Bisulfite genomic sequencing of microdissected cells (original) (raw)
Abstract
Mapping of methylation patterns in CpG islands has become an important tool for understanding tissue-specific gene expression in both normal and pathological situations. However, the inherent cellular heterogeneity of any given tissues can affect the outcome and interpretation of molecular studies. In order to analyse genomic DNA methylation on a pure cell population from tissue sample, we have developed a simple technique of single-cell microdissection from cryostat sections which can be combined with bisulfite-mediated sequencing of 5-methylcytosine. We report here our results on the methylation status of the androgen receptor gene studied by bisulfite genomic sequencing on purified cells isolated from human testis.
INTRODUCTION
To understand biological processes such as regulation of imprinted genes, X chromosome inactivation or gene silencing in human cancer (1), it is now becoming apparent that analysing the global methylation status of a whole tissue is not sufficient, and that refined mapping of individual CpG dinucleotides in genomic DNA has to be known. One major limitation of DNA methylation studies is the purity of the original samples. Recently, microdissection from tissue sections has been used for the isolation of homogeneous, morphologically identified cell populations, thus overcoming the obstacle of tissue complexity (2–5). In conjunction with sensitive analytical techniques, such as the polymerase chain reaction, microdissection allows precise in vivo examination of specific cell populations, which are otherwise inaccessible by conventional molecular studies.
We report here a simple single-cell microdissection process which, combined with bisulfite genomic sequencing, allows the study of the methylation status of pure cell populations isolated from frozen testis samples.
MATERIALS AND METHODS
Microdissection technique
Cryostat sections (Kryostat 1720, Zeiss), 12 µm thick, were fixed for 2 min in 10% formol, before staining with hematoxylin and eosin. Germ cells (spermatids, spermatocytes I, spermatogonia) were visually identified. Individual cells were microdissected and aspirated into a glass micropipette using an inverted microscope (Fig. 1). One to three cells were then injected into a 10 µl drop of molten (80°C) 2% low melting point agarose placed onto a new slide. The agarose/cell beads were immediately solidified on ice and then each bead was transferred into 0.2 ml PCR tube. Control tubes containing all reagents, but no cell, were systematically included.
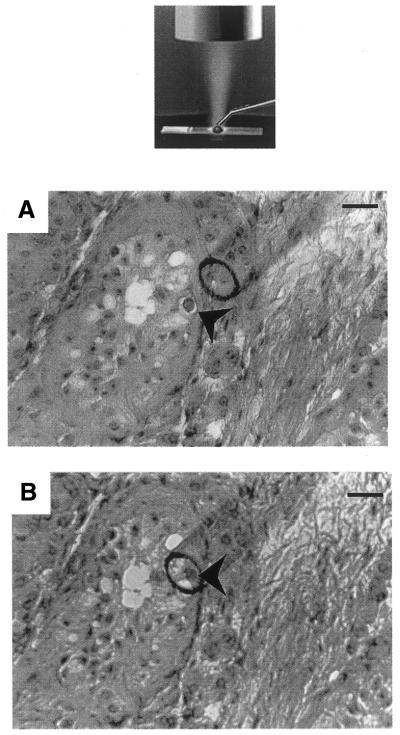
Figure 1.
The microdissection process. Microdissection was performed on cryostat sections using a glass micropipette (tip diameter 70 µm) controlled by a Narishige micromanipulator using an inverted microscope (NT-88, Narishige, Japan), as shown on the schematic representation. Cryostat sections (12 µm) were stained with hematoxylin and eosin. A 10 µl drop of viscoagent (Viscorneal®, Cornéal, France) was deposited on the selected area and the germ cell of interest was microdissected under ×200 magnification. (A) An example of the microdissection of a spermatogonia (arrowhead) from a cryptorchid testis section. The microdissected cell was then aspirated into the micropipette (B) and injected into a 10 µl drop of molten (80°C) 2% low melting point agarose deposited on a new slide. Scale bar, 70 µm.
Bisulfite treatment of agarose beads
Bisulfite treatment of DNA in agarose beads was carried out using a slightly modified version of a published procedure (6,7). 200 µl of lysis solution (0.5 M EDTA, pH 8; 2 mg/ml proteinase K) was deposited over the agarose beads containing a defined number of cells (1–3). After an overnight incubation at 50°C, lysis solution was removed carefully with a pipette and the beads were overlaid with 60 µl of freshly prepared sodium hydroxide (0.3 M NaOH) for 30 min in order to denature DNA. Aliquots (170 µl) of bisulfite solution (4 M sodium bisulfite, Sigma; 125 mM hydroquinone, Sigma; pH 5.0) were directly added to each reaction tube. The reaction mixtures were incubated for at least 4 h at 50°C in the dark. The bisulfite solutions containing the beads were then heated to 80°C for 10 min to melt the agarose. Four to twelve reaction mixtures containing identical cell types were pooled together (corresponding to up to 9–12 cells). Modified DNA was purified using the Wizard® DNA Clean-Up System (Promega) according to the manufacturer’s instructions and eluted into 40 µl of water. Modification was completed by treatment with 4 µl of 3 M NaOH for 15 min at 37°C, followed by addition of 22 µl 6 M ammonium acetate (pH 6). The bisulfite-reacted DNA was precipitated for 30 min at –20°C by 250 µl of cold 100% ethanol. Then, the nucleic acid was washed once with 70% ethanol, resuspended in water (10 µl) and either immediately used or stored at – 20°C.
PCR amplifications, cloning and sequencing of bisulfite treated DNA
The CpG island within exon 1 of the AR gene was amplified by two rounds of PCR using the following primers. In the first round, primers F –1 (5′-AGATTTAGTTAAGTTTAAGGATGGAAGTG-3′) and R +282 (5′-AAAAACCATCCTCACCCTACTACTAC-3′) and, in the second, primers F +34 (5′- GGGTTGGGAAGGGTTTATTTT-3′) and R +282. PCR amplification reactions were carried out using Ampli_Taq_ Gold DNA polymerase (Perkin-Elmer, Norwalk, CT) in a 50 µl reaction containing 10 mM GeneAmp PCR Buffer II; 1.5 mM MgCl2; 0.2 mM dNTPs; 0.5 µM of each primer and 1.25 U of Ampli_Taq_ Gold DNA polymerase. PCR cycles are 5 min of denaturation at 94°C, followed by 40 cycles of 45 s of denaturation at 94°C, 45 s of annealing at 61°C, 1 min of elongation at 72°C and a final 10 min elongation step. The amplified DNA was cloned into the CloneAmp® pAMP1 system (Life Technologies, Gaithersburg, MD) and individual clones are sequenced using an ABI Prism 377 DNA Sequencer (Perkin-Elmer, Norwalk, CT). All experiments were performed in duplicate.
RESULTS
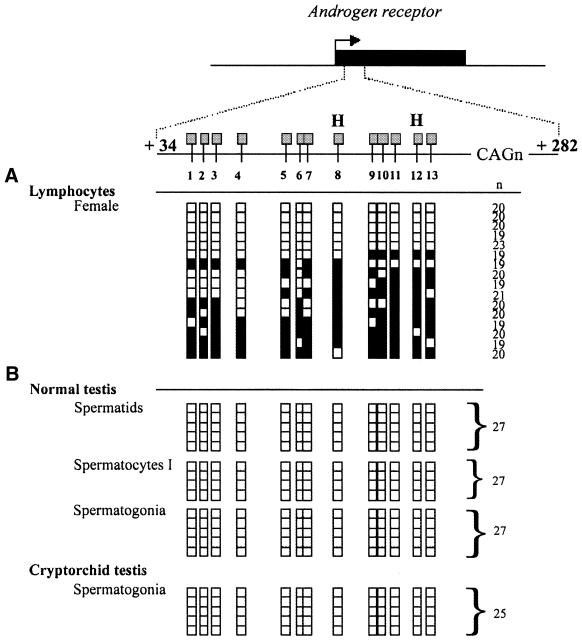
The DNA sequence used to illustrate this method comprised a 248 bp unit including the flanking CpGs sites and the highly polymorphic (CAG)n repeats located in exon 1 of the human androgen-receptor gene (Fig. 2) (8). The close proximity of the polymorphic (CAG)n repeats to the CpGs sites enabled the parental X chromosomes from a female to be distinguished from each other. Analysis of the sequences after bisulfite treatment confirmed the heterozygous status of the female with two major allelic forms of 19 (6/16) and 20 (8/16) (CAG)n repeats respectively (Fig. 2A). Other alleles (n = 23 CAG and n = 21 CAG) possibly corresponded to PCR artefacts during amplification of the repeat (8).
Figure 2.
Analysed region and results of the bisulfite genomic sequencing of the human androgen receptor gene (AR). The region analysed in exon 1 (AR sequence +34 to +282) is shown relative to the AR transcriptional start (0). All CpG sites within the region are indicated and numbered from 1 to 13. The relative position of _Hpa_II sites (H) are shown. Each row of squares represents a single cloned allele, and each square represents a single CpG site (open square, non-methylated cytosines; filled square, methylated cytosines). n, number of repeats (CAGn) of the highly polymorphic CAG repeat sequence. (A) Methylation patterns were analysed from female adult lymphocytes from peripheral blood. Genomic DNA from blood sample (1 µg) was treated with sodium bisulfite as previously described (14). (B) Methylation patterns were analysed from germ cells isolated from a normal testis (spermatids, spermatocytes I, spermatogonia) and from spermatogonia isolated from a cryptorchid testis. Bisulfite modified DNA from 9–12 cells was PCR amplified.
Detailed methylation analysis using bisulfite sequencing was performed on a series of germ cells microdissected from testis samples. The different cell types were identified on the basis of their morphology and relative position within the tubules. Germ cells were microdissected from a normal testis (spermatogonia, spermatocytes I, spermatids) and from a cryptorchid testis (spermatogonia). The overall efficiency of bisulfite treatment and PCR in yielding a product in a limited number (9–12) of picked cells was 68% (15/22 samples). Control tubes were always negative. A constant number of repeats were sequenced in the different germ cell types of the normal testis (27 repeats, 14/14) and the cryptorchid testis (25 repeats, 5/5). As expected, no methylation was found in these microdissected germs cells of normal or abnormal testis.
DISCUSSION
Identifying differences between the methylation profiles of two cell types is clearly affected by the quality of the sample prepared. On one hand, tissue microdissection techniques allow single-cell procurement resulting in pure and homogeneous samples (2–5). On the other hand, the introduction of the bisulfite genomic sequencing technique made it possible to study small amounts of DNA and to identify methylation patterns with single base resolution. We have combined a simple technique of microdissection with bisulfite genomic sequencing. The procedure facilitates bisulfite methylation analysis of very small amounts of microdissected cells. Negative samples may be due to different reasons (9), including partial recovery of cell nuclei, loss of DNA during the experimental process, incomplete DNA modification by bisulfite treatment, or inefficiency of the polymerase.
In summary, bisulfite genomic sequencing of microdissected cells from tissue samples allows us to refine methylation profile of pure cell populations. In the case of the AR gene, it may be revelant in pathological situations such as clonal neoplasm (10), excessive gain of X chromosome (11) or loss of AR expression (12,13), and make the assay complementary to previously published assays that detect X inactivation patterns.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge Andràs Pàldi and Françoise Poirier for fruitful discussions. This work was supported by grants from the ARC (5743).
References
- 1.Robertson K.D. and Wolffe,A.P. (2000) DNA methylation in health and disease. Nature Rev. Genet., 1, 11–19. [DOI] [PubMed] [Google Scholar]
- 2.Curran S., McKay,J.A., McLeod,H.L. and Murray,G.I. (2000) Laser capture microscopy. Mol. Pathol., 53, 64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sirivatanauksorn Y., Drury,R., Crnogorac-Jurcevic,T., Sirivatanauksorn,V. and Lemoine,N.R. (1999) Laser-assisted microdissection: applications in molecular pathology. J. Pathol., 189, 150–154. [DOI] [PubMed] [Google Scholar]
- 4.Schutze K., Posl,H. and Lahr,G. (1998) Laser micromanipulation systems as universal tools in cellular and molecular biology and in medicine. Cell. Mol. Biol., 44, 735–746. [PubMed] [Google Scholar]
- 5.Simone N.L., Bonner,R.F., Gillespie,J.W., Emmert-Buck,M.R. and Liotta,L.A. (1998) Laser-capture microdissection: opening the microscopic frontier to molecular analysis. Trends Genet., 14, 272–276. [DOI] [PubMed] [Google Scholar]
- 6.Olek A., Oswald,J. and Walter,J. (1996) A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res., 24, 5064–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerjean A., Dupont,J.M., Vasseur,C., Le Tessier,D., Cuisset,L., Paldi,A., Jouannet,P. and Jeanpierre,M. (2000) Establishment of the paternal methylation imprint of the human H19 and MEST/PEG1 genes during spermatogenesis. Hum. Mol. Genet., 9, 2183–2187. [DOI] [PubMed] [Google Scholar]
- 8.Allen R.C., Zoghbi,H.Y., Moseley,A.B., Rosenblatt,H.M. and Belmont,J.W. (1992) Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am. J. Hum. Genet., 51, 1229–1239. [PMC free article] [PubMed] [Google Scholar]
- 9.Rein T., DePamphilis,M.L. and Zorbas,H. (1998) Identifying 5-methylcytosine and related modifications in DNA genomes. Nucleic Acids Res., 26, 2255–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang S.J. and Mao,L. (2000) Methylation patterns in human androgen receptor gene and clonality analysis. Cancer Res., 60, 864–866. [PubMed] [Google Scholar]
- 11.McDonald H.L., Gascoyne,R.D., Horsman,D. and Brown,C.J. (2000) Involvement of the X chromosome in non-Hodgkin lymphoma. Genes Chromosomes Cancer, 28, 246–257. [PubMed] [Google Scholar]
- 12.Nakayama T., Watanabe,M., Suzuki,H., Toyota,M., Sekita,N., Hirokawa,Y., Mizokami,A., Ito,H., Yatani,R. and Shiraishi,T. (2000) Epigenetic regulation of androgen receptor gene expression in human prostate cancers. Lab. Invest., 80, 1789–1796. [DOI] [PubMed] [Google Scholar]
- 13.Kinoshita H., Shi,Y., Sandefur,C., Meisner,L.F., Chang,C., Choon,A., Reznikoff,C.R., Bova,G.S., Friedl,A. and Jarrard,D.F. (2000) Methylation of the androgen receptor minimal promoter silences transcription in human prostate cancer. Cancer Res., 60, 3623–3630. [PubMed] [Google Scholar]
- 14.Tremblay K.D., Duran,K.L. and Bartolomei,M.S. (1997) A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol. Cell. Biol., 17, 4322–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]