 μm3 (based on the size of cores using nanoparticle tracking (Jain et al. 2016)), we estimate that there are between 300 and 1500 SG cores in U-2 OS cells after 60’ of arsenite stress if SGs consist of 20% (estimated in Jain et al., 2016) or 100% (a maximal boundary condition) cores, respectively. Since actin mRNA, the most abundant molecule in SGs, has only 160 molecules within SGs/cell and there are at least 300 cores per cell by our most conservative estimate, core RNA composition must be heterogeneous. Moreover, at 1 hour of arsenite stress, we estimate there are 32,000 mRNAs in SGs (Figure 2E). Thus, each core should have approximately 21 to 106 mRNA molecules, and by necessity cores would need to be composed of a heterogeneous mixture of transcripts to explain our transcriptome data.
We identify three parameters that correlate with the partitioning of specific mRNAs into SGs. First, both yeast and mammalian analyses show inefficient translation correlates with higher partitioning of mRNAs into SGs (Figure 4, 5). This is consistent with prior models that mRNAs engaged with ribosomes are restricted from entering SGs and provides additional evidence that mRNAs need to be nontranslating to accumulate within SG (Kedersha et al., 2000) (Kedersha and Anderson, 2002). Second, association with ER or mitochondrial membranes correlates with mRNAs being excluded from SGs, perhaps due to membrane tethering limiting the mRNPs from diffusing into SGs (Figure S6). Third, a strong and predominant metric of mRNA accumulation in SGs is the overall length of the coding region and the 3’UTR in both yeast and mammalian SG transcriptomes and in both organisms is strongly linked to the length of the ORF (Figure 4, 6).
In principle, a longer mRNA could be more efficiently localized to SGs by two potentially overlapping mechanisms. First, a longer coding region could increase SG partitioning by providing multiple binding sites for RNA binding proteins that may target mRNAs into SGs. Although it has been tacitly assumed that mRNAs would enter SGs through the interactions of specific RNA binding proteins, to our knowledge there is no direct evidence in support of this model. Indeed, although actin mRNA has been localized to SG, its association with SG is independent of the ZBP1 RNA binding protein (Stöhr et al., 2006) Moreover, we observe only a very modest bias in the binding of SG RNA binding proteins to mRNAs enriched in SG (Figure 7B). Thus, while mRNA binding proteins can clearly play a role in the overall assembly of SGs (Panas et al., 2016; Protter and Parker, 2016), whether they dictate the specific mRNAs localized to SGs remains to be determined. An alternative model to long ORFs simply providing more RBP sites is that the condensation of mRNAs into SGs would be promoted by RNA-RNA interactions between the exposed coding regions, and any 3’ UTR regions exposed due to the off-rate of mRNA binding proteins and their redistribution into exposed coding regions. In this model, the longer the coding region and/or the 3’ UTR, the more RNA-RNA interactions that could occur. Thus, a key area of future research will be in determining what interactions define the SG transcriptome, and how that affects mRNA function.
STAR METHODS
U-2 OS growth conditions and reagents
Human osteosarcoma U-2 OS cells expressing G3BP1-GFP (Figley et al., 2014), U-2 OS cells and U-2 OS ΔΔG3BP1/2 cells (Kedersha et al., 2016), maintained in DMEM with high glucose, 10% fetal bovine serum, and 1% penicillin/streptomycin at 37°C/5% CO 2, were used in all experiments.
Isolation of RNA from U-2 OS cells and SG Cores for RNA-sequencing
U-2 OS cells expressing G3BP1-GFP were grown to 85% confluency in three 500 cm2 square TC-treated culture dishes (Thermo Fisher Scientific, 07-200-599). One hour prior to stress, cell culture media was exchanged with fresh media. Cells were then treated with 500 μM NaAsO2 for one hour at 37°C/5% CO 2. After stress, cells were washed once with media, transferred to falcon tubes, and pelleted at 1,500 x g for 3 mins. Upon aspirating the media, the pellets were immediately flash-frozen in liquid N2 and stored at -80°C until isolation of mammalian SG cores was performed.
The isolation of SG cores was adapted from two papers (Jain et al., 2016; Wheeler et al., 2016). Briefly, the pellet was thawed on ice for 5 mins, re-suspended in 1 mL SG lysis buffer (50 mM TrisHCl pH 7.4, 100 mM KOAc, 2 mM MgOAc, 0.5 mM DTT, 50 μg/mL Heparin, 0.5% NP40, complete mini EDTA-free protease inhibitor (Sigma Aldrich, 11836170001), 1 U/μL of RNasin Plus RNase Inhibitor (Promega, N2615) and passed through a 25 gauge 5/8 needle attached to a 1 ml syringe 7 times. After lysis, the lysates were spun at 1000 x g for 5 min at 4°C to pellet cell debris. 50 μL and 950 μL of the supernatants were transferred to new microcentrifuge tubes for isolating total and SG RNAs respectively (Figure S1A). For isolating total RNA, Trizol LS reagent (Thermo Fisher Scientific, 10296-028) was added and RNA was extracted following the manufacturers protocol. Following isopropanol precipitation, the RNA pellet was re-suspended in 50 μL RNase-free H2O.
The following steps were performed to isolate mammalian SG cores and extract its RNA: 1) The 950 μL supernatant was spun at 18,000 x g for 20 mins at 4°C to pellet SG cores. 2) The resulting supernatant was discarded, and the pellet was re-suspended in 1 mL SG lysis buffer. 3) Steps 1 and 2 were repeated to enrich for SG cores. 4) The resulting pellet was then re-suspended in 300 μL of SG lysis buffer and spun at 850 x g for 2 mins at 4°C. 5) The supernatant which represents the mammalian SG core enriched fraction was transferred to a new tube. 6) The enriched fraction was pre-cleared twice by adding 60 μL equilibrated DEPC-treated Protein A Dynabeads (Thermo Fisher Scientific, 10002D) and nutating at 4°C for 30 mins. Dynabeads were removed using a magnet. 7) 20 μL of anti-GFP antibody (Life technologies, A1112) was added to the enriched fraction and nutated at 4°C for 1 hr t o affinity purify SG cores. 8) The solution was spun at 18,000 x g for 20 mins at 4°C and the supernatant was discarded to remove any unbound antibody. 9) The pellet was then re-suspended in 500 μL SG lysis buffer and 60 μL equilibrated DEPC-treated Protein A Dynabeads. 10) The sample was nutated for 3 hr at 4°C. 11) The Dynabeads were washed three times with wash buffer 1 (20 mM Tris HCl pH 8.0, 200 mM NaCl, 1 U/μL of RNasin Plus RNase Inhibitor) for 5 mins, once with wash buffer 2 (20 mM Tris HCl pH 8.0, 500 mM NaCl, 1 U/μL of RNasin Plus RNase Inhibitor) for 5 mins, and once with wash buffer 3 (SG lysis buffer + 2M Urea, 1 U/μL of RNasin Plus RNase Inhibitor) for 2 mins at 4°C. 12) The beads were resuspended in 200 μL of 100 μg/mL Protease K solution (1X TE buffer, 2M Urea, 1 U/μL of RNasin Plus RNase Inhibitor) and incubated for 15 mins at 37°C. 13) Trizol LS reagent (Thermo Fisher Scientific, 10296-028) was added to the samples and RNA was extracted following the manufacturers protocol. Following isopropanol precipitation, the RNA pellet was re-suspended in 20 μL RNase-free H2O.
Mammalian library construction and RNA-sequencing
After isolating RNA from cells and SG cores 1 and 20 μL of RNA isolated from cells and SG cores, respectively, were treated with rDNase 1 following manufacturer’s protocol (Thermo Fisher Scientific, AM1906). Immediately, the samples were ribosomal-depleted using Ribo-Zero rRNA removal kit following manufacturer’s protocol (Illumina, MRZH11124). Afterwards, the quality of the RNA was inspected using high sensitivity RNA ScreenTape (Agilent, 5067–5579) on Agilent TapeStation 2200 instrument (University of Colorado-Boulder BioFrontiers Next-Generation Sequencing Facility).
cDNA libraries were prepared following manufacturer’s protocol from three replicates of 10 ng ribosomal-depleted RNA from cells and SG cores using NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina (New England Biolabs, E7420S). The qualities and amounts of the cDNA libraries were assessed using high sensitivity D1000 ScreenTape (Agilent, 5067–5584) on Agilent Tapestation 2200 instrument and the Qubit (Thermo Fisher Scientific), respectively (University of Colorado-Boulder BioFrontiers Next-Generation Sequencing Facility) The cDNA libraries (3 from cells and 3 from SG cores) were pooled and sequenced on the Illumina NextSeq 500 platform using the High Output 150 cycles kit (paired-end reads, single index) (University of Colorado-Boulder BioFrontiers Next-Generation Sequencing Facility).
Isolation and sequencing of RNA from Stress Granules
A 2.4L culture of Pab1-GFP expressing yeast cells and stressed them with sodium azide (NaN3) for 30’, prior to lysis. Total cellular RNA (TotalRNA) and RNA from stress granule cores (SGRNA) were isolated from this culture as described in a previous report (Jain et al., 2016).
Yeast Library Construction
Libraries were constructed using the Bio Scientific NEXTflex Rapid Illumina Directional RNA-Seq Library Prep Kit. We followed the manufacturer supplied directions. Before making the library, the RNA was ribosome depleted using the Ribo-Zero rRNA removal kit, also according to manufacturer's directions. The resulting RNA was incubated with Turbo DNase at 37ºC for 1 hour and then cleaned up using a Zymo Research RNA Clean and Concentrator kit also according to the manufacturer's directions.
Yeast Sequencing Data Analysis
Read quality was assessed using fastqc. Illumina adapters were trimmed using Trimmomatic 0.32 in single end (SE) mode (Bolger et al., 2014). An index genome was built with Bowtie 0.12.7 using the ‘build’ command and the S288C reference genome version (R64-1-1), which was acquired through Saccharomyces Genome Database (SGD)(Langmead et al., 2009). Reads were aligned using Bowtie 0.12.7 using the following options: -S -v 2 -m 3 –best. Reads mapping to each gene were counted using HTSeq with parameters –t gene –I Name –f sam –s reverse, using the R64-1-1 annotation file (Anders et al., 2015). Normalization and differential expression analysis were then performed using DESeq 1.22.1 (Anders et al., 2010).
SUTs and CUTs were analyzed using the same method, but with a different annotation file gene annotation file that contained these transcript annotations (Xu et al., 2009).
Yeast Length, Translation, and Stability analysis
Length: Data for the length analysis was obtained from a previous report (Nagalakshmi et al., 2008). Codon Optimality: The fraction of the optimal codon for each gene was calculated using a custom script based on codon stabilization coefficients presented in a previous report (Presnyak et al., 2016). In brief, the amount of each codon was tallied on a gene by gene basis. Optimal codons were assigned a value of one while non-optimal codons were assigned a value of zero. The total number of optimal codons was then divided by the total number of codons in order to find the fraction of optimal codons. Predicted translation initiation rates and ribosome density data were obtained from a previous study (Siwiak and Zielenkiewicz, 2010) Stability: RNA half-lives were obtained from a previous report (Neymotin et al., 2014). All boxplots were generated using PRISM. Scatterplots were created using PRISM and Tableau.
Mammalian Sequencing Data Analysis
Read quality was assessed using fastqc. Illumina adaptors were trimmed using Trimmomatic 0.32 in paired and (PE) mode (Bolger et al., 2014). An index genome was acquired from GENCODE (Release 19 GRCh37.p13). Reads were aligned using Tophat (version 2.0.6) and Bowtie2 (version 2.0.2) and the following parameters: --b2 –fast –microexon-search (Kim et al., 2013a; Langmead and Salzberg, 2012). Differential expression analysis was performed using Cuffdiff (version 2.2.1) with the default parameters (Trapnell et al., 2013).
Human Length, Translation, and Stability analysis
All length data (5’UTR, ORF, 3’ UTR and total) was acquired using Ensemble’s Biomart tool (Aken et al., 2016; Kinsella et al., 2011). Translational efficiency values were calculated from a previous report (Ribosome protected fragment reads / RNA-Seq reads) (Sidrauski et al., 2015). GC content data was acquired using Ensemble’s Biomart tool. Half-life data was acquired from a previous report (Tani et al., 2012).
eCLIP Data analysis
All eCLIP data was analyzed from a previous report, which examined eCLIP targets of over 80 proteins (Van Nostrand et al., 2016). We examined eCLIP data from known RBPs in the SG proteome as well as non-SG proteins (Jain et al., 2016). In brief, a bed file for each eCLIP experiment was obtained from the ENCODE data repository, which contained the genomic coordinates of all eCLIP peaks. Next, a bed file was created for the human genome using NCBI’s gtf annotation file. The program BEDOPS v2.4.26 was used performed to convert the gtf file into a bed file, using the command ‘gtf2bed’ (Neph et al., 2012). The human genome bed file was then compared to each eCLIP bed file using the ‘intersect’ command in bedtools version 2.16.2, in order to see which genes contained eCLIP peaks (Quinlan and Hall, 2010). The fraction of eCLIP peaks in SG-enriched mRNAs was calculated for a given RBP by dividing the summation of eCLIP peaks aligning to SG-enriched mRNAs by the total number of eCLIP peaks. A similar analysis was done for SG-depleted mRNAs.
Machine learning
A mathematical model for mRNA localization during stress was built using MATLAB’s “Classification Learner” application. For yeast mRNA localization, two parameters (length and fraction optimal codons), and a response (mRNA localization) were imported into the classification learner. The model used 90% of the data for training and held back 10% for testing. This process was performed for multiple iterations until every mRNA had been used for both model building and model testing. The course nearest neighbor model provided the best results. In brief, the course nearest neighbor model makes localization predictions of a given mRNA based on the localization of its 100 nearest neighbors on a scatterplot of length vs. codon optimality. A similar approach was used for modeling mammalian mRNA localization, but instead using ribosome translational efficiency of codon optimality.
Statistical Tests
R2 values were calculated for pairwise comparison of sequencing replicates by squaring MATLAB’s ‘corr’ function. To calculate the statistical significance in boxplots, a 2-sample t-test was performed using PRISM software. A 2-population proportion test was used in R to calculate the overlap of yeast SG enriched and SG depleted mRNA with SRP-localized and mitochondria-localized mRNA.
Stress conditions
To examine mRNA localization during other stresses, we used the following stress conditions in U-2 OS cells. Thapsigargin was added to a final concentration of 1 μM for 1 hour. For heat shock experiments, cells were placed at 42ºC for 1 hour. For osmotic stress, cells were stressed in .4M sorbitol for 2.5 hours.
Sequential immunofluorescence and single molecule FISH
Sequential immunofluorescence and smFISH on fixed U-2 OS and U-2 OS ΔΔG3BP1/2 cells were performed following manufacturer’s protocol (https://biosearchassets.blob.core.windows.net/assets/bti_custom_stellaris_immunofluorescence_seq_protocol.pdf) in Figure 1,5 and Supplemental Figure 1. SmFISH on fixed U-2 OS cells expressing G3BP1-GFP were performed following manufacturer’s protocol (https://biosearchassets.blob.core.windows.net/assets/bti_stellaris_protocol_adherent_cell.pdf) in Figure 2 and 3. smFISH on yeast cells was performed following the manufacturer’s&protocol&(https://biosearchassets.blob.core.windows.net/assets/bti_custom_stellaris_yeast_protocol.pdf).
The primary antibodies used for immunofluorescence include mouse anti-G3BP (5 μg/mL, ab56574(Abcam)), rabbit anti-PABP (1 μg/mL, ab21060(Abcam)), and/or rabbit anti-PUM2 (5 μg/mL, ab10361(Abcam)) and the appropriate secondary antibodies used were goat anti-mouse FITC antibody (1:500, Abcam (ab6785)), and donkey anti-rabbit Alexa Fluor 555 conjugate antibody (1:500, Abcam(ab150062)). Ship ready Stellaris® FISH Probes recognizing TFRC, POLR2A, and GAPDH transcripts labeled with Quasar 570 Dye and GAPDH labeled with Quasar 670 Dye (SMF-2006-1, SMF-2003-1, SMF-2026-1, and SMF-3140-1, respectively, Biosearch Technologies, Inc., Petaluma, CA) and Custom Stellaris® FISH Probes designed against AHNAK, DYNC1H1, PEG3, ZNF704, NORAD, and MDN1 transcripts labeled with Quasar 670 dye by utilizing the Stellaris® RNA FISH Probe Designer (Biosearch Technologies, Inc., Petaluma, CA), available online at www.biosearchtech.com/stellarisdesigner (version 4.2), were used. CDK6 mFISH probes labeled with Atto633 were designed following Gasper et al. (2017). ddUTP-Atto633 were provided to us by Ephrussi lab, and CDK6 22mer reverse complement DNA oligos were purchased from IDT. Finally, oligo d(T)30-Cy3 were purchased from IDT.
Microscopy and image analysis
Fixed U-2 OS, U-2 OS ΔΔG3BP1/2, U-2 OS G3BP1-GFP, and yeast cells stained by immunofluorescence, smFISH and/or DAPI were imaged using a wide field DeltaVision Elite microscope with a 100X objective using a PCO Edge sCMOS camera. At least five images were taken for each experiment comprising of 25 Z-sections each in Figure 1, 2, supplemental Figure 1 and 5. This analysis allows us to capture the entire cell and provide us the means to estimate the number of transcripts in cell and SGs by best fit-line analysis in Figure 2. In Figure 3 and supplemental Figure 2, at least 10 images were taken for each experiment comprising 5 Z-sections. All images shown in the manuscript are one Z-plane and image processed by ImageJ with FIJI plugin and Adobe Photoshop. Minimum and Maximum display values were set in ImageJ for each channel to properly view fluorescence. Quantification of smFISH spots in SGs in cells was determined using the spot and cell functions from Imaris Image Analysis Software (Bitplane) (University of Colorado-Boulder, BioFrontiers Advanced Light Microscopy Core).
Data and Software Availability
Raw sequencing data for both yeast and mammalian SGs and total RNA are available for download GEO:GSE99304.
Supplementary Material
1
Data File S1: Mammalian stress granule transcriptome, Related to Figure 1, 2, 4, and 5
2
Data File S2: Yeast stress granule transctiptome, Related to Figure 6
3
Data File S3: Yeast noncoding RNA stress granule transcriptome, Related to Figure S4
4
Data File S4: eCLIP analysis, related to Figure 7
5
Data File S5: Single molecule FISH probe dataset, Related to Figure 1, 3, 5, S1, S2, and S4
6
Acknowledgments
We would like to thank Nancy Kedersha and Paul Anderson for the ΔΔG3BP1/2 U-2 OS cell line as well as WT U-2 OS cells. We would also like to thank Paul Taylor for the G3BP1-GFP U-2 OS cells. We would like to thank Anne Ephrussi for providing ddUTP-Atto633 and Evan Lester for making CDK6 smFISH probes. We would like to thank Briana Van Treeck for making proportional Venn Diagrams. We thank Carolyn Decker for DeltaVision training. We are grateful to Joe Dragavon for imaging analysis training. The imaging data analysis was performed at the CU Light Microscopy Core Facility and the BioFrontiers Institute Advanced Light Microscopy Core. We also thank Dr. Jamie Kershner and Dr. Meghan Sloan at the BioFrontiers Next Generation Sequencing Facility (University of Colorado Boulder). This work was funded by NIH-F30N2093682 (J.R.W.), NIH-GM05443 (R.P.), and the Howard Hughes Medical Insitute (R.P.).
Footnotes
AUTHOR CONTRIBUTIONS
A.K., T.M., S.J., J.R.W., and S.F.M. conceived and performed experiments, analyzed results and contributed to manuscript preparation. R.P. contributed to project conception and manuscript preparation.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aken BL, Ayling S, Barrell D, Clarke L, Curwen V, Fairley S, Banet JF, Hourlier T, Billis K, Garc C, et al. Database update The Ensembl gene annotation system. 2016:1–19. doi: 10.1093/database/baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W, SAWH Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. Genome analysis HTSeq — a Python framework to work with high-throughput sequencing data. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bley N, Lederer M, Pfalz B, Reinke C, Fuchs T, Glaβ M, Birgit M, Stefan H. Stress granules are dispensable for mRNA stabilization during cellular stress. 2015;43 doi: 10.1093/nar/gku1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Genome analysis Trimmomatic: a flexible trimmer for Illumina sequence data. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Yoon J, Parker R. Stress-specific composition , assembly and kinetics of stress granules in Saccharomyces cerevisiae. 2011:228–239. doi: 10.1242/jcs.078444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis R, Taylor JP, Parker R. Eukaryotic stress granules are cleared by granulophagy and Cdc48 / VCP function. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartron JW, Hunt KCL, Frydman J. Cotranslational signal-independent SRP preloading during membrane targeting. Nature. 2016;536:224–228. doi: 10.1038/nature19309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey CM, Cenik B, Sephton CF, Johnson BA, Herz J, Yu G. TDP-43 aggregation in neurodegeneration: Are stress granules the key? Brain Res. 2012;1462:16–25. doi: 10.1016/j.brainres.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figley MD, Bieri G, Kolaitis R, Taylor JP, Gitler AD. Profilin 1 Associates with Stress Granules and ALS-Linked Mutations Alter Stress Granule Dynamics. 2014;34:8083–8097. doi: 10.1523/JNEUROSCI.0543-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell. 2016;164:487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. Dynamic Shuttling of TIA-1 Accompanies the Recruitment of mRNA to Mammalian Stress Granules 7. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P. Evidence That Ternary Complex ( eIF2-GTP-tRNA iMet )– Deficient Preinitiation Complexes Are Core Constituents of Mammalian Stress Granules. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, et al. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol. 2016;212:845–860. doi: 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding Proteins TIA-1 and TIAR Link the Phoshporylation of eIF-2a to the Assembly of Mammalian Stress Granules. 1999;147:1431–1441. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions , deletions and gene fusions. 2013a:1–13. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NC, Tresse E, Kolaitis R, Molliex A, Thomas RE, Alami NH, Wang B, Joshi A, Smith RB, Ritson GP, et al. Article VCP Is Essential for Mitochondrial Quality Control by PINK1 / Parkin and this Function Is Impaired by VCP Mutations. Neuron. 2013b;78:65–80. doi: 10.1016/j.neuron.2013.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WJ, Kim JH, Jang SK. Anti-inflammatory lipid mediator 15d-PGJ2 inhibits translation through inactivation of eIF4A. 2007;26:5020–5032. doi: 10.1038/sj.emboj.7601920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella RJ, Ka A, Spudich G, Almeida-king J, Staines D, Derwent P, Kerhornou A, Kersey P, Flicek P. Original article Ensembl BioMarts: a hub for data retrieval across taxonomic space. 2011;2011:1–9. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. 2012;9:357–360. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:1–10. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kopp F, Chang T, Yu H, Xie Y, Mendell JT. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell. 2016;164:69–80. doi: 10.1016/j.cell.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. J Cell Biol. 2013;201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Nicholson AM, Sarkar M, Boylan KB, Taylor JP, Rademakers R. TIA1 Mutations in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia Promote Phase Separation and Alter Stress Granule Dynamics Report TIA1 Mutations in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia Promote Phase Separation and Alter Stress Granule Dynamics. 2017:808–816. doi: 10.1016/j.neuron.2017.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, Chen J-J, Anderson P, Kaufman RJ. Heme-regulated Inhibitor Kinase-mediated Phosphorylation of Eukaryotic Translation Initiation Factor 2 Inhibits Translation, Induces Stress Granule Formation, and Mediates Survival upon Arsenite Exposure. J Biol Chem. 2005;280:16925–16933. doi: 10.1074/jbc.M412882200. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science (80- ) 2008:320. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelles DA, Fang MY, O’Connell MR, Xu JL, Markmiller SJ, Doudna JA, Yeo GW. Programmable RNA Tracking in Live Cells with CRISPR/Cas9. Cell. 2016;165:488–496. doi: 10.1016/j.cell.2016.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S, Kuehn MS, Reynolds AP, Haugen E, Thurman RE, Johnson AK, Rynes E, Maurano MT, Vierstra J, Thomas S, et al. BEDOPS: high-performance genomic feature operations. 2012;28:1919–1920. doi: 10.1093/bioinformatics/bts277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neymotin B, Athanasiadou R, Gresham D. Determination of in vivo RNA kinetics using RATE-seq. 2014:1645–1652. doi: 10.1261/rna.045104.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat Meth. 2016;13:508–514. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas MD, Ivanov P, Anderson P. Mechanistic insights into mammalian stress granule dynamics. 2016;215:313–323. doi: 10.1083/jcb.201609081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnyak V, Alhusaini N, Chen Y, Martin S, Kline N, Olson S, Weinberg D, Baker KE, Brenton R, Coller J. Codon optimality is a major determinant of mRNA stability. 2016;160:1111–1124. doi: 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter DSW, Parker R. Principles and Properties of Stress Granules. Trends Cell Biol. 2016;26:668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A, Green R. Connections Underlying Translation and mRNA Stability. J Mol Biol. 2016;428:3558–3564. doi: 10.1016/j.jmb.2016.05.025. [DOI] [PubMed] [Google Scholar]
- Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154:727–736. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhaussser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. :1–8. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Mcgeachy AM, Ingolia NT, Walter P. The small molecule ISRIB reverses the effects of eIF2 α phosphorylation on translation and stress granule assembly. 2015:1–16. doi: 10.7554/eLife.05033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwiak M, Zielenkiewicz P. A comprehensive, quantitative, and genome-wide model of translation. PLoS Comput Biol. 2010;6:4. doi: 10.1371/journal.pcbi.1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WFC, Blackwell TK, Anderson P. prevent stress granule association and ARE-mRNA decay. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöhr N, Lederer M, Reinke C, Meyer S, Hatzfeld M, Singer RH, Hüttelmaier S. ZBP1 regulates mRNA stability during cellular stress. 2006;175:527–534. doi: 10.1083/jcb.200608071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Mizutani R, Salam KA, Tano K, Ijiri K, Wakamatsu A, Isogai T, Suzuki Y, Akimitsu N. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. 2012:947–956. doi: 10.1101/gr.130559.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichon A, Gil N, Lubelsky Y, Solomon TH, Lemze D, Itzkovitz S, Stern-ginossar N, Ulitsky I. A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nat Commun. 2016;7:1–10. doi: 10.1038/ncomms12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotech. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsworth H, Raguz S, Edwards HJ, Higgins CF, Yagüe E. mRNA escape from stress granule sequestration is dictated by localization to the endoplasmic reticulum. FASEB J. 2010;24:3370–3380. doi: 10.1096/fj.09-151142. [DOI] [PubMed] [Google Scholar]
- Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R. Distinct stages in stress granule assembly and disassembly. Elife. 2016;5:1–25. doi: 10.7554/eLife.18413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-mu S, Camblong J, Huber W, Steinmetz LM, Guffanti E. Bidirectional promoters generate pervasive transcription in yeast. 2009:457. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
Data File S1: Mammalian stress granule transcriptome, Related to Figure 1, 2, 4, and 5
2
Data File S2: Yeast stress granule transctiptome, Related to Figure 6
3
Data File S3: Yeast noncoding RNA stress granule transcriptome, Related to Figure S4
4
Data File S4: eCLIP analysis, related to Figure 7
5
Data File S5: Single molecule FISH probe dataset, Related to Figure 1, 3, 5, S1, S2, and S4
6