Toward a Functional Catalog of the Plant Genome. A Survey of Genes for Lipid Biosynthesis (original) (raw)
Abstract
Public databases now include vast amounts of recently acquired DNA sequences that are only partially annotated and, furthermore, are often annotated by automated methods that are subject to errors. Maximum information value of these databases can be derived only by further detailed analyses that frequently require careful examination of records in the context of biological functions. In this study we present an example of such an analysis focused on plant glycerolipid synthesis. Public databases were searched for sequences corresponding to 65 plant polypeptides involved in lipid metabolism. Comprehensive search results and analysis of genes, cDNAs and expressed sequence tags (ESTs) are available online (http://www.canr.msu.edu/lgc). Multiple alignments provided a method to estimate the number of genes in gene families. Further analysis of sequences allowed us to tentatively identify several previously undescribed genes in Arabidopsis. For example, two genomic sequences were identified as candidates for the palmitate-specific monogalactosyldiacylglycerol desaturase (FAD5). A candidate genomic sequence for 3-ketoacyl-acyl-carrier protein (ACP) synthase involved in mitochondrial fatty acid biosynthesis was also identified. Biotin carboxyl carrier protein (BCCP) in Arabidopsis is encoded by at least two genes, but the most abundant BCCP transcript so far has not been characterized. The large number (>165,000) of plant ESTs also provides an opportunity to perform “digital northern” comparisons of gene expression levels across many genes. EST abundance in general correlated with biochemical and flux characteristics of the enzymes in Arabidopsis leaf tissue. In a few cases, statistically significant differences in EST abundance levels were observed for enzymes that catalyze similar reactions in fatty acid metabolism. For example, ESTs for the FatB acyl-ACP thioesterase occur 21 times compared with 7 times for FatA acyl-ACP thioesterase, although flux through the FatA reaction is several times higher than through FatB. Such comparisons may provide initial clues toward previously undescribed regulatory phenomena. The abundance of ESTs for ACP compared with that of stearoyl-ACP desaturase and FatB acyl-ACP thioesterase suggests that concentrations of some enzymes of fatty acid synthesis may be higher than their acyl-ACP substrates.
During the last several years, a large quantity of cDNA and genomic sequences from plants have been produced by several major sequencing projects. As of September 1999, 92.4 Mb of genomic DNA sequences, comprising more than 70% of the Arabidopsis genome, are available in public databases from the Arabidopsis Genome Sequencing Project (Kotani et al., 1997, 1998; Nakamura et al., 1997; Bevan et al., 1998; Kaneko et al., 1998; Sato et al., 1998; http://genome-www.stanford.edu/Arabidopsis/agi.html). The entire genomic sequence will likely be completed by the year 2000 (Ecker, 1998). An important step in gene analysis is the study of corresponding cDNAs. This approach is greatly facilitated by expressed sequence tag (EST) projects in Arabidopsis, rice, and other species aimed at establishing an inventory of expressed genes (Höfte et al., 1993; Newman et al., 1994; Sasaki et al., 1994; Cooke et al., 1996; Yamamoto and Sasaki, 1997).
As of August 5, 1999, 40,737 Arabidopsis ESTs had been deposited in dbEST, which, together with the known full-length cDNA sequences, correspond to approximately 56% of the estimated 21,000 Arabidopsis genes (Bevan et al., 1998). The number of available ESTs provided by the Rice Genome Research Program is comparable to that of Arabidopsis, and recent projects on maize, soybean, and tomato are also accumulating large public EST databases. Smaller but very important plant EST projects have yielded several thousand ESTs from poplar (Sterky et al., 1998), Brassica napus (Park et al., 1993), castor (van de Loo et al., 1995), Brassica campestris (Lim et al., 1996; Kwak et al., 1997), maize (Keith et al., 1993; Shen et al., 1994), and loblolly pine (Allona et al., 1998). Taken together, the number of EST clones for plant species from the dbEST as of September 1999 exceeds 160,000.
One of the major challenges that plant biologists face will be to identify the functions of the thousands of new genes discovered by sequencing. DNA sequence data are accumulating so rapidly that their processing lags behind. For example, on September 9, 1999, more than 27 million bp of completed Arabidopsis genomic sequences were not annotated (http://genome-www.stanford.edu/Arabidopsis/agi.html). Furthermore, almost all annotation and/or gene identification of genomic and EST sequence data is performed automatically by similarity comparisons to previously identified genes. Although such annotations provide an initial clue toward gene identity and function, often the results are incorrect or misleading (Rouze et al., 1999). More complete and valuable information about each gene can be obtained if sequences are examined more thoroughly by a number of additional criteria. Usually these analyses are best performed by researchers who have knowledge of the biology or metabolism underlying a putative gene function.
In the study reported here, we have examined the publicly available sequence databases for information on 65 polypeptides involved in plant glycerolipid synthesis. Our results provide several examples of how additional useful information can be “mined” from detailed considerations of available sequence data and how this approach can extend the value of sequence databases. Extension of such studies to complete plant genomes will eventually result in construction of a sequence/biological database in which the genes, cDNAs, ESTs, and amino acid sequences will be sorted according to their function and linked to metabolic maps and information on expression levels, tissue specificity, and subcellular localization. To our knowledge, no attempts have been made in the public domain to construct such a comprehensive database for plants, although for other organisms, including Escherichia coli and Caenorhabditis elegans, attempts are under way (Karp et al., 1999; http://genome.cornell.edu/cgi-bin/WebAce/webace?db=celegans).
In addition to the identification and classification of new genes, a second major dividend that can emerge from large-scale EST sequencing is information on relative gene expression levels. Because most of the >160,000 plant EST sequences in GenBank are derived from non-normalized cDNA libraries, the number of ESTs for a given gene will in general reflect the abundance of mRNA for that gene in the population used to prepare the library. Comparing the numbers of EST clones for enzymes in a biosynthetic pathway may also provide an insight into possible transcriptional or other control mechanisms. To a large extent, our understanding of the regulation of glycerolipid biosynthesis is based on studying the pools of final lipid products and intermediate metabolites (Browse et al., 1986; for review, see Ohlrogge and Jaworski, 1997). However, in many cases it is difficult or unreliable to quantitate enzyme expression in vivo directly (either activity or concentration), and studies comparing the expression or activity of more than a few enzymes are rare. Although the concentrations of mRNAs (which are reflected in numbers of ESTs) and their protein products can be different due to post-translational regulation, mRNA, and protein turnover, etc., it is reasonable as a first approximation to assume that for the majority of genes, mRNA concentration correlates with protein abundance. In this study, we compared EST abundance values with knowledge of pathway fluxes and catalytic efficiencies. Our survey revealed some unexpected differences between the abundance of ESTs involved in glycerolipid biosynthesis, which may provide new clues concerning genetic control of this primary metabolic pathway.
MATERIALS AND METHODS
The database survey and sequence analysis were accomplished using personal computers and Challenge L multiprocessor (Silicon Graphics Inc., Mountain View, CA). The software used included GCG Sequence Analysis Software (Wisconsin Package Version 10.0, Genetics Computer Group, Madison, WI), Lasergene Sequence Analysis Software for Macintosh and Windows (DNASTAR, Madison, WI), and Clustal X (http://www.eur.nl/FGG/CH1/software.html). Prediction of protein localization sites in cells was based on the program PSORT (http://psort.nibb.ac.jp:8800/). Chloroplast transit peptides were predicted using the ChloroP V1.0 server for chloroplast transit peptide prediction (Center for Biological Sequence Analysis, The Technical University of Denmark, Lyngby; http://www.cbs.dtu.dk/services/). Prediction of gene coding regions in Arabidopsis genomic sequences was done with GENSCAN software, which is available online at http://bioweb.pasteur.fr/seqanal/interfaces/genscan.html.
To retrieve nucleotide and amino acid sequences for glycerolipid biosynthesis genes, we initially did text searches in the GenBank database with enzyme names using the NCBI Internet server (National Center for Biotechnology Information, Bethesda, MD) at www.ncbi.nlm.nih.gov. For most enzymes and proteins we were able to find plant cDNA or genomic sequences that we picked as representative examples and used for further sequence-based searches. For those proteins for which clearly identified plant sequences were unavailable, amino acid sequences of different origin (animal, yeast, or bacterial) were selected. The resulting collection of amino acid sequences was used to search GenBank using the BLASTP and TBLASTN programs at NCBI to retrieve additional plant sequences for glycerolipid biosynthesis genes (Altschul et al., 1997). The genes identified from all searches were grouped according to their biochemical function and for each enzyme or polypeptide the entries were organized alphabetically according to the Latin name of the host plant (links from table IV at the website). The resulting list of annotated entries (GenBank files) was used as a framework for the catalog of plant lipid ESTs that are now available from a number of plants. The results of both EST and GenBank searches are available online (http://www.canr.msu.edu/lgc).
GenBank contains a large and rapidly growing number of genomic sequences from Arabidopsis. Data are organized in large files spanning the sequences of separate BAC (bacterial artificial chromosome) or P1 clones. Because these files are inconvenient to work with, we extracted the segments of sequences and annotations for the lipid biosynthesis genes and linked these abridged files to the table IV list of genes (CATALOG link in the website). Some of these BAC sequence files in GenBank do not have any annotations, many identifications of Arabidopsis genes are only putative, and in a few cases are erroneous. Therefore, we did BLAST searches for each sequence of interest from the Arabidopsis genome sequencing project and saved the top part of the BLAST output in an abridged file together with our identification. In a number of cases for non-annotated genes, we added PSORT, ChloroP, and GENSCAN online software prediction results, as well as alignments of nucleotide and encoded amino acid sequences. All of the errors and experimental artifacts that we detected in the GenBank files were not erased but, rather, were highlighted and annotated at our website.
To obtain “digital northern” analysis, we were particularly interested in accurate comparison of the abundance of ESTs for lipid biosynthesis genes in Arabidopsis and rice and took every possible precaution to avoid potential errors. Several aspects of the available EST data could potentially confound such an analysis. First, although in most cases ESTs are sequenced from the 5′ end of the cDNA clone, some EST clones were sequenced from both ends, and the corresponding sequences are in separate files in dbEST. To identify such cases, the records of individual EST files were inspected, and all clone IDs were retrieved and displayed in the catalog. For the purpose of comparing EST abundance, a clone sequenced from both 5′ and 3′ ends was considered only once. Second, a relevant comparison of EST abundance can be made only if clones originate from non-normalized cDNA libraries and the resulting EST data are not normalized. Unfortunately for our purposes, the consortium of laboratories in France after a certain time chose to select and deposit in dbEST only non-redundant sequences (Cooke et al., 1996). Although we list at our website all lipid biosynthesis ESTs, including those from France, in our “digital northern” analysis we do not include the French ESTs in the considerations of relative EST abundance.
Finally, to avoid redundant sequencing of abundant clones, the cDNA library used for the EST project at the Michigan State University was prescreened with EST clones for photosystem II chlorophyll a/_b_-binding (CAB) protein (T13913, clone 43D8T7), CAB binding protein (T14135, clone 47H3T7), ADP, ATP carrier protein (T14153, clone 48B2T7), heat shock protein (T13873, clone 42F9T7), Fru-bisphosphate aldolase (T04477, clone 36C5T7), elongation factor TU (T04453, clone 34F5T7), catalase (T04280, clone 35F2T7), tonoplast intrinsic protein (T04167, clone 23H5T7), NADH-ubiquinone oxidoreductase (T04342, clone 38C2T7), tonoplast intrinsic protein (T04259, clone 34E6T7), glutathione _S_-transferase (T13961, clone 44C2T7), and Gly rich protein (T13960, clone 44C1T7) (T. Newman, personal communication). We did not search databases with these sequences, so the relative EST abundances estimated in our study are not influenced by the fact that the cDNA library has been prescreened (although absolute levels may be influenced very slightly).
Estimation of Gene Family Size
Approximately 20% of the genes in Arabidopsis are estimated to be members of gene families (Bevan et al., 1998). By examining alignments of EST and genomic sequences, it was often possible for us to assess the number of genes in gene families. In many cases, multiple alignments of ESTs and genes were done in several steps. At the first step, BLAST search results identified the full-length sequences that were most similar to particular ESTs. In many cases, different members of gene families were identified by different top hits in the BLAST searches. Next, these full-length amino acid or cDNA nucleotide sequences (including “cDNA” sequences derived from genomic sequences by intron deletion) were aligned using Clustal X software. Finally, the amino acid sequences deduced from EST sequences were manually aligned to resulting aligned sequences by superimposing the alignments from the BLAST search output. In some cases, additional frame shifts were introduced manually to optimize alignments.
Finally, we took effort in this study not to include sequences that may represent contamination of plant sequencing projects with fungal or bacterial DNA. It is important not to overestimate the significance of new sequences identified by similarity to genes from evolutionarily remote species, particularly from bacteria and fungi. By surveying Arabidopsis genomic sequences, we identified a number of sequences with high similarity to fatty acid biosynthesis genes in fungi and bacteria (e.g. GSSes B78504, B77149, AL094535, and B73815). However, because plants most likely do not have type I fatty acid synthase (FAS) of the fungal type and a sequence in the end of Arabidopsis BAC clone T27J3 (B73815) is 84% identical to Emericella nidulans FAS, we suspect that some of these sequences represent contamination with non-plant DNA.
RESULTS AND DISCUSSION
A Catalog of Sequences for Plant Fatty Acid and Glycerolipid Synthesis
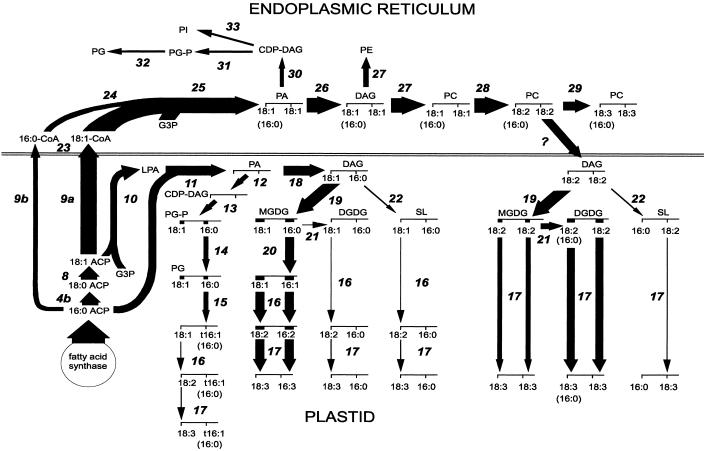
Glycerolipids are essential components of biological membranes in all living organisms. The major reactions of plant fatty acid and glycerolipid synthesis are summarized in Figure 1. This figure represented the starting framework for our analysis of the public DNA and protein sequence databases. Each reaction in Figure 1 has been numbered to provide a reference to Table I and the catalog at the website. In plants, at least 30 enzymatic reactions have been identified that produce the major glycerolipids from acetyl-CoA. Plastidial enzymes are responsible for the biosynthesis of fatty acids from the precursor acetyl-CoA. After fatty acid synthesis, the pathway bifurcates into two different branches, one occurring in the endoplasmic reticulum (“eukaryotic” pathway) and the other in the plastid (“prokaryotic” pathway), leading to different products (Fig. 1). Some reactions are common to both compartments, whereas other are compartment specific. Reviews on this topic have been recently published (Kinney, 1994; Ohlrogge and Browse, 1995; Töpfer et al., 1995; Harwood, 1996).
Figure 1.
Abbreviated diagram of glycerolipid synthesis in Arabidopsis leaves. Widths of the arrows show the relative fluxes through different reactions. Adapted from Browse and Somerville (1991) with permission of Annual Reviews. Numbers of reactions correspond to those in Table I of the present paper and in tables 1 and 4 of the website. Abbreviations for the lipid structures: fatty acids, X:Y, a fatty acyl group containing X carbon atoms and Y cis double bonds; t16:1, hexadec-3trans-enoic acid; G3P, glycerol-3-phosphate; LPA, 1-acyl-glycerol-3-phosphate; PA, phosphatidic acid; DAG, dyacylglycerol; CDP-DAG, cytidine-5′-diphosphate-diacylglycerol; PG, phosphatidylglycerol; PG-P, phosphatidylglycerol-3-phosphate; MGDG, monogalactosyldiacylglycerol; DGDG, digalactosyldiacylglycerol; SL, sulfoguinovosyldiacylglycerol; PC, phosphatidylcholine; PI, phosphatidylinositol; PE, phosphatidylethanolamine.
Table I.
Estimated size of lipid metabolism gene families in Arabidopsis and rice deduced from analysis of available sequencesa
| Gene Product | Arabidopsis | Rice | ||
|---|---|---|---|---|
| Min | Max | Min | Max | |
| Glycerolipid biosynthesis enzymes and proteins | ||||
| 1. ACCases (EC 6.4.1.2) | ||||
| 1a. Homomeric acetyl-CoA carboxylase | 2 | 2 | 1 | 2 |
| 1b. Heteromeric acetyl-CoA carboxylase BC subunit | 1 | 1 | 0 | 0 |
| 1c. Heteromeric acetyl-CoA carboxylase BCCP subunit | 2 | 2 | 0 | 0 |
| 1d. Heteromeric acetyl-CoA carboxylase α-CT subunit | 1 | 1 | 0 | 0 |
| 1e. Heteromeric acetyl-CoA carboxylase β-CT subunit | 1 | 1 | 0 | 0 |
| 2. Acyl carrier proteins (ACP) | ||||
| Plastidial isoforms | 5 | 5 | 5 | 5 |
| Mitochondrial isoforms | 3 | 3 | 1 | 1 |
| 3. Malonyl-CoA:ACP transacylase (EC 2.3.1.39) | 1 | 1 | n.d.b | n.d. |
| 4. Ketoacyl-ACP synthase (KAS) (EC 2.3.1.41) | ||||
| 4a. KAS I | 1 | 1 | 1 | 1 |
| 4b. KAS II | 1 | 1 | 1 | 1 |
| 4c. KAS III | 1 | 1 | 1 | 1 |
| 4d. Putative mitochondrial KAS | 1 | 1 | n.d. | n.d. |
| 5. Ketoacyl-ACP reductase (EC 1.1.1.100) | 1 | 1 | n.d. | n.d. |
| 6. 3-hydroxyacyl-ACP dehydrase (EC 4.2.1.17) | 2 | 2 | n.d. | n.d. |
| 7. Enoyl-ACP reductase (EC 1.3.1.44) | 1 | 1 | n.d. | n.d. |
| 8. Stearoyl-ACP desaturase (EC 1.14.99.6) | 4 | 4 | 4 | 9 |
| 9. Acyl-ACP thioesterase (EC 3.1.2.14) | ||||
| 9a. FatA | 2 | 2 | 1 | 1 |
| 9b. FatB | 1 | 1 | 2 | 3 |
| 10. Glycerol-3-phosphate acyltransferase (EC 2.3.1.15) | 1 | 1 | n.d. | n.d. |
| 11. 1-Acyl-sn-glycerol-3-phosphate acyltransferase (EC 2.3.1.51) | 1 | 1 | n.d. | n.d. |
| 12. Plastidial cytidine-5′-diphosphate-diacylglycerol synthase (EC 2.7.7.41) | Unknown | |||
| 13. Plastidial phosphatidylglycerophosphate synthase (EC 2.7.8.5) | 4 | 4 | 1 | 1 |
| 14. Plastidial phosphatidylglycerol-3-phosphate phosphatase (EC 3.1.3.27) | Unknown | |||
| 15. Phosphatidylglycerol desaturase (palmitate specific) (FAD4) (EC 1.14.99.-) | Unknown | |||
| 16. Plastidial oleate desaturase (FAD6) (EC 1.14.99.-) | 1 | 1 | 1 | 1 |
| 17. Plastidial linoleate desaturase (FAD7/FAD8)(EC 1.14.99.-) | 2 | 2 | n.d. | n.d. |
| 18. Plastidial phosphatidic acid phosphatase (EC 3.1.3.4) | n.d. | n.d. | n.d. | n.d. |
| 19. Monogalactosyldiacylglycerol synthase (EC 2.4.1.46) | 3 | 3 | 1 | 3 |
| 20. Monogalactosyldiacylglycerol desaturase (palmitate-specific) (FAD5) (EC 1.14.99.-) | Unknown | |||
| 21. Digalactosyldiacylglycerol synthase (EC 2.4.1.184) | 1 | 2 | 2 | 2 |
| 22. Sulfolipid biosynthesis protein | 1 | 1 | 1 | 1 |
| 23. Long-chain acyl-CoA synthetase (EC 6.2.1.3) | 18 | 25 | 5 | 18 |
| 24. ER glycerol-3-phosphate acyltransferase (EC 2.3.1.15) | Unknown | |||
| 25. ER 1-acyl-sn-glycerol-3-phosphate acyltransferase (EC 2.3.1.51) | 4 | 4 | 1 | 1 |
| 26. ER phosphatidic acid phosphatase (EC 3.1.3.4) | 2 | 2 | 4 | 4 |
| 27. Diacylglycerol cholinephosphotransferase (EC 2.7.8.2) | 2 | 2 | 1 | 2 |
| 28. ER oleate desaturase (FAD2) (EC 1.14.99.-) | 1 | 1 | 1 | 2 |
| 29. ER linoleate desaturate (FAD3) (EC 1.14.99.-) | 1 | 1 | 1 | 2 |
| 30. ER cytidine-5′-diphosphate-diacylglycerol synthase (EC 2.7.7.41) | 5 | 5 | 2 | 3 |
| 31. ER phosphatidylglycerophosphate synthase (EC 2.7.8.5) | 1 | 5 | n.d. | n.d. |
| 32. ER phosphatidylglycerol-3-phosphate phosphatase (EC 3.1.3.27) | Unknown | |||
| 33. Phosphatidylinositol synthase (EC 2.7.8.11) | 3 | 3 | 1 | 2 |
| 34. Acyl-CoA:diacylglycerol acyltransferase (EC 2.3.1.20) | 1 | 1 | 1 | 1 |
| Other enzymes and proteins involved in lipid metabolism | ||||
| Acyl-ACP desaturases other than stearoyl-ACP desaturase (EC 1.14.99.-) | No indication of presence | |||
| Related to linoleoyl desaturase (EC 1.14.99.-) | No indication of presence | |||
| Δ8 Sphingolipid desaturase (EC 1.14.99.-) | 2 | 2 | n.d. | n.d. |
| Oleate 12-hydroxylase | No indication of presence | |||
| Bifunctional oleate 12-hydroxylase:desaturase | No indication of presence | |||
| Δ12 Fatty acid acetylenase | No indication of presence | |||
| Δ12 Fatty acid epoxygenase | No indication of presence | |||
| Diacylglycerol kinase (EC 2.7.1.107) | 8 | 9 | 1 | 1 |
| Cholinephosphate cytidylyltransferase (EC 2.7.7.15) | 5 | 5 | 1 | 3 |
| Similar to acyl-CoA desaturase (EC 1.14.99.-) | 5 | 6 | n.d. | n.d. |
| Choline kinase (EC 2.7.1.32) | 7 | 9 | 2 | 4 |
| Phospholipase C (EC 3.1.4.11) | 10 | 11 | 3 | 5 |
| Phospholipase D (EC 3.1.4.4) | 17 | 20 | 5 | 9 |
| Phosphatidylserine decarboxylase (EC 4.1.1.65) | 2 | 2 | n.d. | n.d. |
| Phosphatidylinositol-3-kinase (EC 2.7.1.137) | 1 | 1 | 1 | 1 |
| Phosphatidylinositol-4-kinase (EC 2.7.1.67) | 3 | 3 | 2 | 2 |
| Ketoacyl-CoA synthase (KCS) | 21 | 26 | 4 | 8 |
| 3-Ketoacyl reductase (involved in wax biosynthesis) | 3 | 3 | 2 | 3 |
| Wax synthase | 9 | 9 | n.d. | n.d. |
| Possible aldehyde decarbonylase CER1 involved in wax biosynthesis | 2 | 2 | 1 | 1 |
| Putative transcription factor CER2 involved in wax biosynthesis | 2 | 2 | 1 | 1 |
| CER3 protein involved in wax biosynthesis | 1 | 1 | 1 | 2 |
| Oleosin | 7 | 6 | 5 | 6 |
| 3-Ketoacyl-CoA thiolase (EC 2.3.1.16) | 7 | 7 | 5 | 7 |
| Acyl-CoA dehydrogenase (EC 1.3.99.3) | 3 | 3 | 2 | 6 |
| Enoyl-CoA hydratase (EC 4.2.1.17) | 1 | 1 | n.d. | n.d. |
| Acyl-CoA oxidase (EC 1.3.3.6) | 5 | 5 | 2 | 4 |
Our search of public databases with text strings representing each enzyme shown in Figure 1 resulted in a list of 34 enzymes and proteins involved in plant glycerolipid biosynthesis arranged according to reactions in the pathway (Table I). In addition, Table I includes a number of other genes of glycerolipid or fatty acid synthesis that are less central to membrane lipid synthesis or are less well understood. We used this table as a framework for the catalog of genes, cDNAs, and ESTs. The online table summarizes information on over 2,600 clones or sequences for 65 polypeptides involved in plant glycerolipid synthesis.
To retrieve EST sequences, we searched the dbEST database using BLASTN or TBLASTN with nucleotide or amino acid sequences from a number of plant species (Altschul et al., 1997). In the course of this work we observed that searches in dbEST are prone to errors that are mostly due to structural similarities between different genes, experimental errors that remain unnoticed, and the fragmentary nature of EST data. In some cases, only putative identification of an EST is possible, and biological common sense rather than similarity scores can be used to make assignments. To facilitate the handling of many hundreds of annotated sequences identified by GenBank accession numbers, we developed a particular format for the sequence catalog. The core of the catalog is the list of enzymes and proteins involved in metabolic reactions (Table I).
For each enzyme, the sequence entries are organized alphabetically according to the Latin name of the plant species. For each species, all sequences from the GenBank and dbEST are listed with codes indicating the type of sequence (cDNA, genomic, BAC, GSS, or EST). Each entry in the catalog is linked by a hyperlink to a sequence file allowing fast and convenient access (by a single click of the mouse button) without browsing through directory structure, copying and pasting, or memorizing any accession numbers. Although these sequence files are basically GenBank files we found it extremely inconvenient and time consuming to access the sequences by retrieving them directly from GenBank every time. Retrieval time at the plant lipid dedicated website is at least 10-fold faster. Moreover we modified or annotated all retrieved EST files and a number of GenBank files. After retrieval from dbEST, every EST sequence has been compared again with the GenBank sequences using BLASTX or BLASTN search algorithms. This was necessary to unambiguously identify ESTs belonging to gene superfamilies and made it possible to detect and sort out a number of false positives and experimental artifacts. As just one example, several ESTs gave “hits” with the 5′ untranslated region of the homomeric acetyl-CoA carboxylase (ACCase) cDNA in Arabidopsis (D36630, gene _ACC_1). When checked by BLASTN in GenBank, they appeared to be 100% identical to the coding region of an unrelated Arabidopsis gene. Moreover, the corresponding sequence from D36630 is missing from the BAC sequence AC006228, which contains the _ACC_1 gene. Thus, the sequence in D36630 is chimeric and the mentioned ESTs are irrelevant to ACCase. The results of these comparisons and our annotations were saved for each EST in the sequence file containing the original text of the GenBank EST file, the top part of BLAST search output (including the most important alignments), and the date of the BLAST search.
How Complete Is the Database of DNA Sequences for Plant Lipid Pathways?
Examination of the catalog indicates that of the 65 enzymes and proteins of glycerolipid synthesis we examined, DNA sequences or putative DNA sequences are publicly available for 60 of these genes. This high proportion in large part reflects the success of high-throughput DNA sequencing and genomic approaches over the past several years. For the following five reactions: (a) phosphatidylglycerol palmitate desaturase (FAD4), (b) monogalactosyldiacylglycerol palmitate desaturase (FAD5), (c) endoplasmic glycerol-3-phosphate acyltransferase, (d) plastidial and endoplasmic phosphatidylglycerol-3-phosphate phosphatase, and (e) plastidial cytidine-5′-diphosphate-diacylglycerol synthase, no ESTs or genes were identified by our searches. In the case of FAD4 and FAD5, the lack of identified clones is most likely because these genes cannot be distinguished from other desaturases in the database. However, our detailed examination of desaturase sequences has very likely identified strong candidate genomic and EST sequences for the FAD5 desaturase (see below). We were able to find EST representatives for all enzymes and proteins of plant lipid metabolism with known sequences, indicating that the currently available number of EST sequences is large enough to represent essentially all known members of this metabolic pathway.
Discovery of Candidate Genes for FAD5
Genes or clones have previously been identified for six of the eight acyl desaturase reactions shown in Figure 1. However, FAD4 and FAD5 are so far identified only by the existence of putative mutations in these reactions. One of the most intriguing differences in the abundance of ESTs between rice and Arabidopsis concerns a gene family that is most similar to animal and fungal acyl-CoA desaturases. There are 10 independent ESTs in Arabidopsis, four in tomato and two in Brassica, but this sequence class is completely missing from rice. At this time, the precise function of these desaturase-like genes remains unknown. However, further analysis suggests that members of this family encode acyl-lipid desaturases, which likely include palmitate-specific monogalactosyldiacylglycerol desaturase (FAD5).
In animals and yeast, this class of desaturase genes encodes palmitoyl and stearoyl Δ9 desaturases, which use acyl-CoA (or possibly acyl-lipids) as substrate (Thiede et al., 1986; Stukey et al., 1989, 1990). However, desaturation at the Δ9 position in higher plants is catalyzed by stearoyl-ACP desaturases and the currently established pathway of higher plant glycerolipid biosynthesis does not involve any other steps of Δ9 desaturation. Thus, it is possible that enzymes encoded by this gene family are responsible for another reaction(s) similar to desaturation of palmitoyl and stearoyl at the Δ9 position.
The most obvious candidates for these reactions would be desaturation of palmitoyl at the Δ7 position on monogalactosyldiacylglycerol or at the Δ3 trans position on phosphatidylglycerol, reactions that have been associated with the Arabidopsis mutations fad4 (Δ3) and fad5 (Δ7) (Browse et al., 1985; Kunst et al., 1989). Because desaturation of palmitate at Δ3 and Δ7 in higher plants occurs in the chloroplasts, the corresponding enzymes should be synthesized as precursors with chloroplast transit peptides. We did multiple alignment analysis of amino acid sequences for all cDNA, genomic, and EST clones of higher plants similar to Δ9 acyl-CoA desaturases (website: see “Similar to Acyl-CoA Desaturase”).
Surveying genomic Arabidopsis sequences that have not been annotated, we identified a tandem of genes in chromosome 3 that encodes two proteins similar to animal and fungal acyl-CoA and acyl-lipid desaturases (AB017071). When aligned with similar proteins, these sequences, which we named putative FAD5.1 and FAD5.2, have N-terminal extensions recognized as chloroplast transit peptides by ChloroP software. The fad5 mutation has been mapped in chromosome 3 at 28 ± 6 cM from locus gl1 (Hugly et al., 1991). Clone MSJ11 (AB017071) with the putative gene FAD5 is located 27.9 cM from the clone with GL1 and in the same relative position to locus tt5 (http://genome-www3.stanford.edu/cgi-bin/AtDB/SEQmap?chr=3&beg=19&end=24æ). Thus, it is likely that we have identified genomic sequences that are strong candidates for plastidial palmitate-specific monogalactosyldiacylglycerol desaturase.
Putative gene FAD5.1 does not have ESTs in Arabidopsis. However, searching dbEST with the amino acid sequence of the protein encoded by FAD5.1, we found two similar EST sequences in Brassica that overlap with the predicted transit peptide (L38104 and H07631). Thus, in other cruciferous plants, protein products of similar genes are likely to be imported into chloroplasts. Gene FAD5.2 in Arabidopsis is represented by EST clones 145O4 (AI099992 and T76134) and 122A14 (R87006). Both clones appear to be full-length and have the sequence corresponding to the predicted transit peptide. We also identified ESTs similar to animal and fungal acyl-CoA and acyl-lipid desaturases with potential chloroplast transit peptides from tomato and Chlamydomonas reinhardtii.
Finally, we note that occurrence of the transcripts similar to Δ9 acyl desaturases in Arabidopsis, Brassica, tomato, and C. reinhardtii and their apparent absence in rice correlate with our biochemical understanding of desaturation patterns of acyl residues in the sn-2 position of MGDG in these species. All species containing these sequences are considered “16:3 plants” and carry out Δ7 desaturation of palmitate, as opposed to many other angiosperms, including rice (Mongrand et al., 1998), which are “18:3” plants and lack this reaction. The ESTs similar to Δ9 acyl desaturases are clearly associated with the 16:3 phenotype. Thus, based on four criteria: sequence similarity to acyl-CoA desaturases, presence of transit peptides, chromosomal location, and association with the 16:3 phenotype, our survey of public databases has provided strong evidence for the function of one of the previously unidentified members of the desaturase gene family in plants.
Discovery of Candidate 3-Ketoacyl-ACP Synthase Involved in Mitochondrial Fatty Acid Biosynthesis
Fatty acids in plants are synthesized primarily in plastids but also in mitochondria. Our knowledge about the mitochondrial enzymes involved in this pathway is still very limited. Mitochondrial ACP has been characterized from Arabidopsis (Shintani and Ohlrogge, 1994). Biochemical studies of pea leaf mitochondria showed the presence of all enzymes required for de novo fatty acid synthesis from malonate (Wada et al., 1997), but none of the enzymes involved have been cloned or characterized.
By surveying Arabidopsis genomic sequences, we have identified a candidate gene for the previously undescribed mitochondrial 3-ketoacyl-ACP synthase. Gene T1O3.5 in GenBank is annotated as encoding KASII. However, we believe that gene T1O3.5 codes for a mitochondrial KAS based on two criteria. First, the corresponding amino acid sequence does not have a chloroplast transit peptide and is recognized by PSORT algorithm as a mitochondrial protein. Second, in multiple alignments of KAS sequences (website: see reaction 4d), the amino acid sequence of T103.5 groups with bacterial KAS sequences and putative fungal and animal KAS enzymes rather than with the known plant plastidial KAS II. Thus, this is an example where automated annotation has most likely incorrectly identified a gene and more close analysis of the sequence can improve the annotation. Furthermore, it is likely that our survey has identified for the first time a candidate gene for an enzyme of fatty acid biosynthesis in plant mitochondria. Of course, additional experimental work will be needed to verify this hypothesis.
Statistically Significant Differences Occur in EST Abundance for Members of the Glycerolipid Pathway
The very large number (>160,000) of publicly available plant ESTs provides a new opportunity to compare the relative expression levels of large numbers of genes via a “digital northern.” Previous studies of gene expression of members of the glycerolipid biosynthetic pathway (e.g., by northern or western blots) have been limited to examination of one or only a few members. No comprehensive comparative analyses are currently available for the many genes in the glycerolipid synthesis pathway.
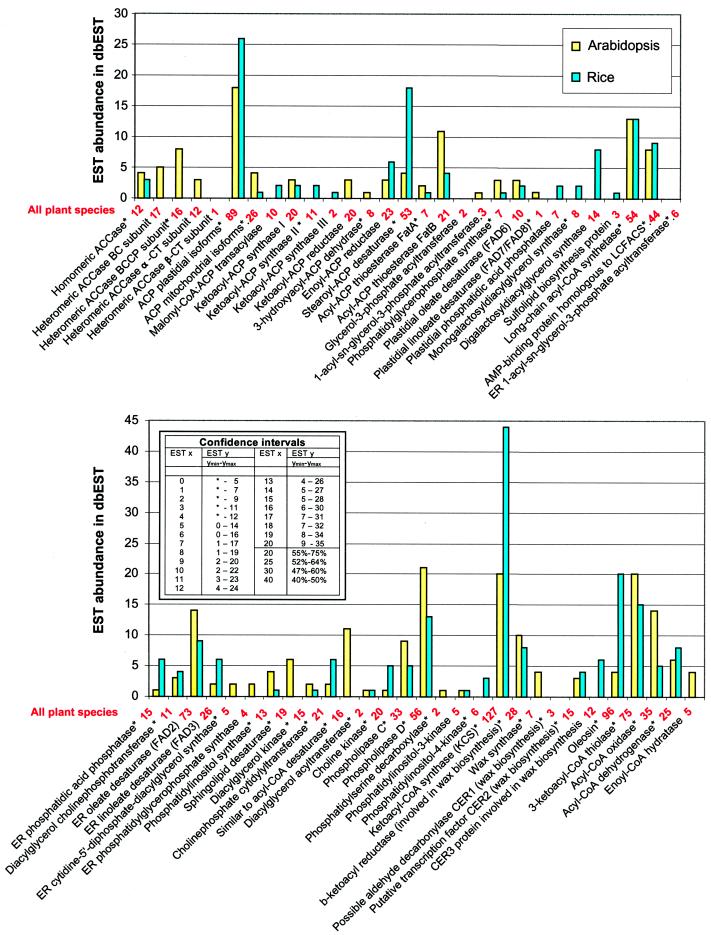
The results of the “digital northern” analysis of EST abundance for 59 genes involved in lipid biosynthesis are presented in Figure 2. Although the data in dbEST represent EST surveys from many species and tissues, most of the reactions of glycerolipid synthesis occur in all tissues and species. Therefore, pooling these data is useful for those reactions that are constitutive or “housekeeping.” However, for some reactions, there can be tissue or species specificity. For example, triacylglycerol synthesis is largely confined to seeds and ESTs for its synthesis might be expected to be underrepresented in the cDNA libraries currently included in dbEST. The histogram (Fig. 2) presents results separately for Arabidopsis and rice and includes the combined results from all plant species reported in dbEST. In our survey, the two genes with the highest number of ESTs found for a single gene in a single plant species are FIDDLEHEAD in rice (30 ESTs) and _FAD_2 in Arabidopsis (14 ESTs). For proteins encoded by gene families, higher numbers are often observed; for example, 26 and 18 in the case of plastidial ACP in rice and Arabidopsis, respectively.
Figure 2.
EST abundance for lipid metabolism enzymes and proteins in Arabidopsis and rice in dbEST as of August 1999. EST numbers for all plant species are shown in red beneath the x axis. Enzymes and proteins encoded by gene families are marked with asterisks. The inset shows 95% confidence intervals in the differences between EST numbers. For a given number of ESTs for a protein shown in the first column, the second column shows the number of ESTs immediately outside the confidence interval (first significantly different values) for the 95% confidence levels (based on table I from Audic and Claverie [1997]). For example, if one protein is represented by 20 ESTs, then for another protein, EST numbers ≤9 or ≥35 are considered different at the 95% confidence level. Proteins listed in Table I but not expected in Arabidopsis and rice, (Legend continues on facing page.)e.g. Δ12 fatty acid epoxydase as well as proteins for which sequences are unknown are not included in this comparison. Putative Arabidopsis wax synthase ESTs (4) may correspond to a hypothetical gene located immediately upstream of the wax synthase gene cluster.
To interpret differences in EST numbers such as those shown in Figure 2, the question of statistical significance is of utmost importance. In a recent study, Audic and Claverie (1997) addressed this issue and established a rigorous significance test for identifying differentially expressed genes by comparing relative abundance of ESTs. The probability that two EST numbers are different due to random variation depends only on the numbers of ESTs themselves and does not depend directly on the number of clones sequenced. Our conclusions concerning differences in expression of genes involved in lipid metabolism are based on the confidence intervals for a 95% significance level (Audic and Claverie, 1997). According to this analysis the surveyed lipid biosynthesis genes shown in Figure 2 can be roughly divided into two classes with respect to their EST abundance. The first class is represented by numerous genes (and gene families) for which the observed numbers of ESTs do not differ significantly from 0. For a 95% confidence level, the first value that is significantly different from 0 is 5. Thus, digital northern analysis does not allow conclusions about differential expression of the genes with numbers of ESTs from zero to four—statistically speaking, they are expressed at the same basal level. For all plant ESTs, 10 of the 59 genes shown in Figure 2 fall into this first class. The second class of genes is represented by five or more ESTs. It can be concluded that many mRNAs of the second class are synthesized at a level higher than those in the first class. For further conclusions, pairwise comparisons are necessary and the significance of such comparisons can be assessed by reference to table I of Audic and Claverie (1997), or the inset of Figure 2. Furthermore, as detailed below, several other conclusions can be made from analysis of these data.
The Abundance of ESTs May Reveal Previously Undescribed Regulation
Many of the reactions of fatty acid and glycerolipid synthesis can be considered a linear sequential metabolic pathway with similar flux through the reactions. If each of these enzymes has a similar catalytic efficiency, then, to a first approximation, the level of expression of each enzyme might be similar. Figure 2 reveals that the expression levels of genes for glycerolipid synthesis enzymes in many cases differ significantly. Some of these differences are easy to rationalize in terms of our understanding of the biochemistry of the pathway. For example, to produce an 18-carbon fatty acid, KAS I must catalyze seven condensation reactions, compared with only one for KAS III or KAS II. Thus, our expectation that KAS I would be more abundant than either KAS II or KAS III was met by the relative abundance of ESTs for these three enzymes (20, 11, and 2, respectively [Fig. 2]).
In other cases we would expect higher levels of expression for enzymes with lower catalytic efficiency. For example, purified stearoyl-ACP desaturase has a specific activity of approximately 1 μmol min−1 mg−1 compared with approximately 2,000 for malonyl-CoA:ACP transacylase. Thus, although flux through malonyl-CoA:ACP transacylase (10 ESTs in all plant species) must be at least 7-fold greater than through stearoyl-ACP desaturase (53 ESTs), the much different catalytic efficiency associated with these enzymes can explain why the relative abundance of ESTs is much higher for the desaturase than for the acyltransferase. These general correlations between EST abundance values and our biochemical expectations validate the concept that ESTs provide a useful first estimate of in vivo mRNA levels. A second type of independent evidence that supports the validity of digital northerns comes from comparison of microarray signal intensity with EST abundance data. When data for microarray signals for genes are compared with their abundance in dbEST, a positive correlation is observed (data not shown; Loftus et al. [1999]).
Despite the overall correlation noted above between EST numbers and biochemical expectations, Figure 2 reveals some statistically significant differences in EST levels that do not correlate with our current understanding of flux or catalytic efficiencies in the glycerolipid pathways. For example, FatA and FatB represent acyl-ACP thioesterases responsible for hydrolysis of primarily oleoyl-ACP and palmitoyl-ACP, respectively (Voelker, 1996). In most plant tissues, and as shown in Figure 1, the flux through the FatA reaction is 4- to 8-fold higher than through FatB. Both enzymes carry out chemically the same hydrolysis reaction and expression of both enzymes in Escherichia coli leads to similar specific activities (Dörmann et al., 1995). However, the abundance of ESTs is significantly higher for FatB (21) than for FatA (7), and these results are the opposite of our expectations based on flux through the pathway. This exception to the general patterns described above may provide the first clue that FatB expression or activity is under regulatory control, such as differential mRNA or protein stability, in addition to gene transcription rates. Of course, the reasons for the discrepancy between enzyme activity/flux data and relative EST numbers can be merely technical; for example, RNA secondary structures might be an obstacle for reverse transcriptase and, therefore, such EST data can be only an indication of a possible regulatory mechanism.
FAS Proteins
Acetyl-CoA Carboxylase
In most plants a multisubunit, heteromeric ACCase is confined to plastids and produces malonyl-CoA used for fatty acid biosynthesis from C4 to C18. Three subunits of the heteromeric ACCase, namely biotin carboxylase (BC), biotin carboxyl carrier protein (BCCP), and the α-subunit of carboxyltransferase (α-CT), are encoded in the nuclear genome, whereas the β-subunit of carboxyltransferase (β-CT) is encoded in the chloroplast genome (Sasaki et al., 1995). The subunits probably occur in stoichiometric amounts in the complex. Therefore, it was of interest to determine if the number of ESTs for each subunit reflects the predicted stoichiometry. We found that, indeed, EST abundance suggests coordinate expression of ACCase subunits in Arabidopsis. The α-CT, BC, and BCCP subunits are represented by 12, 10, and 12 ESTs, respectively, in all plant species. The β-CT subunit is encoded in the chloroplast genome and therefore its message is not polyadenylated. For this reason, β-CT is not found among Arabidopsis ESTs and no conclusion can be made from available EST data concerning its expression.
Closer examination of the BCCP sequences indicates an interesting example of how additional insights can sometimes be mined from dbEST. In Arabidopsis, only one BCCP gene (CAC1) has been detected by Southern hybridization and described in any detail (Choi et al., 1995) (U23155). In Brassica napus developing embryos, six different cDNA sequences were identified that are similar to the Arabidopsis CAC1 gene (Elborough et al., 1996). These sequence data made it possible to assign BCCP function to two previously unidentified Arabidopsis ESTs (T21716 and T43109) (Elborough et al., 1996). We found nine putative BCCP ESTs from Arabidopsis that are different from CAC1. The high sequence identity indicates that they are likely transcribed from a single gene (H37386, T43109, H37396, N38652, R64960, R90694, T21716, AA395831, and H76183). Strikingly, there is only one EST from the _CAC_1 gene (Z25714). The consensus BCCP sequence of the second type encodes a polypeptide that is 72% identical to B. napus BCCP pBP4 (Elborough et al., 1996) and 68% identical to Arabidopsis CAC1.
The ChloroP V1.0 server for chloroplast transit peptide prediction recognizes the polypeptide encoded by the contig for the putative BCCP as a chloroplast protein with a potential transit peptide 55 amino acids long. Significant similarity with B. napus BCCP and the presence of a putative chloroplast transit peptide strongly suggest that these nine ESTs represent a second, more abundant BCCP in Arabidopsis. Northern-blot analysis in B. napus indicated that the BCCP clone isolated from a developing embryo cDNA library and similar to the second type of Arabidopsis BCCP is highly expressed in the embryos but very weakly expressed in the leaves (at least 24 times less according to the number of clones isolated from leaf and embryo libraries). Thus, it is likely that in Arabidopsis there are at least two BCCP genes, one expressed in the green tissues at a low level (CAC1), the other in developing seeds or other tissues at a higher mRNA level. The difference in EST numbers for the two BCCP genes in Arabidopsis is significant at the 95% confidence level. This second type of BCCP remains basically undescribed in Arabidopsis, and it is an intriguing possibility that alternative forms of BCCP are related to the regulation of plastidial ACCase activity.
De Novo Fatty Acid Biosynthetic Enzymes and Acyl-ACP
The relative abundance of the eight to 10 enzymes of plastidial fatty acid biosynthesis has never been assessed by a common technique. Although easily dissociable in vitro, it has been suggested that these enzymes are associated in vivo in some more organized form (Roughan and Ohlrogge, 1996). However, almost no information is available regarding the stoichiometric ratios of the component enzymes. The data in Figure 2 suggest that the mRNA for the enzymes of plastidial fatty acid synthesis are present at much different molar ratios. One of the most abundant classes of ESTs that we observed represents plastidial ACPs with 89 ESTs in all plant species. This finding met our expectations based on its protein abundance (Kuo and Ohlrogge, 1984) and because ACP participates in many reactions. The EST level of plastidial ACP is significantly higher than that of most of the FAS enzymes. However, the ACP pool in plastids represents a complex mixture of dozens of individual acyl-ACP species, and each species constitutes only 0.1% to 5% of the total ACP pool (Post-Beittenmiller et al., 1991).
Consideration of relative ACP and enzyme EST levels leads to the surprising conclusion that in several cases the concentrations of acyl-ACP substrates may be lower than that of the enzymes that act on them. For stearoyl-ACP desaturase and acyl-ACP thioesterase (FatB), the available EST data indicate either higher or similar levels of enzyme relative to its acyl-ACP substrate. We therefore conclude that for a number of FAS enzymes in plants, the concentrations of the corresponding acyl-ACP substrates may be very close to or even lower than the concentrations of enzymes. Such a conclusion can be independently supported in the case of stearoyl-ACP desaturase.
The total ACP pool in chloroplasts has been estimated to be 8 μm (Ohlrogge et al., 1979) and 18:0-ACP is 3% to 5% of the total ACP (or 240–400 nm) (Post-Beittenmiller et al., 1991; Roughan, 1997), while stearoyl-ACP desaturase was estimated to constitute 0.1% to 0.3% of total protein both by enzyme purification data (McKeon and Stumpf, 1982) and by the relative number of clones in a cDNA library (Shanklin and Somerville, 1991). Based on these data and the stearoyl-ACP desaturase molecular mass, the enzyme concentration is 140 to 420 nm. Thus, two independent approaches indicate a similarity in concentration between stearoyl-ACP desaturase and its substrate. An additional independent support of our observation can be found in the RNA hybridization data obtained with microarray technology and publicly available online (Ruan et al., 1998; http://www.monsanto.com/Arabidopsis). In microarray analysis of leaves, flowers, and roots, two plastidial ACP genes were found to be expressed at a similar level compared with stearoyl-ACP desaturase. This observation of a similar concentration of substrates and enzymes is consistent with recent suggestions of channeling of acyl-ACP intermediates in the plastidial FAS machinery (Roughan and Ohlrogge, 1996).
Gene Families
In higher plants, many proteins and enzymes are encoded by gene families, and in Arabidopsis, it has been estimated that 20% of genes belong to members of gene families (Bevan et al., 1998). The existence of gene families can sometimes reflect additional levels of genetic control or isoforms of proteins with specific functions. Therefore, it was of interest to dissect potential gene families in the glycerolipid pathway by comparing genomic sequences and ESTs. We used multiple sequence alignment to determine the potential number of genes encoding individual enzymes of lipid metabolism. Table I presents a summary of results of our multiple sequence analysis, and detailed results for Arabidopsis and rice are presented online in the multiple alignment files linked to the gene catalog. Overall, there are 29 alignments for Arabidopsis and 12 for rice. EST data can either underestimate the gene numbers in families, because some genes are not represented by ESTs, or they can overestimate when short EST sequences from the same gene do not overlap with other partial sequences in a family. In Table I, minimum and maximum numbers are estimated based on these considerations.
Of the 59 proteins surveyed in Arabidopsis, more than half (39) are associated with more than one gene. Thus, for the glycerolipid primary metabolic pathway, gene families are more common than for Arabidopsis genes in general. With one or two exceptions, the most abundant EST classes of the 59 proteins surveyed are represented by gene families. Notable examples of enzymes or proteins with high EST numbers that are transcribed from several genes include: plastidial ACP, stearoyl-ACP desaturase, long-chain acyl-CoA synthetase, phospholipase D, ketoacyl-CoA synthase, and a putative acyl desaturase similar to animal and fungal acyl-CoA desaturases. Thus, within the genes surveyed, there is a general correlation between EST abundance and the existence of gene families. An obvious exception is the endoplasmic reticulum oleate desaturase (FAD2), which in Arabidopsis is the most highly represented single glycerolipid biosynthesis gene (14 ESTs, or 3.7 per 10,000 transcripts). Thus, in agreement with Okuley et al. (1994) mRNA for the FAD2 gene is relatively abundant.
EST abundance can vary significantly for different members of a gene family and may indicate gene specific function or differential expression (spatial, temporal, or inducible). Perhaps the most striking example of a complex gene family is the ketoacyl-CoA synthases (KCS). This large gene family is defined by sequence similarity to the first gene cloned and characterized—_FATTY ACID ELONGATION_1 (_FAE_1) (James et al., 1995). In Arabidopsis, the fae1 mutation results in greatly reduced levels of very long chain fatty acids (VLCFA) in seeds, and the elongation activities from C18 to C22 are reduced (Kunst et al., 1992). Our analysis of KCS sequences indicates that there are at least 21 genes related to _FAE_1 that belong to the KCS family in Arabidopsis. There are 16 full-length sequences available in GenBank, mostly from the Arabidopsis Genome Sequencing Project. Seven of the genes, including _FAE_1, do not have ESTs. Why are there so many KCS genes? VLCFAs play a number of diverse and critical roles in plants, including wax biosynthesis (and are thus involved in cuticle formation and development of epidermis), sphingolipid biosynthesis, and storage lipid synthesis. Although the major function of the plant epidermis is protection against desiccation and environmental stresses, genetic studies in Arabidopsis indicate that cuticular permeability plays a crucial role in developmental signaling between interacting cells, for example, during pollen germination and floral tissue formation (Lolle et al., 1997).
FIDDLEHEAD has been recently identified in Arabidopsis as a gene for putative ketoacyl-CoA synthase (Yephremov et al., 1999). In the Arabidopsis FIDDLEHEAD mutant (fdh-1), the shoot epidermis is changed and pollen germination is promoted on the surface on vegetative organs. In Arabidopsis, FDH-1 is represented by seven ESTs, which is so far the highest number among the KCS class. Not a single full-length KCS sequence is available from rice. However, our EST analysis showed that 30 rice ESTs (75% of rice KCS ESTs) form a contig resulting in a full-length sequence of 539 amino acids that is 72% identical (83% similar) to Arabidopsis FDH-1. Surprisingly, rice FDH-1 ESTs were detected in almost all organs studied. Thus, in rice, the FDH-1 isolog is one of the most highly and ubiquitously expressed lipid biosynthesis genes.
β-Oxidation Transcripts Are Surprisingly Abundant
Although our study has primarily considered lipid biosynthesis rather than degradation pathways, we have observed that ESTs for reactions of fatty acid oxidation are surprisingly abundant. In most plant tissues, fatty acid β-oxidation is considered a minor pathway (Gerhardt, 1992). Based on a recent study with Arabidopsis leaves, we estimated that flux into the fatty acid synthesis pathway was at least 5- to 10-fold higher than the rate of fatty acid degradation (Bao et al., 2000). However, several enzymes of fatty acid oxidation are highly represented in dbEST. For example, in all plant species, acyl-CoA oxidase and 3-ketoacyl-CoA thiolase are represented by 35 and 75 ESTs, respectively. One hypothesis consistent with these data is that β-oxidation enzymes are maintained in plant cells at a high basal level to accommodate potential transient needs (Hooks et al., 1999).
CONCLUSIONS
By surveying GenBank data and ESTs, the “data mining” analyses described above have yielded several new types of information. First, a number of previously undescribed genes for plant glycerolipid synthesis have been putatively identified. Second, the extent to which proteins of the plant lipid synthesis pathway are encoded by gene families and the size of each family has been estimated. Third, more than 160,000 publicly available ESTs have been analyzed to provide a “digital northern” estimate of gene expression levels for 59 plant proteins involved in plant lipid metabolism. With only a few exceptions, the EST abundance patterns for Arabidopsis and rice are very similar (correlation coefficient approximately 0.7), adding support to this method of estimating relative gene expression levels. Such a pathway-wide overview has not been available through previous analyses and has provided new insights regarding the regulation of expression of the pathway. In the near future, with further rapid accumulation of sequence data, such a detailed analysis of gene sequences and ESTs for many metabolic pathways will become an essential approach that will contribute to the development of a functional catalog of plant genes. Of course, such analyses require revision as more information becomes available and the establishment of a website provides users a convenient source of such updates. The database constructed from this survey will be updated as the complete Arabidopsis and rice genome sequences become available.
ACKNOWLEDGMENTS
We thank Toni Voelker, Ralph Dewey, and Tony Hage for bringing to our attention omissions and errors in our catalog, John Browse and Thomas Newman for sharing unpublished results, Christoph Benning and Jay Thelen for critically reading the manuscript, Uwe Rossbach for constructing the website, Curtis Wilkerson for help in automated searches in dbEST, and Natasha Metzler for assistance in editing the numerous files of the web site.
Footnotes
1
This work was supported by grants from the National Science Foundation (no. DCB90–05290) and from the Department of Energy (no. DE–FG02–87ER12729). We also acknowledge the Michigan Agricultural Experiment Station for its support of this research.
LITERATURE CITED
- Allona I, Quinn M, Shoop E, Swope K, Cyr SS, Carlis J, Riedl J, Retzel E, Campbell MM, Sederoff R, Whetten RW. Analysis of xylem formation in pine by cDNA sequencing. Proc Natl Acad Sci USA. 1998;95:9693–9698. doi: 10.1073/pnas.95.16.9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- Bao X, Focke M, Pollard M, Ohlrogge J (2000) Understanding in vivo carbon precursor supply for fatty acid synthesis in leaf tissue. Plant J (in press) [DOI] [PubMed]
- Bevan M, Bancroft I, Bent E, Love K, Goodman H, Dean C, Bergkamp R, Dirkse W, Van Staveren M, Stiekema W, Drost L, Ridley P, Hudson S-A, Patel K, Murphy G, Piffanelli P, Wedler H, Wedler E, Wambutt R, Weitzenegger T, Pohl TM, Terryn N, Gielen J, Villarroel R, De Clerck R, Van Montagu M, Lecharny A, Aubog S, Gy I, Kreis M, Lao N, Kavanagh T, Hempel S, Kotter P, Entian K-D, Rieger M, Schaeffer M, Funk B, Mueller-Auer S, Silvey M, James R, Montfort A, Pons A, Puigdomenech P, Douka A, Voukelatou E, Milioni D, Hatzopoulos P, Piravandi E, Obermaier B, Hilbert H, Düsterhöft A, Moores T, Jones JDG, Eneva T, Palme K, Benes V, Rechman S, Ansorge W, Cooke R, Berger C, Delseny M, Voet M, Volckaert G, Mewes H-W, Klosterman S, Schueller C, Chalwatzis N. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature. 1998;391:485–488. doi: 10.1038/35140. [DOI] [PubMed] [Google Scholar]
- Browse J, Warwick N, Somerville CR, Slack CR. Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the '16:3′ plant Arabidopsis thaliana. Biochem J. 1986;235:25–31. doi: 10.1042/bj2350025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse JA, McCourt PJ, Somerville CR. A mutant of Arabidopsis lacking a chloroplast-specific lipid. Science. 1985;227:763–765. doi: 10.1126/science.227.4688.763. [DOI] [PubMed] [Google Scholar]
- Browse JA, Somerville CR. Glycerolipid metabolism, biochemistry and regulation. Annu Rev Plant Mol Biol. 1991;42:467–506. [Google Scholar]
- Choi JK, Yu F, Wurtele ES, Nikolau BJ. Molecular cloning and characterization of the cDNA coding for the biotin-containing subunit of the chloroplastic acetyl-coenzyme A carboxylase. Plant Physiol. 1995;109:619–625. doi: 10.1104/pp.109.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke R, Raynal M, Laudié M, Grellet F, Delseny M, Morris P-C, Guerrier D, Giraudat J, Quigley F, Clabault G, Li Y-F, Mache R, Krivitzky M, Gy IJ, Kreis M, Lecharny A, Parmentier Y, Marbach J, Fleck J, Clément B, Philipps G, Hervé C, Bardet C, Tremousaygue D, Lescure B, Lacomme C, Roby D, Jourjon M-F, Chabrier P, Charpenteau J-L, Desprez T, Amselem J, Chiapello H, Höfte H. Further progress towards a catalogue of all Arabidopsis genes: analysis of a set of 5000 non-redundant ESTs. Plant J. 1996;9:101–124. doi: 10.1046/j.1365-313x.1996.09010101.x. [DOI] [PubMed] [Google Scholar]
- Dörmann P, Voelker TA, Ohlrogge JB. Cloning and expression in Escherichia coli of a novel thioesterase from Arabidopsis thaliana specific for long-chain acyl-acyl carrier proteins. Arch Biochem Biophys. 1995;316:612–618. doi: 10.1006/abbi.1995.1081. [DOI] [PubMed] [Google Scholar]
- Ecker JR. Genome sequencing: genes blossom from a weed. Nature. 1998;391:438–439. doi: 10.1038/35030. [DOI] [PubMed] [Google Scholar]
- Elborough KM, Winz R, Deka RK, Markham JE, White AJ, Rawsthorne S, Slabas AR. Biotin carboxyl carrier protein and carboxyltransferase subunits of the multi-subunit form of acetyl-CoA carboxylase from Brassica napus: cloning and analysis of expression during oilseed rape embryogenesis. Biochem J. 1996;315:103–112. doi: 10.1042/bj3150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt B. Fatty acid degradation in plants. Prog Lipid Res. 1992;31:417–446. doi: 10.1016/0163-7827(92)90004-3. [DOI] [PubMed] [Google Scholar]
- Harwood JL. Recent advances in the biosynthesis of plant fatty acids. Biochim Biophys Acta. 1996;1301:7–56. doi: 10.1016/0005-2760(95)00242-1. [DOI] [PubMed] [Google Scholar]
- Höfte H, Desprez T, Amselem J, Chiapello H, Rouze P, Caboche M, Moisan A, Jourjon MF, Charpenteau JL, Berthomieu P, Guerrier D, Giraudat J, Quigley F, Thomas F, Yu D-Y, Mache R, Raynal M, Cooke R, Grellet F, Delseny M, Parmentier Y, de Marcillac G, Gigot C, Fleck J, Philipps G, Axelos M, Bardet C, Tremousayue D, Lescure B. An inventory of 1,152 expressed sequence tags obtained by partial sequencing of cDNAs from Arabidopsis thaliana (published erratum appears in Plant J [1994] 5: 611) Plant J. 1993;4:1051–1061. doi: 10.1046/j.1365-313x.1993.04061051.x. [DOI] [PubMed] [Google Scholar]
- Hooks MA, Fleming Y, Larson TR, Graham IA. No induction of β-oxidation in leaves of Arabidopsis that over-produce lauric acid. Planta. 1999;207:385–392. doi: 10.1007/s004250050496. [DOI] [PubMed] [Google Scholar]
- Hugly S, Kunst L, Somerville C. Linkage relationships of mutants that affect fatty acid composition in Arabidopsis. J Hered. 1991;82:484–488. [Google Scholar]
- James DW, Jr, Lim E, Keller J, Plooy I, Ralston E, Dooner HK. Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon activator. Plant Cell. 1995;7:309–319. doi: 10.1105/tpc.7.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Kotani H, Nakamura Y, Sato S, Asamizu E, Miyajima N, Tabata S. Structural analysis of Arabidopsis thaliana chromosome 5. V. Sequence features of the regions of 1,381,565 bp covered by twenty one physically assigned P1 and TAC clones. DNA Res. 1998;5:131–145. doi: 10.1093/dnares/5.2.131. [DOI] [PubMed] [Google Scholar]
- Karp PD, Riley M, Paley SM, Pellegrini-Toole A, Krummenacker M. Eco Cyc: encyclopedia of Escherichia coli genes and metabolism. Nucleic Acids Res. 1999;27:55–58. doi: 10.1093/nar/27.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith CS, Hoang DO, Barrett BM, Feigelman B, Nelson MC, Thai H, Baysdorfer C. Partial sequence analysis of 130 randomly selected maize cDNA clones. Plant Physiol. 1993;101:329–332. doi: 10.1104/pp.101.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney AJ. Genetic modification of storage lipids in plants. Curr Opin Biotechnol. 1994;5:144–151. [Google Scholar]
- Kotani H, Nakamura Y, Sato S, Asamizu E, Kaneko T, Miyajima N, Tabata S. Structural analysis of Arabidopsis thaliana chromosome 5. VI. Sequence features of the regions of 1,367,185 bp covered by 19 physically assigned P1 and TAC clones. DNA Res. 1998;5:203–216. doi: 10.1093/dnares/5.3.203. [DOI] [PubMed] [Google Scholar]
- Kotani H, Nakamura Y, Sato S, Kaneko T, Asamizu E, Miyajima N, Tabata S. Structural analysis of Arabidopsis thaliana chromosome 5. II. Sequence features of the regions of 1,044,062 bp covered by thirteen physically assigned P1 clones. DNA Res. 1997;4:291–300. doi: 10.1093/dnares/4.4.291. [DOI] [PubMed] [Google Scholar]
- Kunst L, Browse J, Somerville C. A mutant of Arabidopsis deficient in desaturation of palmitic acid in leaf lipids. Plant Physiol. 1989;90:943–947. doi: 10.1104/pp.90.3.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L, Taylor DC, Underhill EW. Fatty acid elongation in developing seeds of Arabidopsis thaliana. Plant Physiol Biochem. 1992;30:425–434. [Google Scholar]
- Kuo TM, Ohlrogge JB. Acylation of plant acyl carrier proteins by acyl-acyl carrier protein synthetase from Escherichia coli. Arch Biochem Biophys. 1984;230:110–116. doi: 10.1016/0003-9861(84)90091-2. [DOI] [PubMed] [Google Scholar]
- Kwak JM, Kim SA, Hong SW, Nam HG. Evaluation of 515 expressed sequence tags obtained from guard cells of Brassica campestris. Planta. 1997;202:9–17. doi: 10.1007/s004250050097. [DOI] [PubMed] [Google Scholar]
- Lim CO, Kim HY, Kim MG, Lee SI, Chung WS, Park SH, Hwang I, Cho MJ. Expressed sequence tags of Chinese cabbage flower bud cDNA. Plant Physiol. 1996;111:577–588. doi: 10.1104/pp.111.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus SK, Chen Y, Gooden G, Ryan JF, Birznieks G, Hillard M, Baxevanis AD, Bittner M, Meltzer P, Trent T, Pavan W. Informatic selection of a neural crest-melanocyte cDNA set for microarray analysis. Proc Natl Acad Sci USA. 1999;96:9277–9280. doi: 10.1073/pnas.96.16.9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolle SJ, Berlyn GP, Engstrom EM, Krolikowski KA, Reiter WD, Pruitt RE. Developmental regulation of cell interactions in the Arabidopsis fiddlehead-1 mutant: a role for the epidermal cell wall and cuticle. Dev Biol. 1997;189:311–321. doi: 10.1006/dbio.1997.8671. [DOI] [PubMed] [Google Scholar]
- McKeon TA, Stumpf PK. Purification and characterization of the stearoyl-acyl carrier protein desaturase and the acyl-acyl carrier protein thioesterase from maturing seeds of safflower. J Biol Chem. 1982;257:12141–12147. [PubMed] [Google Scholar]
- Mongrand S, Bessoule J-J, Cabantous F, Cassagne C. The C16:3/C18:3 fatty acid balance in photosynthetic tissues from 468 plant species. Phytochemistry. 1998;49:1049–1064. [Google Scholar]
- Nakamura Y, Sato S, Kaneko T, Kotani H, Asamizu E, Miyajima N, Tabata S. Structural analysis of Arabidopsis thaliana chromosome 5. III. Sequence features of the regions of 1,191,918 bp covered by seventeen physically assigned P1 clones. DNA Res. 1997;4:401–414. doi: 10.1093/dnares/4.6.401. [DOI] [PubMed] [Google Scholar]
- Newman T, de Bruijn FJ, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M, Retzel E, Somerville C. Genes galore: a summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J. Lipid biosynthesis. Plant Cell. 1995;7:957–970. doi: 10.1105/tpc.7.7.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge JB, Jaworski JG. Regulation of fatty acid synthesis. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:109–136. doi: 10.1146/annurev.arplant.48.1.109. [DOI] [PubMed] [Google Scholar]
- Ohlrogge JB, Kuhn DN, Stumpf PK. Subcellular localization of acyl carrier protein in leaf protoplasts of Spinacia oleracea. Proc Natl Acad Sci USA. 1979;76:1194–1198. doi: 10.1073/pnas.76.3.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J. Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell. 1994;6:147–158. doi: 10.1105/tpc.6.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YS, Kwak JM, Kwon OY, Kim YS, Lee DS, Cho MJ, Lee HH, Nam HG. Generation of expressed sequence tags of random root cDNA clones of Brassica napus by single-run partial sequencing. Plant Physiol. 1993;103:359–370. doi: 10.1104/pp.103.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post-Beittenmiller D, Jaworski JG, Ohlrogge JB. In vivo pools of free and acylated acyl carrier proteins in spinach: evidence for sites of regulation of fatty acid biosynthesis. J Biol Chem. 1991;266:1858–1865. [PubMed] [Google Scholar]
- Roughan PG. Stromal concentrations of coenzyme A and its esters are insufficient to account for rates of chloroplast fatty acid synthesis: evidence for substrate channelling within the chloroplast fatty acid synthase. Biochem J. 1997;327:267–273. doi: 10.1042/bj3270267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roughan PG, Ohlrogge JB. Evidence that isolated chloroplasts contain an integrated lipid-synthesizing assembly that channels acetate into long-chain fatty acids. Plant Physiol. 1996;110:1239–1247. doi: 10.1104/pp.110.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouze P, Pavy N, Rombauts S. Genome annotation: which tools do we have for it? Curr Opin Plant Biol. 1999;2:90–95. doi: 10.1016/S1369-5266(99)80019-3. [DOI] [PubMed] [Google Scholar]
- Ruan Y, Gilmore J, Conner T. Towards Arabidopsis genome analysis: monitoring expression profiles of 1400 genes using cDNA microarrays. Plant J. 1998;15:821–833. doi: 10.1046/j.1365-313x.1998.00254.x. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Song J, Koga-Ban Y, Matsui E, Fang F, Higo H, Nagasaki H, Hori M, Miya M, Murayama-Kayano E, Takiguchi T, Takasuga A, Niki T, Ishimaru K, Ikeda H, Yamamoto Y, Mukai Y, Ohta A, Miyadera N, Havukkala I, Minobe Y. Toward cataloguing all rice genes: large-scale sequencing of randomly chosen rice cDNAs from a callus cDNA library. Plant J. 1994;6:615–624. doi: 10.1046/j.1365-313x.1994.6040615.x. [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Konishi T, Nagano Y. The compartmentation of acetyl-coenzyme A carboxylase in plants. Plant Physiol. 1995;108:445–449. doi: 10.1104/pp.108.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Kaneko T, Kotani H, Nakamura Y, Asamizu E, Miyajima N, Tabata S. Structural analysis of Arabidopsis thaliana chromosome 5. IV. Sequence features of the regions of 1,456,315 bp covered by nineteen physically assigned P1 and TAC clones. DNA Res. 1998;5:41–54. doi: 10.1093/dnares/5.1.41. [DOI] [PubMed] [Google Scholar]
- Shanklin J, Somerville C. Stearoyl-acyl-carrier-protein desaturase from higher plants is structurally unrelated to the animal and fungal homologs. Proc Natl Acad Sci USA. 1991;88:2510–2514. doi: 10.1073/pnas.88.6.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Carneiro N, Torres-Jerez I, Stevenson B, McGreery T, Helentjaris T, Baysdorfer C, Almira E, Ferl RJ, Habben JE, Larkins B. Partal sequencing and mapping of clones from two maize cDNA libraries. Plant Mol Biol. 1994;26:1085–1101. doi: 10.1007/BF00040691. [DOI] [PubMed] [Google Scholar]
- Shintani DK, Ohlrogge JB. The characterization of a mitochondrial acyl carrier protein isoform isolated from Arabidopsis thaliana. Plant Physiol. 1994;104:1221–1229. doi: 10.1104/pp.104.4.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterky F, Regan S, Karlsson J, Hertzberg M, Rohde A, Holmberg A, Amini B, Bhalerao R, Larsson M, Villarroel R, Van Montagu M, Sandberg G, Olsson O, Teeri TT, Boerjan W, Gustafsson P, Uhlen M, Sundberg B, Lundeberg J. Gene discovery in the wood-forming tissues of poplar: analysis of 5,692 expressed sequence tags. Proc Natl Acad Sci USA. 1998;95:13330–13335. doi: 10.1073/pnas.95.22.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukey JE, McDonough VM, Martin CE. Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J Biol Chem. 1989;264:16537–16544. [PubMed] [Google Scholar]
- Stukey JE, McDonough VM, Martin CE. The OLE1 gene of Saccharomyces cerevisiae encodes the Δ9 fatty acid desaturase and can be functionally replaced by the rat stearoyl-CoA desaturase gene. J Biol Chem. 1990;265:20144–20149. [PubMed] [Google Scholar]
- Thiede MA, Ozols J, Strittmatter P. Construction and sequence of cDNA for rat liver stearyl coenzyme A desaturase. J Biol Chem. 1986;261:13230–13235. [PubMed] [Google Scholar]
- Töpfer R, Martini N, Schell J. Modification of plant lipid synthesis. Science. 1995;268:681–686. doi: 10.1126/science.268.5211.681. [DOI] [PubMed] [Google Scholar]
- van de Loo FJ, Turner S, Somerville C. Expressed sequence tags from developing castor seeds. Plant Physiol. 1995;108:1141–1150. doi: 10.1104/pp.108.3.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker T. Plant acyl-ACP thioesterases: chain-length determining enzymes in plant fatty acid biosynthesis. Genet Eng. 1996;18:111–133. doi: 10.1007/978-1-4899-1766-9_8. [DOI] [PubMed] [Google Scholar]
- Wada H, Shintani D, Ohlrogge J. Why do mitochondria synthesize fatty acids? Evidence for involvement in lipoic acid production. Proc Natl Acad Sci USA. 1997;94:1591–1596. doi: 10.1073/pnas.94.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Sasaki T. Large-scale EST sequencing in rice. Plant Mol Biol. 1997;35:135–144. [PubMed] [Google Scholar]
- Yephremov A, Wisman E, Huijser P, Huijser C, Wellesen K, Saedler H. Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell. 1999;11:2187–2201. doi: 10.1105/tpc.11.11.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]