PIFs meet Tourists and Harbingers: A superfamily reunion (original) (raw)
Plant genomes host an astounding variety of transposable elements (TEs) (1–7). Among plants, the maize model has been a darling of TE researchers since the classical work of Barbara McClintock (1). Recently, maize was joined by Arabidopsis thaliana, a small plant from the mustard family brought into prominence in biological research because of sequencing of its genome. Studies of TEs from these two plants and a nematode worm, Caenrhabditis elegans, began a story of a new superfamily of DNA transposons. In the part of the story written in this issue of PNAS by Zhang et al. (8), the major superfamily “characters” Tourists and Harbingers are connected with their previously unknown relatives, alive and prosperous in maize. The newly found relatives are P instability factors, or PIFs.
The first Tourist element was discovered as a 128-bp-long insertion mutation in maize. This element became a defining member of the _Tourist_-like family of miniature inverted transposable elements, referred to as MITEs (9). The major characteristics of the _Tourist_-like family are terminal inverted repeats, a 3-bp target site duplication (TSD), and a relatively small size that precludes any protein-coding capabilities. The _Tourist_-MITE family is widespread in plants and is often associated with genes (10). The origin of MITEs remained a mystery although numerous researchers considered them to be short nonautonomous DNA transposons (5, 11–15).
Harbinger, a 5,382-bp-long DNA transposon, was discovered (5) through a systematic survey and analysis of the newly sequenced genome of A. thaliana and deposited in the May 1999 release of Repbase Update, a public database of repetitive elements (16). Harbinger's characteristics include 25-bp terminal inverted repeats, 3-bp target sites, and two ORFs, one of which encodes a transposase-like protein (HARBT). Based on the significant similarity of HARBT to transposases from bacterial insertion elements (IS5, ISL2, IS702, IS493, IS112, and IS470) as well as to putative transposases from C. elegans and Sorghum bicolor, Harbinger was first described as a defining member of a new _Harbinge_r/IS5 superfamily (5). Two diverse C. elegans transposons described in the same study (5), under cryptic names 2746799_CE and 38777883_CE, were deposited as Turmoil1 and Turmoil2 DNA transposons (C. elegans section RU 4.11, http://www.girinst.org) in the November 1999 release of Repbase Update. Together with Harbinger, these are the originally classified autonomous members of the Harbinger/IS5 superfamily. Further DNA sequence studies of the Turmoil1 family in nematodes (C. elegans and Caenrhabditis briggsae) revealed its relationship to _Tourist_-like elements (17).
The existing sequence data do not contain sufficient information to prove that Harbinger or any other element from the Harbinger/IS5 superfamily is currently active in eukaryotes. Indeed, before the paper by Zhang et al. (8) there was no direct evidence for active transposition of any Tourist- or _Harbinger_-like elements. The authors chose to pursue a mysterious class 2 family of transposable elements, PIFs, which were found to actively transpose in maize (18). PIFs were first discovered by studying 5.2-kb and 2.3-kb multiple insertion mutations at the same target in the anthocyanin regulatory gene (R gene) in maize (18). The R gene confers pigmentation to different maize tissues. The insertion of the PIFs into the second intron eliminates the pigmentation and their excision restores it, thus permitting observation of the cut-and-paste activity. The 2.3-kb-long element, named PIF-12, has been sequenced and deposited in public databases but its predicted ORFs failed to match any known protein sequences (18). Apparently, although PIF-12(PIF2.3) and PIF5.2 are actively transposed, they are both nonautonomous elements that evaded broader classification. Classification of the PIF family in relation to other known families of TEs could be done only when Zhang et al. (8) isolated and sequenced a new PIF element associated with active transposition in maize and coding for a transposase-like protein. The authors followed different strains of maize with and without active _PIF_s by monitoring changes in plant pigmentation. Using PCR amplification with a predetermined pull of primers they were able to amplify a 3,728-bp element that cosegregated with _PIF_-active plants. Additional PCR amplifications confirmed that the absence of PIFa correlated with the loss of PIF activity. The authors also have shown that in some active strains PIFa has relocated from the original locus to other site(s). These findings indicate that PIFa itself is mobile and that it is a necessary element involved in active transposition of other PIFs. It is somewhat surprising that the critically important PIFa is shorter than its nonautonomous PIF5.2 relative. It would be interesting to determine whether or not such big nonautonomous companions can encode any additional proteins that coparticipate in the transposition process. In any case, isolating the active PIFa element is a very important contribution that will help to further our understanding of PIFs and related TEs.
The PIFa sequence contains a 331-aa ORF similar to putative transposases from Arabidopsis, sorghum, rice, nematodes (C. elegans and C. briggsae), and a basidiomycete fungus Filobasidiella neoformans. It was also found (8) to be distantly similar to IS5 and other bacterial insertion sequences listed above (5). Similarities to putative transposases from rice and fungus have been reported for the first time. Nematode transposons called CE-PIF1 and CE-PIF2 in this report (8) are the same as Turmoil2 and Turmoil1, respectively. These findings confirm and extend previous reports (refs. 5 and 17; C. elegans section RU 4.11, http://www.girinst.org) and for the first time place the active PIF family in the Harbinger/IS5 superfamily. The authors emphasize the diversity of the superfamily. This diversity is likely to go beyond the Harbinger/IS5 superfamily, which may have a sister IS112-like superfamily present in A. thaliana. This is indicated by a recently discovered putative DNA transposon ATIS112 (A. thaliana section RU 6.01, http://www.girinst.org) flanked by 5-bp TSDs as opposed to the 3-bp TSDs found in Harbinger/IS5 elements (5). Despite the different target sites, the transposase-like ORF from ATIS112 is related to all transposases identified with the Harbinger/IS5 superfamily.
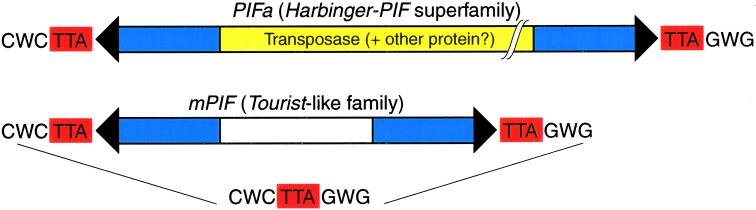
In addition to long PIFs, Zhang et al. (8) studied a small 364-bp _PIF_-like sequence identified in the original report of the PIF family (18). Using Southern blot analysis they found this miniature PIF (or mPIF) to be abundant in maize (around 6,000 copies per haploid genome), but not in sorghum or rice. Based on the 3-bp target site duplications and terminal inverted repeats, mPIF was classified by the authors as a typical _Tourist_-like MITE element. Furthermore, mPIFs share terminal and subterminal regions with other PIF elements. This puts them in a common category of deletion products derived from larger PIFs (Fig. 1). Finally, they appear to use the same 9-bp target as other PIFs, with an approximate consensus CWCTTAGWG (C77W58C77T97T91A97G52W68G61) where W stands for either A or T (Fig. 1). Based on the average frequency of a 9-bp oligonucleotide, the authors estimate that there are about 10,000 such targets in the maize genome. However, given the degenerate character of the target this number is likely to be around four times larger. Moreover, there is additional flexibility in target selection by mPIFs caused by low sequence conservation outside the middle TTA region of the target. Therefore, only a fraction of potential targets may be occupied by the 6,000 or so mPIF elements in maize.
Figure 1.
Harbinger/IS5-like active DNA transposon PIFa and its nonautonomous _Tourist_-like companion mPIF as described by Zhang et al. (8). Black arrows indicate terminal inverted repeats, blue regions show subterminal sequences 70% similar between PIFa and mPIF. Yellow part of PIFa indicates protein coding regions unique to PIFa as well as regions of similarity between PIFa and other long members of the PIF family. The central, white region of mPIF is unique to this nonautonomous element. A consensus sequence of the 9-bp palindromic target is shown on the bottom, and the TTA trinucleotide undergoing duplication upon insertion of PIF elements is marked in red. For further explanation see text.
The 9-bp target resembles an imperfect palindrome, which may be of more general significance. For example, the 8-bp TSDs generated by mammalian hAT transposons are often palindromic (12). However, unlike in haTs, only the internal 3-bp TTA portion of the target undergoes duplication in the case of PIFs. It will be interesting to determine whether TSDs in other DNA transposons are also a part of larger targets.
Classification of mPIFs as _Tourist_-like PIFs brings back the question of whether or not MITEs should be viewed as a distinct group of elements, different from other nonautonomous DNA transposons. The major defining feature of MITEs is their short length, comparable with that of short interspersed nuclear elements. Nevertheless, MITEs include only _Tourist-_like and _Stowaway_-like short elements with 3-bp and 2-bp TSDs, respectively. The short elements with TSDs different from those in Tourist- and _Stowaway_-type elements are arbitrarily excluded by this definition. Following the spirit of the superfamily reunion, MITEs probably should be reunited with other short class II nonautonomous DNA transposons into a single biological category.
Short DNA transposons as well as short interspersed nuclear elements (SINEs) tend to be more abundant in eukaryotes than the long ones. One explanation of this phenomenon is that shorter insertions may have less impact on host phenotype and, for this reason, are more easily tolerated. Indeed, short TEs including MITEs and SINEs appear to be associated with gene-rich rather than with gene-poor regions (10, 19, 20). However, Southern blot analysis of mPIFs (8) indicates that the abundance may be species-specific as much as transposon-specific. Therefore, the abundance of short nonautonomous TEs is a dynamic outcome resulting from a combination of different factors and may, to some extent, be even controlled by the host (8, 20).
Acknowledgments
We thank Alison McCormack, Jolanta Walichiewicz, and Michael Jurka for help with editing the manuscript. Support during preparation of this commentary came from National Institutes of Health Grant 2 P41 LM06252–04A1 (to J.J.).
Footnotes
See companion article on page 12572.
References
- 1.McClintock B. Carnegie Inst Wash Year Book. 1948;47:155–169. [PubMed] [Google Scholar]
- 2.Fedoroff N, Wessler S, Shure M. Cell. 1983;35:235–242. doi: 10.1016/0092-8674(83)90226-x. [DOI] [PubMed] [Google Scholar]
- 3.Bennetzen J L, Swanson J, Taylor W C, Freeling M. Proc Natl Acad Sci USA. 1984;81:4125–4128. doi: 10.1073/pnas.81.13.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flavell A J, Smith D B, Kumar A. Mol Gen Genet. 1992;231:233–242. doi: 10.1007/BF00279796. [DOI] [PubMed] [Google Scholar]
- 5.Kapitonov V V, Jurka J. Genetica. 1999;107:27–37. [PubMed] [Google Scholar]
- 6.Le Q H, Wright S, Yu Z, Bureau T. Proc Natl Acad Sci USA. 2000;97:7376–7381. doi: 10.1073/pnas.97.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapitonov V V, Jurka J. Proc Natl Acad Sci USA. 2001;98:8714–8719. doi: 10.1073/pnas.151269298. . (First Published July 10, 2001; 10.1073/pnas.151269298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Feschotte C, Zhang Q, Jiang N, Eggleston W B, Wessler S R. Proc Natl Acad Sci USA. 2001;98:12572–12577. doi: 10.1073/pnas.211442198. . (First Published October 2, 2001; 10.1073/pnas.211442198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bureau T E, Wessler S R. Plant Cell. 1992;4:1283–1294. doi: 10.1105/tpc.4.10.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bureau T E, Ronald P C, Wessler S R. Proc Natl Acad Sci USA. 1996;93:8524–8529. doi: 10.1073/pnas.93.16.8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oosumi T, Garlick B, Belknap W R. J Mol Evol. 1996;43:11–18. doi: 10.1007/BF02352294. [DOI] [PubMed] [Google Scholar]
- 12.Smit A F A, Riggs A D. Proc Natl Acad Sci USA. 1996;93:1443–1448. doi: 10.1073/pnas.93.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feschotte C, Mouches C. Gene. 2000;30:109–116. doi: 10.1016/s0378-1119(00)00187-6. [DOI] [PubMed] [Google Scholar]
- 14.Feschotte C, Mouches C. Mol Biol Evol. 2000;17:730–737. doi: 10.1093/oxfordjournals.molbev.a026351. [DOI] [PubMed] [Google Scholar]
- 15.Tu Z. Proc Natl Acad Sci USA. 2001;98:1699–1704. doi: 10.1073/pnas.041593198. . (First Published February 6, 2001; 10.1073/pnas.041593198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jurka J. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- 17.Le Q H, Turcotte K, Bureau T. Genetics. 2001;158:1081–1082. doi: 10.1093/genetics/158.3.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker E L, Eggleston W B, Demopulos D, Kermicle J, Dellaporta S L. Genetics. 1997;146:681–693. doi: 10.1093/genetics/146.2.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bureau T, Wessler S R. Proc Natl Acad Sci USA. 1994;91:1411–1415. doi: 10.1073/pnas.91.4.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jurka J, Kapitonov V V. Genetica. 1999;107:239–248. [PubMed] [Google Scholar]