Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis (original) (raw)
Abstract
The ssrA tag, an 11-aa peptide added to the C terminus of proteins stalled during translation, targets proteins for degradation by ClpXP and ClpAP. Mutational analysis of the ssrA tag reveals independent, but overlapping determinants for its interactions with ClpX, ClpA, and SspB, a specificity-enhancing factor for ClpX. ClpX interacts with residues 9–11 at the C terminus of the tag, whereas ClpA recognizes positions 8–10 in addition to residues 1–2 at the N terminus. SspB interacts with residues 1–4 and 7, N-terminal to the ClpX-binding determinants, but overlapping the ClpA determinants. As a result, SspB and ClpX work together to recognize ssrA-tagged substrates efficiently, whereas SspB inhibits recognition of these substrates by ClpA. Thus, dissection of the recognition signals within the ssrA tag provides insight into how multiple proteins function in concert to modulate proteolysis.
Keywords: ClpAP, ClpXP, peptide recognition, protein degradation, SspB
The proteolytic machinery of cells must select the correct protein substrates at the right time and place. Two general mechanisms, degradation tags and regulatory proteins that modulate recognition, help ensure intracellular proteolytic specificity. Degradation signals, which can be present in the protein sequence or added by covalent modification, target substrates to specific proteases. In eukaryotes, for example, proteins can be targeted to the 26S proteosome by posttranslational addition of polyubiquitin (1–3). In bacteria, proteins bearing the ssrA degradation tag, an 11-residue peptide, are recognized and degraded by several different proteases, including ClpXP and ClpAP (4). The ssrA tag is added cotranslationally to the C terminus of polypeptides whose biosynthesis has stalled (5–7). The specificity of proteolysis can be further regulated by protein factors that modulate recognition of degradation signals by the protease. In Escherichia coli, for example, the SspB protein binds specifically to ssrA-tagged substrates and enhances binding of the tagged protein to ClpX (8).
ClpXP and ClpAP are protein machines that promote ATP-dependent degradation. Each of these complexes contains a hexameric Clp/HSP100 family ATPase, ClpA or ClpX, that mediates substrate recognition and catalyzes energy-dependent protein unfolding (9–16). Both Clp ATPases can form a stacked protease complex with ClpP, a double-ring serine peptidase whose active sites face an internal chamber (4, 17–19). The entrance to the inner proteolytic compartment of ClpP is small (18), and before degradation, substrates must be unfolded by ClpX or ClpA and translocated into ClpP. Although similar in function and in their ability to recognize ssrA-tagged proteins, ClpX and ClpA have generally distinct substrate preferences. For example, ClpXP degrades the stationary-phase sigma factor (20) and Mu transposase (21), which are not substrates for ClpAP, whereas ClpAP, but not ClpXP, degrades HemA (22) and MazE (23). Moreover, ClpAP, but not ClpXP, degrades denatured proteins in the absence of a degradation tag (16, 24). Although a few specific recognition sequences for ClpX and ClpA have been identified, general sequence rules governing substrate recognition by either protein have yet to emerge.
In this article, we determine the sequence information within the ssrA degradation tag that is required for efficient recognition by ClpX, ClpA, and SspB. We find that the ssrA tag is rich in signaling information. ClpX and SspB recognize contiguous portions of the ssrA tag and function in concert to bind ssrA-tagged substrates tightly, allowing more efficient degradation of these substrates by ClpXP. In contrast, SspB interacts with sequence determinants that partially overlap those of ClpA, resulting in inhibition of ClpAP-mediated degradation. These results establish that SspB can act as a bifunctional regulator of substrate recognition and that the ssrA tag contains intricate, overlapping recognition signals that allow modulation of proteolysis.
Materials and Methods
Materials.
ClpX (25), ClpP (15), SspB (8), ClpA (26), and GFP-ssrA (27) were purified as described. Polyclonal anti-SspB antibodies were prepared by Covance (Denver, PA), by using SspB purified in our laboratory. PD buffer (pH 7.6) contains 25 mM Hepes-KOH/5 mM MgCl2/5 mM KCl/15 mM NaCl/0.032% (vol/vol) Nonidet P-40/10% (vol/vol) glycerol. HO buffer (pH 7.5) contains 25 mM Hepes-KOH/20 mM MgCl2/300 mM NaCl/10% (vol/vol) glycerol/0.5 mM DTT.
Green Fluorescent Protein (GFP) Mutants.
A gene encoding GFP-ssrA with S6G and S72A mutations in the GFP coding sequence (GFPmut3-ssrA) (28), a gift of A. J. Anderson (The Technical University of Denmark), was cloned into the _Not_I site of pACYC184 to create pMS30. Mutant ssrA tags were introduced by ligating the _Stu_I and _Hin_dIII cleaved backbone fragment of pMS30 to synthetic oligonucleotide cassettes. DNA sequences were determined for all GFP-ssrA variants to confirm the expected sequence. The molecular weights of GFP-ssrA (A10D) and GFP-ssrA (A11D) were confirmed by mass spectrometry.
Degradation Assays.
ClpX6 (0.3 μM), ClpP14 (0.8 μM), ATP (4 mM), and an ATP regeneration system (50 μg/ml creatine kinase and 2.5 mM creatine phosphate) were mixed in PD buffer and incubated for 2 min at 30°C. GFP-ssrA or variants were then added, and the mixture was transferred to a 50-μl cuvette; fluorescence readings were begun within 10 s. In some reactions, SspB was added, in concentrations indicated in the figure legends (as monomer equivalents), following the 2-min incubation at 30°C but before addition of substrate. Changes in GFP fluorescence (excitation 467 nm, emission 511 nm) were monitored in a Fluoromax-2 instrument (ISA, Jobin-Yvon, Longjumeau, France). Degradation of GFP-ssrA or variants by ClpAP was performed as above except by using ClpA6 (0.05 μM) and ClpP14 (0.1 μM) in HO buffer. Reaction solution conditions for ClpXP and ClpAP were different to optimize the activity observed for each enzyme.
Peptide⋅SspB Binding.
A cellulose filter containing 220 synthetic ssrA peptide variants was prepared by the Massachusetts Institute of Technology Biopolymers facility using an Abimed instrument. Each peptide contained two additional alanines, C-terminal to the end of the ssrA sequence, and was covalently attached to the filter via a polyethylene glycol linker. The filter was blocked for 3 h in TBST (50 mM Tris, pH 7.5/125 mM NaCl/0.1% Tween 20) plus 10% milk, incubated with 10 μg/ml SspB in TBST plus 0.1% milk, washed three times in TBST, incubated with polyclonal rabbit anti-SspB antibody for 1 h, washed three times in TBST, and incubated for 30 min with secondary goat anti-rabbit IgG horseradish peroxidase-conjugated antibody (Amersham Pharmacia). Three final washes with TBST were performed, the filter was incubated with ECL substrate (NEN), and binding was visualized on film. Attempts to probe ClpX or ClpA binding to the peptide filter were unsuccessful.
Results
Mutant Derivatives of the ssrA Tag.
To identify the residues within the 11-aa ssrA tag that are important for recognition by ClpX and ClpA, we constructed a set of mutant tags fused to the C terminus of GFP (GFP-ssrA). Each non-alanine residue in the tag sequence was mutated to alanine, and each alanine was changed to aspartic acid (Fig. 1A). Because GFP-ssrA (L9A) (numbering relative to the N terminus of the tag) was a relatively conservative mutation, we also constructed the GFP-ssrA (L9D) mutant. In total, 12 single GFP-ssrA mutants were constructed and purified.
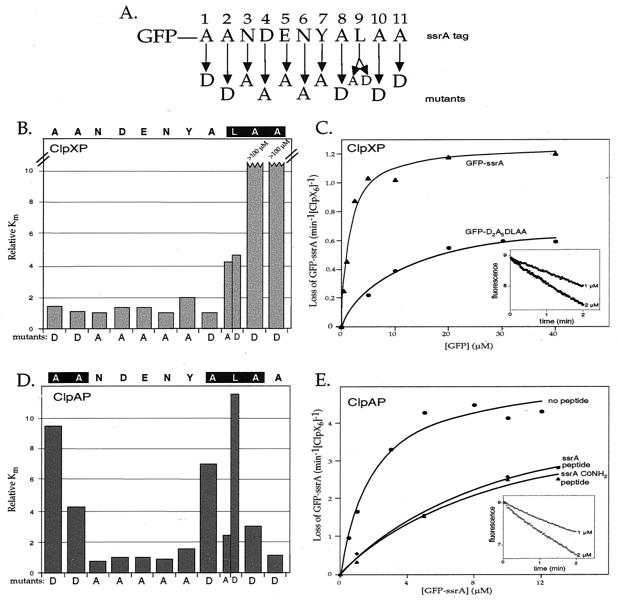
Figure 1.
Degradation of GFP-ssrA variants by ClpXP and ClpAP. (A) SsrA-tag sequence and identity of single-residue substitutions. (B) Relative _K_m values for ClpXP degradation of GFP-ssrA mutants. Rates of ClpXP-mediated degradation of GFP-ssrA variants, determined by the loss of native fluorescence, were determined at different substrate concentrations (see Materials and Methods) and fit to a Michaelis–Menten model. The _K_m values plotted were normalized by dividing by _K_m for ClpXP degradation of wild-type GFP-ssrA (1.5 μM). _V_max values for mutants 1–9 were within 2-fold of the wild-type value (1.2 min−1 ClpX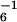 ) except for Y7A, which had a _V_max of 0.45 min−1 ClpX
) except for Y7A, which had a _V_max of 0.45 min−1 ClpX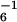 ). (C) Michaelis–Menten plots of ClpXP degradation of GFP-ssrA and GFP-D2A5DLAA. The solid lines are fits to the Michaelis–Menten equation for GFP-ssrA (_K_m = 1.5 μM, _V_max = 1.2 min−1) and GFP-D2A5DLAA (_K_m = 10.1 μM, _V_max = 0.8 min−1). The decrease in _V_max for the consensus mutant is probably caused by the decreased _V_max of the Y9A substitution. (Inset) The change in fluorescence at 511 nm of 1 μM GFP-ssrA and 2 μM GFP-ssrA following incubation with ClpXP. (D) Relative _K_m values for ClpAP degradation of GFP-ssrA mutants. _K_m values were normalized by dividing by the _K_m value (1.5 μM) for ClpAP degradation of wild-type GFP-ssrA. See legend to B for other details. (E) Inhibition of ClpAP degradation of GFP-ssrA by ssrA peptides. Michaelis–Menten plots for ClpAP degradation of GFP-ssrA in the absence of peptide (_K_m = 1.5 ± 0.4 μM, _V_max = 4.9 ± 0.3 μM/min−1) or presence of the wild-type ssrA peptide (_K_m apparent = 10.4 ± 1.6 μM, _V_max = 5.1 ± 0.4 μM/min−1, _K_I = 16.9 μM) or the carboxamide ssrA peptide (_K_m apparent = 10.7 ± 1.2 μM, _V_max = 4.9 ± 0.3 μM/min−1, _K_I = 16.4 μM). _K_I values were calculated from _K_m apparent = [1 + ([I]/_K_I)] * _K_m. (Inset) The change in fluorescence at 511 nm of 1 μM GFP-ssrA and 2 μM GFP-ssrA following incubation with ClpAP.
). (C) Michaelis–Menten plots of ClpXP degradation of GFP-ssrA and GFP-D2A5DLAA. The solid lines are fits to the Michaelis–Menten equation for GFP-ssrA (_K_m = 1.5 μM, _V_max = 1.2 min−1) and GFP-D2A5DLAA (_K_m = 10.1 μM, _V_max = 0.8 min−1). The decrease in _V_max for the consensus mutant is probably caused by the decreased _V_max of the Y9A substitution. (Inset) The change in fluorescence at 511 nm of 1 μM GFP-ssrA and 2 μM GFP-ssrA following incubation with ClpXP. (D) Relative _K_m values for ClpAP degradation of GFP-ssrA mutants. _K_m values were normalized by dividing by the _K_m value (1.5 μM) for ClpAP degradation of wild-type GFP-ssrA. See legend to B for other details. (E) Inhibition of ClpAP degradation of GFP-ssrA by ssrA peptides. Michaelis–Menten plots for ClpAP degradation of GFP-ssrA in the absence of peptide (_K_m = 1.5 ± 0.4 μM, _V_max = 4.9 ± 0.3 μM/min−1) or presence of the wild-type ssrA peptide (_K_m apparent = 10.4 ± 1.6 μM, _V_max = 5.1 ± 0.4 μM/min−1, _K_I = 16.9 μM) or the carboxamide ssrA peptide (_K_m apparent = 10.7 ± 1.2 μM, _V_max = 4.9 ± 0.3 μM/min−1, _K_I = 16.4 μM). _K_I values were calculated from _K_m apparent = [1 + ([I]/_K_I)] * _K_m. (Inset) The change in fluorescence at 511 nm of 1 μM GFP-ssrA and 2 μM GFP-ssrA following incubation with ClpAP.
To assay recognition by ClpX and ClpA, we measured degradation of the GFP-ssrA variants by ClpXP and ClpAP in vitro.
The initial rate of degradation of each mutant was determined by measuring the loss of GFP-ssrA fluorescence. To determine _K_m values, degradation rates were determined at a series of substrate concentrations. Consistent with previous reports, _K_m for ClpXP degradation of GFP-ssrA was 1.5 ± 0.3 μM (8, 15) (see Fig. 1C). _K_m for ClpAP degradation of GFP-ssrA (1.5 ± 0.4 μM) was found to be similar (see Fig. 1E).
SsrA-Tag⋅ClpX Recognition.
Of the 12 GFP-ssrA mutants tested, only those with substitutions at tag positions 9, 10, and 11 caused greater than 2-fold increases in _K_m for ClpXP degradation relative to the wild-type value (Fig. 1B). Ala-10 and Ala-11 were found to be critical determinants for recognition by ClpX. GFP-ssrA with either the A10D or A11D substitution had a _K_m for ClpXP degradation that was increased by at least a factor of 100 (no degradation was observed at substrate concentrations of 100 μM). Mutation of Leu-9 to either alanine or aspartic acid also weakened productive interaction of the substrate with ClpX, increasing the _K_m about 4-fold (L9A _K_m = 6.2 ± 0.6 μM; L9D _K_m = 6.9 ± 1.1 μM). In contrast, residues 1–8 of the ssrA tag did not play major roles in ClpX recognition, as judged by _K_m values similar to wild type. Furthermore, _V_max values for the mutants with detectable degradation rates were similar to the wild-type value of 1.2 ± 0.1 min−1 ClpX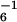 (data not shown, but see legend to Fig. 1). Our finding that Ala-10 and Ala-11 play the largest role in ClpX recognition is consistent with previous studies showing that replacing both residues with aspartic acids greatly reduces degradation by ClpXP of a tagged version of the N-terminal domain of λ repressor (4).
(data not shown, but see legend to Fig. 1). Our finding that Ala-10 and Ala-11 play the largest role in ClpX recognition is consistent with previous studies showing that replacing both residues with aspartic acids greatly reduces degradation by ClpXP of a tagged version of the N-terminal domain of λ repressor (4).
To determine whether the Leu-9–Ala-10–Ala-11 sequence motif was sufficient to mark a protein as a substrate for ClpX, we constructed two additional variants. In one protein, residues 1–8 of the tag were mutated to the same amino acids shown in Fig. 1A to generate GFP-D2A5DLAA. In the other, residues 1–8 of the tag were changed to glycines resulting in GFP-G8LAA. The GFP-D2A5DLAA protein was a substrate for ClpXP degradation (Fig. 1C), although with an increased _K_m value (10.1 ± 1.4 μM). This change in _K_m probably results from the cumulative minor effects of the eight single mutations. The glycine-rich GFP-ssrA variant was resistant to degradation by ClpXP at concentrations of 50 μM and below (data not shown). We conclude that a C-terminal Leu-Ala-Ala tripeptide is sufficient to allow ClpX recognition and ClpXP-dependent degradation in some but not all sequence contexts. Because of its flexibility, the glycine-rich linker may not allow the terminal Leu-Ala-Ala residues to adopt a conformation appropriate for ClpXP recognition.
SsrA-Tag⋅ClpA Recognition.
Degradation of the GFP-ssrA mutants by ClpAP (Fig. 1D) revealed that ClpA relies on a different set of residues than ClpX to recognize the ssrA tag. The mutations that caused the largest increases in _K_m for ClpAP degradation (wild-type value 1.5 μM) were A1D (14.3 ± 1.5 μM), A2D (6.4 ± 1.5 μM), A8D (10.1 ± 1.7 μM), L9D (17.1 ± 1.2 μM), and A10D (4.5 ± 0.4 μM). These results show that ClpA recognizes information in both the N-terminal and C-terminal regions of the ssrA tag.
Because mutation of the C-terminal alanine of the ssrA tag to aspartic acid had no effect on ClpAP degradation, we suspected that the free α-carboxyl group—a unique chemical signature of the C-terminal residue—might also be dispensable. To investigate this question, we compared the ability of peptides with either a normal α-carboxyl group (ssrA peptide) or a terminal carboxamide group (ssrA-CONH2) to inhibit degradation of GFP-ssrA by ClpAP. As shown in Fig. 1E, the ssrA-CONH2 peptide (_K_i = 16.9 μM) was as effective as the ssrA peptide (_K_i = 16.4 μM) in inhibiting degradation of GFP-ssrA by ClpAP. These results suggest that ClpA may be able to recognize an ssrA-like signal in any exposed region of a protein without restriction to the C-terminal end. Previous studies have shown that the α-carboxyl group is an important determinant of ClpX recognition of the ssrA tag, with the ssrA-CONH2 peptide being 10-fold less effective as an inhibitor than the normal ssrA peptide (15).
SsrA-Tag⋅SspB Recognition.
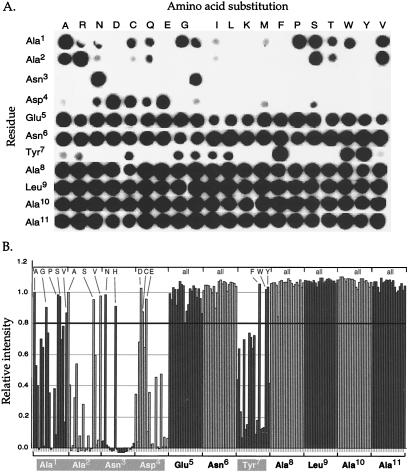
SspB binds to ssrA-tagged proteins and enhances recognition of these proteins by ClpX. Previous studies showed that SspB binds specifically to the tag, that the N3A tag mutation abrogates this binding, and that deletion of the last three amino acids from the tag does not prevent binding (8). To define further the interaction between SspB and the ssrA tag, we synthesized an immobilized peptide library in which each residue of the ssrA peptide was individually changed to each of the other 19 aa, whereas the rest of the sequence remained unchanged. These peptides, which contained two additional C-terminal alanines, were covalently attached via their C termini to a cellulose filter by a polyethylene glycol linker. The filter contained 220 “spots,” with each spot corresponding to one peptide sequence.
Interaction with the peptides was measured by incubating the filter with SspB and subsequently detecting bound SspB with anti-SspB antibody (Fig. 2A). Inspection of the filter showed that SspB bound poorly to many of the peptides with substitutions at tag positions 1, 2, 3, 4, and 7. At position 3, for example, only peptides with Asn or His were efficiently bound. In contrast, at tag positions 5, 6, 8, 9, 10, and 11, SspB had no significant sequence preferences. Fig. 2B quantifies the efficiency of the SspB⋅peptide interactions. Using an arbitrary cut-off value of 80% of wild-type binding produced the consensus [AGPSV]1-[ASV]2-[NH]3-[DCE]4-X5-X6-[FWY]7 for SspB recognition. These results suggest that SspB and ClpX interact with discrete sets of residues in the ssrA tag, whereas SspB and ClpA interact with some of the same residues.
Figure 2.
Effects of ssrA-peptide mutations on SspB recognition. (A) A library consisting of 220 ssrA peptide variants was used to assay SspB binding via an “indirect” Western. The filter containing covalently bound peptides was first incubated in 10 μg/ml SspB, and bound SspB was detected with anti-SspB antibody followed by horseradish peroxidase-conjugated goat anti-rabbit IgG antibody and the ECL substrate. (B) The filter in A was digitally scanned, and the number of pixels in each spot was quantified by using imagequant. These values are presented relative to the intensity of the wild-type ssrA peptide. Substitutions that show 80% or more of wild-type binding are indicated above the graph.
Requirement for Dual Recognition of the ssrA Tag by SspB and ClpX.
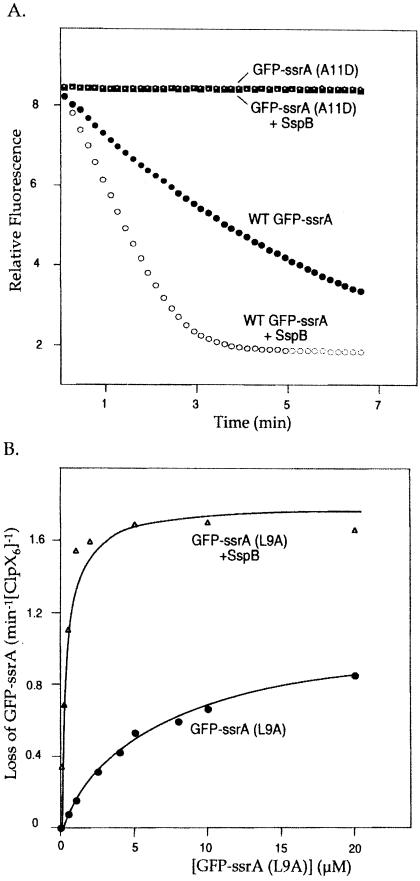
Previous studies established that SspB decreases _K_m for ClpXP degradation of GFP-ssrA from 1.5 μM to less than 0.3 μM (8). In principle, binding of SspB to the ssrA tag might be sufficient to target a tagged protein to ClpX without requiring independent recognition of the tag by ClpX. To test this possibility, we monitored degradation in the presence of SspB of the three GFP-ssrA mutants defective in ClpX recognition (L9A, A10D, and A11D). Even with SspB, the GFP-ssrA (A10D) and GFP-ssrA (A11D) proteins remained refractory to ClpXP degradation, indicating that binding by SspB does not bypass the requirements for ClpX recognition of these two residues (Fig. 3A and data not shown). SspB did, however, enhance recognition of the GFP-ssrA (L9A) mutant by ClpX. In the presence of 0.24 μM SspB, _K_m for degradation of GFP-ssrA (L9A) was reduced from 6.2 μM to less than 0.3 μM (Fig. 3B). Thus, SspB can compensate for decreased interactions with ClpX caused by this mutation. However, the GFP-ssrA (A10D) and GFP-ssrA (A11D) results clearly establish that SspB-regulated degradation of ssrA-tagged proteins depends on both sets of binding determinants, those for ClpX and those for SspB.
Figure 3.
(A) Degradation of GFP-ssrA (A11D) in the presence of SspB. ClpXP degradation, assayed by loss of fluorescence at 511 nm of 1 μM GFP-ssrA with or without SspB and 1 μM GFP-ssrA (A11D) with or without SspB. When present, the SspB concentration was 1 μM. (B) Degradation of L9A in the presence of SspB. Michaelis–Menten plots for ClpXP degradation of GFP-ssrA (L9A) in the absence (_K_m = 6.2 μM, _V_max = 1.1 min−1) or presence of saturating amounts of SspB (_K_m = 0.34 μM, _V_max = 1.8 min−1). The _K_m represents an upper limit because of the relatively high enzyme concentration (0.3 μM ClpX6) used in the experiment. The solid lines are fits to the Michaelis–Menten equation.
SspB Inhibits Degradation of GFP-ssrA by ClpAP.
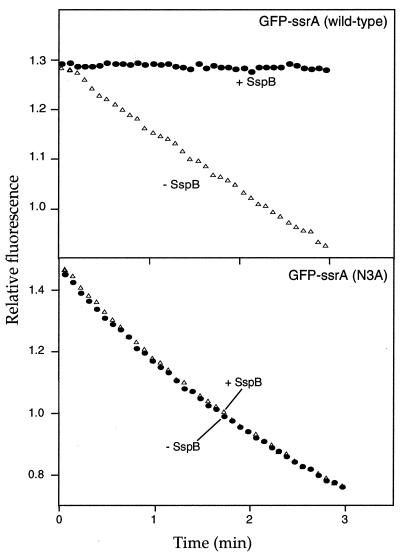
Because SspB and ClpA both interact with Ala-1 and Ala-2 in the ssrA tag, it seemed likely that their binding would be mutually exclusive and thus that SspB could inhibit ClpAP degradation of ssrA-tagged proteins. The results shown in Fig. 4 confirm this prediction. ClpAP degradation of GFP-ssrA was completely inhibited in the presence of a 2-fold excess of SspB. To ensure that SspB inhibits ClpAP degradation of GFP-ssrA by binding to the ssrA tag, we measured ClpAP degradation of GFP-ssrA (N3A). This mutation prevents binding of SspB to ssrA-tagged GFP (Fig. 2 and ref. 8) but does not affect ClpA recognition (Fig. 1D). SspB did not inhibit GFP-ssrA (N3A) degradation by ClpAP (Fig. 4), indicating that specific interaction of SspB with the ssrA tag is required to inhibit ClpAP degradation of the tagged protein. Thus, SspB binds specifically to the ssrA tag and seems to mask sequence elements important for ClpA interactions.
Figure 4.
SspB inhibits degradation by ClpAP. ClpAP degradation of 1 μM GFP-ssrA or GFP-ssrA (N3A), assayed by loss of fluorescence at 511 nm, without SspB or with SspB (2 μM).
Discussion
Binding Determinants for ClpX and ClpA in the ssrA Tag.
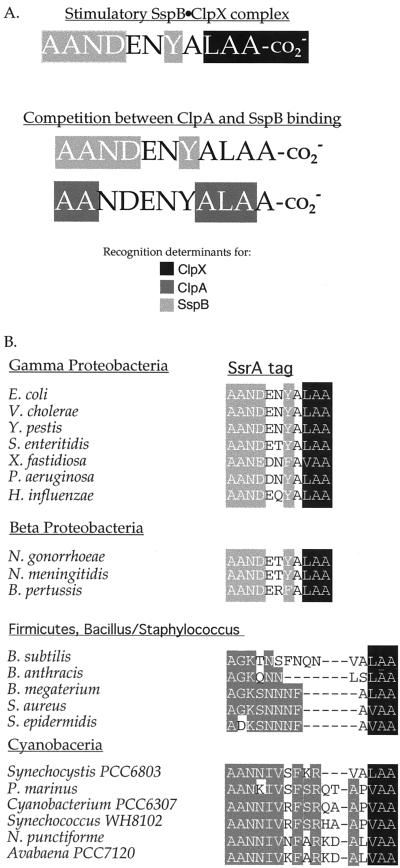
In E. coli, addition of the ssrA degradation tag to a protein is a signal to destroy the resulting polypeptide, and ssrA-tagged proteins are degraded by ClpXP, ClpAP, FtsH (HflB), and Tsp (Prc) (4, 29, 30). The Clp proteases are cytoplasmic, FtsH is a membrane protease, and Tsp is a periplasmic protease, ensuring that tagged proteins are degraded in all cellular compartments. In addition, SspB binds ssrA-tagged proteins in the cytoplasm and enhances their binding to and degradation by ClpXP (8). Thus, the 11-residue ssrA tag must encode sufficient information to mediate at least five sets of protein–protein interactions. Here, we dissected the sequence elements within the tag that are recognized by SspB, ClpX, and ClpA, the three proteins principally responsible for degradation of ssrA-tagged proteins in the cytoplasm. Our results show that the ssrA tag contains contiguous binding sites for ClpX and SspB but overlapping binding sites for ClpA and SspB (Fig. 5A).
Figure 5.
(A) Recognition determinants within the ssrA tag for ClpX, ClpA, and SspB. Recognition determinants for ClpX are highlighted in black, those for ClpA in dark gray, and those for SspB in light gray. (B) SsrA-degradation tags from different bacteria. The conserved SspB-binding determinants in the γ- and β-proteobacteria are highlighted in light gray. Shown are the predicted ssrA tag sequences from representative members of various families of bacteria. The conserved residues in the N-terminal regions of the ssrA tag in the other families are highlighted in dark gray. All sequenced γ and β proteobacteria have a predicted ssrA tag sequence that contains an acceptable SspB binding site with the exception of Buchnera sp., strain APS [tag sequence (A)ANNKQNYALAA]. Interestingly, this bacterium does not have a detectable ortholog of SspB. Of the bacteria listed, the following appear to have a ClpA ortholog, in addition to a ClpX: E. coli, Vibrio cholerae, Xylella fastidiosa, and Pseudomonas aeruginosa.
The ClpX-binding determinants in the ssrA tag are highly localized, composed of the α-carboxyl group and C-terminal residues, Leu-9–Ala-10–Ala-11. Within this set, however, aspartic-acid substitutions at Ala-10 or Ala-11 completely blocked substrate recognition by ClpX and were far more deleterious than substitutions at Leu-9 or the α-carboxyl group. Interestingly, Tsp recognizes ssrA-tagged polypeptides (31) and non-ssrA-tagged substrates that end with Leu-Ala-Ala (30), indicating that this protease interacts with the same portion of the ssrA tag as ClpX. Although the ClpX determinants are highly localized at the C-terminal end of the ssrA tag, it is important to note that GFP-G8LAA, which has the terminal Leu-Ala-Ala sequence, was not degraded by ClpXP. These data suggest that the sequence context or structure of a peptide containing this terminal tripeptide can influence ClpX interactions.
About half of the known ClpX substrates are similar to ssrA-tagged proteins in having nonpolar side chains at the penultimate and C-terminal residues (13). This group includes MuA (Ala-Ile), Mu repressor (Ala-Val), Mu repressor vir 3061 (Val-Leu), and CtrA (Ala-Ala). In several cases, these nonpolar residues have been implicated in ClpX recognition (25, 32, 33). It seems likely that ClpX uses the same substrate-binding site to interact with each of these substrates and with the ssrA tag. In contrast, other ClpX substrates—λO (34), UmuD′ (35), TrfA (36), Phd (37), and σs (38)—lack nonpolar residues at their C termini. Furthermore, where determined, the sequences responsible for protease targeting in these proteins have been localized to regions other than the C terminus. Thus, it is an attractive model that these proteins are recognized by ClpX using a different binding surface than the one that recognizes the ssrA tag.
Rules governing substrate recognition by ClpA are currently poorly defined. Our mutational analysis reveals that the most important residues of the ssrA tag for recognition by ClpA were Ala-1, Ala-2, Ala-8, Leu-9, and Ala-10, with the substitutions A1D, A8D, and L9D being especially deleterious. Thus, the ClpA recognition determinants, like those of ClpX, involve aliphatic side chains. Unlike the ClpX determinants, however, those for ClpA are not highly localized. It is unclear whether the five-residue spacing between the Ala-1–Ala-2 and Ala-8–Leu-9–Ala-10 determinants is important for ClpA recognition. Surprisingly, GFP-D2A5DLAA was found to be efficiently degraded by ClpAP in vitro (_K_m ≈ 2 μM, unpublished observations). The tag of this substrate does not contain several important ClpA-recognition determinants, nor does it contain a ΦΦX5ΦΦΦ motif (where Φ represents an aliphatic side chain). This tag does, however, contain ΦΦX4ΦΦΦ, ΦΦX3ΦΦΦ, and ΦΦX2ΦΦΦ motifs, suggesting that ClpA might recognize short clusters of aliphatic residues with variations in spacing.
ClpX and ClpA are related proteins that both recognize the ssrA tag. Thus, it was a reasonable hypothesis that they might share homologous substrate-binding pockets responsible for this common substrate recognition. However, we find that these ATPases achieve common recognition of the ssrA tag by interacting with different sequences in the peptide (Fig. 5A). This finding clearly favors the idea that the ssrA tags are recognized by these two proteins using substrate-binding pockets with substantially different recognition characteristics. Consistent with this conclusion, ClpX and ClpA generally recognize distinct proteins.
SspB Is a Bifunctional Regulator of Substrate Recognition.
SspB exhibits strong preferences for specific side chains at positions 1, 2, 3, 4, and 7 of the ssrA tag. These SspB-binding determinants are adjacent to those recognized by ClpX, allowing both proteins to bind to the same ssrA tag. Mutual binding, in this instance, is required for SspB to stimulate ClpXP degradation of ssrA-tagged substrates. Disruption of either SspB or ClpX recognition of the ssrA tag abolishes efficient degradation of ssrA-tagged substrates by ClpXP (see Fig. 3 and ref. 8). Consistent with this substrate-docking mechanism, ClpX, SspB, and an ssrA-tagged substrate form stable ternary complexes (8). In contrast, the SspB-binding determinants in the ssrA tag overlap those for ClpA recognition, and SspB, as a consequence, inhibits ClpAP degradation of ssrA-tagged substrates. Hence, SspB binding to the ssrA-tagged substrates enhances their degradation by ClpXP but inhibits proteolysis by ClpAP. The ability of SspB to divert ssrA-tagged substrates from ClpAP to ClpXP helps explain the observation that both proteases degrade ssrA-tagged proteins similarly in vitro, whereas these substrates are preferentially degraded by ClpXP in vivo (4, 8).
Is SspB-mediated channeling of ssrA-tagged substrates from ClpAP to ClpXP biologically important? The answer to this question is uncertain, but the different activities of the two proteases toward certain substrates provides an opportunity for speculation. For example, ClpAP but not ClpXP degrades unfolded proteins without targeting signals (24, 39), an activity that is probably most important during heat shock or other types of environmental stress. Up-regulation of SspB in response to stress could redirect ssrA-tagged substrates to ClpXP, leaving ClpAP free to degrade unfolded substrates.
Conservation of ClpX- and SspB-Recognition Modules Within the ssrA Tag.
The C-terminal tripeptide of the ssrA tag from a variety of bacterial species is highly conserved (LAA or VAA; Fig. 5B), consistent with the observation that ClpX and Tsp orthologs, which are likely to recognize these positions, are present in these bacteria. SspB orthologs are only found in the γ- and βproteobacteria (8). Alignment of the ssrA tags from these bacteria (Fig. 5B) reveals a consensus for the first seven tag residues, [A]1-[A]2-[N]3-[DE]4-[SDE]5-[TNRQ]6-[YF]7, that is a subset of the E. coli SspB consensus, [AGPSV]1-[ASV]2-[NH]3-[DCE]4-X5-X6-[FWY]7, determined here. The N-terminal portions of ssrA-tag sequences from other bacterial families are still highly conserved (Fig. 5B), although clearly distinct from the sequence bound by SspB. These observations suggest either that these bacteria contain an SspB-like regulator or that these regions are conserved because they mediate interactions with other proteases.
Acknowledgments
We thank members of the Sauer and Baker labs for advice and help. This work was supported by National Institutes of Health Grant AI-16892 and the Howard Hughes Medical Institute. T.A.B. is an employee of the Howard Hughes Medical Institute.
Abbreviation
GFP
green fluorescent protein
References
- 1.Hochstrasser M. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 2.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 3.Voges D, Zwickl P, Baumeister W. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 4.Gottesman S, Roche E, Zhou Y, Sauer R T. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tu G F, Reid G E, Zhang J G, Moritz R L, Simpson R J. J Biol Chem. 1995;270:9322–9326. doi: 10.1074/jbc.270.16.9322. [DOI] [PubMed] [Google Scholar]
- 6.Keiler K C, Waller P R, Sauer R T. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 7.Roche E D, Sauer R T. EMBO J. 1999;18:4579–4589. doi: 10.1093/emboj/18.16.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levchenko I, Seidel M, Sauer R T, Baker T A. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- 9.Gottesman S, Clark W P, de Crecy-Lagard V, Maurizi M R. J Biol Chem. 1993;268:22618–22626. [PubMed] [Google Scholar]
- 10.Wojtkowiak D, Georgopoulos C, Zylicz M. J Biol Chem. 1993;268:22609–22617. [PubMed] [Google Scholar]
- 11.Wawrzynow A, Wojtkowiak D, Marszalek J, Banecki B, Jonsen M, Graves B, Georgopoulos C, Zylicz M. EMBO J. 1995;14:1867–1877. doi: 10.1002/j.1460-2075.1995.tb07179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenney K, Maurizi M R. Proc Natl Acad Sci USA. 1994;91:12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levchenko I, Smith C K, Walsh N P, Sauer R T, Baker T A. Cell. 1997;91:939–947. doi: 10.1016/s0092-8674(00)80485-7. [DOI] [PubMed] [Google Scholar]
- 14.Weber-Ban E U, Reid B G, Miranker A D, Horwich A L. Nature (London) 1999;401:90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- 15.Kim Y I, Burton R E, Burton B M, Sauer R T, Baker T A. Mol Cell. 2000;5:639–648. doi: 10.1016/s1097-2765(00)80243-9. [DOI] [PubMed] [Google Scholar]
- 16.Hoskins J R, Singh S K, Maurizi M R, Wickner S. Proc Natl Acad Sci USA. 2000;97:8892–8897. doi: 10.1073/pnas.97.16.8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ortega J, Singh S K, Ishikawa T, Maurizi M R, Steven A C. Mol Cell. 2000;6:1515–1521. doi: 10.1016/s1097-2765(00)00148-9. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Hartling J A, Flanagan J M. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 19.Grimaud R, Kessel M, Beuron F, Steven A C, Maurizi M R. J Biol Chem. 1998;273:12476–12481. doi: 10.1074/jbc.273.20.12476. [DOI] [PubMed] [Google Scholar]
- 20.Schweder T, Lee K H, Lomovskaya O, Matin A. J Bacteriol. 1996;178:470–476. doi: 10.1128/jb.178.2.470-476.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levchenko I, Luo L, Baker T A. Genes Dev. 1995;9:2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, Elliott M, Elliott T. J Bacteriol. 1999;181:1211–1219. doi: 10.1128/jb.181.4.1211-1219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engelberg-Kulka H, Glaser G. Annu Rev Microbiol. 1999;53:43–70. doi: 10.1146/annurev.micro.53.1.43. [DOI] [PubMed] [Google Scholar]
- 24.Katayama Y, Gottesman S, Pumphrey J, Rudikoff S, Clark W P, Maurizi M R. J Biol Chem. 1988;263:15226–15236. [PubMed] [Google Scholar]
- 25.Levchenko I, Yamauchi M, Baker T A. Genes Dev. 1997;11:1561–1572. doi: 10.1101/gad.11.12.1561. [DOI] [PubMed] [Google Scholar]
- 26.Maurizi M R, Thompson M W, Singh S K, Kim S H. Methods Enzymol. 1994;244:314–331. doi: 10.1016/0076-6879(94)44025-5. [DOI] [PubMed] [Google Scholar]
- 27.Yakhnin A V, Vinokurov L M, Surin A K, Alakhov Y B. Protein Expression Purif. 1998;14:382–386. doi: 10.1006/prep.1998.0981. [DOI] [PubMed] [Google Scholar]
- 28.Andersen J B, Sternberg C, Poulsen L K, Bjorn S P, Givskov M, Molin S. Appl Environ Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herman C, Thevenet D, Bouloc P, Walker G C, D'Ari R. Genes Dev. 1998;12:1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keiler K C, Sauer R T. J Biol Chem. 1996;271:2589–2593. doi: 10.1074/jbc.271.5.2589. [DOI] [PubMed] [Google Scholar]
- 31.Beebe K D, Shin J, Peng J, Chaudhury C, Khera J, Pei D. Biochemistry. 2000;39:3149–3155. doi: 10.1021/bi992709s. [DOI] [PubMed] [Google Scholar]
- 32.Laachouch J E, Desmet L, Geuskens V, Grimaud R, Toussaint A. EMBO J. 1996;15:437–444. [PMC free article] [PubMed] [Google Scholar]
- 33.Domian I J, Quon K C, Shapiro L. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 34.Gonciarz-Swiatek M, Wawrzynow A, Um S J, Learn B A, McMacken R, Kelley W L, Georgopoulos C, Sliekers O, Zylicz M. J Biol Chem. 1999;274:13999–14005. doi: 10.1074/jbc.274.20.13999. [DOI] [PubMed] [Google Scholar]
- 35.Gonzalez M, Rasulova F, Maurizi M R, Woodgate R. EMBO J. 2000;19:5251–5258. doi: 10.1093/emboj/19.19.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konieczny I, Helinski D R. Proc Natl Acad Sci USA. 1997;94:14378–14382. doi: 10.1073/pnas.94.26.14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lehnherr H, Yarmolinsky M B. Proc Natl Acad Sci USA. 1995;92:3274–3277. doi: 10.1073/pnas.92.8.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y, Gottesman S, Hoskins J R, Maurizi M R, Wickner S. Genes Dev. 2001;15:627–637. doi: 10.1101/gad.864401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoskins J R, Kim S Y, Wickner S. J Biol Chem. 2000;275:35361–35367. doi: 10.1074/jbc.M006288200. [DOI] [PubMed] [Google Scholar]