CR16 forms a complex with N-WASP in brain and is a novel member of a conserved proline-rich actin-binding protein family (original) (raw)
Abstract
The Neuronal Wiskott–Aldrich syndrome protein (N-WASP) has emerged as a central regulator of the actin cytoskeleton with abilities to integrate multiple upstream signal inputs and transmit them to the Arp2/3 complex. Here, we demonstrate that native N-WASP is present in a tight complex with a proline-rich protein, CR16, which shares ≈25% identity with WASP interacting protein. CR16 is encoded by a gene previously cloned as a glucocorticoid-regulated mRNA from a rat hippocampal cDNA library. Although N-WASP is expressed ubiquitously, full-length CR16 protein is found predominately in the brain. CR16 and N-WASP colocalize in primary hippocampal neurons and at the tips of their growth cone filopodia. In vitro, CR16 directly binds both monomeric and filamentous actin but does not affect the kinetics of actin polymerization mediated by N-WASP and the Arp2/3 complex. Sequence homologues of CR16 are found not only in other vertebrates but also in the invertebrate_Caenorhabditis elegans_ and in yeast. Thus, CR16 and WASP interacting protein belong to a family of N-WASP-binding proteins.
During the past few years, members of the Wiskott–Aldrich syndrome protein (WASP) family that include WASP, N-WASP, Scar/WAVE, and Bee1 have emerged as key intermediates that link upstream signals from small GTPases, perhaps tyrosine kinases and phosphoinositides to the actin cytoskeleton (1–4). For instance, N-WASP and WASP directly bind to and are activated by Cdc42, phosphoinositides, and adaptor proteins like Nck and Grb2 (4–6). Scar/WAVE proteins seem to be indirect downstream effectors of Rac signaling (7). All WASP family proteins regulate the actin cytoskeleton by activating a common effector, the Arp2/3 complex (8, 9), which accelerates the de novo nucleation and side branching of actin filaments (10).
N-WASP can exist in an inactive or an Arp2/3 interaction competent activated state (8). We have previously demonstrated that binding of Cdc42 and phosphotidylinositol-4,5-bisphosphate [PI(4,5)P2] to the N terminus of N-WASP disrupts an intramolecular inhibitory interaction between its N terminus and its Arp2/3-binding C-terminal VCA (forverprolin homology, cofilin homology, andacidic region) domain, allowing the VCA domain to activate the Arp2/3 complex (11). N-WASP is also an essential component of a Cdc42 and PI(4,5)P2-dependent actin assembly pathway in Xenopus egg and bovine brain extracts (8,12).
Although the mechanism of N-WASP regulation has been studied intensively, the precise cellular function of N-WASP remains largely a mystery. Thus, we have used bovine brain extracts to identify novel N-WASP interacting proteins in an attempt to gain insight into the biological role of the N-WASP pathway. Here, we report the identification and characterization of a novel N-WASP-binding protein, CR16.
Materials and Methods
Purification of the N-WASP/CR16 Complex from Bovine Brain Extracts.
Throughout the purification, the N-WASP complex was followed by immunoblotting with an anti-N-WASP antibody (8). All chromatography media were purchased from Amersham Pharmacia.
Preparation of bovine brain high-speed supernatant was described in ref. 8. The supernatant was sequentially fractionated on butyl Sepharose, Resource S, Superose 6, MonoQ, and a 5–20% wt/vol sucrose gradient. The sucrose gradient fractions containing N-WASP were used for immunoaffinity purification. Immunoprecipitation of N-WASP was performed as described (8). The bound N-WASP complex was eluted from the antibody beads by using 200 mM glycine, pH 2.0, neutralized and analyzed by SDS/PAGE.
Mass Spectrometry.
Sequence analysis of the 67-kDa N-WASP associated protein was performed at the Harvard Microchemistry Facility by microcapillary reverse-phase HPLC nano-electrospray tandem mass spectrometry.
Molecular Biology.
The portion of the CR16 ORF extending from the beginning of exon 7 to the end was PCR amplified from rat brain Marathon cDNA (CLONTECH). The fragment was then fused to the rest of CR16 (obtained from Jeffery Masters, Abbott) by PCR and subcloned into the pCS2+ vector (untagged) and the pCS2+HA vector [N-terminal hemagglutinin (HA)-tagged]. The reconstructed CR16+Ex7 was checked by sequencing. CR16−Ex7 was reconstructed by PCR, cloned into pCS2+, and checked by sequencing.
Baculoviruses were constructed and used for infections according to the Bac-to-Bac baculovirus expression system (GIBCO/BRL). Recombinant proteins were purified on an anti-HA antibody column (Roche Biochemicals) and eluted with HA peptide (Roche Biochemicals). The proteins were further purified on a Mono S column. The recombinant CR16/N-WASP complex was synthesized by coinfecting Sf9 cells with the HA-CR16+Ex7 and N-WASP viruses. Glutathione _S_-transferase (GST)-N-WASP and GST-WH1 proteins were described previously (11).
Binding Assays.
GST pull-down was performed as described (11).
Antibodies.
Anti-N-WASP antibody was previously described (8). A C-terminal fragment (amino acids 370–485) of CR16 with an N-terminal hexahistidine tag was expressed in Escherichia coli and purified on an Ni–NTA column. This fragment was used to raise antisera in rabbits (Zymed). The antibodies were affinity purified as described (8).
Tissue Blot and Fractionation.
Mouse tissues were Dounce homogenized in the presence of protease inhibitors. Five micrograms of total protein from each tissue were immunoblotted with an α-CR16 antibody. A homogenized mouse brain was centrifuged at 3,500 × g for 15 min to separate nuclei and cell debris from the postnuclear supernatant. The postnuclear supernatant was centrifuged at 100,000 × g for 30 min to separate membranes and cytoskeleton from the high-speed supernatant. The high-speed pellet was washed in lysis buffer with 1 M NaCl. Fractions were normalized by original volumes and analyzed by immunoblotting.
Immunofluorescence Microscopy.
Preparation of primary rat hippocampal neurons and immunocytochemistry followed (13). For single-staining experiments, cells were incubated with affinity-purified α-N-WASP antibody (final concentration 7 μg/ml) or α-CR16 antibody (final concentration 4 μg/ml), followed by incubation with Texas Red-conjugated donkey α-rabbit secondary antibody (1:250 dilution, Jackson ImmunoResearch). After washing, cells were stained with Alexa-488 conjugated phalloidin (1:100 dilution, Molecular Probes).
For double staining of CR16 and N-WASP, an affinity-purified α-CR16 antibody was directly labeled by using the Alexa-488 protein coupling kit (Molecular Probes). After first staining with α-N-WASP and Cy5-conjugated secondary antibody (Jackson ImmunoResearch) and then washing extensively, cells were stained with Alexa-488-conjugated α-CR16 antibody at 70 μg/ml. Cells were then stained with rhodamine-conjugated phalloidin.
Images were viewed and acquired by using the DeltaVision fluorescence microscopy system (Applied Scientific) without deconvolution. Images were processed by using the ADOBE PHOTOSHOP or CANVAS 5 software.
Results
Purification of an N-WASP Complex from Bovine Brain Extracts.
To isolate native protein complexes containing N-WASP, we compared the hydrodynamic properties of N-WASP in crude bovine brain extracts to recombinant, purified N-WASP expressed in insect cells. We observed that endogenous bovine N-WASP fractionated at a considerably higher molecular weight than the recombinant protein. Specifically, the purified recombinant rat N-WASP has a Stokes radius of 4.4 nm by gel filtration and an uncorrected sedimentation coefficient of ≈2.0 S by sucrose gradient zonal sedimentation (data not shown). In the brain extracts, native N-WASP migrated with a Stokes radius of 8 nm and an uncorrected S value of 3.21 S (data not shown). Thus, N-WASP is likely either a dimer or is associated with other proteins in the brain extracts.
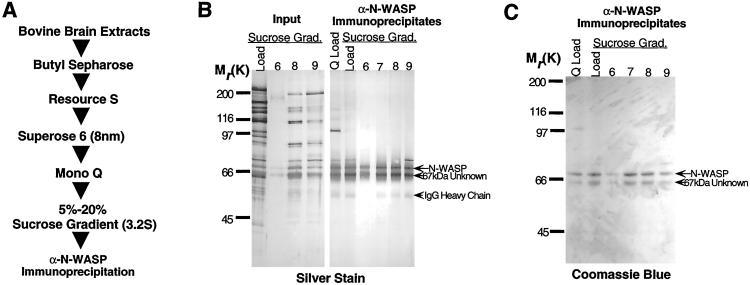
To purify the N-WASP containing complex from brain extracts, we used a purification scheme that combined five conventional fractionation steps, followed by an immunoaffinity step using an affinity-purified polyclonal anti-N-WASP antibody (Fig.1A). The N-WASP complex was followed by α-N-WASP immunoblotting. Analysis of the purified complex by SDS/PAGE and silver staining revealed that the complex consisted of two proteins, N-WASP (70 kDa by SDS/PAGE) and a second protein that migrated at 67 kDa by SDS/PAGE (Fig. 1B).
Figure 1.
Purification of a N-WASP complex from bovine brain extracts. (A) Scheme used for purification of the N-WASP complex. (B) A 10% silver-stained SDS/polyacrylamide gel and (C) an identical Coomassie-stained gel used to fractionate acid eluates of α-N-WASP immunoprecipitates from material loaded on the Mono Q column, material loaded on sucrose gradient, and from the sucrose gradient fractions (6–9) containing the N-WASP peak by immunoblotting. Also shown (B) are the total complement of protein (input) in various sucrose gradient fractions before immunoprecipitation.
Based on Coomassie dye binding in SDS gels (Fig.1C) followed by densitometry, the stoichiometry of the two proteins in the complex is ≈2:1 (N-WASP:67-kDa band). However, the native molecular mass of the complex, based on the diffusion coefficient from gel filtration and the sedimentation coefficient from sucrose gradient sedimentation, is 110–120 kDa, more consistent with a 1:1 stoichiometry. It is likely that N-WASP and the 67-kDa protein bind Coomassie blue with different efficiencies.
Identification and Cloning of the 67-kDa N-WASP-Associated Protein.
Analysis of 5 μg (70 pmol) of the 67-kDa N-WASP-associated protein by trypsin digestion and mass spectrometry identified four peptides that exactly matched the sequence of the CR16 protein from rat (amino acid sequence shown in Fig. 2). CR16 was originally cloned as a glucocorticoid-regulated mRNA from a rat hippocampal cDNA library (14, 15). The CR16 ORF predicts a 49-kDa protein with an unusually high proline content (32%), which likely contributes to its aberrant migration at 67 kDa on SDS/PAGE and perhaps its reduced dye binding.
Figure 2.
Sequence comparison of CR16, WIP, and related proteins. (A) Alignment of rat CR16 (RNcr16), human CR16 (hsCR16), and human WIP (hsWIP1). Identical residues among the three proteins are shaded, and similar residues are boxed. The blue and red lines demarcate the verprolin homology segment and the alternatively spliced exon 7 (CR16 only), respectively. (B) Phylogenetic tree showing the phylogenetic relationship of CR16, WIP, and related proteins from selected species.
The CR16 gene exists in two alternatively spliced forms (16). The less abundant form includes exon7 (the penultimate exon), whereas the more abundant form excludes this exon. Exon7 adds 102 bp in the CR16 cDNA sequence and replaces the Gly at amino acid position 419 with 35 additional amino acids at the C terminus (Figs. 2 and3) (16).
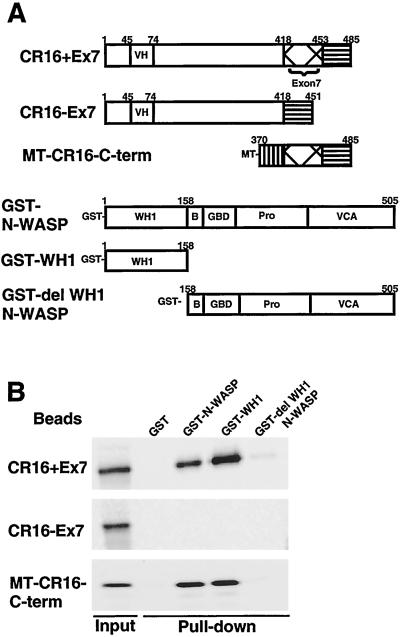
Figure 3.
Sequence determinants of the interaction between CR16 and N-WASP. (A) A schematic illustration and amino acid boundaries of the various CR16 (rat) and N-WASP (bovine) constructs used in this study. (B) Recombinant GST–N-WASP, GST-WH1 domain, and GST-del WH1 N-WASP (and GST alone as a negative control) were immobilized on glutathione-Sepharose beads and tested for their ability to pull down full-length CR16 (CR16+Ex7), the alternatively spliced CR16 (CR16−Ex7), and a Myc-tagged C-terminal fragment of CR16 (MT–CR16–C-term). Ten percent of the input and 20% of the pulled down material was analyzed on an SDS/5–15% polyacrylamide gel.
Sequence comparison revealed that a human CR16 sequence shares 72% identity with the rat CR16 protein. In addition, CR16 is ≈25% identical to WASP interacting protein (WIP, see Fig.2A for an alignment of CR16 and WIP) (17). However, CR16 is clearly a different protein from WIP because human and_Xenopus_ WIP are more closely related to each other than to human or rat CR16 (Fig. 2B). The alignment in Fig.2A reveals two highly homologous regions between CR16 and WIP. One region, near the N terminus of the proteins, overlaps the actin-binding verprolin homology region of WIP (underlined blue in Fig. 2A). The second region, in the C terminus, coincides with the alternatively spliced exon 7 (underlined red in Fig.2A). This C-terminal region of WIP mediates interaction with the WASP and N-WASP (17).
A database search revealed that CR16 and WIP-related sequences are found not only in vertebrates but also in the invertebrate_Caenorhabditis elegans_ (T16755). CR16 is also distantly related to the yeast actin-binding protein verprolin, based on primary amino acid sequences (18). Thus, CR16 and WIP represent members of a proline-rich protein family conserved through evolution (Fig. 2B).
Defining the Sequence Elements Important for the CR16–N-WASP Interaction.
A GST pull-down method was used to map the sequence elements in CR16 and N-WASP required for their interaction. The proteins and protein fragments used in these experiments are summarized in Fig.3A. Briefly, CR16+Ex7, CR16−Ex7, and a Myc-tagged C-terminal fragment of CR16 containing exon7 (MT–CR16–C-term), were synthesized by in vitro translation in the presence of [35S]methionine. The proteins were tested for their abilities to interact with GST fusions of full-length N-WASP (GST–N-WASP), the isolated WH1 domain of N-WASP (GST-WH1), and N-WASP lacking the WH1 domain (GST-del WH1 N-WASP) immobilized on glutathione-Sepharose beads. CR16+Ex7 interacts with both GST–N-WASP and GST-WH1 but not with GST-del WH1 N-WASP or the control GST (Fig.3B). Thus, the WH1 domain of N-WASP is necessary and sufficient to interact with CR16. CR16−Ex7 lacks the ability to bind GST–N-WASP or GST-WH1 completely, suggesting that sequences in the exon 7 of CR16 are necessary for high affinity binding to N-WASP. MT–CR16–C-term can bind both GST–N-WASP and GST-WH1 with similar affinity when compared with CR16+Ex7. Thus, the C-terminal 115 aa of CR16+Ex7 contain all of the sequence elements required for high affinity binding to N-WASP. Whereas exon7 is required for the CR16–N-WASP interaction, it alone is not sufficient. These binding results also confirm that CR16 is indeed the N-WASP-associated protein we purified from bovine brain extracts.
Expression of the Recombinant CR16/N-WASP Complex in Insect Cells.
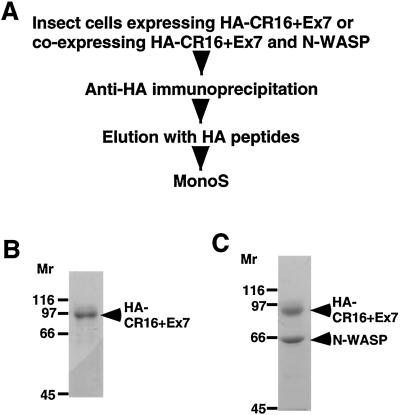
We have generated a recombinant baculovirus expressing CR16+Ex7 with an N-terminal HA epitope tag (HA-CR16+Ex7) and overexpressed this protein in insect cells. The recombinant HA-CR16+Ex7 protein was purified as shown in Fig. 4 A and_B_. We have also successfully expressed the CR16/N-WASP complex in insect cells by coinfection of Sf9 cells with both HA-CR+Ex7 and N-WASP (wild type, untagged) expressing viruses. The recombinant complex was purified as described above for HA-CR16+Ex7. HA-CR16+Ex7 and N-WASP cofractionate on both anti-HA and Mono S columns (Fig.4C), suggesting the two proteins form a complex when coexpressed in insect cells. HA-CR16+Ex7 expressed in insect cells sometimes migrates as a doublet band in SDS/PAGE (see Fig.7A) and is likely because of phosphorylation, as treatment with lambda phosphatase causes the doublets to collapse into the lower band (data not shown). Frequently, native CR16 protein also appears as a doublet in SDS/PAGE (Fig. 5).
Figure 4.
Expression of recombinant CR16 protein and CR16/N-WASP complex in insect cells. (A) Scheme used for the expression and purification of recombinant HA-CR16+Ex7 protein and HA-CR16+Ex7/N-WASP complex from insect cells. (B) A Coomassie-stained SDS/polyacrylamide gel showing the purified HA-CR16+Ex7 protein from the peak Mono S fraction. (C) A Coomassie-stained SDS/polyacrylamide gel showing the purified recombinant HA-CR16+Ex7/N-WASP complex from the peak Mono S fraction.
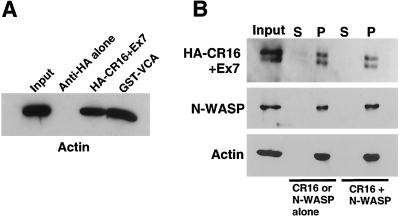
Figure 7.
CR16 directly interacts with both G-actin and F-actin. (A) HA-CR16 immobilized on α-HA antibody-coated beads was tested for its ability to pull down G-actin from solution. GST-VCA of N-WASP immobilized on glutathione-Sepharose and α-HA beads were used as positive and negative controls, respectively. Thirty-three percent of input and material pulled down were analyzed by immunoblotting with α-actin. (B) The ability of CR16 to interact with F-actin was determined in an F-actin cosedimentation assay. Recombinant HA-CR16+Ex7 and N-WASP, either alone or together, were incubated with F-actin and then subject to ultracentrifugation. The supernatants (S) and pellets (P) containing F-actin and proteins associated with F-actin were normalized by original volume and resolved on a 5–15% SDS/polyacrylamide gel, followed by immunoblotting with α-HA, α-N-WASP, and α-actin. The appearance of recombinant HA-CR16+Ex7 as a doublet is likely because of protein phosphorylation (see Results for details).
Figure 5.
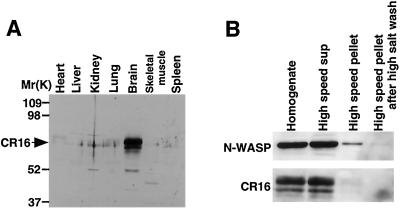
Tissue distribution and subcellular fractionation of the CR16 protein. (A) Tissue distribution of CR16 protein. Five micrograms of total protein from each indicated mouse tissue was resolved by SDS/PAGE and immunoblotted for CR16. (B) Mouse brain homogenates were fractionated into cytoplasmic, membrane, and cytoskeletal fractions by centrifugation. Sample fractions were normalized by volume, resolved by SDS/PAGE, and analyzed by immunoblotting with α-N-WASP and α-CR16.
Tissue and Subcellular Distribution of CR16 Protein.
The CR16 mRNA was previously reported to be expressed in brain, heart, lung, and testis but not kidney, liver, and spleen (15). We sought to determine the tissue and subcellular distribution of CR16 protein. We raised a polyclonal rabbit antibody against a C-terminal fragment of rat CR16 (amino acids 370–485) that includes the region encoded by exon 7. The antibody is specific for CR16 and recognizes CR16 in rat, mouse, and bovine brain lysates (data not shown). In agreement with the mRNA expression pattern, CR16 protein is found mainly in the brain (Fig. 5A), but it is also present in the lung and the heart at lower levels (data not shown). Thus, even though N-WASP is expressed ubiquitously, the expression of the CR16 protein is limited to the nervous system and perhaps a few other tissues.
To determine the subcellular distribution of CR16 and N-WASP, fresh mouse brain was fractionated into postnuclear supernatant, insoluble membrane/cytoskeleton, and soluble cytosolic fractions by differential centrifugation. The majority of CR16 and N-WASP (>95%) was found in the soluble cytosolic fraction (Fig. 5B). This result suggests that the CR16/N-WASP complex is not associated or is only very weakly associated with the membrane or the cytoskeleton.
CR16 and N-WASP Colocalize in Embryonic Hippocampal Neurons.
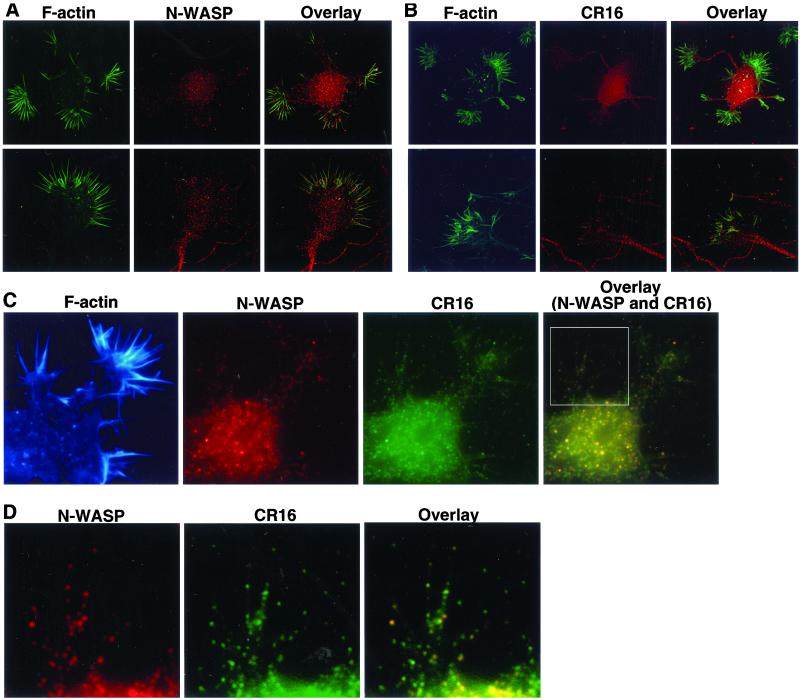
We localized the CR16/N-WASP complex in embryonic hippocampal neurons by immunofluorescence microscopy. Primary cultures of embryonic hippocampal neurons are advantageous for this study because these cells not only express CR16 and N-WASP endogenously but also exhibit elaborate growth cones with extensive filopodial and lamellipodial structures. We first examined the localization patterns of CR16 and N-WASP individually. Both CR16 and N-WASP displayed a punctate staining pattern throughout the cell body, the axons, and the growth cones (Fig. 6). The puncta are unlikely to be associated with vesicles, as most CR16 and N-WASP is found in the soluble cytosol (Fig. 5B). In the growth cones, N-WASP and CR16 are found at the tips (distal ends) of many filopodia (Fig. 6). Double staining of CR16 and N-WASP showed that CR16 and N-WASP colocalize significantly in the cell body, axons, and growth cones, including many of the filopodial tips (Fig. 6 C and_D_). These results also support the in vivo interaction of CR16 and N-WASP.
Figure 6.
Localication of CR16 and N-WASP in embryonic hippocampal neurons by immunofluorescence. Primary embryonic hippocampal neurons were fixed 10 or 24 h after plating and subject to immunostaining. (A) Cells were costained with Alexa 488-phalloidin (green) to visualize F-actin and an affinity-purified α-N-WASP antibody (red). (B) Cells were costained with Alexa 488-phalloidin (green) and an affinity-purified α-CR16 antibody (red). Shown in both A and B are the whole neuron (Upper) and the growth cone of a neuron (Lower). (C) Cells were triple stained with an α-N-WASP antibody (red), an α-CR16 antibody (green), and rhodamine-conjugated phalloidin (false colored blue). (D) A close-up view of the growth cone boxed in_C_. N-WASP and CR16 stainings were overlaid to show colocalization of the two antigens (C and_D_, yellow signals).
Biochemical Activity of the CR16/N-WASP Complex.
The N-terminal verprolin homology (also known as WASP homology 2, or WH2) domain of CR16 contains a conserved G-actin-binding motif (RLRK) that is also found in WIP (KLKK), verprolin (KLKK), N-WASP (QLKK, QLKS), WASP (QLKS), and Scar1/WAVE (QLRK). Thus, we wanted to test the ability of CR16 to interact with G-actin. As shown in Fig.7A, recombinant HA-CR16 immobilized on agarose beads coated with anti-HA monoclonal antibody efficiently pulled down G-actin (below the critical concentration of actin polymerization). Anti-HA beads alone did not pull down detectable amounts of G-actin, whereas the VCA fragment of N-WASP used as a positive control pulled down G-actin with similar efficiency.
Because WIP has recently been reported to bind F-actin directly (19), we asked whether CR16 also interacts with F-actin. Using an F-actin copelleting assay, we have found that CR16 alone, or in the presence of N-WASP, cosediments with F-actin (Fig. 7B). N-WASP alone cosediments with F-actin, as reported (20), and is not affected by the presence of CR16 (Fig. 7B). Thus, we conclude that CR16 is a G-actin and F-actin-binding protein.
We next compared the effects of CR16 on actin assembly kinetics in a purified component system. In this system, guanosine 5′-[γ-thio]triphosphate-loaded Cdc42 and PI(4,5)P2 are used to activate N-WASP, which in turn stimulates the actin nucleating activity of the Arp2/3 complex (8). Actin polymerization is initiated by the addition of purified actin and pyrene-labeled actin to the reaction mixture, and the rate of assembly is monitored by the change in pyrene fluorescence. Addition of recombinant HA-CR16+Ex7 protein (up to 0.5 μM final concentration) to the polymerization reaction (containing 100 nM N-WASP and 30 nM Arp2/3) did not affect the kinetics of the assembly or the total mass of F-actin polymerized (data not shown). HA-CR16+Ex7 also has no effect on actin polymerization when only guanosine 5′-[γ-thio]triphosphate-loaded Cdc42 [but no PI(4,5)P2] was added (data not shown). Recombinant complexes (HA–CR16+Ex7/N-WASP) coexpressed and copurified from insect cells were indistinguishable from N-WASP alone (data not shown).
Discussion
We have purified a complex of N-WASP and a protein of unknown function, CR16, from bovine brain extracts. The interaction was confirmed in vitro by pull-down assay, coexpression in insect cells, and in vivo by double-staining immunofluorescence microscopy in primary hippocampal neurons. We have also shown that the interaction is directly mediated by the N-terminal WH1 domain of N-WASP and by a C-terminal region of CR16, which includes the alternatively spliced exon 7. Alternative splicing of CR16 presents a potential regulatory mechanism for the interaction between the CR16/N-WASP complex and its interacting partners, as only CR16+Ex7 and not CR16−Ex7 has the ability to bind N-WASP. Both splice variants of CR16 contain multiple putative recognition sites for SH3 domains, as well as several potential phosphorylation sites (16). It is interesting to note that only a small fraction of total CR16 mRNA has the exon 7 required for binding to N-WASP (16), and the physiological significance of this observation is currently unknown.
CR16 and WIP share ≈25% protein sequence identity, and both proteins interact with the WH1 domain of N-WASP through their conserved C-terminal regions. Like WIP, CR16 directly interacts with both G-actin and F-actin. Thus, CR16 and WIP belong to a novel family of N-WASP/WASP-binding proteins. Remarkably, CR16 is highly conserved throughout evolution. Sequence homologues of CR16 are found not only in vertebrates but also in the invertebrate C. elegans and the hemichordate acorn worm, Saccoglossus kowalesky (M.W.K., unpublished data).
The biological function of CR16 remains to be elucidated. WIP has recently been implicated in the recruitment of N-WASP to the surface of Vaccinia virus during actin comet tail motility, suggesting its role in the spatial control of actin assembly machinery (21). It is also important for filopodia formation in fibroblasts (19). In the absence of evidence suggesting CR16 can directly affect the actin polymerization machinery, we speculate that CR16 may target N-WASP to particular subcellular locations where positive signals for actin assembly are received. Colocalization of the CR16/N-WASP complex to neuronal growth cones, particularly the tips of filopodia, is consistent with this idea. The localization of the CR16/N-WASP complex at the tips of growth cone filopodia is particularly interesting because N-WASP is important for filopodia formation induced by microinjection of Cdc42 or treatment with bradykinin in cultured cell (2). Further study is required to understand the physiological function of the CR16/N-WASP complex in neurons.
The richness of putative SH3 domain protein-binding sites along the sequence of CR16 also suggests that CR16 may be a signaling intermediate that links N-WASP to upstream pathways. In addition, the WH1 domain of the actin cytoskeleton regulatory Ena/VASP proteins has been implicated in protein localization, raising the possibility that the WH1 domain functions as a general protein-targeting module (22). Nevertheless, further biochemical and cellular studies are required before the roles of CR16 in the regulation of actin cytoskeleton become clear. Understanding the cellular function of CR16 will no doubt shed light on the yet obscure physiological functions of the Cdc42–N-WASP–Arp2/3 pathway, and our identification and characterization of the CR16/N-WASP complex is an important step in that direction.
Acknowledgments
We thank Jeff Masters (Abbott) for providing a cDNA clone for CR16−Ex7 and Sharon Eden for preparing the α-CR16 rabbit polyclonal antibody. We especially thank Maria Morabito and Li-Huei Tsai (Harvard Medical School) for providing primary hippocampal neuron and the use of their DeltaVison microscopy system. We thank Rong Li (Harvard Medical School) for critically reading the manuscript. R.R. is a member of the Medical Scientist Training Program at Harvard Medical School. This work was supported by National Institutes of Health Grant NIHGM 5R01 GM26875-24 (to M.W.K.).
Abbreviations
PI(4,5)P2
phosphotidylinositol-4,5-bisphosphate
HA
hemagglutinin
N-WASP
neuronal Wiskott–Aldrich syndrome protein
GST
glutathione_S_-transferase
WIP
WASP interacting protein
References
- 1.Bear J E, Rawls J F, Saxe C L., III J Cell Biol. 1998;142:1325–1335. doi: 10.1083/jcb.142.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miki H, Sasaki T, Takai Y, Takenawa T. Nature (London) 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- 3.Li R. J Cell Biol. 1997;136:649–658. doi: 10.1083/jcb.136.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Symons M, Derry J M, Karlak B, Jiang S, Lemahieu V, McCormick F, Francke U, Abo A. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- 5.Miki H, Miura K, Takenawa T. EMBO J. 1996;15:5326–5335. [PMC free article] [PubMed] [Google Scholar]
- 6.Carlier M F, Nioche P, Broutin-L'Hermite I, Boujemaa R, Le Clainche C, Egile C, Garbay C, Ducruix A, Sansonetti P, Pantaloni D. J Biol Chem. 2000;275:21946–21952. doi: 10.1074/jbc.M000687200. [DOI] [PubMed] [Google Scholar]
- 7.Miki H, Suetsugu S, Takenawa T. EMBO J. 1998;17:6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner M W. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 9.Machesky L M, Insall R H. Curr Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- 10.Mullins R D, Heuser J A, Pollard T D. Proc Natl Acad Sci USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rohatgi R, Ho H Y, Kirschner M W. J Cell Biol. 2000;150:1299–1310. doi: 10.1083/jcb.150.6.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma L, Cantley L C, Janmey P A, Kirschner M W. J Cell Biol. 1998;140:1125–1136. doi: 10.1083/jcb.140.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zukerberg L R, Patrick G N, Nikolic M, Humbert S, Wu C L, Lanier L M, Gertler F B, Vidal M, Van Etten R A, Tsai L H. Neuron. 2000;26:633–646. doi: 10.1016/s0896-6273(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 14.Nichols N R, Masters J N, Finch C E. Brain Res Bull. 1990;24:659–662. doi: 10.1016/0361-9230(90)90004-j. [DOI] [PubMed] [Google Scholar]
- 15.Masters J N, Cotman S L, Osterburg H H, Nichols N R, Finch C E. Neuroendocrinology. 1996;63:28–38. doi: 10.1159/000126932. [DOI] [PubMed] [Google Scholar]
- 16.Weiler M C, Smith J L, Masters J N. J Mol Neurosci. 1996;7:203–215. doi: 10.1007/BF02736841. [DOI] [PubMed] [Google Scholar]
- 17.Ramesh N, Anton I M, Hartwig J H, Geha R S. Proc Natl Acad Sci USA. 1997;94:14671–14676. doi: 10.1073/pnas.94.26.14671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donnelly S F, Pocklington M J, Pallotta D, Orr E. Mol Microbiol. 1993;10:585–596. doi: 10.1111/j.1365-2958.1993.tb00930.x. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Quiles N, Rohatgi R, Anton I M, Medina M, Saville S P, Miki H, Yamaguchi H, Takenawa T, Hartwig J H, Geha R S, Ramesh N. Nat Cell Biol. 2001;3:484–491. doi: 10.1038/35074551. [DOI] [PubMed] [Google Scholar]
- 20.Egile C, Loisel T P, Laurent V, Li R, Pantaloni D, Sansonetti P J, Carlier M F. J Cell Biol. 1999;146:1319–1332. doi: 10.1083/jcb.146.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreau V, Frischknecht F, Reckmann I, Vincentelli R, Rabut G, Stewart D, Way M. Nat Cell Biol. 2000;2:441–448. doi: 10.1038/35017080. [DOI] [PubMed] [Google Scholar]
- 22.Gertler F B, Niebuhr K, Reinhard M, Wehland J, Soriano P. Cell. 1996;87:227–239. doi: 10.1016/s0092-8674(00)81341-0. [DOI] [PubMed] [Google Scholar]