Supermolecular structure of the enteropathogenic Escherichia coli type III secretion system and its direct interaction with the EspA-sheath-like structure (original) (raw)
Abstract
Enteropathogenic Escherichia coli (EPEC) secretes several Esp proteins via the type III secretion system (secreton). EspA, EspB, and EspD are required for translocation of the effector proteins into host cells, in which EspB and EspD are thought to form a pore in the host membrane. Recent study has shown that EspA forms a filamentous structure that assembles as a physical bridge between bacteria and host cell surfaces, which then functions as a conduit for the translocation of bacterial effectors into host cells. To investigate the supermolecular structure of the type III secreton in EPEC, we partially purified it from the bacteria membrane and observed it via transmission electron microscopy. The EPEC type III secreton was composed of a basal body and a needle part and was similar to those of_Salmonella_ and Shigella, except for a sheath-like structure at the tip of the needle. The length of sheath-like structures varied; it extended more than 600 nm and was 10 times longer than the Shigella needle part. The putative major needle component, EscF, was required for both secretion of Esp proteins and needle complex formation. Interestingly, elongation of the sheath-like structure was observed under constitutive expression of EspA but not of EscF. Furthermore, the transmission electron microscopy view with immunogold labeled anti-EspA antibodies clearly showed that EspA is a component of the sheath-like structure. This study revealed, to our knowledge for the first time, the supermolecular structure of the EPEC type III secreton and its direct association with the EspA-sheath-like structure.
Enteropathogenic_Escherichia coli_ (EPEC) is a major cause of diarrhea in young children (1). Related pathogens, which cause disease by using similar mechanisms (2), include enterohemorrhagic E. coli O157:H7, rabbit enteropathogenic E. coli, and_Citrobacter rodentium_, which is found in mice. These pathogens induce a characteristic histopathological lesion, termed an attaching/effacing (A/E) lesion, which is defined by the intimate attachment of bacteria to the epithelial surface and the effacement of host cell microvilli (3). The A/E lesion is mediated by bacteria–host cell interactions, including triggering of host signal transduction pathways and cytoskeletal arrangements (4), and is required for full virulence in vivo (5).
Factors responsible for the A/E lesion formation are encoded by a 35-kilobase pair (kbp) locus termed the LEE (6), which encodes (i) the type III secretion system (the secreton) (7), (ii) the translocated intimin receptor (Tir) (8) and intimin (9), and (iii) Esp proteins. To elicit A/E lesion formation, Tir must be translocated into the host cell membrane via the type III secreton, and then the bacteria adhesion molecule, intimin, directly associates with the translocated Tir (8). Tir–intimin interaction is required for A/E lesion formation and induces accumulations of actin and other cytoskeletal components beneath the attached bacteria (8).
In addition to Tir, three other additional secreted proteins, EspA, EspB, and EspD, are required for formation of A/E lesions and are secreted via the EPEC/enterohemorrhagic E. coli type III secreton. EspA is a structural protein and the major component of a filamentous surface organelle termed “EspA filament” (10). The EspA filament forms a physical bridge between bacteria and the host cell, and it then functions as a conduit for the translocation of bacterial effectors into the host cell. EspB and EspD are thought to be delivered to the host cell membrane, and both proteins showed homology to Yersinia YopB/D proteins, which are believed to form a pore complex in the host membrane and correlate with the ability to induce contact-dependent hemolysis of red blood cells (RBCs) (11, 12). Recent study has shown that EPEC induces contact-independent hemolysis to RBCs, and EspA filament and EspD are required for this event (13,14). On the other hand, the espB mutant still caused weak hemolysis, indicating that EspD may be the major component of a translocation pore into the host cell membrane (14). However, the contradictory finding has been presented that the espB mutant showed the nonhemolytic phenotype (13).
Type III secretons are found in many other Gram-negative bacteria species, and they are the mechanism for translocation of bacterial proteins into host cells. Many components of type III secretons show sequence similarities with those of flagellar basal bodies (15). Kubori_et al._ identified the supermolecular structure termed the “needle complex” (NC) from the Salmonella type III secreton (16), and a similar structure was also identified from_Shigella_ (17, 18), indicating that the type III secreton appears to have universal form. The size of each part of the_Shigella_ NC has been characterized and measured (17); the needle part is 8 nm wide and 45 nm long, and the basal body consists of upper and lower doublet rings with diameters of 15 and 26 nm, respectively, and resembles those of flagella. The height of the basal body is 32 nm, which presumably allows it to traverse both bacterial membranes and the peptidoglycan. The protein components of the NC have been proposed and characterized in both Salmonella (16, 19,20) and Shigella (17, 18). These studies have revealed that in Salmonella, the basal body is composed of PrgH/K and InvG, and the needle part is composed of PrgI/J. In_Shigella_, the basal body and needle part are composed of MxiD/G/J and MxiH/I. MxiG, MxiJ, MxiD, MxiH, and MxiI show sequence similarity to PrgH, PrgK, InvG, PrgI, and PrgJ, respectively.
Although extensive knowledge has been accumulated about the EPEC type III secreton and EspA filament, the supermolecular structure of the type III secreton and its relationship to EspA filament are poorly understood. Interestingly, no EspA homologue was found in components of type III secretons in Salmonella, Shigella, and other pathogens, except for enterohemorrhagic E. coli. In the EspA defect strain, EspB and EspD are still secreted into the culture supernatant (21), but they are not able to translocate both proteins into host cells (10, 14), suggesting that the assembly of the EPEC type III secreton may be somewhat different from those of_Salmonella_ or Shigella, and that EspA may be associated with the type III secreton. We report here the identification and characterization of the EPEC type III secreton and its relationship to EspA.
Materials and Methods
Bacterial Strains and Growth Media.
EPEC was grown in LB broth or DMEM at 37°C. Shigella was grown in LB broth at 37°C. For details of strains and its phenotypes, see Table 1, which is published as supporting information on the PNAS web site, www.pnas.org.
Cloning and Construction of the Nonpolar Mutant.
EPEC escF and espA were cloned into pTrc99A (Amersham Pharmacia) to obtain p99-escF_s and p99-espA, which constitutively produce EspA and EscF and which were used for complementation of espA and_escF mutants. Details of clones and construction of EPEC Δ_escF_ (strain KILS001) can be found in the supporting_Methods_, which are published on the PNAS web site.
Purification of the NCs from EPEC and Shigella.
EPEC and its mutant strains were grown in LB broth overnight at 37°C without shaking, and then overnight cultures were diluted 1:25 in DMEM and incubated for 5 h in a CO2 incubator.Shigella or EPEC was grown in LB broth overnight at 37°C with shaking, and then overnight culture was diluted 1:100 in LB broth and incubated for 2.5 h at 37°C with shaking. Bacteria were then harvested, and the NC was purified by protocols for the partial purification of the Shigella NC (17).
Preparation of Secreted Proteins, SDS/PAGE, and Immunoblotting.
Bacteria grown in DMEM or LB were removed by centrifugation (18,000 × g, 10 min), and proteins in the supernatant were precipitated by the addition of ice-cold trichloroacetic acid at a final concentration of 10% and then incubated on ice for 1 h. After centrifugation, the pellets were resuspended in Laemmli sample buffer and analyzed by SDS/PAGE. For immunoblotting, the proteins were resolved by SDS/PAGE and transferred to polyvinylidene difluoride membranes. EspA, EspB, and EscC were detected with guinea pig polyclonal antibodies against their respective recombinant proteins. Antibodies were purified by affinity chromatography by using respective antigen-immobilized columns.
Infection of Cultured Cells and Immunofluorescence Microscopy.
HeLa cells (105) were seeded and grown overnight on 12-mm round glass coverslips, then infected with bacteria for 3 h. Cells were washed three times with PBS and fixed with 3.0% paraformaldehyde in PBS (pH 7.2), then washed three times with PBS. The fixed cells were permeabilized with 20 μl 0.1% Triton X-100 in PBS in the presence of phalloidin–Texas red (to stain filamentous actin) or antiphosphotyrosine antibodies (4G10, Upstate Biotechnology). Alexa-conjugated anti-mouse IgG and IgM (Molecular Probes) were used as the secondary antibody for antiphosphotyrosine. Stained samples were visualized and photographed as described elsewhere (22).
Hemolysis Assay.
Bacteria were grown in LB broth overnight at 37°C without shaking, and then overnight cultures were diluted 1:25 into DMEM and incubated for 4 h in a CO2 incubator. They were collected by centrifugation and resuspended in fresh DMEM corresponding to 1/10 volume of the culture media. Rabbit blood cells (RBCs) were sedimented, washed three times in PBS, and resuspended with DMEM at 109/ml. Equal volumes of bacteria and RBC suspension were mixed together, and each 100-μl aliquot was poured into round-bottom 96-well plates and incubated at 37°C for 90 min in the CO2 incubator. The bacteria–RBC suspensions were gently resuspended with an additional 150 μl of PBS, and then plates were centrifuged. Supernatants (100 μl) were transferred to a fresh plate, where optical density at 550 nm was measured.
Electron Microscopy.
Samples were negatively stained with 2% phosphotungstic acid, pH 7.3, that contained 0.2% (wt/vol) sucrose on Butval-98 grids and observed under a JEM 1010 transmission electron microscope (JEOL). For immunolabeling of the NC, samples were applied to Butval-98 grids, fixed with 1% formaldehyde in physiological salt solution, immunolabeled with the affinity-purified anti-EspA polyclonal antibody and with 6-nm colloidal gold-conjugated antibodies against guinea pig IgG (Aurion, Wageningen, The Netherlands) at room temperature for 20 min. After further thorough washing, the NC were fixed and stained as described above.
Results and Discussion
Supermolecular Structure of the EPEC Type III Secreton.
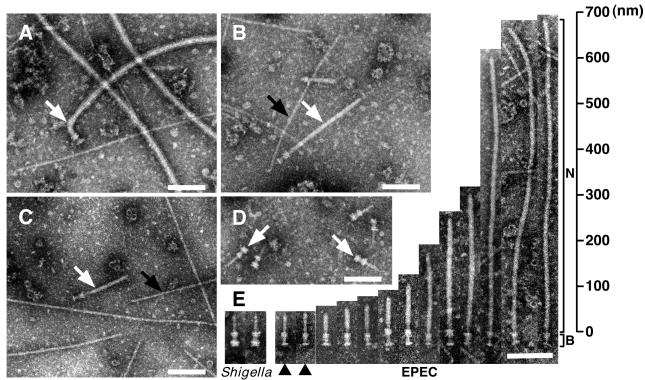
To identify the supermolecular structure of the EPEC type III secreton, EPEC was grown in LB broth, and the NC fraction was prepared, negatively stained, and then observed by transmission electron microscopy (TEM). Although we could detect a supermolecular structure similar to that of the Shigella NC, the major components were flagella complexes (Fig.1A). It has been reported that expression of Esp proteins is affected by environmental conditions such as culture media (23), host body temperature (24), and Congo red (25). Indeed, expression of Esp proteins was induced when EPEC was grown in DMEM but not in LB broth (Fig. 3C). Therefore, the NC fraction was prepared from EPEC wild type (WT) grown in DMEM and observed by TEM (Fig. 1B). On induction in DMEM, the number of flagella complexes was greatly reduced, and the number of NCs with cylindrical symmetry was predominantly increased (white arrow in Fig.1B). Although the basal body of the NC was reminiscent of that of Shigella (arrows in Fig. 1D), the needle was extraordinarily long and thick (Fig. 1B). To exclude the possibility that the long needle may be specific to the E2348/69 strain (serotype O127), NCs were also prepared from B171–8 (serotype O111) and rabbit enteropathogenic E. coli (REPEC) (serotype O103) strains (Fig. 1C and summarized in Table 1). The TEM view clearly showed that NCs from B171–8 and REPEC had the same shape as that of E2348/69. Our preparation techniques were also confirmed by using Shigella flexneri M94 strain (17), and we found the typical Shigella NCs (arrows in Fig. 1D) that had already been characterized elsewhere (17, 18). To further characterize the needle parts, TEM images of 13 particles were aligned, and these structures were compared with those of Shigella (Fig.1E). In contrast to Shigella, the length of the needle varied, and some of the needles were more than 600 nm long (Fig. 1E). Needle elongation has not been observed in Shigella WT, unless the major needle component, MxiH, was overexpressed (17). In this study, we also observed pilus-like structures from different serotypes of EPEC (black arrows in Fig. 1 B and C), and these structures were not observed in Shigella NC fraction (Fig. 1D), indicating that EPEC possesses additional surface appendages that were different from NCs.
Figure 1.
Electron micrographs of negatively stained NC fractions from EPEC and_Shigella_. The NCs were partially purified from EPEC E2348/69 strain grown in LB broth (A), in DMEM (B), and from EPEC B171–8 grown in DMEM (C). (D) Electron micrograph of purified_Shigella_ NC. (E) Alignment of EPEC NCs and comparisons to Shigella. N and B indicate the needle and basal body of EPEC NCs, respectively. Black arrowheads indicate putative immature NCs. White arrows indicate flagellar complexes (A) and NCs (B, C, and_D_). Black arrows indicate pilus-like structures. (Bars = 100 nm.)
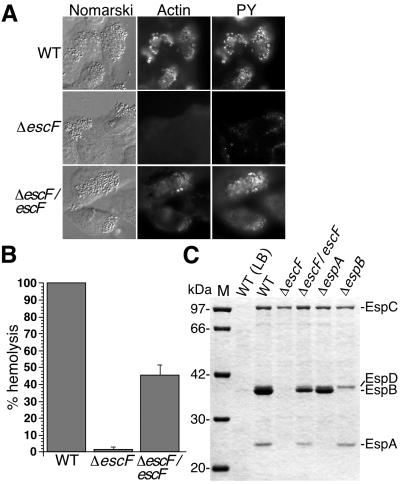
Figure 3.
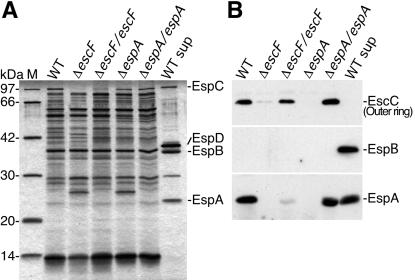
The effect of escF mutation on A/E lesion formation, hemolytic activity, and secretion of Esp proteins. (A) Infected HeLa cells with WT or escF mutant were fixed, and then actin and tyrosine-phosphorylated proteins (PY) were detected by immunofluorescence microscopy. (B) Contact-independent hemolytic activity. EPEC WT and the_escF_ mutant strains were assayed for their ability to hemolyze RBCs. Results from three independent experiments are shown. Error bars, standard deviation. (C) Secreted protein profiles of EPEC WT and escF mutant strains. Bacteria were grown in DMEM or LB broth, and secreted proteins were resolved by 10% SDS/PAGE and stained with Coomassie blue.
EPEC drastically changed the supermolecular structure on the bacterial cell surface when the environmental condition was shifted to an_in vivo_ conditional medium such as DMEM from an in vitro rich medium such as LB media (Fig. 1 A and_B_). These observations may indicate that EPEC has two different phases: (i) a planktonic phase where the flagella complex is used to aid in bacterial swimming; and (ii) a virulent phase where the production of flagella complexes is repressed, and instead the type III secreton is produced to use for the translocation of bacterial effectors into the host cells.
EscF Is Required for A/E Lesion Formation, Hemolytic Activity, and Secretion.
To investigate whether the NC was an EPEC type III secreton, we decided to disrupt a gene encoding EscF. EscF showed homology to the major needle parts of type III secretons in S. typhimurium SPI1 PrgI (24% identity) (19) and Shigella flexneri MxiH (25% identity) (17). In addition, EscF shared homology with other putative type III needle parts of S. typhimurium SPI2 SsaH (35% identity), Pseudomonas aeruginosa PscF (25% identity) (Fig.2A). The escF located on the LEE4 transcriptional unit in EPEC LEE (26,27) (Fig. 2C) was amplified by PCR, and the escF mutant strain was constructed as described in the supporting_Methods_. To analyze whether EscF is involved in A/E lesion formation, HeLa epithelial cells were infected with WT and Δ_escF_, and then cytoskeletal actin and phosphorylated proteins were labeled with phalloidin–Texas red and fluorescently labeled antiphosphotyrosine antibody, respectively (Fig.3A). Infection with WT induced the accumulation of actin and tyrosine-phosphorylated proteins beneath the adherent bacteria. In contrast, infection with the escF mutant did not elicit A/E lesion formation. Complementation of Δ_escF_ with the cloned escF in trans restored the ability to form A/E lesions. These results indicate that the lack of cytoskeletal rearrangements could be attributed to the disruption of_escF_.
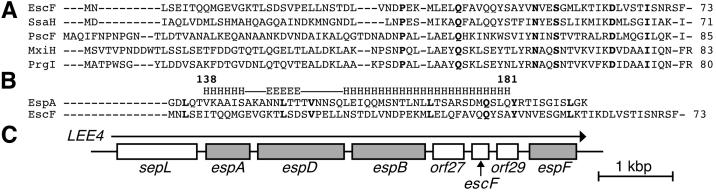
Figure 2.
Sequence analyses of EscF and EspA and a genetic map of the_LEE4_. (A) The sequence analyses of needle parts were performed with blast (National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/). (B) The N terminus of EscF showed homology (13% identity) to the C terminus of EspA (135–190 aa) that contains the coiled-coil region (138–181 aa). The EspA coiled-coil region predicted elsewhere (30) is illustrated (E, β sheet; H, α helix). Note that the leucine-rich heptad repeat of EscF at position Leu3-Leu38 was overlapped with the heptad repeat in EspA coiled-coil region that is required for the A/E lesion formation and EspA-EspA interaction (30). Bold letters indicate identical amino acids. (C) Organization of_esp_ genes and escF in the_LEE4_ transcriptional unit. An arrow indicates the direction of transcription, and gray boxes indicate genes encoding secreted proteins.
A recent study showed that EPEC induces contact-independent hemolysis in RBCs, and that this hemolytic activity depends on the type III secreton (14). To confirm the contribution of EscF to the EPEC-induced hemolysis, we measured the hemolytic activity of the escF mutant and compared its activity to that of WT (Fig. 3B). Hemolytic activity was observed in WT without close contact to RBCs by centrifugation. In contrast, this activity was completely abolished by the escF mutant, and it was partially restored when cloned_escF_ was reintroduced into the escF mutant_in trans_, indicating that EscF is involved in the hemolysis.Yersinia (11, 12) and Shigella (28, 29) induce hemolytic activities, but centrifugation is required for to evoke hemolysis in RBCs. These findings indicate that the contact-independent hemolysis in EPEC infection can probably be attributed to the extraordinary length of the needle indicating the formation of an EspA filament (10). Indeed, EPEC needle parts varied and were longer than those of Shigella (Fig. 1E), thus permitting the elongated needle to be in direct contact with RBCs without centrifugation.
To further investigate the defect caused by escF mutation, EPEC-secreted proteins were analyzed by 12% SDS/PAGE. As shown in Fig. 3C, secretion of EspA, EspB, and EspD was completely blocked by the escF mutation. Complementation of the_escF_ mutant with cloned escF in trans restored secretion of all type III secreted proteins. These results indicate that EscF is required for the secretion of Esp proteins, and this observation agrees with secretion-defect phenotypes of mutant strains of Shigella mxiH (17, 18) and Salmonella prgI (19, 20) that encode major needle components.
Characterization and Assembly of the EPEC NC.
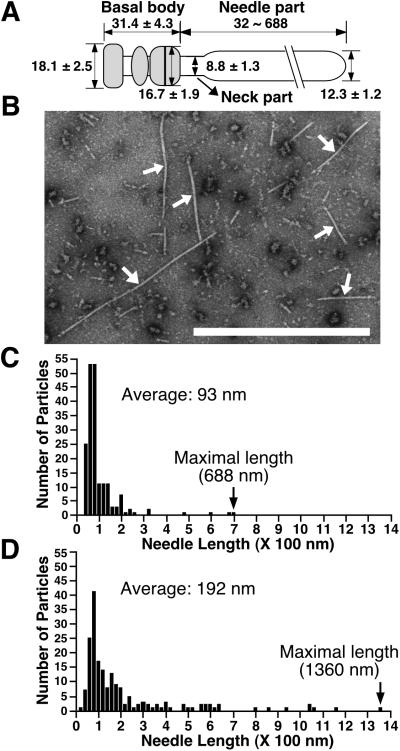
We prepared and measured 34 conserved NCs from EPEC WT, and each part was estimated, as shown in Fig.4A. Although we could not figure out exact sizes of lower and upper rings of the basal body because of the instability in the structure, both rings appeared to be doublet. The widths of the upper and lower rings were estimated to be 16.7 ± 1.9 and 18.1 ± 2.5 nm, respectively, and the height of the basal body was 31.4 ± 4.3 nm. These observations indicate that diameters of the upper and lower rings in EPEC type III secretons are somewhat smaller than those of Shigella (Fig.1E), whereas the height of the EPEC basal body is nearly identical to that of Shigella. In contrast to_Shigella_ and Salmonella, the EPEC needle is associated with unknown components that seemed to form a sheath-like structure (12.3 ± 1.2 nm in width, Fig. 1E). The measured value was wider than the width of the Shigella needle [8.0–8.5 nm; Tamano _et al._ (17) and this study]. Furthermore, minor particles, less than 1% of the total particles, of EPEC NCs appear to have thinner needles (arrowheads in Fig.1E). Indeed, the NCs possessing the thin needles (termed the neck part in this study), which were located between the sheath-like structure and the basal body, are similar in widths to those of Shigella. These findings suggest that the sheath-like structure is associated with the surface of the needle part and that the needle structure is extended from the basal body of the EPEC type III secreton.
Figure 4.
(A) Presumed model of the EPEC NC with the sheath-like structure. Each size is represented in nanometers. The gray area indicates the basal body. (B) Electron micrographs of the negatively stained NC prepared from the espA mutant containing cloned espA. Arrows indicate NCs. (Bar = 1 μm.) Distribution of the needle length in the NCs purified from EPEC WT (C) and the espA mutant containing espA (D).
Next, we prepared NCs from various mutants and observed them by TEM (summarized in Table 1). As expected, the espB mutant did not affect the NC formation. In contrast, NCs were not detected in the type III secretion-defective strain, CVD452 (escN mutant strain). As in the case of CVD452, NCs were not detected in the_escF_ mutant, but NCs were restored when cloned_escF_ was reintroduced into the escF mutant_in trans_. Although the Shigella needle was elongated up to 1 μm when MxiH was overexpressed, we did not observe needle elongation in the escF mutant containing cloned_escF,_ which allows constitutive expression (data not shown). Interestingly, the NC formation was completely abolished by the_espA_ mutant; even secretion of EspB and EspD was not affected by this mutation (Fig. 3C). In contrast, the NC was completely restored when cloned espA was introduced into the_espA_ mutant in trans. To our surprise, the sheath-like structure in this strain appears to be longer than that of the EPEC WT (Fig. 4B). To confirm this observation, we examined the distribution of needle length in 187 randomly chosen NCs from WT (Fig. 4C) and Δ_espA_ containing cloned_espA_ (Fig. 4D). Needle lengths of WT varied, ranging from 32 to 688 nm (average, 93 nm) with a peak at 40–140 nm. Although the peak of the needle length in Δ_espA_ containing cloned espA was similar to that of WT, the needle length was distributed over a wider range, from 32 to 1,360 nm (average, 192 nm), with the maximal size nearly corresponding to the whole bacterial size. For complementation studies of escF and espA mutants, we used the same expression vector and growth conditions. We determined that EspA, but not EscF, may be a major component of the sheath-like structure, because the length of the sheath-like structure appears to be controlled by the amount of EspA.
Component of the NC with the Sheath-Like Structure.
To further analyze the requirement of EspA for NC formation, we analyzed NC fractions by silver staining 12% SDS/PAGE (Fig.5A). The NC fraction prepared from WT, Δ_escF_, and Δ_espA_ appears to contain other components, because unknown pilus-like structures were also observed in this NC fraction (Fig. 1B), and we could not detect drastic changes of NC fractions between WT and mutants. Next, EspA was detected by immunoblotting by using anti-EspA antibodies. Although we detect EspA in the bacterial culture supernatant (Fig.3C), we also detected it in the NC fraction prepared from WT (Fig. 5B). To eliminate the possibility of contamination of bacterial culture supernatants into the NC fraction, immunoblotting was carried out by using antibodies against EspB secreted into culture supernatants. As expected, EspB was not detected in the NC fraction. These results indicate that EspA is specifically localized in the NC fraction. Furthermore, EspA was not detected in the NC fraction prepared from the escF mutant, and the localization of EspA in the NC fraction was partially restored by reintroduction of cloned escF into the escF mutant in trans, indicating that the EspA may be associated with EscF.
Figure 5.
The effect of escF and espA mutations on the NC formation. (A) NC fractions were prepared and analyzed by 12% SDS/PAGE followed by silver staining. (B) Immunoblot analysis of NC fractions from EPEC WT and its mutants. Affinity-purified anti-EspA, -EspB, and -EscC antibodies were used for immunodetection. M and WT sup indicate the size marker and secreted proteins prepared from the EPEC WT, respectively.
Although basal bodies, type III secretons without needles, have been observed in Shigella mxiH (17, 29) and Salmonella prgI (20) mutants, we could not detect basal body structure in_escF_ and espA mutants. To analyze the basal body, we tried to detect EscC by immunoblotting (Fig. 5B). EscC is thought to be one of components of the basal body and showed homology to the Shigella MxiD outer ring. Although EscC was detected in the NC fraction from WT, this protein was greatly decreased in both of the escF and espA mutants, and even MxiD has been detected in the NC prepared from the mxiH mutant (17). Both deficient phenotypes were restored in the reintroduction of cloned_escF_ or espA into their respective strains. These results agreed with our TEM views and explained why we could not detect basal bodies from escF and espA mutants. Collectively, EscF and EspA may affect the assembly and stability of the basal body.
EspA Directly Associates with the NC and Is an Essential Component of the Sheath-Like Structure.
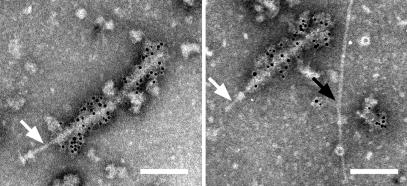
The study cited above suggests that EspA may interact directly with the type III secreton. To confirm this fascinating hypothesis, NCs purified from EPEC WT were stained with immunogold-labeled anti-EspA antibodies and observed by TEM (Fig. 6). The TEM view clearly showed that the sheath-like structure was attached to the tip of the needle part (an arrow in the left column), and the needle beyond the upper ring is observed from the incomplete NC lacking the lower base (an arrow in the right column). To our surprise, only the sheath-like structure was stained with the anti-EspA antibodies. In contrast, the needle, which was observed as the neck part (Fig.1E and an arrow in the left column in Fig. 6), and the needle beneath the upper ring (an arrow in the right column in Fig.6) were not labeled with anti-EspA antibodies. Our TEM view was strongly supported by the finding of Shaw et al. (14), who showed that about 50 nm of the EspA filament close to the bacterial surface was not stained with anti-EspA antibodies. Although we observed an unknown pilus-like structure (black arrow in Fig. 6; Fig. 1 B and C), this did not react to the anti-EspA antibodies. These results clearly indicate that EspA is the major component of the sheath-like structure, which is supported by elongation of the sheath-like structure under constitutive expression of EspA (Fig. 4B).
Figure 6.
Direct association of EPEC type III secreton with EspA. The NC was partially purified from EPEC WT, and EspA was detected with the immunogold-labeled anti-EspA antibodies. Sheath-like structures coated with 6-nm gold particles were observed. Note that the neck (white arrow, Left), the stem beneath the upper ring (white arrow, Right), and the pilus-like structure (black arrow, Right) were not stained with the anti-EspA antibodies.
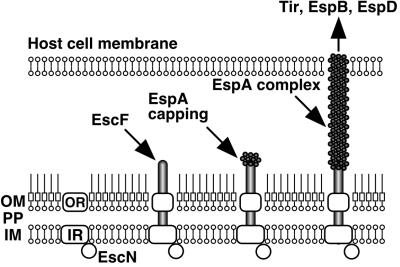
In conclusion, we have shown, to our knowledge for the first time, a direct association of the EPEC type III secreton with EspA. A hypothetical model of the EPEC type III secreton and the sheath-like structure is illustrated in Fig. 7. First, the inner and outer ring components are assembled; a_sec_-dependent pathway probably requires this step, because the typical signal peptide exists in the outer ring component, EscC. Second, EscF is polymerized and assembled into outer and inner rings, and then EspA is secreted via the type III secreton and attached to the tip of the needle. Recently, Delahay et al. revealed that a coiled-coil domain with heptad periodicity is located at the C terminus of EspA (Fig. 2B) (30), and that this domain is required for EspA–EspA interaction. Interestingly, we found that the N terminus of EscF showed homology to the coiled-coil domain of EspA (Fig. 2B). Probable interaction between the EspA-coiled-coil domain and EscF helps the stability of the type III secreton, because the NC was not detected in the espA mutant. Finally, EspA initiates the polymerization from the tip of the needle and assembles the sheath-like structures. The sheath-like structure is expandable, and its elongation is controlled by the amount of EspA (Fig. 4B). The sheath-like structure builds a physical bridge between the bacterium and the host cell membrane, and Tir and other effectors can translocate into host cells.
Figure 7.
Schematic model of the assembly of the EPEC type III secreton and the sheath-like structure. First, inner and outer ring components are assembled, and then EscF is polymerized and assembled into the outer and inner rings. EspA is secreted via the type III secreton and attached to the tip of the needle. Finally, EspA initiates the polymerization and assembles the sheath-like structure. The sheath-like structure is expandable and is controlled by the amount of EspA (Fig.4B). The sheath-like structure builds a physical bridge between the bacterium and the host cell membrane, and Tir and other effectors can be translocated into host cells. OM, outer membrane; PP, periplasm; IM, inner membrane; OR, outer ring; IR, inner ring.
Supplementary Material
Supporting Information
Acknowledgments
We thank Brett Finlay (Biotechnology Laboratory, University of British Columbia, Vancouver, BC, Canada), Jim Kaper (Center for Vaccine Development and Department of Microbiology and Immunology, University of Maryland School of Medicine, Baltimore, MD), and Michael Donnenberg (Division of Infectious Diseases, Department of Medicine, University of Maryland School of Medicine, Baltimore, MD) for providing the E2348/69 derivatives. We also thank Asaomi Kuwae for critical reading for the manuscript and helpful discussions. This work was supported by operating grants from the Research for the Future Program of the Japanese Society for the Promotion of Science and the All Kitasato Project Study.
Abbreviations
EPEC
enteropathogenic E. coli
NC
needle complex
TEM
transmission electron microscopy
WT
wild type
Tir
translocated intimin receptor
A/E
attaching/effacing
References
- 1.Donnenberg M S, Kaper J B. Infect Immun. 1992;60:3953–3961. doi: 10.1128/iai.60.10.3953-3961.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeVinney R, Gauthier A, Abe A, Finlay B B. Cell Mol Life Sci. 1999;55:961–976. doi: 10.1007/PL00013202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moon H W, Whipp S C, Argenzio R A, Levine M M, Giannella R A. Infect Immun. 1983;41:1340–1351. doi: 10.1128/iai.41.3.1340-1351.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenshine I, Donnenberg M S, Kaper J B, Finlay B B. EMBO J. 1992;11:3551–3560. doi: 10.1002/j.1460-2075.1992.tb05438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abe A, Heczko U, Hegele R G, Finlay B B. J Exp Med. 1998;188:1907–1916. doi: 10.1084/jem.188.10.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDaniel T K, Jarvis K G, Donnenberg M S, Kaper J B. Proc Natl Acad Sci USA. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jarvis K G, Giron J A, Jerse A E, McDaniel T K, Donnenberg M S, Kaper J B. Proc Natl Acad Sci USA. 1995;92:7996–8000. doi: 10.1073/pnas.92.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kenny B, Devinney R, Stein M, Reinscheid D J, Frey E A, Finlay B B. Cell. 1997;91:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 9.Jerse A E, Yu J, Tall B D, Kaper J B. Proc Natl Acad Sci USA. 1990;87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knutton S, Rosenshine I, Pallen M J, Nisan I, Neves B C, Bain C, Wolff C, Dougan G, Frankel G. EMBO J. 1998;17:2166–2176. doi: 10.1093/emboj/17.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hakansson S, Schesser K, Persson C, Galyov E E, Rosqvist R, Homble F, Wolf-Watz H. EMBO J. 1996;15:5812–5823. [PMC free article] [PubMed] [Google Scholar]
- 12.Neyt C, Cornelis G R. Mol Microbiol. 1999;33:971–981. doi: 10.1046/j.1365-2958.1999.01537.x. [DOI] [PubMed] [Google Scholar]
- 13.Warawa J, Finlay B B, Kenny B. Infect Immun. 1999;67:5538–5540. doi: 10.1128/iai.67.10.5538-5540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaw R K, Daniell S, Ebel F, Frankel G, Knutton S. Cell Microbiol. 2001;3:213–222. doi: 10.1046/j.1462-5822.2001.00105.x. [DOI] [PubMed] [Google Scholar]
- 15.Hueck C J. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galan J E, Aizawa S I. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 17.Tamano K, Aizawa S, Katayama E, Nonaka T, Imajoh-Ohmi S, Kuwae A, Nagai S, Sasakawa C. EMBO J. 2000;19:3876–3887. doi: 10.1093/emboj/19.15.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blocker A, Jouihri N, Larquet E, Gounon P, Ebel F, Parsot C, Sansonetti P, Allaoui A. Mol Microbiol. 2001;39:652–663. doi: 10.1046/j.1365-2958.2001.02200.x. [DOI] [PubMed] [Google Scholar]
- 19.Kubori T, Sukhan A, Aizawa S I, Galan J E. Proc Natl Acad Sci USA. 2000;97:10225–10230. doi: 10.1073/pnas.170128997. . (First Published August 15, 2000; 10.1073/pnas.170128997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimbrough T G, Miller S I. Proc Natl Acad Sci USA. 2000;97:11008–11013. doi: 10.1073/pnas.200209497. . (First Published September 12, 2000; 10.1073/pnas.200209497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kenny B, Lai L C, Finlay B B, Donnenberg M S. Mol Microbiol. 1996;20:313–323. doi: 10.1111/j.1365-2958.1996.tb02619.x. [DOI] [PubMed] [Google Scholar]
- 22.Finlay B B, Ruschkowski S, Dedhar S. J Cell Sci. 1991;99:283–296. doi: 10.1242/jcs.99.2.283. [DOI] [PubMed] [Google Scholar]
- 23.Kenny B, Abe A, Stein M, Finlay B B. Infect Immun. 1997;65:2606–2612. doi: 10.1128/iai.65.7.2606-2612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abe A, Kenny B, Stein M, Finlay B B. Infect Immun. 1997;65:3547–3555. doi: 10.1128/iai.65.9.3547-3555.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abe A, Nagano H. Microbiol Immunol. 2000;44:857–861. doi: 10.1111/j.1348-0421.2000.tb02574.x. [DOI] [PubMed] [Google Scholar]
- 26.Mellies J L, Elliott S J, Sperandio V, Donnenberg M S, Kaper J B. Mol Microbiol. 1999;33:296–306. doi: 10.1046/j.1365-2958.1999.01473.x. [DOI] [PubMed] [Google Scholar]
- 27.Friedberg D, Umanski T, Fang Y, Rosenshine I. Mol Microbiol. 1999;34:941–952. doi: 10.1046/j.1365-2958.1999.01655.x. [DOI] [PubMed] [Google Scholar]
- 28.Sansonetti P J, Ryter A, Clerc P, Maurelli A T, Mounier J. Infect Immun. 1986;51:461–469. doi: 10.1128/iai.51.2.461-469.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blocker A, Gounon P, Larquet E, Niebuhr K, Cabiaux V, Parsot C, Sansonetti P. J Cell Biol. 1999;147:683–693. doi: 10.1083/jcb.147.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delahay R M, Knutton S, Shaw R K, Hartland E L, Pallen M J, Frankel G. J Biol Chem. 1999;274:35969–35974. doi: 10.1074/jbc.274.50.35969. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information