NanoJ-SQUIRREL: quantitative mapping and minimisation of super-resolution optical imaging artefacts (original) (raw)
. Author manuscript; available in PMC: 2018 Aug 19.
Published in final edited form as: Nat Methods. 2018 Feb 19;15(4):263–266. doi: 10.1038/nmeth.4605
Abstract
Most super-resolution microscopy techniques depend on steps that can contribute to the formation of image artefacts, leading to misinterpretation of biological information. We present NanoJ-SQUIRREL, an ImageJ-based analytical approach that provides quantitative assessment of super-resolution image quality, capable of guiding researchers in optimising imaging parameters. By comparing diffraction-limited images and super-resolution equivalents of the same acquisition volume, this approach generates a quality score and quantitative map of super-resolution defects.
The quality and resolution of super-resolution images is largely dependent on factors including the photophysics of fluorophores used, the chemical environment of the sample, imaging conditions, and analytical approaches used to produce final images [1–5] (Supplementary Note 1). Thus far super-resolution data quality assessment relies on subjective comparison of the data relative to prior knowledge of the expected structures [6,7] or benchmarking of the data against other high-resolution imaging methods like electron microscopy [8]. An exception exists in the Structured Illumination Microscopy (SIM) field [9], where analytical frameworks exist for quantitative evaluation of image quality [10,11].
The simplest, most robust way to visually identify defects in super-resolution images is the direct comparison of diffraction-limited and super-resolved images of the sample. For images that represent the same focal volume, the super-resolution version should provide an improved resolution representation of the reference diffraction-limited one. When this analysis is performed empirically it is subject to human bias and interpretation. Here we present a new analytical approach named SQUIRREL (super-resolution quantitative image rating and reporting of error locations), which allows for quantitative mapping of local image errors thereby providing a framework to assist in their reduction. This is implemented as an easy-to-use open-source ImageJ and Fiji [12] plugin (named NanoJ-SQUIRREL), exploiting high-performance GPU-enabled computing.
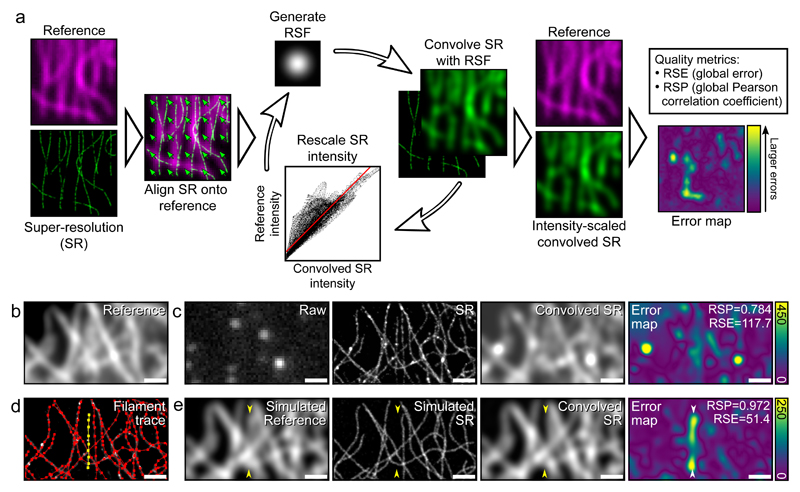
SQUIRREL is formed solely on the premise that a super-resolution image should be a high-precision representation of the underlying nanoscale position and photon emission of the imaged fluorophores. Although based on the principle of comparing conventional and super-resolution images, in contrast to other approaches it requires no prior knowledge of the expected properties of the sample or label. Assuming an imaged field-of-view has a spatially-invariant point spread function (PSF), application of a resolution rescaling transfer function to the super-resolution image should produce an image with a high degree of similarity to the original diffraction-limited one. Variance between these images beyond a noise floor can be used as a quantitative indicator of local macro-anomalies in the super-resolution representation (Fig. 1, Supplementary Fig. 1). The algorithm requires three inputs: a reference image (generally diffraction-limited), a super-resolution image and a representative resolution scaling function (RSF) image. The RSF can be provided by the user or automatically estimated through optimisation (Supplementary Note 2, Supplementary Fig. 2).
Figure 1. Overview of quantitative error mapping with SQUIRREL.
a) Representative workflow for SQUIRREL error mapping. b) Fixed microtubules labelled with Alexa Fluor 647 imaged in TIRF. c) Raw - single frame from raw dSTORM acquisition of structure in b, SR - super-resolution reconstruction of dSTORM data set, Convolved SR - super-resolution image convolved with appropriate RSF, Error map - quantitative map of errors between the reference and convolved SR images. d) SuReSim [13] filament tracing used to generate e, yellow filament is made to be present in reference image but absent in super-resolution image. e) Simulated reference image, super-resolution image, and super-resolution image convolved with RSF and error map. Yellow arrowheads indicate position of yellow filament seen in d. Scale bars = 1 μm. b-d represents data from 1 of 5 independent experiments showing similar results.
The stages involved in error mapping are: 1) Correcting for any analytical or optical spatial offsets between the super-resolution and reference images; 2) iterative estimation of the RSF and linear rescaling coefficients to convert the super-resolution image into its diffraction-limited equivalent (the ‘resolution-scaled image’); 3) calculation of the pixel-wise absolute difference between the reference and resolution-scaled image to generate the final error map (Fig. 1a). In addition to local quality assessment, two global image quality metrics are calculated: the RSE (Resolution Scaled Error), representing the root-mean-square-error between the reference and resolution-scaled image; and the RSP (Resolution Scaled Pearson coefficient), which is the Pearson correlation coefficient between the reference and resolution-scaled images with values truncated between -1 and 1. The RSE is more sensitive to differences in contrast and brightness than the RSP, whereas RSP provides a score of image quality that can be compared across different super-resolution imaging modalities. A full description of the SQUIRREL algorithm is provided in Supplementary Note 3.
To demonstrate the capacity of SQUIRREL to identify defects in super-resolution images, we acquired total internal reflectance fluorescence (TIRF) microscopy images of immunolabelled microtubules (Fig 1b) and a corresponding dSTORM [4] dataset. From these we produced an error map indicating regions of high dissimilarity (Fig. 1c). Regions surrounding filament junctions and overlapping filaments, where the increased local density of fluorophores limits the capacity for single-molecule localisations, were particularly dissimilar. Based on this we generated two simulated optical and photophysical realistic datasets using the SuReSim software [13] (Fig. 1d): a diffraction-limited reference dataset containing all the traced filaments; and a structural artefact dSTORM dataset in which a filament was removed. SQUIRREL analysis of the reference and artefactual super-resolution images produced an error map that clearly highlights the absence of the selected filament (Fig. 1d). These results exemplify the power of SQUIRREL to identify large-scale image artefacts in instances where subjective comparison of the widefield (i.e. Simulated Reference) and super-resolution (i.e. Simulated SR) images would be insufficient.
SQUIRREL is not only sensitive to disappearance of structures. It can also identify common super-resolution artefacts including merged structures and bright aggregates (Supplementary Note 1, Supplementary Fig. 1). The software is not limited to single molecule localisation microscopy (SMLM), and we have demonstrated that for SIM images it provides complementary information to SIMcheck [10] (Supplementary Note 4, Supplementary Fig. 3). Although SQUIRREL cannot highlight errors in the axial direction, Supplementary Note 5 and Supplementary Figs. 4-6 explore how out-of-focus information affects SQUIRREL metrics. For example, using widefield references of thick samples compromises the metrics’ fidelity, which can be minimized by using optical-sectioning systems such as TIRF (Fig. 1, Supplementary Fig. 6), confocal and lattice light sheet microscopes. We estimate that SQUIRREL is capable of accurately identifying 2D image artefacts within a ~600 nm focal volume. The major limitation of SQUIRREL is that small-scale artefacts cannot be identified due to the diffraction-limited reference image. To define this limit we carried out simulations of an 8-molecule ring structure of varying diameter; for typical signal-to-noise ratios encountered in super-resolution microscopy SQUIRREL can quantify image anomalies as small as 150 nm (Supplementary Note 6, Supplementary Fig. 7). This limit is set by the resolution of the reference image, and so using a higher resolution image as the reference (acquired using another super-resolution modality) can provide smaller-scale artefact detection and cross-validation. To demonstrate this, correlative SMLM, SIM, Super-Resolution Radial Fluctuations (SRRF) [14] and Stimulated Emission Depletion (STED) microscopy were performed on vaccinia virus (VACV) lateral bodies [15], structures separated by < 200 nm. Using SQUIRREL to cross-validate different super-resolution techniques, we found that artefacts not discernable using a diffraction-limited reference image were highlighted (Supplementary Note 7, Supplementary Fig. 8).
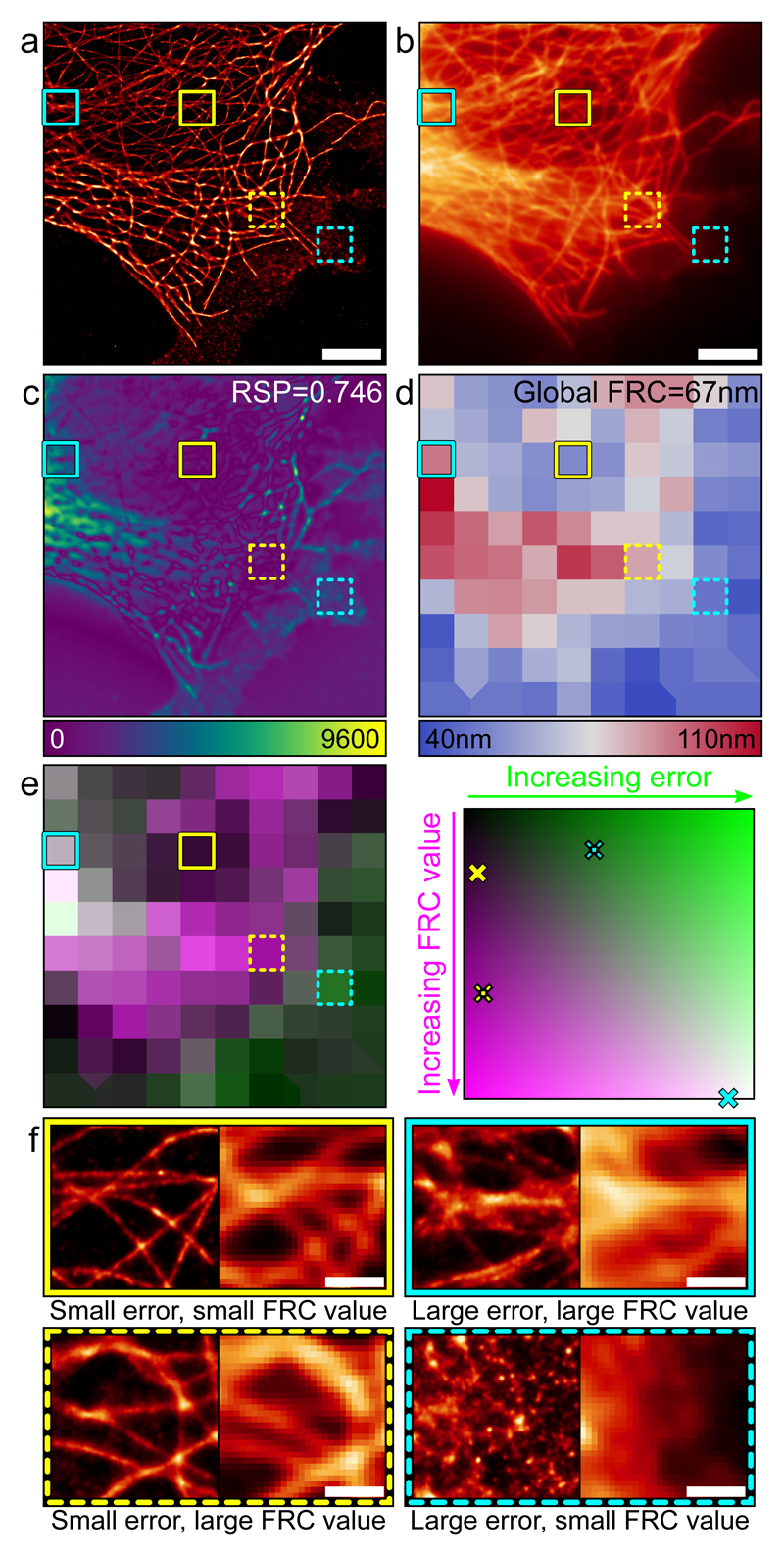
Image resolution is commonly used as a reporter of image quality, although in super-resolution studies these factors weakly correlate [6,7,16]. One popular method for quantifying image resolution in super-resolution and electron microscopy is Fourier Ring Correlation (FRC) [16]. Conventional FRC measurements represent the global resolution of an image; within the NanoJ-SQUIRREL package we have implemented block-wise FRC resolution mapping to provide local resolution measurements (Supplementary Fig. 9). In Fig. 2 we map the local FRC-estimated resolution of a dSTORM dataset and compare it against the SQUIRREL error map. Highlighting various regions of the dataset (Fig. 2a-b), we show that high resolution does not necessarily correlate with low error (Fig. 2c-f). Thus SQUIRREL error mapping allows for direct visual detection of structural anomalies without coupling quality to a description of resolution.
Figure 2. Error mapping and FRC analysis.
a) Super-resolution image of fixed Alexa Fluor 647-labelled microtubules reconstructed via MLE. Scale bar = 5 μm. b) Corresponding TIRF image. Scale bar = 5 μm. c) Error map for super-resolution image in a using b as the reference. d) Local mapping of FRC values for the super-resolution image in a. e) Left: Merge of FRC map (magenta) and error map (green, binned to match FRC map). Right: Map of error-resolution space showing where the four boxed regions are located. f) Enlargements of the super-resolution (left) and widefield (right) boxed regions indicated on panels a-e. Scale bars = 1 μm. Figure represents data from 1 of 5 independent experiments showing similar results.
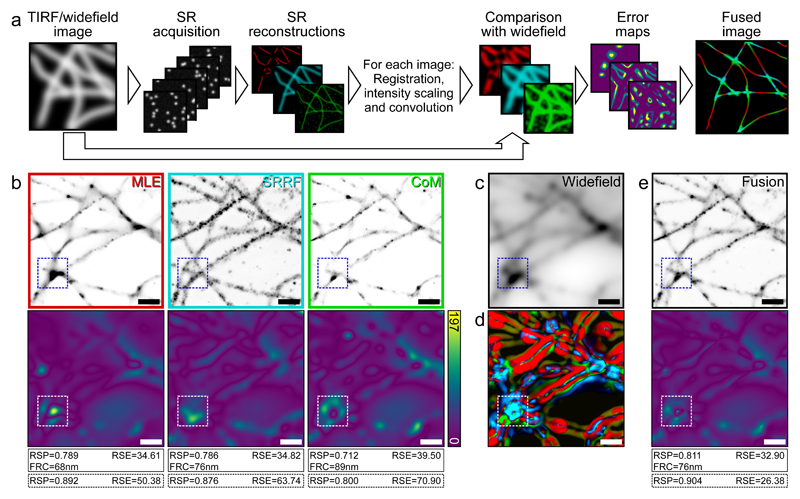
By providing an assessment of image quality, SQUIRREL can be used as a tool to improve various aspects of super-resolution image acquisition. One of these is the choice of analytical method for SMLM image reconstruction. As dozens of high-performing algorithms are available [5] it can be challenging to determine which will be most appropriate for a given dataset. We acquired a dSTORM dataset of immunolabelled microtubules and analysed it using three distinct algorithms: ThunderSTORM using a multi-emitter maximum likelihood estimator engine [17], SRRF, and QuickPALM [18]. SQUIRREL error maps and quality metrics were generated for these three super-resolution images (Fig. 3a,b) using the same diffraction-limited reference image for each (Fig. 3c). In addition to providing the means to ‘rank’ the quality of each reconstruction, the error maps provide spatial details on the local accuracy of each algorithm. By converting these into weights (Fig. 3d, Supplementary Note 8) a new composite image with minimal defects can be generated using the lowest error features of each reconstruction (Fig. 3e). SQUIRREL can also be used to empirically optimise super-resolution images, as exemplified by determining the optimal DNA-PAINT imaging conditions for clathrin-coated pits (Supplementary Note 9, Supplementary Fig. 10), and number of frames for dSTORM imaging of neuronal actin rings along axons (Supplementary Note 10, Supplementary Figs. 11-12).
Figure 3. Image fusion of SMLM data using SQUIRREL.
a) Workflow for generating fused images from different super-resolution images from the same SMLM data set. b) Top row: Three super-resolution images generated from the same dSTORM dataset using different algorithms. MLE = maximum likelihood estimator with multi-emitter fitting, SRRF = super-resolution radial fluctuations, CoM = centre of mass. Bottom row: Corresponding error maps with the widefield image shown in c used as the reference. d) Contributions of different images to the final fused images, colour coded as in the top row of b. e) Top: Fused image, Bottom: Error map of fused image with c again used as the reference image. Values in solid line boxes indicate the quality metrics of the whole images, values in dashed boxes represent quality values from highlighted inset region only. Scale bars = 1 μm. b-e represents data from 1 of 5 independent experiments showing similar results.
In conclusion, SQUIRREL is a quick and easy approach to immediately improve super-resolution data acquisition and quality. Being an open-source ImageJ plugin, NanoJ-SQUIRREL is highly accessible to super-resolution users. We envisage that eventually SQUIRREL will be implemented for continual monitoring of super-resolution image quality during acquisition. By pairing such a feedback approach with automated adaptation of acquisition parameters users could ensure optimal image quality, shorten acquisition times, and reduce data storage requirements.
Online Methods
Super-resolution image simulation with SuReSim
In order to simulate disappearance of a filament from a realistic microtubule network, a real super-resolution image of microtubules (Fig. 1c) was used as a support for SuReSim data simulation. Raw data of blinking Alexa 647-labelled microtubules imaged using TIRF were reconstructed using ThunderSTORM maximum likelihood multi-emitter fitting and then loaded into the SuReSim software and all filaments were traced using the editor function and the WIMP file saved. SuReSim was used to generate a simulated super-resolution reconstruction of all filaments, which was then convolved by a Gaussian PSF to generate a simulated reference image. The object in the WIMP file corresponding to the filament highlighted in Fig. 1d-e was deleted, and SuReSim was used again to render a simulated super-resolution reconstruction, except this time missing a filament. SuReSim was also used for the simulations in Sup. Fig. 1.
Cell lines and primary cells
HeLa cells (Figs. 1-2) were kindly provided by Prof Mark Marsh, MRC LMCB, UCL and cultured in phenol-red free DMEM (Gibco) supplemented with 2 mM GlutaMAX (Gibco), 50 U/ml penicillin, 50 µg/ml streptomycin and 10% fetal bovine serum (FBS; Gibco). CHO cells (Fig. 3) were cultured in phenol red-free Minimum Essential Medium Alpha (MEMα; Gibco) supplemented with 10% FBS (Gibco) and 1% penicillin/streptomycin (Sigma).
Rat hippocampal neurons and glial cells (Sup. Figs. 10-11) were harvested from embryonic day 18 pups, following established guidelines of the European Animal Care and Use Committee (86/609/CEE) and approval of the local ethics committee (agreement D13-055-8), and cultured in Neurobasal medium (Gibco) supplemented with 2 mM GlutaMAX-I (Gibco) and B27 supplement (Gibco). All cells were grown at 37°C in a 5% CO2 humidified incubator.
Sample preparation for fixed microtubules
For TIRF-SMLM imaging of microtubules (Figs. 1-2), 13 mm diameter, thickness #1.5 coverslips were washed overnight in 1:1 HCl:methanol and washed 5 times in ddH2O and twice in 90% isopropyl alcohol. Coverslips were then incubated overnight in poly-L-lysine (0.01%) (Sigma Aldrich) and rinsed twice in PBS. HeLa cells were seeded on these coverslips and grown overnight in 12-well plates. Cells were fixed with 4% PFA in cytoskeleton buffer (10 mM MES, pH 6.1, 150 mM NaCl, 5 mM EGTA, 5 mM glucose, 5 mM MgCl2) for 15 min at 37°C, washed 3x with PBS, then permeabilised with 0.1% Triton X-100 in PBS for 10 min and blocked in 2.5% BSA in PBS for a further 30 min. Samples were then labelled with 2 µg/ml anti-α-tubulin (DM1A mouse monoclonal, Sigma Aldrich) in 2.5% BSA in PBS for 1 hour, followed by 3x washes with PBS and labelling with Alexa Fluor 647-labelled goat anti-mouse secondary antibody (Invitrogen) (2µg/ml in 2.5% BSA in PBS) for 1 hour. Samples were washed 3x with PBS and fixed again in 4% PFA in cytoskeleton buffer for 10 min, before being washed 3x with PBS. Samples were mounted on a parafilm-formed gasket [3] in STORM buffer (150 mM TRIS, pH 8, 10 mM NaCl, 1 % glycerol, 1 % glucose, 1 % BME), sealed with clear nail varnish (Maybelline) and imaged within 3 hours of mounting.
For imaging in different focal volumes (Sup. Fig. 6) COS cells were fixed with glutaraldehyde and labelled with two monoclonal mouse anti-alpha tubulin antibodies (DM1A and B-5-1-2, both from Sigma) and a goat anti-mouse Alexa Fluor-647-labelled secondary antibody (Thermo Fisher Scientific). Samples were mounted in Smart Buffer (Abbelight) for imaging.
For widefield super-resolution imaging of microtubules (Fig. 3), CHO cells were seeded on ultra-clean [3] 8 mm diameter thickness #1.5 coverslips (Zeiss) at a density of 0.1×106 per 35 mm dish. Fixation was performed with 4% PFA in a modified version of cytoskeleton stabilising buffer (CSB) (5 mM KCl, 0.1 mM NaCl, 4 mM NaHCO3, 11 mM Na2HPO4, 2 mM MgCl2, 5 mM PIPES, 2 mM EGTA, pH 6.9) for 15 min at 37°C, followed by washing with the same CSB (without PFA). Additional permeabilization was performed (0.05% Triton X-100 in CSB) for 5 min followed by three washing steps using 0.05% Tween-20 in the modified version of CSB and blocking in 5% BSA (Sigma) for 40 min. Microtubules were stained and submitted to a secondary fixation step as described above. 100 nm TetraSpeck beads (Life Technologies) were added at a dilution of 1:1000 in PBS for 10 min to each coverslip. Coverslips were mounted on clean microscope slides [3] in 100 mM mercaptoethylamine (Sigma) at pH 7.3 and all imaging was performed within 3 hours of mounting.
Fixed microtubule imaging
Fixed microtubule samples were imaged by TIRF-SMLM (Figs. 1-2) on a N-STORM microscope (Nikon Instruments), using a 100× TIRF objective (Plan-APOCHROMAT 100×/1.49 Oil, Nikon) with additional 1.5× magnification. A reference TIRF image was acquired with 5% power 647 nm laser illumination and 100 ms exposure time, before SMLM data acquisition of 40 000 frames at 100% power 647 nm illumination with 405 nm stimulation and an exposure time of 30 ms per frame.
Imaging with different illumination regimes (Sup. Fig. 6) was performed on an N-STORM microscope using a 100x, 1.49NA objective as above, but with no additional magnification and an exposure time of 15 ms. Prior to dSTORM imaging a reference image was acquired using a high-pressure mercury lamp (Intensilight, Nikon) with a Cy5 filter cube (Nikon); the filter cube was then switched and the laser illumination set to either vertical (i.e. widefield), HILO, or TIRF. A second reference image was then acquired, this time with laser illumination. A cylindrical lens was inserted into the detection path and 60,000 frame dSTORM dataset acquired at this angle.
Widefield and super-resolution imaging for fusion (Fig. 3) was carried out on a Zeiss Elyra PS.1 inverted microscope system, using a 100× TIRF objective (PlanAPOCHROMAT 100×/1.46 Oil, Zeiss) and additional 1.6× magnification. The sample was illuminated with a 642 nm laser operating at 100% laser power. 45000 frames were acquired with a 20 ms exposure time per frame.
Reconstruction algorithms for dSTORM data
The freely available software packages ThunderSTORM [17] (Figs. 1-3, Sup. Figs. 6, 8, 10), SRRF (Fig. 3, Sup. Figs. 8, 10) and QuickPALM [18] (Fig. 3, Sup. Fig. 10) were used for super-resolution image reconstruction. Images labelled ‘MLE’ were reconstructed with ThunderSTORM with the integrated PSF method with maximum likelihood fitting and multi-emitter fitting enabled. Drift correction was performed post-localization and images were rendered using a normalized 20 nm Gaussian. Images labelled ‘SRRF’ were analysed with the most appropriate parameters for each individual data set and drift corrected during analysis. Images labelled ‘CoM’ were reconstructed using QuickPALM with the default parameters, following drift correction of the raw data using the NanoJ-SRRF package. The particle tables from QuickPALM were then loaded into ThunderSTORM for rendering using a normalized 20 nm Gaussian.
SIM imaging
For SQUIRREL analysis of SIM images (Sup. Fig. 3), FluoCells prepared slide #2 (Invitrogen) with BPAE cells stained with Texas Red-X phalloidin and Alexa Fluor 488-tubulin was imaged on a Zeiss Elyra PS.1 system, using a 63x NA 1.4 objective with additional 1.6x magnification for SIM and widefield acquisition. For actin imaging, ‘Low SNR’ images were acquired with a 561 nm laser at 0.05 % laser power, using 100 ms exposure time, and 5 grid rotations. ‘High SNR’ images were acquired with a 561 nm laser at 5 % laser power, 100 ms exposure, 5 grid rotations. Widefield images were acquired with a 561 nm laser at 0.2 % laser power, 100 ms exposure time. SIM reconstructions were generated with the Zeiss Elyra Zen software using automatic settings. For microtubule imaging, raw SIM data was acquired with a 488 nm laser at 10 % laser power using 100 ms exposure time and 3 grid rotations. The SIM reconstruction was generated using FairSIM [19].
Generation and analysis of synthetic data at different z-positions
The bead images used in Sup. Fig. 4 were obtained from the open source dataset ‘z-stack-Bead-2D-Exp-as-stack’ available to download from the SMLMS Challenge 2016 website, http://bigwww.epfl.ch/smlm/challenge2016/datasets/Beads/Data/data.html (data used here was downloaded on 4th September 2017). This dataset comprises 151 slices of an image of six fluorescent beads covering the z-range -750 nm to 750 nm (step size = 10 nm). The central x,y location of each of the six beads in this was determined at the central plane of the z-stack, and this was used to define the centre of a 3.3 μm x 3.3 μm region about each bead. For generation of the dataset containing PSFs from all z-positions in each frame, these regions were pasted into an image where the x,y coordinates mapped to a specific z-position from the bead image stack. The target x,y coordinates for pasting the images were spaced such that there was 5 μm between adjacent bead centres, and regions were randomly from the six original bead images. This was repeated 1000 times to generate a 1000-frame dataset. Gaussian-Poisson noise was then added to the image stack to mask the edges of the pasted bead images. This dataset was then analysed with SRRF and ThunderSTORM (default software settings in both cases) to produce a single super-resolution image for each algorithm. The reference image was an average projection of all 1000 frames. For generation of the dataset containing constant z-positions in each frame, regions from the bead z-stack were again selected and pasted but this time z was varied between slices as opposed to the x,y position within each frame. 10 frames were produced for each z-position, and noise was added again as above. ThunderSTORM and SRRF analyses (default settings) were then run on this image stack 10 slices at a time to generate a single super-resolution image for each z-position. The reference was the average of the 10 frames corresponding to z=0 nm.
For assessing the impact of out-of-focus fluorescence on defect detection (Sup. Fig. 5), a test structure was simulated consisting of three semicircles of radius 500 nm and axial tilt ranging from -750 nm to +750 nm. A widefield representation of this structures was produced via convolution with a 3D PSF generated using the ImageJ PSF Generator plugin [20] with the following settings: Born and Wolf optical model, numerical aperture 1.4, wavelength 640 nm, z-range 1500 nm, z-step size 2 nm. Single molecule blinking data sets were generated with custom-written simulation software with the same PSF used for rendering molecule appearances, and were binned into 100 nm ‘camera’ pixels with Gaussian-Poisson noise. This was performed for both the defect-free structure and an artefactual equivalent where 100 nm stretches of the structure had been deleted. These data sets were analysed using weighted-least-squares fitting in ThunderSTORM.
VACV sample preparation and imaging
2.5 x 106 VACV particles (WR strain, EGFP-F18 in tk locus [21]) were diluted in 100 μl 1 mM TRIS, pH 8, sonicated for 3x 30 s and incubated on gridded #1.5 glass-bottom petri dishes (Zell-Kontakt GmbH) for 1 hour at room temperature and fixed with 4 % PFA in PBS for 15 min. Samples were quenched with 50 mM NH4Cl in PBS for 10 min, washed in PBS, and incubated in permeabilization/blocking buffer (1% Triton X-100, 5% BSA, 1% FBS in PBS) for 30 min. Samples were labelled in blocking/permeabilisation buffer overnight at 4 °C or 2 hours at room temperature with anti-GFP nanobodies (Chromotek), labelled in-house with Alexa Fluor 647 NHS-ester (Life Technologies) with a dye-to- protein ratio of approximately 1, as previously described [22]. Samples were then washed 3x with PBS, fixed in 4% PFA in PBS for 10 min, quenched with 50 mM NH4Cl in PBS for 10 min and washed in PBS.
VACV samples were imaged in STORM buffer on a Zeiss Elyra PS.1 system, using a 100x TIRF objective with additional 1.6x magnification (as above) for SIM, SRRF and SMLM acquisition (Sup. Fig. 5). Buffer was exchanged to PBS and STED images were acquired on a Leica SP8, re-localising the same region of interest based on the grid. SMLM data acquisition parameters were 30,000 frames at 100% laser power 647 nm illumination with 405 nm stimulation and an exposure time of 33 ms per frame.
Clathrin coated pits sample preparation and imaging
Rat glial cells (from embryonic day 18 pups) were cultured on 18-mm coverslips at a density of 4000/cm2, respectively. After 9 days in culture, samples were fixed using 4% PFA in PEM (80 mM PIPES, 2 mM MgCl2, 5 mM EGTA, pH 6.8) for 10 min. For PAINT imaging [23] of clathrin coated pits (CCPs) in glial cells, fixed samples were incubated with a rabbit anti-clathrin primary antibody (abCam, catalogue #21679) overnight at 4 °C, then with an anti-rabbit DNA-conjugated secondary antibody (Ultivue) for 1 hour at room temperature.
DNA-PAINT imaging of CCPs in glial cells (Sup. Fig. 10) was performed on a N-STORM microscope using a 100x objective as above. The same glial cell (present in low numbers in hippocampal cultures) was imaged in serial dilutions of Imager-650 (2 mM stock, from lower to higher concentration) in Imaging Buffer (Ultivue). The sample was illuminated at 647 nm (50% laser power) and a sequence of 20,000 images at 33 Hz was acquired for each Imager-650 dilution, before switching to a higher concentration of Imager-650 in Imaging Buffer.
Actin rings sample preparation and imaging
Rat hippocampal neurons (from embryonic day 18 pups) were cultured on 18-mm coverslips at a density of 10,000 After 9 days in culture, samples were fixed using 4% PFA in PEM (80 mM PIPES, 2 mM MgCl2, 5 mM EGTA, pH 6.8) for 10 min. Preparation of actin-stained neurons for SMLM was performed similarly to the protocol described in [24], with minor modifications. After blocking, neurons were incubated with a mouse anti-map2 primary antibody (Sigma Aldrich, catalogue #M4403) for 1h30 at RT, then with a Alexa Fluor 488 labelled donkey anti-mouse secondary antibody (Thermo Fisher) for 45 min at RT, then with 0.5 mM phalloidin-Alexa Fluor 647 (Thermo-Fisher) overnight at 4 °C. Neurons were mounted in a modified STORM buffer (50 mM Tris, pH 8, 10 mM NaCl, 10% glucose, 100 mM mercaptoethylamine, 3.5 U/ml pyranose oxidase, 40 μg/mL catalase) complemented with 0.05 mM phalloidin-Alexa Fluor 647, to mitigate phalloidin unbinding during acquisition and imaged immediately.
Neuron samples were imaged on a N-STORM microscope using a 100x objective as above (Sup. Fig. 11). The sample was illuminated at 100% laser power at 647 nm. A sequence of 60,000 images at 67 Hz was acquired. Images were rendered with ThunderSTORM using a normalized 20 nm Gaussian from particle tables generated with SMAP, a MATLAB based software package developed by the Ries group at the EMBL, Heidelberg. Localizations were determined using a probability based method after background subtraction by wavelet filtering and lateral drift was corrected by cross-correlation.
Visibility analysis
To quantify the quality of the super-resolution reconstructions of parallel actin rings (Sup. Fig. 11i), a normalized visibility similar to that described in Geissbuehler et al. [25] was calculated as follows. Average intensity profiles were plotted for a 0.5 x 1 μm stretch of axon containing 5 actin rings (region shown in Sup. Fig. 11h) for each of the 120 reconstructed images. The MATLAB function findpeaks was used to find the 5 peak positions in the average profile measured from the 60,000 frames reconstruction, and mean pairwise visibility was calculated as follows.
v¯=12∑i=14(Imax,i−Imin,i→i+1Imax,i+Imin,i→i+1+Imax,i+1−Imin,i→i+1Imax,i+1+Imin,i→i+1)
_I_max,i and _I_max,i+1 are the intensities at peak positions i and i + 1 respectively, where i denotes the index of the actin ring in the sampled regions and _I_min,i_→_i+1 is the intensity at the midpoint of two adjacent peaks. Higher visibilities correspond to a greater ability to differentiate between two structures up to a maximum value of v̄ = 0.5.
Colour maps
Colour maps used for displaying images (‘NanoJ-Orange’), error maps (‘SQUIRREL-errors’) and FRC maps (‘SQUIRREL-FRC’) are provided in the NanoJ-SQUIRREL software package.
Software Availability
NanoJ-SQUIRREL can be downloaded and installed in ImageJ and Fiji automatically by following the instructions in the manual, available here: https://bitbucket.org/rhenriqueslab/nanoj-squirrel. Source code is also available at the same website.
Data Availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request. Sample datasets can be downloaded from https://bitbucket.org/rhenriqueslab/nanoj-squirrel.
Supplementary Material
Reporting summary
Supplementary figures
Supplementary notes and figures
Acknowledgements
We thank A. Knight and S. Holden for critical reading of the manuscript. We thank J. Ries at European Laboratory for Molecular Biology (EMBL) Heidelberg for provision of customised MATLAB software and critical reading of the manuscript. K. Tosheva at University College London (UCL) for critical reading of the manuscript and beta testing the software. We thank B. Baum at UCL for reagents. Many of the look-up tables used here are based on the open-source repository of D. Williamson at King's College London. This work was funded by grants from the UK Biotechnology and Biological Sciences Research Council (BB/M022374/1; BB/P027431/1; BB/R000697/1) (R.H. and P.M.P), MRC Programme Grant (MC_UU12018/7) (J.M.), the European Research Council (649101—UbiProPox) (J.M.), the UK Medical Research Council (MR/K015826/1) (R.H. and J.M.), the Wellcome Trust (203276/Z/16/Z) (S.C. and R.H.) and the Centre National de la Recherche Scientifique (CNRS ATIP-AVENIR program AO2016) (C.L.). D.A. is presently a Marie Curie fellow (Marie Sklodowska-Curie grant agreement No 750673). C.J. funded by a Commonwealth scholarship, funded by the UK government.
Footnotes
Author Contributions
S.C. and R.H. devised the conceptual framework and derived theoretical results. S.C., D.A., C.L., J.M. and R.H. planned experiments. S.C. and R.H. wrote the algorithm. Simulations were done by S.C. Experimental data sets were acquired by S.C. (Fig. 1), D.A. (Fig. 2, Sup. Figs. 3, 5, 8), C.J. (Fig. 2, Sup. Fig. 9), P.M.P. (Fig. 3) and C.L. (Sup. Figs. 6, 10, 11). Data analysed by S.C. and D.A. while C.L., J.M. and R.H. provided research advice. The paper was written by S.C., D.A., J.M. and R.H. with editing contributions of all the authors.
Competing Financial Interests
The authors declare no competing financial interests.
References
- [1].Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat Methods. 2011;8:1027–1036. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Almada P, Culley S, Henriques R. PALM and STORM: Into large fields and high-throughput microscopy with sCMOS detectors. Methods. 2015;88:109–21. doi: 10.1016/j.ymeth.2015.06.004. [DOI] [PubMed] [Google Scholar]
- [3].Pereira PM, Almada P, Henriques R. High-content 3D multicolor super-resolution localization microscopy. Methods Cell Biol. 2015;125:95–117. doi: 10.1016/bs.mcb.2014.10.004. [DOI] [PubMed] [Google Scholar]
- [4].van de Linde S, Löschberger A, Klein T, Heidbreder M, Wolter S, Heilemann M, Sauer M. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat Protoc. 2011;6:991–1009. doi: 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]
- [5].Sage D, Kirshner H, Pengo T, Stuurman N, Min J, Manley S, Unser M. Quantitative evaluation of software packages for single-molecule localization microscopy. Nat Methods. 2015;12:1–12. doi: 10.1038/nmeth.3442. [DOI] [PubMed] [Google Scholar]
- [6].Pengo T, Olivier N, Manley S. Away from resolution, assessing the information content of super-resolution images. 2015:1–22. http://arxiv.org/abs/1501.05807.
- [7].Fox-Roberts P, Marsh R, Pfisterer K, Jayo A, Parsons M, Cox S. Local dimensionality determines imaging speed in localization microscopy. Nat Commun. 2017;8 doi: 10.1038/ncomms13558. 13558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–5. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- [9].Gustafsson MG. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc. 2000;198:82–7. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- [10].Ball G, Demmerle J, Kaufmann R, Davis I, Dobbie IM, Schermelleh L. SIMcheck: a Toolbox for Successful Super-resolution Structured Illumination Microscopy. Sci Rep. 2015;5 doi: 10.1038/srep15915. 15915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Förster R, Wicker K, Müller W, Jost A, Heintzmann R. Motion artefact detection in structured illumination microscopy for live cell imaging. Opt Express. 2016;24:22121. doi: 10.1364/OE.24.022121. [DOI] [PubMed] [Google Scholar]
- [12].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Venkataramani V, Herrmannsdörfer F, Heilemann M, Kuner T. SuReSim: simulating localization microscopy experiments from ground truth models. Nat Methods. 2016;13:319–21. doi: 10.1038/nmeth.3775. [DOI] [PubMed] [Google Scholar]
- [14].Gustafsson N, Culley S, Ashdown G, Owen DM, Pereira PM, Henriques R. Fast live-cell conventional fluorophore nanoscopy with ImageJ through super-resolution radial fluctuations. Nat Commun. 2016;7 doi: 10.1038/ncomms12471. 12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cyrklaff M, Risco C, Fernández JJ, Jiménez MV, Estéban M, Baumeister W, Carrascosa JL. Cryo-electron tomography of vaccinia virus. Proc Natl Acad Sci U S A. 2005;102:2772–7. doi: 10.1073/pnas.0409825102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nieuwenhuizen RPJ, Lidke Ka, Bates M, Puig DL, Grünwald D, Stallinga S, Rieger B. Measuring image resolution in optical nanoscopy. Nat Methods. 2013;10:557–62. doi: 10.1038/nmeth.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ovesný M, Křížek P, Borkovec J, Švindrych Z, Hagen GM. ThunderSTORM: A comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30:2389–2390. doi: 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Henriques R, Lelek M, Fornasiero EF, Valtorta F, Zimmer C, Mhlanga MM. QuickPALM: 3D real-time photoactivation nanoscopy image processing in ImageJ. Nat Methods. 2010;7:339–340. doi: 10.1038/nmeth0510-339. [DOI] [PubMed] [Google Scholar]
- [19].Müller M, Mönkemöller V, Hennig S, Hübner W, Huser T. Open-source image reconstruction of super-resolution structured illumination microscopy data in ImageJ. Nat Commun. 2016;7 doi: 10.1038/ncomms10980. 10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kirshner H, Aguet F, Sage D, Unser M. 3-D PSF fitting for fluorescence microscopy: implementation and localization application. J Microsc. 2013;249:13–25. doi: 10.1111/j.1365-2818.2012.03675.x. [DOI] [PubMed] [Google Scholar]
- [21].Schmidt FI, Bleck CKE, Reh L, Novy K, Wollscheid B, Helenius A, et al. Vaccinia virus entry is followed by core activation and proteasome-mediated release of the immunomodulatory effector VH1 from lateral bodies. Cell Rep. 2013;4:464–76. doi: 10.1016/j.celrep.2013.06.028. [DOI] [PubMed] [Google Scholar]
- [22].Albrecht D, Winterflood CM, Sadeghi M, Tschager T, Noé F, Ewers H. Nanoscopic compartmentalization of membrane protein motion at the axon initial segment. J Cell Biol. 2016;215 doi: 10.1083/jcb.201603108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jungmann R, Avendaño MS, Woehrstein JB, Dai M, Shih WM, Yin P. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat Methods. 2014;11:313–8. doi: 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ganguly A, Tang Y, Wang L, Ladt K, Loi J, Dargent B, et al. A dynamic formin-dependent deep F-actin network in axons. J Cell Biol. 2015;210:401–17. doi: 10.1083/jcb.201506110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Geissbuehler S, Sharipov A, Godinat A, Bocchio NL, Sandoz PA, Huss A, et al. Live-cell multiplane three-dimensional super-resolution optical fluctuation imaging. Nat Commun. 2014 doi: 10.1038/ncomms68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reporting summary
Supplementary figures
Supplementary notes and figures
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request. Sample datasets can be downloaded from https://bitbucket.org/rhenriqueslab/nanoj-squirrel.