Mapping of Chromosome 1p Deletions in Myeloma Identifies FAM46C at 1p12 and CDKN2C at 1p32.3 as being Genes in Regions Associated with Adverse Survival (original) (raw)
. Author manuscript; available in PMC: 2018 Jan 3.
Abstract
Purpose
Regions on 1p with recurrent deletions in presenting myeloma patients were examined with the purpose of defining the deletions and assessing their survival impact.
Experimental Design
Gene mapping, gene expression, FISH and mutation analyses were performed on patient samples from the MRC Myeloma IX trial and correlated with clinical outcome data.
Results
1p32.3 was deleted in 11% of cases, and deletion was strongly associated with impaired overall survival (OS) in patients treated with autologous stem cell transplant (ASCT). In patients treated less intensively, del(1)(p32.3) was not associated with adverse progression free survival (PFS) or OS. The target of homozygous deletions was CDKN2C, however its role in the adverse outcome of cases with hemizygous deletion was less certain. 1p22.1-21.2 was the most frequently deleted region and contained the candidate genes MTF2 and TMED5. No mutations were identified in these genes. 1p12 was deleted in 19% of cases, and deletion was associated with impaired OS in univariate analysis. The target of homozygous deletion was FAM46C, which was mutated in 3.4% of cases. When cases with FAM46C deletion or mutation were considered together, they were strongly associated with impaired OS in the intensive treatment setting.
Conclusion
Deletion of 1p32.3 and 1p12 were associated with impaired OS in myeloma patients receiving ASCT. FAM46C was identified as a gene with potential pathogenic and prognostic significance based on the occurrence of recurrent homozygous deletions and mutations.
Keywords: Myeloma, 1p12, 1p32.3, FAM46C, CDKN2C, Prognosis, FISH
INTRODUCTION
High risk genetic lesions can be divided into primary genetic events, namely translocations involving the IGH gene at 14q32, and secondary genetic events which usually constitute chromosomal gains or deletions (1). IGH translocations that have been associated with short survival in myeloma include t(4;14), t(14;16) and t(14;20), whilst structural chromosomal abnormalities that have been shown to be important include deletion of 17p, involving the TP53 gene, and gain of 1q (2-7). A prognostic model incorporating these lesions with the International Staging System (ISS) has been described which identified patients with poor prognosis at diagnosis (8).
Deletion of 1p has been identified as a common recurrent genetic event in myeloma that has prognostic significance when detected by conventional cytogenetics (9-10). Several minimally altered regions on 1p have been identified, including 1p32.3, 1p31.3, 1p22.1-1p21.3 and 1p12 (11). We have previously used an approach of focusing on regions with recurrent homozygous deletions, as these are usually small, pathogenically relevant, and allow for the definition of critically de-regulated genes (12). We described recurrent homozygous deletions of 1p32.3 which incorporate two genes, CDKN2C and FAF1, with homozygous deletion being associated with abrogation of expression of both of these genes (13-14). We found deletion of 1p32.3 to be associated with impaired survival in 510 newly presenting patients treated intensively (14). In another study, genome wide correlation of CGH-defined genomic lesions with survival identified 1p31-32 as the only factor independently associated with prognosis in 131 patients treated with high dose melphalan and autologous stem cell transplantation (ASCT) (15). Two data sets have, therefore, found loss of 1p32 to be prognostically significant in patients treated with ASCT, although its significance outside of this clinical context is unknown. We have previously identified recurrent homozygous deletions of FAM46C at 1p12, identifying it as a gene with potential pathogenic relevance (11). The next generation sequencing of 38 myeloma tumours recently found FAM46C to be frequently mutated, highlighting the potential significance of this gene (16). It is not known whether abnormalities of 1p12 / FAM46C impact prognosis. We have also previously identified a region from 1p22.1-21.3 as being a region of potential significance as, although it did not harbour homozygous mutations, it was the most frequently deleted region (11). Two candidate genes with down-regulated expression within this region (MTF2 and TMED5) were identified.
In this study we conducted an analysis of the survival associations of genetic lesions of 1p32.3, 1p22.1 and 1p12 in patients treated intensively and non-intensively, with conventional and thalidomide-based chemotherapy regimens, and have integrated gene expression and mutational analysis to define de-regulated genes on 1p.
MATERIALS AND METHODS
Patients
The MRC Myeloma IX trial (ISRCTN68454111) enrolled 1960 patients with newly diagnosed multiple myeloma requiring treatment, and received ethical approval from the MRC Leukaemia Data Monitoring and Ethics Committee (MREC 02/8/95). The design and results of which have been reported elsewhere, but in summary treatment followed two pathways, an intensive or a non-intensive pathway, based on patient age and performance status (17-18). Patients in the intensive pathway underwent an initial randomization to CVAD (cyclophosphamide, vincristine, doxorubicin and dexamethasone) or CTD (cyclophosphamide, thalidomide and dexamethasone) followed by ASCT. After the autograft procedure there was a second randomization to maintenance thalidomide versus no ongoing therapy. In the non-intensive pathway patients were randomized to either MP (melphalan and prednisolone) or CTDa (attenuated cyclophosphamide, thalidomide and dexamethasone) to maximum response, followed by the same maintenance randomization. Irrespective of treatment pathway, all patients underwent a bisphosphonate randomization to receive zoledronic acid or clodronic acid. Median follow up was 3.7 years. An extended data set for 1p12 FISH testing included patients randomized in the Myeloma IX trial along with other diagnostic myeloma samples referred to the Wessex Regional Cytogenetic laboratory.
Genetic Test Data Sets
Bone marrow aspirate and peripheral blood samples were taken from patients after informed consent. Bone marrow was purified for plasma cells using CD138 magnetic microbead cell selection (Miltenyi Biotec). 1140 / 1960 randomized patients provided a bone marrow aspirate sample that was suitable for analysis. A variety of genetic tests were carried out on the CD138 selected cell fraction. The number of samples tested by each method was decided by the quality and quantity of the provided samples, resulting in several overlapping data sets: 1140 had FISH performed, including 859 with results for 1p32.3 and 378 with results for 1p12; 272 had global gene expression arrays performed; 114 had SNP-based gene mapping performed; 147 had mutation analysis of FAM46C and MTF2. 124 had a complete data set of deletional annotation of 1p by mapping or FISH, plus gene expression and mutation analysis (Table 1). These various data sets were representative of the overall trial population (supplementary data, Table 1)
Table 1.
Genetic Test Data Sets
| Data Set | Number of Patients |
|---|---|
| Myeloma IX clinical trial entrants | 1960 |
| Any FISH performed | 1140 |
| FISH results for 1p32.3 | 859 |
| FISH results for 1p12 | 378 |
| Global gene expression arrays | 272 |
| Gene mapping arrays | 114 |
| FAM46C/MTF2 mutation analysis | 147 |
Gene Mapping and Expression Analysis
CD138 selected cells were stored in RLT buffer (Qiagen) at −80 °C immediately after purification until extraction. DNA and RNA were extracted using commercial kits as previously described(19). 100 ng of tumor RNA was amplified using the 2-cycle target labeling kit and hybridized to Human Genome U133 Plus 2.0 arrays (Affymetrix), washed and scanned following the manufacturer’s protocol. 500 ng of tumor and 500 ng of germ line DNA were used for mapping purposes. DNA was digested, amplified, labeled and hybridized to GeneChip mapping 500K arrays (Affymetrix) as previously described (19-20). SNP genotypes were derived as previously described using Affymetrix GCOS and GTYPE software (19). Regions of gain or loss of heterozygosity were identified using dChip 2006 (www.dchip.org), and expression data were normalized using dChip. Microarray data are accessible through Gene Expression Omnibus Series accession number GSE15695.
FISH
FISH was performed on CD138 selected bone marrow samples. FISH analysis of 1p32.3 used two bespoke probes (RP11-278J17 and RP11-116M11) grown and labeled in the laboratory, as previously described (13). FISH for 1p12 utilized the RP11-418J17 probe. A mixture of commercially available and bespoke probes were used as previously described to detect the presence of an immunoglobulin heavy chain (IGH) translocation, the common IGH translocation partners (MMSET at 4p16, CCND3 at 6p21, CCND1 at 11q13, MAF at 16q23 and MAFB at 20q12), hyperdiploid status using the iFISH ploidy classification, deletion of 13q14, 16q23, 17p13, 22q11 and gain of 1q21 (21-22).
Mutation Analysis
Genomic DNA underwent whole genome amplification using the REPLI-g commercial kit (Qiagen) following the manufacturer’s protocol. High resolution melt (HRM) analysis was used to screen tumor DNA samples for genetic variants of FAM46C, using the Type It HRM commercial kit (Qiagen) and the Rotorgene Q real-time cycler (Qiagen). Primers were designed to span segments of approximately 250 bases (supplementary data, Table 2). The PCR reaction comprised 7 pmol of combined forward and reverse primer mix, 20 ng of DNA, 5 μl of 2x HRM Master Mix and 3.3 μl of RNAse free water. PCR conditions were a 95 °C hold for 5 minutes, followed by 40 cycles of 95 °C for 10 seconds, 55 °C for 30 seconds and 72 °C for 10 seconds. High resolution melt analysis was performed from 65 °C to 95 °C at 0.1 °C increments. Data were analyzed using the Rotorgene Q series software version 1.7. Samples identified as variants by HRM underwent purification using the QIAquick PCR purification kit (Qiagen) and were directly sequenced with BigDye terminators v3.1 (Applied Biosytems) on a 3500 Genetic Analyzer (Applied Biosystems). Variants that were not labeled as SNPs on Ensembl had constitutional DNA sequenced using DNA extracted from peripheral blood samples.
Statistical Methods
Statistical analysis was performed using R and SPSS version 19.0 (SPSS Inc., Chicago, USA). Survival curves were plotted using the Kaplan-Meier method. Differences between curves were tested for statistical significance using the log-rank test, with P<0.05 taken as the level of significance. Variables associated with survival in univariate analysis at the P<0.05 level were entered into the model for multivariate testing. Multivariate analysis of variables associated with survival used a backwards elimination Cox proportional hazards model, with factors retained if they were significant at P<0.05. The test for interaction used a Cox regression model, with chi square values derived from the difference of the log-likelihood values. Comparison of categorical variables used the Fisher exact or chi-squared tests. Comparison of continuous variables used the Wilcoxon rank sum test. The gene expression of groups was compared using a t-test.
RESULTS
Survival Impact and Molecular Biology of Specific Regional Loss
1p32.3
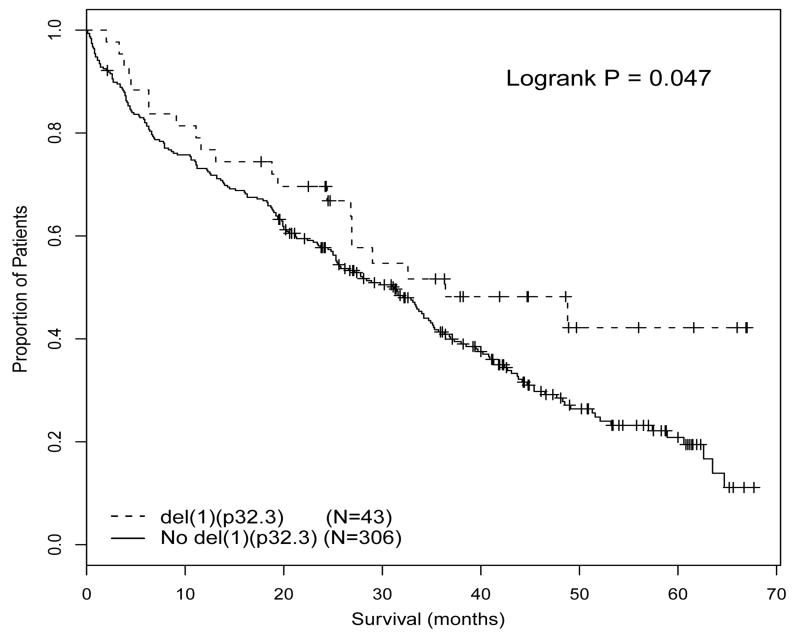
FISH for 1p32.3 was performed on 859 patients, of which 96 (11.2%) had del(1)(p32.3). As previously reported, intensively treated patients with this deletion had very short survival (median overall survival (OS) 34.5 months vs median not reached, P<0.001) (11). In contrast, patients with del(1p)(32.3) treated with a non-intensive approach had improved survival compared to patients without deletion (median OS 36.4 months vs 31.2 months, P=0.047) (Figure 1); del(1)(p32.3) was not associated with progression free survival (PFS) in either treatment pathway. A formal test for interaction supported the finding that the prognostic impact of del(1)(p32.3) varied by treatment pathway (chi-square 13.69, df 1, P<0.001). We examined whether other variables associated with survival were balanced between the intensive and non-intensive arms in patients with del(1)(p32.3), in an attempt to explain this paradoxical result. There was no significant bias in frequency of other high risk genetic lesions including t(4;14), t(14;16), t(14;20), +1q or del(17p). Similarly, there were no significant differences comparing intensively and non-intensively treated patients with del(1)(p32.3) in ISS stratification, length of 1p deletion, rate of homozygous 1p32.3 deletion or rate of co-segregation of 1p12 deletion (supplementary data, Table 3). In a multivariate analysis including these genetic variables, del(1)(p32.3) was significantly associated with adverse OS in the intensive arm (HR1.68 (1.11 – 2.56), p=0.015). Surprisingly, the association of del(1)(p32.3) with favorable survival in the patients treated non-intensively remained weakly significant in multivariate analysis (HR 0.63 (0.39 – 1.00), p=0.050). These data suggest that the paradoxical survival effect in the two arms is not due to a confounding variable but is associated with the 1p32.3 deletion.
Figure 1.
OS of patients undergoing non-intensive treatment comparing patients with del(1)(p32.3) to those without deletion.
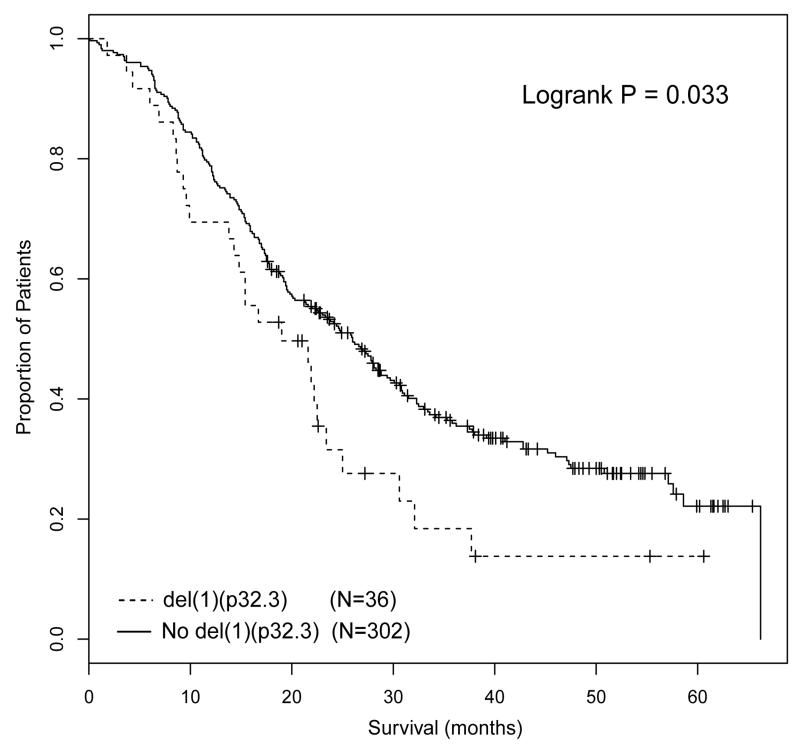
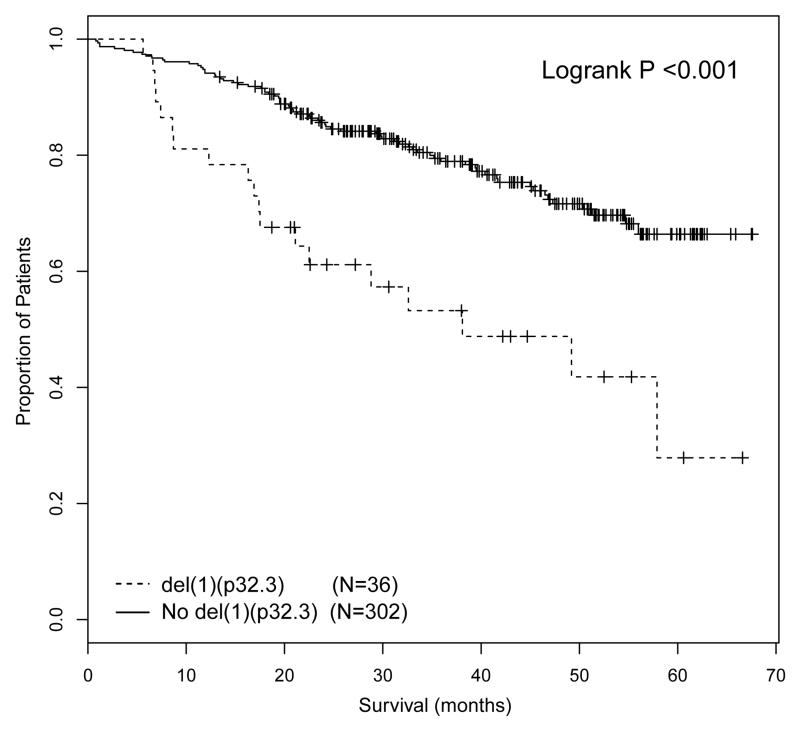
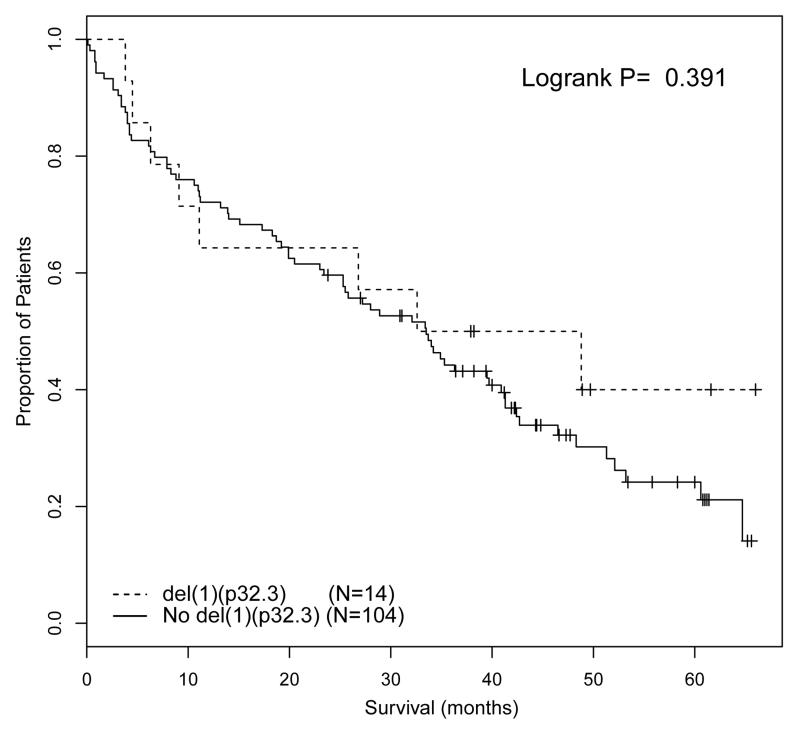
The major differences between the intensive and non-intensive arms were treatment and patient age. The intensive pathway incorporated treatment with ASCT, whereas the non-intensive pathway did not. Whether a patient entered the intensive or non-intensive pathway was decided by the treating physician based on patient suitability for ASCT, with no defined age limit. However, in practice, the majority of patients under the age of 70 were treated intensively, whilst the majority of patients over the age of 70 were treated non-intensively. This raised the question as to whether del(1)(p32.3) was associated with adverse prognosis in the context of ASCT or young age, and taking this question further we investigated age and ASCT as factors in subgroup analyses. We examined the impact of del(1)(p32.3) in patients that received autograft and in young patients treated in the non-intensive pathway. Although there was an intention-to-treat with ASCT in the intensive pathway, only 346/503 (68.6%) analyzable patients in the intensive arm received ASCT. The analysis of the association of del(1)(p32.3) with survival in the intensive pathway was landmarked from time of ASCT, thus excluding patients that did not proceed to transplant. In this landmarked analysis del(1)(p32.3) was associated with impaired PFS and OS (median PFS from time of ASCT 19.0 months vs 26.0 months, p=0.033; median OS from time of ASCT 38.1 months vs median not reached, P<0.001) (Figure 2A, B). 118 patients in the non-intensive pathway were less than 70 years of age, including 14 with del(1)(p32.3). Del(1)(p32.3) in these patients was not associated with impaired OS (median 40.7 months del(1)(p32.3) vs 33.5 months (no del(1)(p32.3), P=0.391) (Figure 3). From this we conclude that del(1)(p32.3) is only associated with impaired survival in the context of ASCT. The fact that del(1)(p32.3) was not associated with OS in young patients that were not autografted suggests that this may be a treatment-specific effect.
Figure 2.
PFS (2A) and OS (2B) of patients treated in the intensive pathway, landmarked at time of ASCT and comparing cases with del(1)(p32.3) with patients without deletion.
Figure 3.
OS of patients ≤70 years of age in the non-intensive pathway, comparing patients with del(1)(p32.3) with patients without deletion.
To investigate the biological basis of this effect, the deletions were defined in more detail. Of the 114 patients with gene mapping data, 6 had a homozygous deletion of this region, with a further 12 having a hemizygous deletion. In our data set all the homozygous deletions affected two genes, CDKN2C and FAF1, and in order to try and define the key deleted gene we examined the mapping of an additional 565 publically available myeloma mapping cases (available through www.broad.mit.edu/mmgp (incorporating the Mayo clinic, Carrasco and MMRC reference collections) and the IFM collection at www.ncbi.nlm.nih.gov/geo GSE 12896). These data identified a further 15 cases with homozygous deletion of 1p32.3. In the majority of cases the deletions incorporated both CDKN2C and FAF1, but in 3 cases CDKN2C appeared to be the sole target of deletion.
The relationship of deletion and gene expression was examined. Although homozygous deletion of 1p32.3 abrogates expression of CDKN2C, hemizygous deletion was not associated with decreased expression. Moreover, although the homozygously deleted cases with abrogated CDKN2C expression were associated with short survival, low CDKN2C expression was not associated with impaired survival in other cases. In fact, the converse was true; patients with top quartile CDKN2C expression were associated with significantly shorter survival than those with bottom quartile expression (median OS 54.8 months low expression vs 36.1 months high expression, p=0.001). This is consistent with previously reported data linking high CDKN2C expression with a high proliferation rate (13, 23). We have previously not found CDKN2C to be inactivated through mutation or methylation (13). As hemizygous deletion was not associated with low expression, low expression was not associated with adverse survival, and other mechanisms of allelic inactivation have not been demonstrated, it is difficult to link the adverse survival associated with hemizygous deletion of 1p32.3 to modulation of CDKN2C. We examined FAF1, and found that hemizygous deletion of 1p32.3 was associated with decreased expression of this gene (218080_x_at fold change −1.48 (−1.29 to −1.73), P<0.001; 224217_s_at fold change −1.66 (−1.37 to −2.07), P<0.001). However, there was no association of low expression of these probes with short survival.
1p22.1
In a previous report we identified 1p22.1-21.3 as being the most frequently deleted of 1p, and the meta-analysis of the additional 565 publically available mapping cases confirmed this, with 22.5% of cases having a deletion of 1p22.1 (11). We previously identified MTF2 and TMED5 as candidate genes within this region as both had significantly down-regulated expression in cases with deletion (11). We now report that low expression of MTF2 was not associated with impaired OS or PFS. Moreover, no mutations of MTF2 were identified by HRM analysis in 147 newly presenting patients. Low TMED5 expression was weakly associated with short survival (median OS 29.9 months low expression vs 51.2 months, high expression, p=0.050). However, no mutations were identified in 20 newly presenting patients.
1p12
Of 114 cases with mapping data, 1p12 was homozygously deleted in 2 cases and hemizygously deleted in a further 20 cases. In addition, uniparental disomy (UPD) was a mechanism of loss of heterozygosity in 4 cases. In an extended data set of 378 cases with FISH for 1p12, using a probe that was not specific for FAM46C, the rate of 1p12 deletion was 19%. When mapping of human myeloma cell lines (HMCLs) was examined, small homozygous deletions of FAM46C were identified in KMS11, LP1, and MM1s, with further hemizygous deletions in H929, JIM1, JIM3, KMS26, KMS28 and MM1R. The fact that 2/114 presenting cases had homozygous deletion of this region, compared to 3/18 HMCLs suggest that this lesion is associated with disease progression. Mutations of FAM46C were screened for in 147 patients, and 5 (3.4%) were found to harbor mutations (Table 2). All mutations were single base substitutions, resulting in either an amino acid change or a stop codon. Two of the mutations occurred in samples with hemizygous deletion of 1p12, whilst three did not co-segregate with either deletion or UPD.
Table 2.
Annotated FAM46C Mutations
| Sample no. | Base Change | Amino Acid Change | 1p12 Mapping |
|---|---|---|---|
| 323 | c357 C>G | F119L | No deletion |
| 1527 | c463 A>T | I155F | Hemizygous deletion |
| 245 | c537 C>A | F179C | No deletion |
| 326 | c872 A>G | Y291C | No deletion |
| 127 | c1068 C>G | Y356X | Hemizygous deletion |
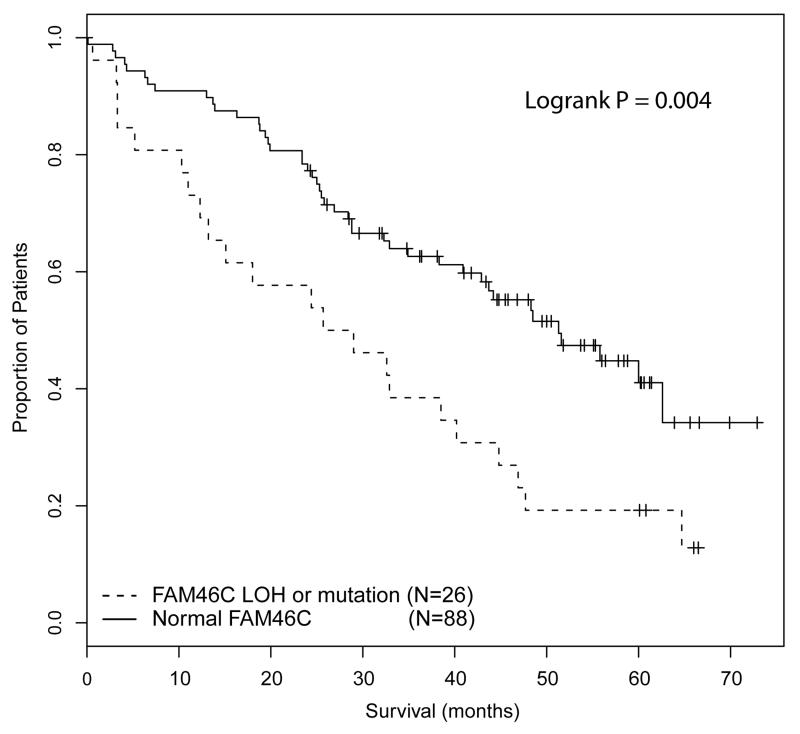
The survival of patients with any abnormality of FAM46C (incorporating loss of heterozygosity or mutation) was compared to patients with normal FAM46C, and we show that abnormal FAM46C was associated with impaired survival (median OS 25.7 months vs 51.3 months, P=0.004) (Figure 4). Although this effect was most marked in patients treated intensively (median OS 24.4 months vs not reached, P=0.005), a formal test for interaction of pathway and del(1)(p12) was not significant (chi square 3.08; df 1; P=0.078) which does not support there being the same differential survival impact in patients treated intensively and non-intensively as was evident in the analysis of 1p32.3. The survival impact of del(1)(p12) was extended to the FISH data set of 378 patients and deletion was found to be weakly associated with impaired OS (median OS 44.5 months vs 53.0 months, P=0.0497). This association was not significant in multivariate analysis when +1q, del17p and adverse IGH translocations (t(4;14), t(14;16) and t(14;20)) were included as co-variates.
Figure 4.
OS of cases with any abnormality of FAM46C (incorporating loss of heterozygosity and mutation) compared to cases with normal FAM46C.
The Relative Survival Impact of del(1)(p32.3) and del(1)(p12)
Hemizygous deletion are often large, so that 1p deletions often encompass both 1p12 and del1p32.3. In order to assess the relative contributions of these two regions to the adverse prognostic signal, 372 patients with FISH results for both 1p12 and 1p32.3 were examined. 38 (10.2%) had del(1)(p12) only, 18 (4.8%) had del(1)(p32.3) only and 30 (8.1%) cases had deletion of both regions. The 38 patients with only del(1)(p12) were weakly associated with impaired survival (median OS 40.9 months vs 53.0 months, p=0.045), whilst there was a stronger association with short survival in patients with only del(1p)(32.3) (median OS 21.6 months vs 51.6 months, P<0.001). This suggests that when deletion alone is being considered, 1p32.3 has the largest prognostic impact.
DISCUSSION
In this analysis we identify genes and regions of 1p with prognostic impact and potential pathogenic relevance. 1p32.3 underwent homozygous deletion in 5.3% of cases, with the deletions affecting CDKN2C and also FAF1, although the mapping data of other data sets suggest that CDKN2C may be the key deleted gene (23). Deletions detected by FISH were associated with impaired survival in patients treated in the intensive arm of the Myeloma IX trial, but surprisingly the converse was true in patients treated non-intensively, with deletion being associated with a marginal improvement in OS. Both these associations were true in multivariate analyses including other genetic lesions, and we found no bias in confounding variables to explain this effect. We examined this paradox in more detail to assess whether this was related to patient therapy or patient age, and the association of deletion with impaired survival was only observed in patients receiving ASCT, suggesting that del(1)(p32.3) may specifically impair the response to this therapy. However, the low numbers of young patients not autografted in the analysis means that age cannot be completely discounted as a factor.
Whilst CDKN2C is likely to be the key deregulated gene in cases with homozygous deletion, it’s involvement in the outcome of cases with hemizygous deletion was less certain, as in these cases CDKN2C was not under-expressed, and we have previously reported no evidence of allelic silencing through methylation or mutation (13). Moreover, low expression was not associated with short survival but the converse was true, with high expression being linked to impaired survival. This is consistent with previously reported data showing that myeloma with high proliferation defined by a gene expression signature was associated with high CKDN2C expression (23). It is possible, then, that in patients with hemizygous deletion of 1p32.3, deletion of other genes may play a role in the clinical outcome. Whilst homozygous deletions were small and focal, mapping analysis showed that hemizygous deletions are often large and involve other regions, including 1p22.1 and 1p12.
1p22.1 was the most frequently deleted region in our mapping data set and in the extended analysis of all publically available mapping data sets. However, the two candidate genes that we had previously identified within this region (MTF2 and TMED5) did not have convincing evidence that down-regulation adversely affected survival. Moreover, we did not identify any mutations in these genes. Whilst it remains likely that this region has pathogenic relevance, a key de-regulated gene within this region has not been identified.
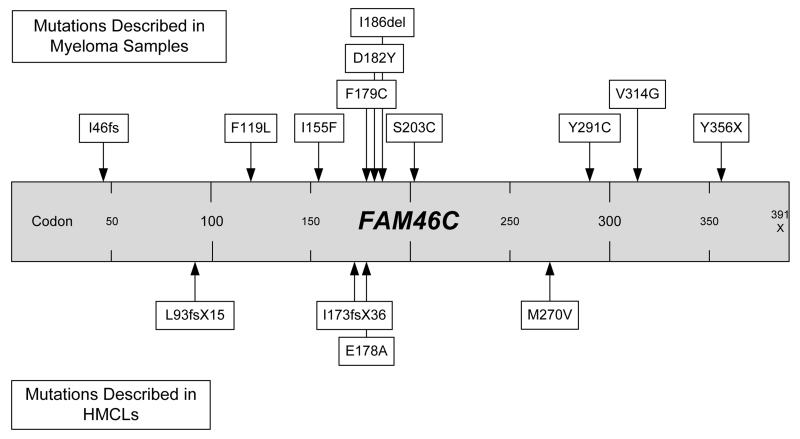
Homozygous deletions of 1p12 affecting only FAM46C were observed in two presenting myeloma patients and three HMCLs. In our initial gene mapping and expression data set we identified the potential importance of FAM46C, and this was confirmed in the report of the initial genome sequencing of 38 myeloma tumors which found FAM46C mutations in 5/38 patient samples and 4/17 HMCLs, and suggested a role for this functionally uncharacterized gene in regulation of translation by acting as an mRNA stability factor (11, 16). We screened presenting myeloma tumors for variants of FAM46C by high resolution melt analysis, and found mutations in 5 samples. The mutation rate was 3.4%, which is significant for presenting myeloma samples, but is lower than the 13% reported in the next-generation sequencing analysis. All our mutations were mis-sense mutations, as were 5/9 of the mutations reported previously (16). FAM46C consists of two exons, with only part of exon 2 being translated, and is made up of 391 codons. It is notable that 35.7% (5/14) of mutations described in FAM46C in myeloma are in a small region between codon 173 and 186, suggesting that this may constitute a mutation hotspot (Figure 5). As FAM46C shows occasional homozygous deletion in presenting myeloma cases, and has a reported mutation rate of between 3.4% and 13%, it likely represents a gene with pathogenic significance.
Figure 5.
Mutations of FAM46C described to date in myeloma.
We examined the prognostic significance of abnormalities of this gene and region. The association of deletion of 1p12 with adverse survival was confirmed by FISH in 379 patients, although the association was of borderline significance, and was not significant in multivariate analysis. Moreover, when cases with del(1)(p12) alone were compared to patients with del(1)(p32.3) alone, the association of del(1)(p32.3) with short survival was much stronger. A caveat of this extended analysis was that the FISH probe for 1p12 did not target FAM46C and may, therefore, have missed some FAM46C deleted cases. These analyses also did not consider FAM46C mutation events, which were frequent, and when we examined survival within patients with complete annotation of deletion and mutation of FAM46C, patients with any LOH or mutation of FAM46C were associated with impaired survival, although the numbers of patients involved in this analysis means that these results should be treated as preliminary. Taken together, these data suggest that when deletion alone is being considered, del(1)(p32.3) is the more prognostically important lesion. However, if deletion and mutation events are considered together, FAM46C may still be prognostically important and future studies incorporating next generation sequencing data will illuminate this further.
In summary, mapping of homozygous deletions in newly presenting myeloma patients identified two regions of potential importance, 1p32.3 and 1p12. Deletion of 1p32.3 was strongly associated with adverse prognosis, but only in patients treated with ASCT, whilst in patients treated non-intensively its impact was neutral, or even slightly favorable. Whilst CDKN2C appeared to be the target of homozygous deletions of 1p32.3, it remains unclear if the adverse survival seen in patients with hemizygous deletion was mediated through CDKN2C dysregulation, or if other genes played a role in the observed clinical outcome. In comparison, the target of 1p12 deletion was more apparent, as the homozygous deletions targeted only one gene, FAM46C, and this gene was found to be frequently mutated. Del(1)(p12) was not associated with adverse survival to the same extent as del(1)(p32.3), but if deletion and mutation events were both considered, there may be a prognostic effect. Taken together, these data highlight the importance of genetic events affecting 1p in myeloma, and show that the clinical relevance of these events needs to be individualized based on the site of deletion and treatment context.
Supplementary Material
1
2
3
STATEMENT OF TRANSLATIONAL RELEVANCE.
Cytogenetic lesions that are associated with impaired survival have been identified in myeloma, and there is an aim to use such lesions to define high risk disease in a risk-adapted therapeutic approach. The lesions generally accepted to constitute high risk lesions are t(4;14), t(14;16), t(14;20), del(17p) and +1q. We identified two regions of 1p that have prognostic impact when deleted: 1p32.3 and 1p12. Deletion of 1p32.3 targets CDKN2C, and it only impacts prognosis in patients treated with ASCT. Deletion of 1p12 targets FAM46C, which is also frequently mutated. Cases with deletion or mutation of FAM46C also had impaired survival. Deletion of 1p has prognostic significance in patients treated with ASCT, and could be incorporated into the definition of high risk disease in risk adapted treatment strategies.
Table 3.
Multivariate analysis of genetic variables associated with OS in the intensive pathway
| Genetic Variable | Hazard Ratio | 95% CI | P= |
|---|---|---|---|
| +(1)(q21) | 1.79 | 1.33 – 2.42 | <0.001 |
| del(17)(p13) | 1.90 | 1.21 – 2.98 | 0.005 |
| del(1)(p32.3) | 1.68 | 1.11 – 2.56 | 0.015 |
Table 4.
Multivariate analysis of genetic variables associated with OS in the non-intensive pathway
| Genetic Variable | Hazard Ratio | 95% CI | P= |
|---|---|---|---|
| Adverse IGH group | 2.12 | 1.52 – 2.95 | <0.001 |
| del(1)(p32.3) | 0.63 | 0.39 – 1.00 | 0.050 |
ACKNOWLEDGEMENTS
We thank Rebecca Protheroe, David Stockley, Tim Parker, Hazel Robinson, Elisabet Dachs Cabanas and Christina Rudduck of the LLR UKMF Cytogenetics group for probe preparation and FISH scoring. We also thank staff from the Clinical Trials Research Unit, University of Leeds, for trial co-ordination and data management. The University of Leeds acted as the trial sponsor. We also acknowledge the support of the National Institute for Health Research, through the National Cancer Research Network.
Supported by: Main financial support for the MRC Myeloma IX clinical trial was from the UK Medical Research Council, with additional funding in the form of unrestricted educational grants from Novartis, Schering Health Care, Chugai, Pharmion, Celgene and Ortho Biotech. The LLR UKMF Cytogenetics group was supported by Leukemia and Lymphoma Research. The Institute of Cancer Research received financial support from Myeloma UK and the Biological Research Centre of the National Institute for Health Research at the Royal Marsden Hospital.
Footnotes
C.O.I – The authors have no relevant conflicts of interest to declare
REFERENCES
- 1.Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol. 2005;23:6333–8. doi: 10.1200/JCO.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 2.Fonseca R, Blood E, Rue M, Harrington D, Oken MM, Kyle RA, et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood. 2003;101:4569–75. doi: 10.1182/blood-2002-10-3017. [DOI] [PubMed] [Google Scholar]
- 3.Avet-Loiseau H, Attal M, Moreau P, Charbonnel C, Garban F, Hulin C, et al. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myelome. Blood. 2007;109:3489–95. doi: 10.1182/blood-2006-08-040410. [DOI] [PubMed] [Google Scholar]
- 4.Ross FM, Chiecchio L, Dagrada G, Protheroe RK, Stockley DM, Harrison CJ, et al. The t(14;20) is a poor prognostic factor in myeloma but is associated with long-term stable disease in monoclonal gammopathies of undetermined significance. Haematologica. 2010;95:1221–5. doi: 10.3324/haematol.2009.016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanamura I, Stewart JP, Huang Y, Zhan F, Santra M, Sawyer JR, et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood. 2006;108:1724–32. doi: 10.1182/blood-2006-03-009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fonseca R, Bergsagel PL, Drach J, Shaughnessy J, Gutierrez N, Stewart AK, et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009;23:2210–21. doi: 10.1038/leu.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd KD, Ross FM, Tapper WJ, Chiecchio L, Dagrada G, Konn ZJ, et al. The clinical impact and molecular biology of del(17p) in multiple myeloma treated with conventional or thalidomide-based therapy. Genes, Chromosomes and Cancer. 2011 doi: 10.1002/gcc.20899. In press GCC 11-0042.R2 (20899) [DOI] [PubMed] [Google Scholar]
- 8.Boyd KD, Ross FM, Chiecchio L, Dagrada G, Konn ZJ, Tapper WJ, et al. A novel prognostic model in myeloma based on co-segregating adverse FISH lesions and the ISS: analysis of patients treated in the MRC Myeloma IX trial. Leukemia. 2011 doi: 10.1038/leu.2011.204. in press, SC11-LEU-0152R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu KL, Beverloo B, Lokhorst HM, Segeren CM, van der Holt B, Steijaert MM, et al. Abnormalities of chromosome 1p/q are highly associated with chromosome 13/13q deletions and are an adverse prognostic factor for the outcome of high-dose chemotherapy in patients with multiple myeloma. Br J Haematol. 2007;136:615–23. doi: 10.1111/j.1365-2141.2006.06481.x. [DOI] [PubMed] [Google Scholar]
- 10.Qazilbash MH, Saliba RM, Ahmed B, Parikh G, Mendoza F, Ashraf N, et al. Deletion of the short arm of chromosome 1 (del 1p) is a strong predictor of poor outcome in myeloma patients undergoing an autotransplant. Biol Blood Marrow Transplant. 2007;13:1066–72. doi: 10.1016/j.bbmt.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 11.Walker BA, Leone PE, Chiecchio L, Dickens NJ, Jenner MW, Boyd KD, et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood. 2010;116:e56–65. doi: 10.1182/blood-2010-04-279596. [DOI] [PubMed] [Google Scholar]
- 12.Dickens NJ, Walker BA, Leone PE, Johnson DC, Brito JL, Zeisig A, et al. Homozygous deletion mapping in myeloma samples identifies genes and an expression signature relevant to pathogenesis and outcome. Clin Cancer Res. 2010;16:1856–64. doi: 10.1158/1078-0432.CCR-09-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leone PE, Walker BA, Jenner MW, Chiecchio L, Dagrada G, Protheroe RK, et al. Deletions of CDKN2C in multiple myeloma: biological and clinical implications. Clin Cancer Res. 2008;14:6033–41. doi: 10.1158/1078-0432.CCR-08-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker BA, Wardell CP, Chiecchio L, Smith EM, Boyd KD, Neri A, et al. Aberrant global methylation patterns affect the molecular pathogenesis and prognosis of multiple myeloma. Blood. 2010 doi: 10.1182/blood-2010-04-279539. [DOI] [PubMed] [Google Scholar]
- 15.Chng WJ, Gertz MA, Chung TH, Van Wier S, Keats JJ, Baker A, et al. Correlation between array-comparative genomic hybridization-defined genomic gains and losses and survival: identification of 1p31-32 deletion as a prognostic factor in myeloma. Leukemia. 2010;24:833–42. doi: 10.1038/leu.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–72. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan GJ, Davies FE, Gregory WM, Cocks K, Bell SE, Szubert AJ, et al. First-line treatment with zoledronic acid as compared with clodronic acid in multiple myeloma (MRC Myeloma IX): a randomised controlled trial. Lancet. 2010;376:1989–99. doi: 10.1016/S0140-6736(10)62051-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morgan GJ, Davies FE, Gregory WM, Russell NH, Bell SE, Szubert AJ, et al. Cyclophosphamide, thalidomide, and dexamethasone (CTD) as initial therapy for patients with multiple myeloma unsuitable for autologous transplantation. Blood. 2011 doi: 10.1182/blood-2011-02-338665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker BA, Leone PE, Jenner MW, Li C, Gonzalez D, Johnson DC, et al. Integration of global SNP-based mapping and expression arrays reveals key regions, mechanisms, and genes important in the pathogenesis of multiple myeloma. Blood. 2006;108:1733–43. doi: 10.1182/blood-2006-02-005496. [DOI] [PubMed] [Google Scholar]
- 20.Jenner MW, Leone PE, Walker BA, Ross FM, Johnson DC, Gonzalez D, et al. Gene mapping and expression analysis of 16q loss of heterozygosity identifies WWOX and CYLD as being important in determining clinical outcome in multiple myeloma. Blood. 2007;110:3291–300. doi: 10.1182/blood-2007-02-075069. [DOI] [PubMed] [Google Scholar]
- 21.Chiecchio L, Protheroe RK, Ibrahim AH, Cheung KL, Rudduck C, Dagrada GP, et al. Deletion of chromosome 13 detected by conventional cytogenetics is a critical prognostic factor in myeloma. Leukemia. 2006;20:1610–7. doi: 10.1038/sj.leu.2404304. [DOI] [PubMed] [Google Scholar]
- 22.Chiecchio L, Dagrada GP, Ibrahim AH, Dachs Cabanas E, Protheroe RK, Stockley DM, et al. Timing of acquisition of deletion 13 in plasma cell dyscrasias is dependent on genetic context. Haematologica. 2009;94:1708–13. doi: 10.3324/haematol.2009.011064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dib A, Peterson TR, Raducha-Grace L, Zingone A, Zhan F, Hanamura I, et al. Paradoxical expression of INK4c in proliferative multiple myeloma tumors: bi-allelic deletion vs increased expression. Cell Div. 2006;1:23. doi: 10.1186/1747-1028-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3