Biosynthesis of a fluorescent cyanobacterial C-phycocyanin holo-α subunit in a heterologous host (original) (raw)
Abstract
The entire pathway for the synthesis of a fluorescent holophycobiliprotein subunit from a photosynthetic cyanobacterium (Synechocystis sp. PCC6803) was reconstituted in Escherichia coli. Cyanobacterial genes encoding enzymes required for the conversion of heme to the natural chromophore 3_Z_-phycocyanobilin, namely, heme oxygenase 1 and 3_Z_-phycocyanobilin:ferredoxin oxidoreductase, were expressed from a plasmid under control of the hybrid _trp_-lac (trc) promoter. Genes for the apoprotein (C-phycocyanin α subunit; cpcA) and the heterodimeric lyase (cpcE and cpcF) that catalyzes chromophore attachment were expressed from the trc promoter on a second plasmid. Upon induction, recombinant E. coli used the cellular pool of heme to produce holo-CpcA with spectroscopic properties qualitatively and quantitatively similar to those of the same protein produced endogenously in cyanobacteria. About a third of the apo-CpcA was converted to holo-CpcA. No significant bilin addition took place in a similarly engineered E. coli strain that lacks cpcE and cpcF. This approach should permit incisive analysis of many remaining questions in phycobiliprotein biosynthesis. These studies also demonstrate the feasibility of generating constructs of these proteins in situ for use as fluorescent protein probes in living cells.
The phycobiliproteins are a family of light-harvesting proteins found in cyanobacteria, red algae, and the cryptomonads. These proteins absorb strongly in the visible region of the spectrum because they carry various covalently attached linear tetrapyrrole prosthetic groups (bilins). Phycobiliproteins are tightly associated αβ heterodimers in which each subunit carries bilin(s) thioether-linked to particular cysteinyl residues (1–3).
Steps involved in bilin biosynthesis and bilin addition to apophycobiliprotein subunits have been inferred from diverse studies (e.g., refs. 4–6). However, the entire pathway from heme to a particular holophycobiliprotein subunit has not hitherto been reconstituted either in vitro or in vivo. This achievement is reported here for a pathway from endogenous heme to phycocyanin α subunit-linked phycocyanobilin (PCB) in an engineered Escherichia coli strain.
The studies described here are important as prototypes for experiments directed to exploring many unanswered questions in the enzymology and chemistry of phycobiliprotein synthesis. They are also of great interest in opening the way to the use of phycobiliproteins as in vivo fluorescent protein probes. Purified native phycobiliproteins and their subunits fluoresce strongly and, since 1982, have been widely used as external labels for cell sorting and analysis and a wide range of other fluorescence-based assays (7, 8). Fluorescent protein probes expressed in vivo, such as Aequorea victoria green fluorescent protein and its variants, have proved to be of great value in all fields of cell biology (9). Probes generated by the spontaneous addition of exogenously supplied phycoerythrobilin to recombinant apophytochrome fragments in living cells are also very promising (10). Phycobiliprotein constructs represent a broad array of spectroscopically distinctive proteins with photophysical properties superior to those of probes currently available (11). Hence, the ability to express them conveniently in functional fluorescent form in diverse cell types would greatly expand the repertoire of tools available for examining a broad array of questions in cell biology.
Materials and Methods
Materials.
Enzymes for DNA manipulation were obtained from New England Biolabs and Life Technologies (Rockville, MD), and antibiotics were obtained from Sigma. Agar and organic nutrients for LB were obtained from Difco, and other chemicals were from Sigma or Fisher Scientific. “Superflow” Ni2+-nitrilotriacetic acid agarose for isolation of His-tagged proteins was purchased from Qiagen (Chatsworth, CA).
Cultures and Strains.
E. coli strain DH5α (Life Technologies) grown in LB was used in all experiments. Plasmids conferring resistance to spectinomycin (Sp) were selected with 100 μg⋅ml−1 Sp dihydrochloride, and those conferring resistance to kanamycin (Km) were selected with 50 μg⋅ml−1 Km sulfate. For expression of _P_trc-controlled genes, isopropyl β-d-thiogalactoside was added to a final concentration of 0.5 mM to exponentially growing cells. Induced cells were grown for 3.5 h at 37°C with shaking. Cultures were harvested and pellets were stored at −20°C until they were used.
Cloning of Relevant Synechocystis sp. PCC6803 Genes.
Standard procedures were used for most DNA manipulations. Gene sequences were obtained from the Kazusa DNA Research Institute CyanoBase (refs. 12 and 13; http://www.kazusa.or.jp/cyano/index.html), and all accession numbers given below refer to that database. Using primers described below, all genes were amplified from PCC6803 genomic DNA by the PCR. The fidelity of all PCR-generated fragments was verified by direct nucleotide sequencing. A more typical E. coli ribosomal binding site was engineered upstream of the cpcE, cpcF, and hox1 ORFs. DNA sequence analysis was performed with the program editbase (Purdue Research Foundation and U.S. Department of Agriculture/Agricultural Research Service), and predicted amino acid sequences were deduced with lasergene (DNAstar, Madison, WI).
Cloning of the Gene Encoding Synechocystis sp. PCC6803 Phycocyanin α Subunit.
Primers used to amplify the cpcA gene (sll1578) were 5′-CGG GAG ATA CCA GAC ATA TGA AAA CCC CTT TAA CTG-3′ and 5′-TAG AAT TCG AAG ACT AGC TCA GAG CAT TGA TGG CG-3′. The resulting 0.5-kb product was digested with restriction enzymes _Nde_I and _Eco_RI and cloned into _Nde_I- and _Eco_RI-digested cloning vector, pBS350V (described in ref. 11), giving plasmid pBS405V. The α subunit of C-phycocyanin (CpcA) construct expressed from pBS405V consists of CpcA fused at the N terminus to a 24-amino acid sequence that includes a His6-tag (11).
Cloning of the Genes Encoding the Synechocystis sp_._ PCC6803 Phycocyanin α Subunit Phycocyanobilin Lyase.
Primers used to amplify the cpcE gene (slr1878) were 5′-ATT GAC GTC GAC AAG GAG CTT GCA TAT GAG TGA ACC TAA CCT CAA CCC CG-3′ and 5′-GTG CGG CCG CGA CGC TTT TAG AGT AAA CTA TCC ATT AAT TCC AAT ATT TGT CG-3′. The resulting 0.85-kb PCR fragment was digested with restriction enzymes _Sal_I and _Not_I and cloned into _Sal_I- and _Not_I-digested pBS350V, giving plasmid pBS406V. Primers used to amplify the cpcF gene (sll1051) were 5′-ATT AGC GGC CGC TAG GAG GGC TAA CAT ATG GAG GGT AAT AGC GTC GTA ACT CC-3′ and 5′-GAG GGA TCC TAG AAG ACT AGA TTG GGC CGA TGT TTT CCA GG-3′. The resulting 0.7-kb product was digested with restriction enzymes _Eag_I and _Bam_HI and cloned into _Eag_I- and _Bam_HI-digested pBS350V, giving plasmid pBS407V.
Cloning of the Gene Encoding Synechocystis sp. PCC6803 Heme Oxygenase 1 (HO1).
Primers used to amplify the hox1 gene (sll1184; ref. 14) were 5′-CAT TGG ATC CAA GGT ACC CAG GGA TTT TTC ATA TGA GTG TCA ACT TAG CTT CCC AGT TGC-3′ and 5′-GTT GGC GCG CCA GGG AAG ACT AGC CTT CGG AGG TGG CGA G-3′. The resulting 0.75-kb product was digested with restriction enzymes _Bam_HI and _Asc_I and cloned into _Bam_HI- and _Asc_I-digested pBS350V, yielding plasmid pBS421V.
Cloning of the Gene Encoding Synechocystis sp. PCC6803 3_Z_-Phycocyanobilin:Ferredoxin Oxidoreductase.
Primers used to amplify the pcyA gene (slr0116) were 5′-CTT GGC GCG CCA TAG GAG TAT TTC ATA TGG CCG TCA CTG ATT TAA GTT TGA CC-3′ and 5′-TTC GCT GCA TGC TGG AGA GTC TTT TAT TGG ATA ACA TCA AAT AAG ACT TGG CTC ATA TAC-3′. The resulting 0.8-kb product was digested with restriction enzymes _Asc_I and _Sph_I and cloned into the _Asc_I- and _Sph_I-digested pBS350V, giving the plasmid pBS422V.
Design of Expression Vectors.
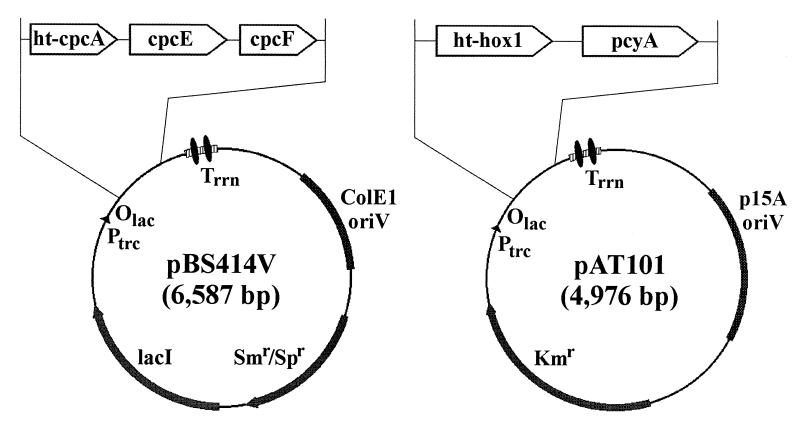
Two expression vectors were designed, each containing a cassette of two or more Synechocystis sp. PCC6803 genes. The aim was to introduce them together into E. coli cells and thereby produce all of the catalytic functions and components required for the formation of phycocyanin holo-α subunit in vivo. The cassette in one vector, pBS414V, contained cpcA along with cpcE and cpcF, and provides all of the components (apo-CpcA, CpcE, and CpcF) known to be both necessary and sufficient for the correct addition of PCB to apo-CpcA (15). The cassette in the other vector, pAT101, contained hox1 and pcyA (6), to provide enzymes required for the conversion of heme to PCB, the proximal precursor to the polypeptide-bound bilin (15). Construction of these cassettes is described below (see also Fig. 1).
Figure 1.
Physical maps of the pBS414V and pAT101 expression vectors used for in vivo bilin addition to Synechocystis sp. PCC6803 His6-tagged phycocyanin apo-α subunit (HT-CpcA) in E. coli. Vector pBS414V carries the phycocyanin α subunit phycocyanobilin lyase genes cpcE and cpcF and ht-cpcA, the gene encoding HT-CpcA. Plasmid pBS414V contains the ColE1 oriV for replication in E. coli and the aadA gene conferring resistance to Sp and streptomycin (Sm). Expression is controlled by the trc promoter, the lac operator, and the lacI q gene encoding the Lac repressor. Two strong, bidirectional, ρ-independent transcriptional terminator structures from the E. coli rrnB gene are located downstream of the expression cassette. Vector pAT101 expresses the genes involved in phycocyanobilin biosynthesis from heme: HO1 (hox1) and 3_Z_-phycocyanobilin:ferredoxin oxidoreductase (pcyA). Plasmid pAT101 contains the p15A oriV for replication in E. coli and the Km-resistance gene. The use of compatible origins of replication and different selectable markers allows for both plasmids to be maintained within the same E. coli cell. Expression of the genes carried by pAT101 is under the same control elements as in pBS414V. Plasmid pBS405V (see text) was equivalent to plasmid pBS414V, except that the insert contained only ht-cpcA.
Plasmid pBS414V Containing the cpcA-cpcE-cpcF Cassette.
The cpcF gene, as a 0.75-kb _Bam_HI–_Eag_I fragment from pBS407V, was cloned into the _Bam_HI- and _Eag_I-digested pBS405V containing ht-cpcA, giving plasmid pBS412V. Subsequently, the cpcE gene, as a 0.85-kb _Sal_I–_Not_I fragment from pBS406V, was cloned into the _Sal_I- and _Not_I-digested pBS412V containing ht-cpcA and cpcF, giving the final cassette plasmid, pBS414V.
Construction of Plasmid pAT101 Containing the PCB Biosynthesis Cassette.
The hox1 gene, as a 0.7-kb _Nde_I–_Asc_I fragment from pBS421V, was cloned into the _Nde_I- and _Asc_I-digested pBS350V, giving plasmid pBS425V. Unlike pBS421V, pBS425V allows for expression of an N-terminal His6-tagged HO1 (HT-HO1) fusion protein. Subsequently, the pcyA gene, as a 0.8-kb _Asc_I–_Sph_I fragment from pBS422V, was cloned into the _Asc_I- and _Sph_I-digested pBS425V containing hox1, giving plasmid pBS426V. As noted above, a more typical E. coli ribosomal binding site is present upstream of each of the ORFs in this cassette and for the ORFs in plasmid pBS414V.
Because plasmids pBS414V and pBS426V, containing the two distinct gene cassettes, were derived from within the same parental pBS350V with the ColE1 origin of replication, they are incompatible within the same E. coli cell. To maintain and express these two cassettes within the same E. coli cell, the use of different but compatible origins of replication was required. To achieve this goal, the 2.2-kb fragment generated from pBS426V by _Ssp_I (partial) and _Bsp_HI digestion, containing the trc promoter, the lac operator, and the genes encoding HO1 and 3_Z_-phycocyanobilin:ferredoxin oxidoreductase (PcyA), was cloned into the 2.77-kb _Bsp_HI- (partial) and _Psi_I-digested plasmid pACYC177 (GenBank accession no. X06402), which contains the p15A origin of replication (16) and confers resistance to Km. The β-lactamase gene in pACYC177 was completely removed in the cloning. The resulting pACYC177 derivative, pAT101, carries the genes required for PCB synthesis (_ht_-hox1 and pcyA) and is compatible with pBS414V, which carries the genes encoding the heterodimeric phycocyanin α subunit PCB lyase (cpcE and cpcF) and HT-CpcA (ht-cpcA) (Fig. 1).
Isolation of His-Tagged Proteins by Immobilized Metal Affinity Chromatography.
Cell pellets were thawed and resuspended in 20 ml of cold (0–4°C) buffer 0 (20 mM Tris⋅HCl, pH 8.0/50 mM NaCl/50 mM KCl). Phenylmethylsulfonyl fluoride and 2-mercaptoethanol were added to final concentrations of 1 mM and 10 mM, respectively, immediately before breakage of cells by passage through a French pressure cell three times at 18,000 psi (1 psi = 6.89 kPa). Cell debris was removed by centrifugation at 4°C in a Beckman JA20 rotor at 30,000 × g for 1 h. The supernatant solution was mixed with 2–3 ml of Ni2+-nitrilotriacetic acid agarose at 4°C for 15 min before the agarose was loaded onto a column. The agarose then was washed with 10 column volumes each of cold buffer A1 (20 mM Tris⋅HCl, pH 8.0/50 mM NaCl/50 mM KCl/20 mM imidazole/5% vol/vol glycerol), buffer B (20 mM Tris⋅HCl, pH 8.0/500 mM NaCl/500 mM KCl), and buffer A2 (20 mM Tris⋅HCl, pH 8.0/50 mM NaCl/50 mM KCl/30 mM imidazole). His-tagged proteins were eluted from the agarose with buffer C (20 mM Tris⋅HCl, pH 8.0/50 mM NaCl/50 mM KCl/200 mM imidazole) and then were dialyzed overnight with 50 mM sodium phosphate (pH 7.0).
Absorbance and Fluorescence Spectrometry.
Absorbance spectra were acquired on a computer-controlled, dual-beam λ6 UV/VIS spectrophotometer (Perkin–Elmer). Corrected fluorescence spectra were obtained on a FP-750 spectrofluorometer (Jasco, Easton, MD). Excitation and emission slits were set at 5 nm for all measurements.
Results
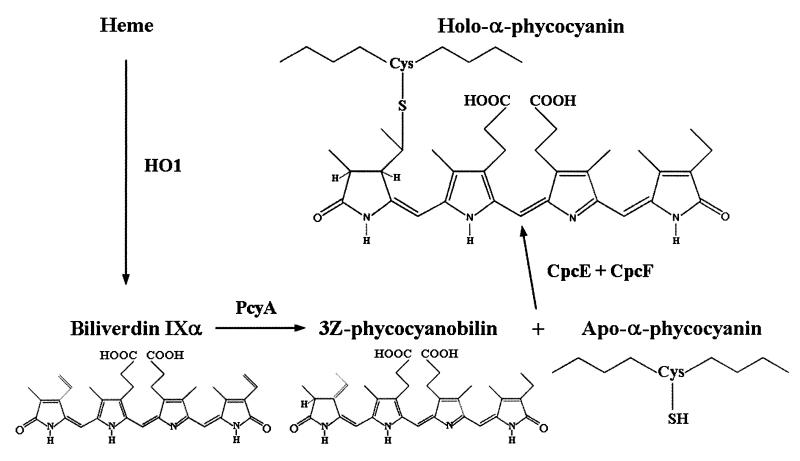
A minimal biosynthetic pathway in cyanobacteria leading from heme to the formation of the cysteinyl adduct of PCB with CpcA (Fig. 2) is consistent with substantial in vivo and in vitro data (4, 6, 14, 15, 17–21).
Figure 2.
Minimal biosynthetic pathway for the production of phycocyanobilin from heme and its addition to the C-phycocyanin apo-α subunit.
To test the predictions of this scheme, two strains of E. coli, each carrying two expression vectors, were generated. To maintain the two plasmids simultaneously, double transformants were selected for resistance to both Sp and Km and were grown with both Sp and Km. E. coli strain DH5α(414V,101) carries plasmids pBS414V (_ht_-cpcA-cpcE-cpcF cassette, Spr) and pAT101 (_ht_-hox1-pcyA cassette, Kmr) and expresses all of the known genes believed to be involved in PCB biosynthesis and its addition to apo-CpcA (Fig. 2). The other E. coli strain, DH5α(405V,101), carries plasmid pBS405V (expressing CpcA but lacking CpcE and CpcF, Spr) and plasmid pAT101. Comparison of the properties of the His-tagged CpcA proteins recovered from these two E. coli strains tests the stringency of the requirement for CpcE and CpcF for PCB addition in vivo. Previous studies have shown that HT-CpcA behaves just like untagged CpcA in cyanobacteria (11).
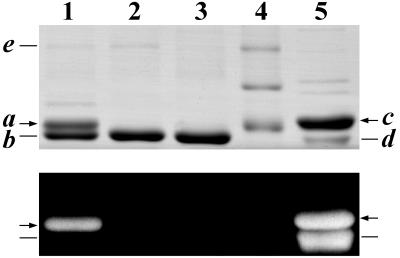
Expression of the gene cassettes in these two strains of E. coli led to two very different phenotypes. Upon induction with isopropyl β-d-thiogalactoside, the E. coli strain DH5α(414V,101) culture acquired a pronounced blue tint. This color change was not seen in the culture of DH5α(405V,101). HT-CpcA was purified from strain DH5α(414V,101) by affinity chromatography. Analysis of Coomassie blue-stained gels after SDS/PAGE of the purified protein showed two distinct bands in a ratio of ≈1:2, of 21.1 kDa (band a) and 20.5 kDa (band b), corresponding to the calculated molecular masses of His-tagged holo- and apo-CpcA, respectively (Fig. 3, lane 1). Upon exposure to Zn2+ (22) and UV illumination, only band a was fluorescent, indicating that it contained covalently attached bilin (Fig. 3, lane 1, Lower).
Figure 3.
SDS/PAGE analysis of Synechocystis sp. PCC6803 His-tagged proteins expressed in various E. coli strains and purified by immobilized metal affinity chromatography. Proteins were visualized by Coomassie blue stain (Upper) and by the UV-excited fluorescence of bilin-bearing polypeptides in the presence of Zn2+ (ref. 22; Lower). Lane 1, HT-CpcA (holo- and apo-HT-CpcA, bands a and b, respectively) expressed in E. coli strain DH5α(414V,101), which also expresses CpcE and CpcF, HT-HO1 (band e), and PcyA (see Fig. 1). Lane 2, HT-CpcA expressed in strain DH5α(405V,101), which also expresses HT-HO1 (band e) and PcyA, but not CpcE or CpcF. Lane 3, HT-CpcA expressed in strain DH5α(pBS405V), which does not express HO1, PcyA, CpcE, or CpcF. Lane 4, molecular mass standards (from the top): 30, 25, and 20 kDa. Lane 5, holo-HT-CpcA (21.9 kDa; band c) and the phycocyanin holo-β subunit (19.5 kDa; band d), both from Anabaena sp. PCC7120 (11). The difference in the relative intensities of the Coomassie blue-stained HT-α and β bands and of the fluorescence of these bands in the presence of Zn2+ is due to the fact that Coomassie blue stains the β subunit less strongly than the His-tagged α subunit, and that the β subunit carries two phycocyanobilins, whereas the α carries one.
The ratio of bands a:b was unaltered in HT-CpcA preparations from DH5α(414V,101) cultures grown in medium supplemented with 5 μM δ-aminolevulinic acid (a precursor of heme biosynthesis), with 0.5 μM ferric ammonium citrate (enhances heme biosynthesis in E. coli), or with both (data not shown). The yield of holo-HT-CpcA, calculated from the 625-nm (ɛM = 127,600 M−1⋅cm−1; see below) absorbance was ≈0.4 mg/g wet weight of E. coli cells.
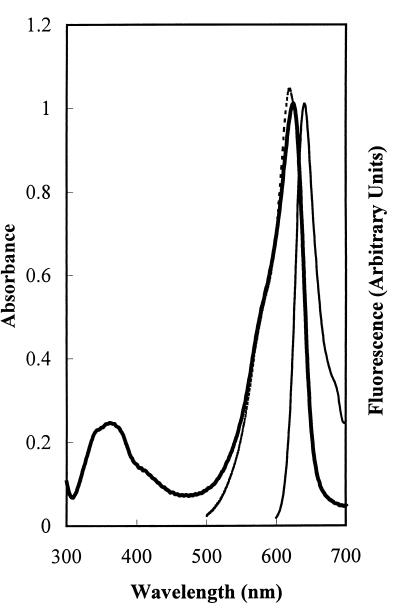
Because apo-HT-CpcA does not absorb light above 320 nm, the spectroscopic properties of holo-HT-CpcA were determined without fractionation of the apo- and holo-HT-CpcA mixture. The absorbance spectrum of holo-HT-CpcA had a λmax at 625 nm and a ratio of _A_625 nm:_A_369 nm of 4.14 (Fig. 4). The corresponding values for Synechococcus sp. PCC7002 holo-CpcA are 620 nm and 4.22 (see figure 3 in ref. 5). The λmax for Anabaena sp. PCC7120 holo-HT-CpcA is at 622 nm with ɛM = 109,000 M−1⋅cm−1 (Y.A.C., J. T. Murphy, G. J. Wedemayer, and A.N.G., unpublished data). The absorbance properties of native holo-CpcA preparations from other cyanobacteria are similar (e.g., ref. 23). The holo-HT-CpcA produced in E. coli, denatured in 8 M urea at pH 2, had a λmax of 661 nm and a ratio of _A_352:_A_661 of 1.03, values characteristic of polypeptide-bound PCB in the denatured cyanobacterial C-phycocyanin α subunit (24). From the acid urea spectrum, an ɛM of 127,600 M−1⋅cm−1 at 625 nm in 50 mM sodium phosphate (pH 7) was calculated for the native holo-HT-CpcA. The fluorescence emission maximum of the holo-HT-CpcA produced in E. coli was at 641 nm (Fig. 4), as compared with 642 nm for Synechococcus sp. PCC7002 holo-CpcA (12), and 637 nm for Anabaena sp. PCC7120 holo-HT-CpcA (Y.A.C., J. T. Murphy, G. J. Wedemayer, and A.N.G., unpublished data). The excitation spectrum of the holo-HT-CpcA produced in E. coli corresponded well to the absorbance spectrum (see Fig. 4), indicating that only one red fluorescence-emitting species was present. The absorbance spectrum was determined at a holo-HT-CpcA concentration of 9.2 × 10−6 M, whereas the excitation spectrum was measured at 4.6 × 10−7 M; the small red shift in the peak of the absorbance spectrum relative to that of the excitation spectrum reflects the concentration-dependent red shift that accompanies dimerization of holo-CpcA (24). The fluorescence quantum yield of holo-HT-CpcA produced in E. coli was 0.31, as compared with 0.23 for Anabaena sp. PCC7120 holo-HT-CpcA (Y.A.C., J. T. Murphy, G. J. Wedemayer, and A.N.G., unpublished data).
Figure 4.
Spectroscopic properties of Synechocystis sp. PCC6803 holo-HT-CpcA expressed in E. coli. Shown are the absorbance (λmax 625 nm; thick solid line), fluorescence emission (λmax 641 nm; λexc 580 nm; thin solid line), and fluorescence excitation (λem 645 nm; fluorescence excitation λmax 619 nm; dashed line) spectra for HT-CpcA purified from E. coli by affinity chromatography. The fluorescence excitation spectrum was normalized to the absorbance spectrum at 625 nm.
Upon induction, DH5α(405V,101) expressed a level of HT-CpcA similar to that of DH5α(414V,101). SDS/PAGE analysis of HT-CpcA purified from DH5α(405V,101) showed a single major band with a molecular weight corresponding to apo-HT-CpcA, and exposure of the gel to UV illumination after exposure to Zn2+ indicated the complete absence of any fluorescent bands (Fig. 3, lane 2). When 10 times as much protein was loaded on the gel, a very weak fluorescence was detected in the corresponding band (data not shown). Spectroscopic analysis of the affinity-purified HT-CpcA fraction from DH5α(405V,101) indicated an upper limit of <1% to any bilin-bearing CpcA in the preparation (data not shown). The analysis of HT-CpcA purified from an E. coli strain expressing only CpcA is shown for comparison in Fig. 3 (lane 3). Thus, overall, the results show at most a trace of bilin addition to HT-CpcA in the absence of CpcE and CpcF.
Of the proteins produced upon induction of the E. coli strains described here, both CpcA and HO1 were His-tagged (see Fig. 1). HO1 was tagged in this study solely to check for protein expression from pAT101. It was anticipated that HT-HO1 (calculated mass 29.9 kDa) would copurify on the Ni2+ affinity column along with HT-CpcA, which indeed is the case. A weak band e with a mass of 30 kDa is seen in Fig. 3, lanes 1 and 2, where the purified His-tagged protein preparation was derived from strains carrying the hox1 and cpcA genes, but is absent in lane 3, where the protein was obtained from an E. coli strain expressing solely the cpcA gene.
Discussion
The entire genome of Synechocystis sp. PCC6803 has been sequenced (12). This organism is particularly convenient for exploratory studies on phycobiliprotein biosynthesis, because PCB is the sole bilin attached to the phycobiliproteins in this cyanobacterium. The Synechocystis sp. PCC6803 genes encoding the phycocyanin α subunit, cpcA; both of the genes required for the conversion of heme to PCB, hox1 and pcyA; and the two genes required for bilin attachment, cpcE and cpcF, encoding the heterodimeric phycocyanin α subunit PCB lyase, have all been identified (6, 12–14). These advances allowed the design described here of an E. coli strain wherein endogenous heme could be converted to PCB that would then be attached to apo-CpcA (Fig. 2). The ability to generate PCB within heterologous cells is critical. Earlier studies with yeast expressing CpcA, CpcE, and CpcF showed that even though soluble and active proteins were produced, no holo-CpcA formed upon PCB addition to the cells under a wide variety of conditions (ref. 25; B. Schroeder, J. T. Thorner, and A.N.G., unpublished observations). Evidently, intracellular PCB concentrations adequate for bilin addition to CpcA were not achieved.
We show here that upon induction, E. coli strain DH5α(414V,101) produces HT-HO1, PcyA, CpcE, and CpcF in active form and converts about one-third of the soluble apo-HT-CpcA produced into holo-HT-CpcA with endogenous heme. By SDS/PAGE and spectroscopic analyses, the holo-HT-CpcA is in all respects similar to the holo-CpcA produced in wild-type cyanobacteria. Because the addition of δ-aminolevulinic acid and/or iron did not affect the extent of PCB addition, the yield of holo-CpcA appears not to be limited by the availability of heme. Rather, the lack of complete conversion may be due in part to codon usage (26) unfavorable for E. coli to generate large amounts of HO1, PcyA, CpcE, and CpcF. Codon usage does not seem to be a problem for the expression of cpcA in either E. coli or yeast (25). Another likely contributing factor is that in strain DH5α(414V,101) the copy number of the plasmid pAT101 is significantly lower than that of plasmid pBS414V (data not shown). Yet another unexplored issue is the fraction of each HO1, PcyA, CpcE, and CpcF that aggregates into inclusion bodies. Earlier studies of the expression of CpcA, CpcE, and CpcF in E. coli showed that a substantial amount of these proteins is found in the cell pellet (15). In a more complete list, there may yet be other, unidentified concerns.
The evidence that CpcE and CpcF are needed for the correct addition of PCB to apo-CpcA, both in vivo in various cyanobacteria (17–19) as well as in vitro (5, 15), is very strong. In vitro, nonenzymic addition of PCB to CpcA leads to the formation of an unnatural, oxidized adduct, mesobiliverdin-CpcA, with distinctive spectroscopic properties (20, 21); however, in the presence of CpcE and CpcF, normal holo-CpcA is produced (5, 15). The E. coli strain DH5α(405V,101) produces HO1, PcyA, and CpcA, but not CpcE and CpcF. In this strain no significant bilin addition of any kind takes place. Here, CpcE and CpcF are absolutely required for PCB addition to CpcA. Little, if any, of the spontaneous mesobiliverdin-CpcA adduct (20, 21) is formed, presumably because of the highly reducing conditions within the E. coli cell (e.g., ref. 27).
Two other bilin lyases with specificities different from those of CpcE and CpcF have been described to date. PecE plus PecF catalyzes the addition of PCB to phycoerythrocyanin apo-α subunit and the isomerization of the bound bilin to phycobiliviolin (28, 29). CpeY plus CpeZ has been reported to catalyze the addition of phycoerythrobilin to one of the bilin attachment sites on the α subunit of C-phycoerythrin (30). Additional phycobiliprotein-bilin lyases remain to be discovered. In addition to PcyA, which converts biliverdin to PCB, Frankenberg et al. (6) have described PebA, which converts biliverdin to 15,16-dihydrobiliverdin, and PebB, which converts 15,16-dihydrobiliverdin to 3_Z_-phycoerythrobilin.
Phycobiliprotein subunits are highly versatile as fusion partners (11). Consequently, the expression in prokaryotic or eukaryotic cells of an appropriate selection of genes encoding enzymes and apophycobiliprotein subunit-containing fusion proteins will lead to the intracellular production of constructs carrying specific bilins at unique locations, with broad utility in addressing a variety of questions in cell biology.
Acknowledgments
We thank J. Clark Lagarias for sharing information with us on the cyanobacterial ferredoxin-dependent bilin reductases in advance of publication. We are grateful to Jeremy Thorner and J. Clark Lagarias for their helpful comments on the manuscript and Cynthia Voong for her assistance. This research was supported in part by grants from the Lucille P. Markey Charitable Trust, the W. M. Keck Foundation, and the Director, Office of Energy Research, Office of Health and Environmental Research of the U.S. Department of Energy under Contract DE-FG-91ER61125.
Abbreviations
CpcA
α subunit of C-phycocyanin
CpcE and CpcF
subunits of the heterodimeric phycocyanin α-subunit phycocyanobilin lyase
HO1
heme oxygenase 1
HT
His6-tag
PCB
phycocyanobilin
PcyA
3_Z_-phycocyanobilin:ferredoxin oxidoreductase
Sp
spectinomycin
Km
kanamycin
References
- 1.Glazer A N. In: Advances in Molecular and Cell Biology. Bittar E E, Barber J, editors. Greenwich, CT: JAI; 1994. pp. 119–149. [Google Scholar]
- 2.Sidler W A. In: The Molecular Biology of Cyanobacteria. Bryant D A, editor. Dordrecht, The Netherlands: Kluwer; 1994. pp. 139–216. [Google Scholar]
- 3.Glazer A N. Methods Enzymol. 1988;167:291–303. doi: 10.1016/0076-6879(88)67034-0. [DOI] [PubMed] [Google Scholar]
- 4.Beale S I. In: The Molecular Biology of Cyanobacteria. Bryant D A, editor. Dordrecht, The Netherlands: Kluwer; 1984. pp. 519–558. [Google Scholar]
- 5.Fairchild C D, Glazer A N. J Biol Chem. 1994;269:8686–8694. [PubMed] [Google Scholar]
- 6.Frankenberg N, Mukougawa K, Kohchi T, Lagarias J C. Plant Cell. 2001;13:965–972. doi: 10.1105/tpc.13.4.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oi V T, Glazer A N, Stryer L. J Cell Biol. 1982;93:981–986. doi: 10.1083/jcb.93.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glazer A N. In: Chemicals from Microalgae. Cohen Z, editor. London: Taylor & Francis; 1999. pp. 261–280. [Google Scholar]
- 9.Heim R, Tsien R Y. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 10.Murphy J T, Lagarias J C. Curr Biol. 1997;7:870–876. doi: 10.1016/s0960-9822(06)00375-7. [DOI] [PubMed] [Google Scholar]
- 11.Cai Y A, Murphy J T, Wedemayer G J, Glazer A N. Anal Biochem. 2001;290:186–204. doi: 10.1006/abio.2000.4979. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura Y, Kaneko T, Hirosawa M, Miyajima N, Tabata S. Nucleic Acids Res. 1998;26:63–67. doi: 10.1093/nar/26.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamura Y, Kaneko T, Tabata S. Nucleic Acids Res. 2000;28:72. doi: 10.1093/nar/28.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornejo J, Willows R D, Beale S I. Plant J. 1998;15:99–107. doi: 10.1046/j.1365-313x.1998.00186.x. [DOI] [PubMed] [Google Scholar]
- 15.Fairchild C D, Zhao J, Zhou J, Colson S E, Bryant D A, Glazer A N. Proc Natl Acad Sci USA. 1992;89:7017–7021. doi: 10.1073/pnas.89.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. , Sect. 1.3, pp. 3–5. [Google Scholar]
- 17.Zhou J, Gasparich G E, Stirewalt V L, De Lorimier R, Bryant D A. J Biol Chem. 1992;267:16138–16145. [PubMed] [Google Scholar]
- 18.Swanson R V, Zhou J, Leary J A, Williams T, De Lorimier R, Bryant D A, Glazer A N. J Biol Chem. 1992;267:16146–16154. [PubMed] [Google Scholar]
- 19.Cai Y A, Schwartz S H, Glazer A N. Photosynth Res. 1997;53:109–120. [Google Scholar]
- 20.Arciero D M, Bryant D A, Glazer A N. J Biol Chem. 1988;263:18343–18349. [PubMed] [Google Scholar]
- 21.Arciero D M, Dallas J L, Glazer A N. J Biol Chem. 1988;263:18350–18357. [PubMed] [Google Scholar]
- 22.Berkelman T R, Lagarias J C. Anal Biochem. 1986;156:194–201. doi: 10.1016/0003-2697(86)90173-9. [DOI] [PubMed] [Google Scholar]
- 23.Glazer A N, Fang S, Brown D M. J Biol Chem. 1973;248:5679–5685. [PubMed] [Google Scholar]
- 24.Glazer A N, Fang S. J Biol Chem. 1973;248:659–662. [PubMed] [Google Scholar]
- 25.Schroeder B G. Doctoral dissertation. Berkeley: Univ. of California; 1997. [Google Scholar]
- 26.Nakamura Y, Gojobori T, Ikemura T. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Printz W A, Aslund F, Holmgren A, Beckwith J. J Biol Chem. 1997;272:15661–15667. doi: 10.1074/jbc.272.25.15661. [DOI] [PubMed] [Google Scholar]
- 28.Jung L J, Chan C F, Glazer A N. J Biol Chem. 1995;270:12877–12884. doi: 10.1074/jbc.270.21.12877. [DOI] [PubMed] [Google Scholar]
- 29.Zhao K-H, Deng M-G, Zheng M, Zhou M, Parbel A, Storf M, Meyer M, Strohmann B, Scheer H. FEBS Lett. 2000;469:9–13. doi: 10.1016/s0014-5793(00)01245-x. [DOI] [PubMed] [Google Scholar]
- 30.Kahn K, Mazel D, Houmard J, Tandeau de Marsac N, Schaefer M R. J Bacteriol. 1997;179:998–1006. doi: 10.1128/jb.179.4.998-1006.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]