In Situ Structure of Neuronal C9ORF72 Poly-GA Aggregates Reveals Proteasome Recruitment (original) (raw)
. Author manuscript; available in PMC: 2018 Jul 7.
Summary
Protein aggregation and dysfunction of the ubiquitin-proteasome system are hallmarks of many neurodegenerative diseases. Here we address the elusive link between these phenomena employing cryo-electron tomography to dissect the molecular architecture of protein aggregates within intact neurons at high resolution. We focus on the poly-Gly-Ala (GA) aggregates resulting from aberrant translation of an expanded GGGGCC repeat in C9orf72, the most common genetic cause of amyotrophic lateral sclerosis and frontotemporal dementia. We find that poly-GA aggregates consist of densely packed twisted ribbons that recruit numerous 26S proteasome complexes, while other macromolecules are largely excluded. Proximity to poly-GA ribbons stabilizes a transient substrate-processing conformation of the 26S proteasome, suggesting stalled degradation. Thus, poly-GA aggregates may compromise neuronal proteostasis by driving the accumulation and functional impairment of a large fraction of cellular proteasomes.
Keywords: ALS, FTD, dipeptide-repeat proteins, UPS, cryo-electron microscopy, cryo-EM, cryo-ET, cryo-FIB
Introduction
The ubiquitin-proteasome system (UPS) is the main cellular pathway for targeted protein degradation (Collins and Goldberg, 2017; Hershko et al., 2000). UPS alterations have been implicated in many human diseases, including multiple neurodegenerative disorders (Dantuma and Bott, 2014; Hipp et al., 2014; Schmidt and Finley, 2014). In particular, frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS) have been associated with mutations in UPS components (Deng et al., 2011; Johnson et al., 2010; Watts et al., 2004) and altered UPS function (Cheroni et al., 2009; Tashiro et al., 2012). However, the contribution of UPS dysfunction to neurodegeneration and its underlying mechanisms are not yet well understood.
UPS impairment has also been linked to C9orf72 mutations, the most common genetic cause of ALS/FTD (Edbauer and Haass, 2016; Freibaum and Taylor, 2017; Gendron and Petrucelli, 2017; Lin et al., 2017). A massive expansion of a GGGGCC (G4C2) repeat in a non-coding region of the C9orf72 gene to up to several thousand copies is found in 10–50% of familial ALS/FTD cases and in 5–7% of patients with sporadic disease (DeJesus-Hernandez et al., 2011; Majounie et al., 2012; Renton et al., 2011; van der Zee et al., 2013). Three non-mutually exclusive mechanisms have been suggested to mediate the toxicity of the G4C2 repeat expansion: 1) loss of native function of the C9orf72 protein due to reduced transcription of the mutant allele, 2) aberrant RNA interactions, and 3) production of toxic translation products and aggregates via repeat-associated non-ATG (RAN) translation (Zu et al., 2011).
Although the G4C2 repeat is found in a non-coding region of the C9orf72 gene, sense and anti-sense transcripts are unconventionally translated in all reading frames into five dipeptide-repeat proteins (Ash et al., 2013; Gendron et al., 2013; Mori et al., 2013a; Mori et al., 2013b; Zu et al., 2013): poly-GA, poly-GR, poly-GP, poly-PR and poly-PA. While all five proteins form TDP-43-negative, p62-positive inclusions in ALS/FTD patient brain, the vast majority of these aggregates contains poly-GA (Mackenzie et al., 2015; Mori et al., 2013b; Zhang et al., 2014).
Poly-GA expression leads to toxicity in heterologous cells, primary neuron cultures and mice (Jovicic et al., 2015; May et al., 2014; Schludi et al., 2017; Yamakawa et al., 2015; Zhang et al., 2016; Zhang et al., 2014). Similar to other toxic aggregating proteins (Olzscha et al., 2011; Park et al., 2013), poly-GA aggregates sequester critical cellular factors including Unc119 and multiple UPS components (May et al., 2014; Zhang et al., 2016). UPS impairment is critically involved in poly-GA-mediated toxicity (Yamakawa et al., 2015; Zhang et al., 2016; Zhang et al., 2014), but our understanding of the underlying mechanisms remains incomplete. This is aggravated by the limited structural information currently available on poly-GA aggregates, especially within an unperturbed cellular context.
Here we address these challenges using state-of-the-art cryo-electron tomography (cryo-ET) technologies, which allow 3D imaging of the cell interior in close-to-native conditions and at molecular resolution (Beck and Baumeister, 2016). We reveal the structure and cellular interactions of poly-GA aggregates within intact neurons to an unprecedented level of detail. Interestingly, we find that poly-GA aggregates consist of densely packed twisted ribbons that recruit large numbers of 26S proteasome complexes. Structural analysis of these proteasome complexes by subtomogram averaging and classification into functional states provides mechanistic insights into proteasomal dysfunction in C9orf72 ALS/FTD.
Results
Poly-GA Aggregates Contain Densely Packed Twisted Ribbons
To study neuronal poly-GA aggregates without interference from C9orf72 loss-of-function and RNA-mediated toxicity, we transduced primary rat neuronal cultures with a GFP-tagged codon-modified synthetic construct expressing (GA)175-GFP and containing an ATG start codon (May et al., 2014). We have previously shown that lentiviral poly-GA expression results in inclusions of similar size and poly-GA intensity as in C9orf72 patient tissue (May et al., 2014). Neurons were transduced at day in vitro (DIV) 5 and allowed to express the protein for another 5 days (DIV 5 + 5). The cultures were then vitrified and subsequently imaged by cryo-light microscopy to locate cellular poly-GA inclusions (Figure S1A). Correlative microscopy allowed the production of 100–200 nm-thick lamellas at the location of these aggregates using cryo-focused ion beam milling (Bauerlein et al., 2017; Rigort et al., 2012) (Figure S1B–D). Lastly, the samples were transferred to a cryo-transmission electron microscope for high resolution 3D imaging by cryo-ET (Figure S1E, F).
Poly-GA aggregate cross-sections were typically ~3 μm in diameter, and consisted of a dense network of elongated polymorphic ribbons (Figure 1A, B). Whereas the thickness of the ribbons was well defined (13 – 15 nm), their length (100 nm – 1 μm) and width (20 – 80 nm) varied considerably (Figure 1D top). Our measurements likely underestimate ribbon length, as parts of ribbons oriented perpendicular to the electron beam were not reliably detected due to missing information along this direction (Lucic et al., 2005). The ribbons were twisted along their axis with a variable helical pitch, and often bifurcated and/or associated laterally with neighboring ribbons (Figure 1D bottom). This polymorphism contrasts with the uniform fibrils forming polyQ-expanded huntingtin exon 1 aggregates in mammalian cells (Bauerlein et al., 2017). Poly-GA ribbons were also more densely packed than polyQ fibrils, which occupied a lower fraction of the inclusion volume (poly-GA, ≥ 10 %; polyQ, ≤ 4 %). Thus, different amyloids adopt different morphologies in situ.
Figure 1. In Situ Neuronal Poly-GA Aggregates Form Twisted Ribbons.
(A) Tomographic slice of an aggregate within a (GA)175-GFP-transduced neuron (DIV 5 + 5). Colored boxes show macromolecules magnified in (C). (B) 3D rendering of the aggregate shown in (A). Selected poly-GA ribbons (red) magnified in (D) are indicated. (C) Series of higher magnification tomographic slices of representative protein complexes detected in the tomogram shown in (A). Yellow and magenta boxes show the typical smaller (yellow) and larger (magenta) ring-like structures found in the aggregate region. Blue and orange boxes show side views of single-capped (blue) and double-capped (orange) 26S proteasomes. (D) Selected ribbons from (B) rotated and magnified for visualization. Note the variable width of the ribbons (a–c). Some ribbons show bifurcations (d–e). (E, F) Higher magnification tomographic slices of aggregates within neurons transduced with (GA)175-GFP, DIV 5 + 5 (E) or untagged (GA)175, DIV 5 + 5 (F). Yellow boxes mark similar small ring-like structures as in (A). Note that (GA)175-GFP ribbons (red arrowheads) are decorated by additional densities (green arrowheads), which are missing from untagged (GA)175 ribbons. Tomographic slices are 5 nm thick. Scale bars: 200 nm (A, B), 50 nm (C–F). See also Figure S1.
However, similarly to GFP-tagged polyQ fibrils, GFP-labeled poly-GA ribbons were decorated by additional densities (Figure 1E). To investigate the nature of these densities, neurons were co-transduced with untagged poly-GA and tagRFP-p62, which co-localizes with poly-GA aggregates (May et al., 2014; Mori et al., 2013b; Yamakawa et al., 2015; Zhang et al., 2014) and allows targeting untagged poly-GA by correlative microscopy. As for polyQ fibrils (Bauerlein et al., 2017), the decorating densities were absent from untagged poly-GA ribbons (Figure 1F), demonstrating that these additional densities require GFP for their formation and that the ribbons consisted indeed of poly-GA aggregates. Thus, poly-GA forms amyloid-like ribbons in neurons.
26S Proteasomes Are Recruited to Poly-GA Aggregates
Unlike polyQ fibrils (Bauerlein et al., 2017), poly-GA ribbons did not visibly interact with cellular endomembranes. However, both the aggregate interior and periphery were densely populated by macromolecular complexes (Figure 1A, C, E, F). Ribosomes were abundant around poly-GA aggregates but largely absent from their interior. In contrast, the space between poly-GA ribbons was densely populated with macromolecules that appeared as ~10 nm rings in tomographic cross-sections. Larger (~20 nm) cross-sectioned rings were also found at the aggregate periphery and occasionally in the interior. To investigate the identity of these macromolecules, we performed unbiased subtomogram averaging (Figure S2A). A small set of particles were hand-picked from the tomogram, aligned and averaged. The resulting average was used as a template to search the tomogram for additional occurrences of the same structure. These additional particles were then visually inspected, aligned, classified and averaged again to produce a higher resolution average. The iterative application of this procedure yielded an average structure unequivocally corresponding to the 26S proteasome (Chen et al., 2016a; Huang et al., 2016; Schweitzer et al., 2016; Wehmer et al., 2017) for the smaller ring-like structures, and to the TRiC/CCT chaperonin (Leitner et al., 2012; Zang et al., 2016) for the larger ones (Figure 2, Figure 3, Figure S3A–C, Movie S1). Other large UPS components such as p97/VCP did not appear abundant at poly-GA aggregates.
Figure 2. Mapping Macromolecules within Poly-GA Aggregates Shows a Substantial Recruitment of 26S Proteasomes.
(A) 3D rendering of an aggregate within a neuron transduced with (GA)175-GFP (DIV 5 + 5) showing different macromolecules found either within or at the periphery of the aggregate. Poly-GA ribbons (red), 26S proteasomes (green), ribosomes (yellow), TRiC/CCT chaperonins (purple). The macromolecules are mapped in their original locations and orientations, computationally determined by template matching and subtomogram averaging. (B–E) Maximum intensity projection heat maps of the molecular species shown in (A). Note that the proteasomes are mostly found in between poly-GA ribbons, whereas ribosomes occur almost exclusively outside of the aggregate. TRiC/CCT molecules mostly populate the aggregate periphery, but some can also be found between poly-GA ribbons. See also Figure S2A, Figure S3, Figure S4, Figure S5 and Movie S1.
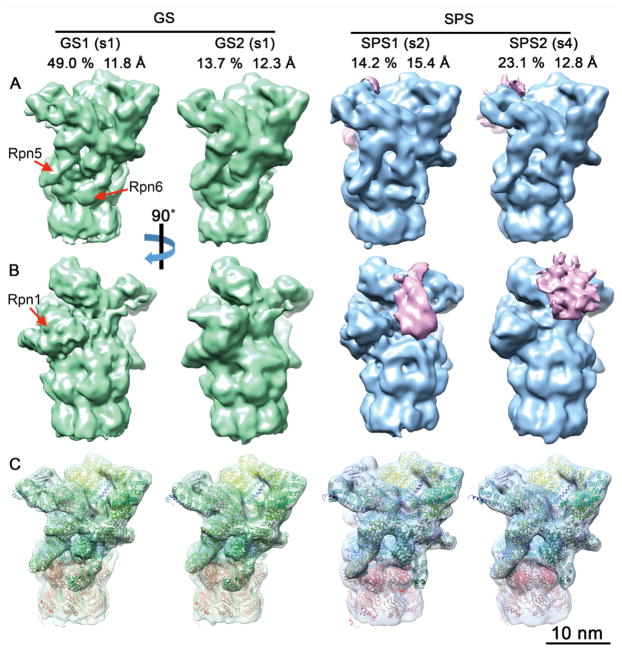
Figure 3. Subtomogram Classification of 26S Proteasomes Reveals Enrichment of Substrate Processing Conformations.
To analyze the functional state of proteasome regulatory particles single- and double-capped proteasomes were cut by the half of the CP. The resulting half proteasomes were classified according to RP conformation into ground or substrate processing states (Asano et al., 2015), yielding two ground states (GS1, GS2) and two substrate processing classes (SPS1, SPS2). (A, B) The four density maps are displayed in solid surface representation in two different views. The positions of the Rpn1, Rpn5 and Rpn6 subunits are indicated. Prominent densities in the substrate binding region of SPS1 and SPS2 are colored in pink. For each class, the percentage of the total number of classified particles and the global resolution are indicated. (C) Same view as (A), with semi-transparent maps superimposed with the atomic models generated by MDFF. The classes respectively represent the s1 state with different Rpn1 positions (GS1, GS2), the s2 state (SPS1) and the s4 state (SPS2). Atomic models are colored by subunits: Rpn1 (brown), Rpn2 (yellow), Rpn9/5/6/7/3/12 (different shades of green), Rpn8/Rpn11 (light/dark magenta), Rpn10 and Rpn13 621 (purple), AAA-ATPase hexamer (blue), and CP (red). See also Figure S2B, Figure S3 and Table S1.
The abundance of TRiC/CCT complexes was not significantly different around poly-GA aggregates compared to the cell body of control neurons (untransduced or transduced with GFP only). However, the estimated concentration of proteasomes within the aggregate (~7 μM) was approximately 30-fold higher than in the cell body (Figure S4B) or the processes (Asano et al., 2015) of control cells. Given that poly-GA expression did not increase overall proteasome expression levels (Figure S4C), these data suggest that proteasomes are removed from other regions of the cell to accumulate at poly-GA aggregates. This is consistent with immunofluorescence staining (Figure S4A) and biochemical fractionation experiments showing reduced levels of Triton-soluble neuronal proteasomes (Figure S4C). Furthermore, our tomograms showed that 26S proteasomes accumulated almost exclusively within the aggregate interior (Figure 2, Figure S5A–D). Taken together, these results show that a substantial fraction of neuronal 26S proteasomes is sequestered into poly-GA aggregates.
To test the influence of the poly-GA expression level on proteasome recruitment, we analyzed aggregates formed in neurons at an earlier time point after transduction (DIV 5 + 3). Although these aggregates were smaller, they contained a similar concentration of proteasomes in their interior (Figure S5E). Importantly, analogous observations were made for poly-GA aggregates generated from a RAN-translated (G4C2)73 construct, which more closely mimics the C9orf72 patient situation (Figure S4E, Figure S5F). Thus, poly-GA aggregate morphology and proteasome recruitment were comparable in all the experimental conditions tested.
Poly-GA Aggregation Alters Proteasome Structure
The 26S proteasome consists of a barrel-shaped 20S core particle (CP) that harbors the catalytic activity, bound to one or two 19S regulatory particles (RP). Regulatory particles are responsible for substrate recognition, unfolding and translocation into the CP for proteolysis. Recent single particle cryo-EM studies have revealed how RP conformational dynamics are coupled to the functional cycle of the 26S complex (Chen et al., 2016a; Unverdorben et al., 2014; Wehmer et al., 2017). Initial binding of substrates to the 26S proteasome presumably occurs in a low energy ground state (s1) (see also Lu et al., 2015). Bound substrates are committed for degradation (s2 state), and then translocated into the CP (s3-s4 states). In the s4 state the gate of the 20S CP is open, allowing the substrates to access the proteolytic chamber. We took advantage of the large number of proteasome complexes recruited to poly-GA aggregates to investigate their functional states in situ by subtomogram averaging and classification (Figure S2B).
We first sorted 26S proteasomes according to the number of regulatory particles (one or two) bound per CP. Previous structural (Asano et al., 2015) and biochemical (Tai et al., 2010) data indicated that in control neurons the large majority of 26S proteasomes contain only one RP (single-capped 26S). In striking contrast, 76% of poly-GA-associated 26S proteasomes were double-capped (Figure S3D, F). Therefore, the labile interaction between the proteasome core and regulatory particles (Kleijnen et al., 2007) is apparently stabilized within poly-GA aggregates.
We further classified 26S proteasomes according to RP conformation (Asano et al., 2015; Unverdorben et al., 2014). This yielded four well-defined classes, two of which (GS1, GS2) were consistent with the RP ground state conformation (s1), whereas the other two (SPS1, SPS2) corresponded to substrate processing states (s2-s4) (Figure 3). Interestingly, 37% of all 26S proteasomes belonged to substrate processing classes, almost twice the number than in control neurons (Asano et al., 2015) (Figure S3E, G). Thus, poly-GA aggregates recruit a large number of 26S proteasome regulatory particles, a substantial fraction of which adopts substrate processing conformations.
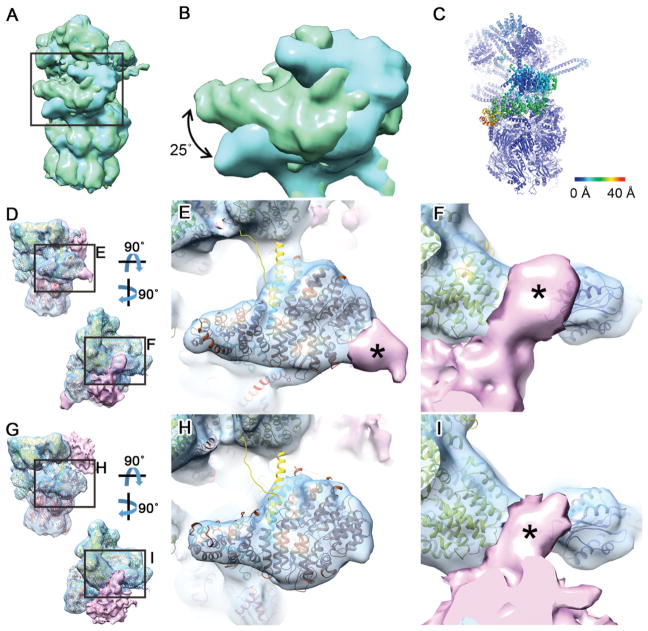
The relatively high resolution of the classes (11.8–15.4 Å, Figure 3, Figure S3H) enabled us to assign each class to a functional state. To this end we employed molecular dynamics flexible fitting (MDFF; Trabuco et al., 2009) initiated through the atomic models of the s1–4states of the yeast 26S proteasome (Figure 3C) (Wehmer et al., 2017). The s1 state was clearly the best fit for the GS1 and GS2 classes (Table S1). The yeast s1 structure fitted GS1 (49% of the total number of particles) without large discrepancies except for the position of the Rpn1 subunit, which in GS1 was similar to that observed in the human 26S proteasome (Chen et al., 2016a; Huang et al., 2016; Schweitzer et al., 2016). Also, in agreement with these studies, no prominent density was visible in our data for the Rpn13 subunit. The GS2 class (13.7% of particles) was overall similar to GS1, but the Rpn1 subunit pivoted 25° on its N-terminal region to shift its C-terminus towards the CP with respect to GS1 (Figure 4A–C). This is a novel conformation of Rpn1, a particularly dynamic subunit (Asano et al., 2015; Huang et al., 2016; Schweitzer et al., 2016; Wehmer et al., 2017) that serves as binding hub for 26S regulatory cofactors containing ubiquitin-like (UBL) domains (Elsasser et al., 2002; Leggett et al., 2002) and was recently identified as an ubiquitin receptor (Shi et al., 2016).
Figure 4. Detailed Structural Differences between 26S Proteasome Conformations Highlight its In Situ Structural Dynamics and Interactions.
(A) Superimposition of GS1 (green) and GS2 (cyan) density maps aligned by their CP. Both classes are consistent with the s1 state and differ only in the position of Rpn1. (B) Magnified view of the region boxed in (A) showing a 25° rotation of the Rpn1 subunit in the GS2 map. (C) Atomic model of the GS2 class colored according to the root mean square deviation (RMSD) from the GS1 model. Note that the only substantial differences are found in the Rpn1 region. (D) Two views of the SPS1 map (consistent with the s2 state) shown in surface representation superimposed with its atomic model. A prominent density in the substrate binding region is colored in pink. (E, F) Magnified view of the regions boxed in (D). The atomic models of Rpn1 (E), Rpn2 and Rpn10 (F) are shown in brown, yellow and purple respectively. Parts of the additional density denoted by asterisks may correspond to proteasome-bound ubiquitin or UBL domain proteins. (G) Two views of the SPS2 map (consistent with the s4 state) shown in surface representation superimposed with its atomic model. A prominent density in the substrate binding region is colored in pink. (H, I) Magnified view of the regions boxed in (G). Note that the density in the Rpn2/10 region is similar in the SPS1 and SPS2 class averages (I), whereas no additional density was found on the Rpn1 region of the SPS2 map (H).
SPS1 proteasomes (14.2% of particles) were most similar to the substrate commitment state s2 (Table S1). In contrast, the SPS2 class (23.1% of particles) was best fitted by the s4 model of actively translocating proteasomes (Table S1). This is remarkable because, in vitro, the s4 state was only recently detected in proteasomes incubated with non-hydrolysable nucleotide analogs (Wehmer et al., 2017), suggesting that it is normally a highly transient conformation. Thus, the interaction of poly-GA aggregates with the proteasome appears to stall its s4 conformation.
Both substrate processing classes showed prominent additional densities in the substrate binding region of the proteasome, in contact with the ATPase ring (Figure 3). In SPS1 the density was well defined (Figure 3), indicating a relatively stable interaction with the 26S complex that may in part correspond to bound UBL domain proteins (Aufderheide et al., 2015; Bashore et al., 2015). The density contacted Rpn1 (Figure 3, Figure 4D, E), consistent with the bound UBL domains of Rad23 or the ubiquitin C-terminal hydrolase 6 (Ubp6)/USP14 (Chen et al., 2016b; Shi et al., 2016), and extended to another binding site of Rad23 at the Rpn10 subunit (Hiyama et al., 1999; Mueller and Feigon, 2003; Walters et al., 2002) (Figure 4D, F). Interestingly, similar to our SPS1 class, Ubp6-bound proteasomes have been shown to mainly adopt the s2 conformation (Aufderheide et al., 2015). Because the Rpn1 and Rpn10 UBL binding sites also interact with ubiquitin, proteasome-bound ubiquitinated substrates may also contribute to the extra density. Consistent with this notion, the estimated molecular mass of the density (~ 70 kDa) was larger than Ubp6 (56 kDa) or Rad23 (40/44 kDa for Rad23A/B). For the SPS2 class, the additional density contacted the Rpn10 subunit but not Rpn1 (Figure 4G–I). The density was overall less well defined than for SPS1, indicating a more dynamic interaction, perhaps involving a more extensive participation of substrates (see below). Therefore, both substrates and cofactors may contribute to the additional densities found on substrate processing proteasomes.
Direct Interactions with Poly-GA Aggregates Impair Proteasome Function
To address the physiological role of the different proteasome conformations observed, we investigated their cellular distribution by mapping the particles back into the tomograms (Figure 5). We found that proteasome conformation correlated with the distance to poly-GA ribbons (Figure 5B, C, E, p < 0.001, Chi-square test, N = 6080 regulatory particles from 4 tomograms). For proteasomes directly touching (Figure 5D) or very close to ribbons the SPS2 class was overrepresented (36% vs 23% of the total proteasomes; Figure 5E), whereas the fraction of GS1 proteasomes was smaller than within the total (40% vs 49%; Figure 5E). The fraction of GS1 proteasomes increased with their distance to ribbons, whereas SPS2 proteasomes followed the opposite trend. For SPS2 proteasomes associated with poly-GA ribbons, the contact interface was consistent with the location of the additional density observed in this class (Figure 3, Figure 4G–I). Only small variations were found in the fractions of GS2 and SPS1 particles with respect to the distance to poly-GA ribbons (Figure 5E). These results indicate that association with poly-GA aggregates modifies the functional state of the 26S proteasome.
Figure 5. Spatial Mapping of Proteasomes within Poly-GA Aggregates Reveals Poly-GA Influence on Proteasome Conformation.
(A) 3D rendering of an aggregate within a neuron transduced with (GA)175-GFP (DIV 5 + 5). Poly-GA ribbons (red), proteasomes in ground state (green), proteasomes in substrate processing states (blue). Proteasome location and orientation were determined by template matching and subtomogram averaging. (B, C) Magnification of the region boxed in (A) showing only proteasomes less than 15 nm away from poly-GA ribbons (B) or 30 – 45 nm away (C). Poly-GA ribbons are shown as a transparent red surface. Note that substrate processing proteasomes are more abundant close to poly-GA ribbons. (D) Examples of SPS2 proteasomes directly touching poly-GA ribbons in the tomogram shown in (A). The additional density in the substrate binding region overlaps with the poly-GA ribbons. (E) Plot of proteasome conformation vs. distance to poly-GA ribbons. The influence of the distance to poly-GA ribbons in proteasome conformation was statistically significant (p < 0.001, Chi-square test, N = 6080 regulatory particles from 4 tomograms). Scale bars: 200 nm (A), 100 nm (B, C). See also Figure S4.
In agreement with this notion, functional measurements show that poly-GA expression impairs proteasome function (Figure S4D) (Yamakawa et al., 2015; Zhang et al., 2014). Interestingly, GA-rich sequences have been reported to slow or even stall proteasomal substrate processing in the context of the Epstein-Barr virus-encoded nuclear antigen 1 protein (Hoyt et al., 2006; Kraut, 2013; Levitskaya et al., 1997). Whereas in vitro the s4 state was only observed in the presence of non-hydrolysable nucleotide analogs (Wehmer et al., 2017), 23% of all proteasomes within neuronal poly-GA aggregates, and 36% of the proteasomes located in the immediate vicinity of poly-GA ribbons adopted the s4 conformation in situ (SPS2 class). Therefore, our results suggest that proteasomal degradation is slowed down by poly-GA-mediated stalling of the otherwise highly transient s4 state. This may play an important role in the proteostasis impairment observed in poly-GA models.
Discussion
Previous studies using classical EM reported that cellular poly-GA inclusions consist of a network of filaments 15–17 nm in diameter (Zhang et al., 2016; Zhang et al., 2014). Our 3D imaging of unstained fully hydrated neurons shows that rather than filaments, poly-GA forms twisted ribbons similar to those observed in vitro for (GA)15 (Chang et al., 2016). Whereas most amyloids are believed to be largely unbranched (Knowles et al., 2014), poly-GA ribbons bifurcated extensively. Together with their variable width, this suggests that in situ poly-GA ribbons are formed by different numbers of laterally stacked protofilaments. Furthermore, the similar morphology of (GA)15 (Chang et al., 2016) and (GA)175 (this study) ribbons is consistent with a molecular arrangement in which stacked GA-repeats give rise to the long axis of the protofilament.
Poly-GA aggregates recruited striking numbers of 26S proteasomes, whereas other macromolecules were excluded from the aggregate interior. This is remarkably different from our recent observations on polyQ inclusions, which interact with and may disrupt the membranes of the endoplasmic reticulum and other organelles, but do not harbor substantial numbers of 26S proteasomes or other large macromolecules (Bauerlein et al., 2017). This difference is surprising as proteasomes were also reported to colocalize with polyQ aggregates (Bennett et al., 2005; Waelter et al., 2001). Thus, different aggregating proteins may trigger UPS dysfunction and cellular toxicity by distinct mechanisms, and how proteasomes associate with other disease-related aggregates remains to be elucidated (Deriziotis et al., 2011; Myeku et al., 2016). Future work should also address the extent of proteasome recruitment by aggregates of poly-GA proteins expressed at endogenous levels, as well as by other C9orf72 dipeptide-repeat proteins.
UPS impairment is known to play an important role in poly-GA induced toxicity (Yamakawa et al., 2015; Zhang et al., 2016; Zhang et al., 2014). The proteasome and other UPS components are major poly-GA interactors in neurons (May et al., 2014), and poly-GA expression leads to reduced proteasome activity (Yamakawa et al., 2015; Zhang et al., 2014 and our results). Our data provides mechanistic insights into these phenomena. First, poly-GA aggregates sequester a large fraction of cellular proteasomes, depleting them from other cellular functions critical for proteostasis maintenance, such as ER-associated degradation (Zhang et al., 2014). This may be particularly damaging to neurons given their extended morphology. Second, many of the poly-GA associated proteasomes may be functionally impaired. The fraction of double-capped proteasomes and proteasome RPs in substrate processing conformations was much higher within poly-GA aggregates than in control cells. Given that CP/RP interactions are stabilized during substrate degradation (Kleijnen et al., 2007), these data suggest that many poly-GA-associated proteasomes are processing substrates or stalled in the process. Consistently, our analysis shows that contact with poly-GA ribbons causes the proteasomes to adopt the so-called s4 conformation, an otherwise highly transient intermediate state of substrate translocation (Wehmer et al., 2017). This is in line with previous reports that GA-rich sequences slow proteasomal translocation or even stall it (Kraut, 2013; Levitskaya et al., 1997). Interestingly, despite the strong association between proteasomes and poly-GA aggregates observed here, inhibiting proteasomal degradation does not affect poly-GA levels (Yamakawa et al., 2015). Therefore, the recruitment of proteasomes to poly-GA aggregates may be unproductive and may not lead to poly-GA degradation. The mechanisms driving such recruitment, which may involve ubiquitination of poly-GA and/or of associated factors, require further investigation.
Our data may also provide insights into the cellular mechanisms of proteasome regulation in the presence of protein aggregates. Poly-GA-associated proteasomes in substrate processing states showed additional densities that may correspond to bound ubiquitin and/or UBL domain-containing cofactors such as the deubiquitinating enzyme Ubp6/USP14 or the substrate shuttle factor Rad23 (Aufderheide et al., 2015; Bashore et al., 2015; Chen et al., 2016b; Shi et al., 2016). Although the binding of these factors to poly-GA-associated proteasomes remains to be conclusively demonstrated, several UBL domain proteins (Rad23, ubiquilin2 or Bag6) were highly enriched in the poly-GA interactome (May et al., 2014), and Rad23 is an important regulator of poly-GA induced toxicity (Zhang et al., 2016). UBL domain proteins strongly modulate proteasome activity (Finley et al., 2016) and have recently been implicated in neurodegeneration and aggregate clearance (Deng et al., 2011; Hjerpe et al., 2016). Our results are consistent with the notion that UBL domain cofactors regulate the interactions of the proteasome with protein aggregates.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Wolfgang Baumeister (baumeist@biochem.mpg.de).
Experimental Model and Subject Details
Cell Culture
HEK293 cells (female) stably expressing UbG76V-GFP (Dantuma et al., 2000; De Smet et al., 2017) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Biochrom) supplemented with 10 % (v/v) fetal bovine serum (Gibco), 2 mM L-glutamine (Gibco), penicillin-streptomycin (Thermo Fisher) and non-essential amino acids (Gibco). Transfection was carried out using FuGENE 6 (Promega).
Rats (IGS background, Charles River; RRID:RGD_734476) were housed in a pathogen-free facility with 12:12 h light/dark cycle and food/water available ad libitum. All animal experiments were performed in compliance with institutional policies approved by the government of Upper Bavaria following §11 Ab.1 TierSchG for the DZNE animal facility (Inst.-Nr. 04-26). Primary cortical neurons were prepared from embryonic day 19 animals of both sexes. Neocortex and hippocampus were dissected in ice-cold dissection media (HBBS, penicillin-streptomycin, 10 mM HEPES pH 7.3). The tissue was enzymatically dissociated at 37 °C (for cortices 20 min in 0.25 % trypsin, 0.7 mg/ml DNase I; for hippocampi 15 min in 0.15 % trypsin in dissection media) followed by gentle trituration.
For EM analysis, 250,000 cells/ml cortical neurons were plated on EM grids and cultured in Neurobasal medium containing 2 % B27 (Thermo Fisher), penicillin-streptomycin, 0.5 mM L-glutamine (Thermo Fisher). For biochemical analysis, 250,000 cells/ml cortical neurons were plated 771 on 12-well plates (Thermo Fisher) and cultured in Neurobasal medium. For immunofluorescence experiments, 85,000 cells/ml hippocampal neurons were plated on 12-well plates containing glass coverslips (VWR) coated with poly-D-lysine and cultured in Neurobasal medium supplemented with 12.5 μM glutamate.
Method Details
Lentivirus Packaging
HEK293FT cells (female; from Thermo Fisher) of low passage number were plated in three 10 cm dishes (5,000,000 cells/dish) and cultured in DMEM (Thermo Fisher), penicillin-streptomycin, 1 % non-essential amino acids (Thermo Fisher) and 10 % fetal bovine serum (Sigma). A transfection mix was set up as follows: 18.6 μg transfer vector (FhSynW-(GA)175-GFP, FhSynW (GA)149, FU3a-tagRFP-p62 or FhSynW-GFP), 11 μg pSPAX2 and 6.4 μg pVSVg in 4.5 ml Opti-MEM were combined with 108 μl Lipofectamine 2000 in 4,5 ml Opti-MEM (Thermo Fisher) and incubated for 20 minutes. Cell media was replaced with 5 ml Opti-MEM and 3 ml of transfection mix were added per dish. The transfection media was replaced after 6 h by plating media supplemented with 13 mg/ml bovine serum albumin, and the supernatant was collected after additional 24 h. Lentiviral particles were harvested by ultracentrifugation using a Sw28 rotor (22,000 rpm, 2 h), resuspended in 150 μl Neurobasal media on a rocking platform overnight and stored in aliquots at −80 °C.
Generation of pcDNA3.1 – STOP-GA(G4C2)73-GFP
The method to generate the construct was adapted from Lee et al. (Lee et al., 2013). Complementary G4C2 repeat oligonucleotides with phosphorylated ends (5′-PHOS-(G4C2)5-3′ and 5′-PHOS-CC(G2C4)4GGCC-3′) were energetically optimized to maximize heterodimer formation. After initial denaturation for 5 min at 95°C in T4 ligase buffer (New England Biolabs), the complementary oligonucleotides were allowed to anneal by slow cool down to room temperature during 2 h. Hybridized oligonucleotides were then self-ligated for 1 h to extend the G4C2 DNA sequence. Adapter oligonucleotides containing NheI (5′) and BamHI (3′) restriction sites (5′ adapter oligonucleotides containing NheI: 5′-GCCGTCAAGGCCGCATCTAGTAGCTAGC-3′ and 5′-PHOS-CCGCTAGCTACTAGATGCGGCCTTGACGGC-3′; 3′ adapter oligonucleotides containing BamHI: 5′-PHOS-GGGATCCCTAGTACTGGGCCTCATGGGC-3′ and 5′-GCCCATGAGGCCCAGTACTAGGGATC-3′) were separately hybridized and sequentially added in excess to the ligation reaction mix to stop G4C2 elongation. Ligation products were separated in a 1.25% agarose gel, and bands running at the desired molecular size were excised. The DNA was then purified with the Zymolclean Gel DNA recovery kit (Zymo Research) according to the manufacturer’s guidelines. The resulting blunt-end fragments were phosphorylated and cloned into a dephosphorylated SmaI-restricted pUC18 plasmid for amplification. Colonies were screened for G4C2 repeat length, and a construct with 73 G4C2 repeats was chosen for further experiments. The selected sequence was subcloned into the mammalian expression vector pcDNA3.1(+)/myc-His between NheI and BamHI in front of a GFP-encoding region. Finally, multiple stop codons were added by site-directed mutagenesis in every reading frame at the 5′ of the sequence of interest, thereby removing all initiator codons between the T7 promoter and the sequence of interest. The sequence was verified by multiple sequencing reactions as well as restriction digest.
Immunofluorescence and Cellular Fractionation
Immunofluorescence stainings were performed on primary hippocampal neurons 5 days after infection with (GA)175-GFP or GFP lentivirus on day 5, or 3 days upon transfection with G4C2 on day 3 using Lipofectamine 2000 (Thermo Fischer). Cells were fixed for 10 min at room temperature (4 % paraformaldehyde, 4 % sucrose in PBS). Anti-PSMC4 (Bethyl Laboratories, 1:250, RRID:AB_2620201) or anti-GA (Mackenzie et al., 2013, 1:200) primary antibodies as well as secondary antibodies (Alexa 555, Alexa 647, Thermo Fisher, 1:400, RRID:AB_2535850, RRID:AB_141780, RRID:AB_2535813) were diluted in staining buffer (0.1 % gelatin, 0.3 % Triton X-100, 450 mM NaCl, 16 mM sodium phosphate pH 7.4). After mounting the coverslips, images were taken using a LSM710 confocal microscope (Carl Zeiss, Jena) with a 63× oil immersion objective.
Biochemical experiments were performed on primary cortical neurons 10 days after infection with either (GA)175-GFP or GFP lentivirus on day 3. Cells were lysed (PBS with 1 % Triton-X-100, 15 mM MgCl2, 0.2 mg/ml DNase with protease inhibitor cocktail). Upon centrifugation (18,000 x_g_, 4 °C for 30 min) the soluble fraction was collected from the supernatant. The pellets were washed twice with lysis buffer yielding the insoluble fraction. After adding 4x Laemmli buffer (Bio-Rad), samples were denaturated (95 °C for 10 min) and loaded on Tricine gradient gels (Thermo Fisher). The following antibodies were used: Anti-PSMC4 (Bethyl Laboratories, 1:1000), anti-GFP (NeuroMab, 1:5000, RRID:AB_10671445) and anti-Calnexin (Enzo Life Sciences, 1:7000, RRID:AB_10616095). The data in Figure S4C was normalized to the total proteasome levels in GFP-transduced control neurons.
Flow Cytometry
HEK293 cells stably expressing UbG76V-GFP were harvested 72 h after transient transfection with (GA)175-tagRFP or tagRFP as control and analyzed with a BD FACSAriaIII flow cytometer, as described previously (Hipp et al., 2012a; Hipp et al., 2012b). In brief, to ensure a sufficient number of cells with elevated levels of the transfected proteins >100,000 events were analyzed per condition. To plot the level of the reporter protein versus the level of the transfected protein, a set of gates was established in the tagRFP channel. HEK293 cells transfected with tagRFP were used as single-color control to compensate the bleed-through between GFP and tag-RFP individually for each gate, and to correct for effects due to high expression of tagRFP. The compensated mean fluorescence of the 855 reporter protein UbG76V-GFP in each of these gates was plotted on the y-axis and the gate number (corresponding to the log of fluorescence intensity of the transfected protein) was plotted on the x-axis. The data shown in Figure S4D are from a single representative experiment out of three independent repeats. Cells expressing low levels of tagRFP (gates 1–9) and gates with < 1000 events were not included in the analysis. Raw flow cytometry data were analyzed using FlowJo software (version 9.9; Tree Star).
Cryo-EM Sample Preparation
Quantifoil grids (R2/1, Au 200 mesh grid, Quantifoil Micro Tools, Germany) were coated with an additional carbon layer (~20 nm thick) using a carbon evaporator (MED 020, BAL-TEC). Before use, the grids were glow discharged using a plasma cleaner (PDC-3XG, Harrick) for 20 seconds and neurons were seeded as described above. Neurons were vitrified 5 days after transduction with GFP or tagRFP-p62/untagged (GA)175 on day 5, 3 or 5 days after transduction with (GA)175-GFP on day 5, and 3 days after transfection with 881 (G4C2)73 on day 3. Untransduced and untransfected control neurons were vitrified on DIV 10.
For vitrification, the grids were blotted for 10 seconds from the back side using filter paper and immediately plunged into a liquid ethane/propane mixture (Tivol et al., 2008) using a manual plunge freezer. Grids were transferred to sealed boxes and stored in liquid nitrogen until usage.
Cryo-Fluorescent Light Microscopy and Cryo-FIB Microscopy
EM grids were mounted onto modified Autogrid (FEI, Hillsboro, OR, USA) sample carriers, allowing subsequent FIB milling with a shallow incident ion beam (Rigort et al., 2012), and then transferred to the cryo-stage of an FEI CorrSight microscope for cryo-light microscopy. Overview images of the grid and poly-GA-GFP/tagRFP-p62 signal were respectively acquired in transmitted light and the spinning disk confocal modes using a 20x lens (air, N.A. 0.8). Image acquisition was done with FEI MAPS software. The samples were then transferred into a FEI Scios dual beam cryo-FIB/scanning electron microscope (SEM) using a cryo-transfer system.
To improve the sample conductivity, a layer of organometallic platinum was deposited onto the grid using the in situ gas injection system with the following parameters: 10 mm working distance, 26°C pre-heating temperature, and 8 s gas injection time. MAPS software allowed correlation between cryo-light microscopy and SEM images via the 3-point alignment method. Thin lamellas were prepared in the regions of poly-GA-GFP/tagRFP-p62 fluorescence signal using the Ga2+ ion beam at 30 kV under a 20° stage tilt angle. 0.5 nA beam current was used for rough milling, followed by sequentially lower currents during the thinning procedure. A current of 30 pA was used for the final polishing step to reach a final lamella thickness of 150–200 nm. SEM imaging was used to monitor the milling progress.
Cryo-Electron Tomography and Reconstruction
The specimens were examined at liquid nitrogen temperature in an FEI Titan Krios cryo-electron microscope operated at 300 kV and equipped with a field emission gun and a Gatan post column energy filter. Lamellas were loaded vertical to the tilt axis, and were precisely aligned by adjusting the β angle of the cryo-stage. Images were collected using a 4 k x 4 k K2 Summit (Gatan) direct detector camera operated in dose fractionation mode (0.4 s, 0.3 electrons/Å2 per frame). Tilt series were recorded using SerialEM software (Mastronarde, 2005) at a nominal magnification of 42,000 X, resulting in a pixel size of 3.42 Å at the specimen level. Unidirectional tomographic tilt series were recorded from −50° to +70° with an increment of 2°. On average, 6 frames were collected for each image resulting in a total dose between 100 e−/Å2 to 110 e−/Å2 per tilt series. K2 frames were aligned using in house software (K2Align) based on previous work (Li et al., 2013). Tilt series were aligned using fiducial-less patch tracking, and the tomograms were reconstructed by weighted back projection using the IMOD software package (Kremer et al., 1996). For tomograms of lamellas superficially contaminated by ice crystals, a surface cleaning procedure was employed after alignment (Fernandez et al., 2016). The resulting tilt series were then aligned and reconstructed again to obtain the final reconstructions.
Template Matching and Subtomogram Averaging
To identify the macromolecules found in the tomograms, a de novo subtomogram averaging procedure without any external structural information was developed (Figure S2A). The MATLAB (Mathworks) TOM toolbox (Nickell et al., 2005) was used as general platform for image processing. Firstly, all the tomograms were binned twice (13.68 Å3 per voxel) for processing. In one tomogram, several identical small ring-like structures were hand-picked, aligned and averaged to obtain a first tube-like average structure. This was used as an initial template for template matching in all the binned tomograms using PyTom (Hrabe et al., 2012). The resulting subtomograms were cropped, CTF-corrected and classified using Relion (Bharat and Scheres, 2016). The resulting structure clearly showed a 20S proteasome core complex, but because of the missing wedge and the preferred orientation induced by the initial template, the 19S regulatory particles (RP) were not well resolved. This structure was used to perform a new round of template matching and classification, which clearly resolved proteasomes either single- or doubled-capped. The single-capped proteasome were low-pass filtered to 40 Å and used as a reference for template matching again to produce the final dataset. In total, 10,367 proteasome subtomograms of 1803 pixel volume were picked for further analysis from 9 tomograms containing large (GA)175-GFP aggregates (DIV 5 + 5). The same template matching procedure was applied to tomograms of control neurons, either untransduced (16 tomograms) or transduced with GFP only (17 tomograms).
The subtomograms were then 3D classified using Relion (Figure S2B). They were firstly divided into single-capped and double-capped proteasomes. To further rule out reference bias, a single-capped proteasome, a double-capped proteasome and a mirrored single-capped proteasome were used as references for classification. The results were similar, indicating negligible reference bias during classification. To further analyze the conformational status of the 19S regulatory particles, all subtomograms were cut in silico between the β-rings of the 20S, resulting in two independent particles for the double-capped proteasomes (Asano et al., 2015). All cut-out half proteasomes were merged into a new dataset for another round of classification. Ground state and substrate processing state structures were distinguished by the relative orientation of the Rpn5 and Rpn6 subunits (Asano et al., 2015; Unverdorben et al., 2014). Ground state and substrate processing state structures were further classified by applying a soft sphere mask in the RP region only. Identical subclasses were merged resulting in two ground state and two substrate processing state classes, which were further refined to achieve the final structures. A similar analysis could not be applied to control neurons (GFP-transduced or untransduced) due to the low numbers of proteasomes found in these samples. Therefore, the results from untransduced neurons from a previous study (Asano et al., 2015) were used as reference. Visualization of the subtomogram averages was performed in UCSF Chimera (Pettersen et al., 2004). Resolution was determined using the 0.143 criterion according to the gold standard Fourier Shell Correlation (Scheres and Chen, 2012).
To identify the larger ring-like structures found in the tomograms, a similar template matching procedure was employed. A spherical structure was generated after processing, which showed a clear eight-fold symmetry by rotational correlation coefficient analysis (Figure S3B). Furthermore, the size of the average fitted well with the crystal structure of TRiC/CCT (PDB 4V94) (Leitner et al., 2012), indicating that these larger rings correspond to the TRiC/CCT chaperonin. A total of 1,366 TRiC subtomograms were used for averaging. For ribosome template matching, a human 80S ribosome structure (EMDB 2938) (Khatter et al., 2015) was filtered to 40 Å and used as a template.
Atomic Model Fitting
The S. cerevisiae proteasome models of the states s1 (PDB 5MP9, 5MPD), s2 (PDB 5MPA, 5MPE), s3 (PDB 5MPB), and s4 (PDB 5MPC) (Wehmer et al., 2017) were used as initial models. The core particles of the models were fitted into each of the groups obtained by classification of the poly-GA proteasome dataset using rigid body docking within UCSF Chimera. For each class, MDFF was performed to refine the s1–4models according to the density maps. Then, the RMSD between the best fitting atomic models and the initial s1–4state models was calculated (Table S1). This showed that the GS1 class reflected the s1 state with the Rpn1 positioning of the human s1 state (PDB 5L5K) (Schweitzer et al., 2016), GS2 reflected the s1 state with a rotated Rpn1 position, SPS1 reflected the s2 state and SPS2 reflected the s4 state. To obtain the final atomic rat models, the best fitting yeast structures for each class were used as templates for comparative modeling and real-space structure refinement (Goh et al., 2016). MDFF simulations were prepared using QwikMD (Ribeiro et al., 2016), analyzed with VMD (Humphrey et al., 1996), and carried out with NAMD (Phillips et al., 2005).
Segmentation of Poly-GA Aggregates and Distance Measurement
Poly-GA ribbons are locally planar and were consequently segmented using a filter based on tensor voting, which distinguishes planar-like from line- and blob-like structures (Martinez-Sanchez et al., 2014). The filter outputs a scalar map, where voxel intensity value is proportional to the local similarity with a plane. The final segmentation was generated by thresholding the filter output combined with a manually generated mask to discard other locally planar structures like membranes and correct minor artifacts. Due to the missing information along the electron beam direction (Lucic et al., 2005), poly-GA ribbons were not clearly visible when oriented within the xy plane. 984 Segmentations were visualized using Amira (Thermo Fischer, RRID:SCR_014305).
For Figure 5, proteasome regulatory particles and poly-GA ribbons were represented by an isosurface, properly placed and oriented in their original tomogram. For regulatory particles, surfaces were obtained by applying the marching cubes algorithm, implemented in the VTK Open Source library (www.vtk.org; RRID:SCR_015013; Schroeder et al., 2006), on the corresponding subtomogram average. Isosurface threshold was set manually for every case with the criterion of choosing the minimum value avoiding noisy features. For poly-GA ribbons, isosurfaces were generated from the output of the planar filter using the marching cubes algorithm. The final ribbon surfaces were obtained by masking the isosurface with the corresponding ribbon segmentation from the tomogram. To calculate the fraction of the volume of poly-GA inclusions occupied by poly-GA ribbons, total ribbon volume was measured from the segmentations and divided by the total tomogram volume. For this calculation, tomograms (N = 4, (GA)175-GFP transduced neurons at DIV 5 + 5) were selected in which the large majority of the volume contained poly-GA aggregates.
985 RP to poly-GA ribbon distance was defined as the shortest Euclidean distance between the center of the RP to any surface point of any ribbon in the same tomogram. A custom Python software package was developed to measure RP-to-ribbon distance for every RP in all tomograms. The results were grouped by particle class to facilitate statistical analysis. The shortest distance among surfaces was computed with the help of a VTK library (Schroeder et al., 2006).
Quantification and Statistical Analysis
To measure proteasome concentration, for each tomogram (N = 9, (GA)175-GFP transduced neurons, DIV 5 + 5; N = 17, GFP transduced neurons; N = 16, untransduced neurons) the number of proteasomes found by template matching and subtomogram averaging was divided by the total tomogram volume. Because other cellular structures (such as poly-GA aggregates, other macromolecules or cellular organelles) were also present in the tomogram volume, this calculation underestimates the cytosolic concentration of proteasomes. For poly-GA aggregates, only tomograms with more than half volume occupied by aggregates were considered for the concentration calculation. In Figure S4B, the top and bottom boundaries of the boxes indicate ± 2x standard error, and whiskers extend to the maximum and minimum values. Statistical analysis was performed by the non-parametric Mann-Whitney test, as not all data was normally distributed according to the Shapiro-Wilk test.
For the analysis of proteasome conformation as a function of the distance to poly-GA ribbons (Figure 5B, C, E), 6080 regulatory particles from 4 tomograms of (GA)175-GFP transduced neurons (DIV 5 + 5) were analyzed. The data were divided in a 4x4 table (4 RP states x 4 distance bins) resulting in 9 degrees of freedom. Statistical analysis was performed by Chi-square test.
The proteasome levels in the total, soluble and insoluble fractions of (GA)175-GFP transduced neurons were quantified from western blots by measuring the grey levels using Fiji (Schindelin et al., 2012). Error bars in Figure S4C indicate standard error. All data was normally distributed according to the Shapiro-Wilk test (95 % confidence level). Statistical analysis was performed by two-sided paired t-test (N = 6 replicates from 4 independent experiments). All graphs were plotted using OriginPro (OriginLab).
Data and Software Availability
The Python scripts used to calculate the distance between proteasomes and poly-GA ribbons are available at https://github.com/anmartinezs/poly-GA.git
The tomogram analyzed in Figure 2 has been deposited at the Electron Microscopy Data Base (EMDB) with accession number EMD-4191. The structures of the proteasome and TRiC/CCT obtained by in situ subtomogram averaging have been deposited at EMDB with the following accession numbers: EMD-3916 (GS1 proteasome), EMD-3913 (GS2 proteasome), EMD-3914 (SPS1 proteasome), EMD-3915 (SPS2 proteasome), EMD-3917 (TRiC/CCT). The fitted atomic models of the different proteasome states have been deposited at the Protein Data Base with the following accession numbers: 6EPF (GS1), 6EPC (GS2), 6EPD (SPS1), 6EPE (SPS2).
Supplementary Material
S2
Figure S1. Correlative Cryo-Light and Electron Microscopy Workflow, Related to Figure 1. (A) Rat cortical neurons were cultured on EM grids and transduced with (GA)175-GFP on day 5. Upon vitrification by plunge freezing 5 days after transfection, the grids were imaged by cryo-light microscopy. (A) shows an overlay of phase and GFP channels. (B) EM grids were transferred to a cryo-FIB/SEM microscope. To locate cells of interest (boxed), SEM images were aligned and superimposed with the GFP signal from the cryo-LM image. (C) FIB-induced secondary electron image of the cell shown in (B), superimposed with the GFP signal. Yellow boxes mark the regions of the cell removed by FIB to produce a lamella containing the GFP-positive region. (D) SEM image of the lamella resulting from FIB milling, with a final thickness of ~ 200 nm, superimposed with the GFP signal. (E) Cryo-TEM low magnification image of the lamella shown in (D). (F) 5 nm-thick tomographic slice of a tomogram recorded in the area marked by a white square in (E), containing the GFP signal. A dense network of poly-GA-GFP ribbons (red arrowheads) is visible. The inset shows a high magnification of a poly-GA-GFP ribbon decorated by GFP-associated additional densities (green arrowheads). Scale bar: 20 nm.
Figure S2: Subtomogram Averaging and Classification Workflow, Related to Figure 2 and Figure 3. (A) Unbiased template matching procedure. Several identical structures were first hand-picked from one tomogram, aligned and averaged. The resulting average was used as a template to search more tomograms for additional occurrences of the same structure. These additional particles were then visually inspected, aligned, classified and averaged again to produce a higher resolution average. This average was low-pass filtered and used as a template to search all the tomograms to produce the final subtomogram dataset. (B) All proteasome subtomograms were first classified into single-capped or double-capped. To further analyze the conformational status of the regulatory particles, all the subtomograms were cut in silico between the β-rings of the core particle (Asano et al., 2015), resulting in two independent particles for double-capped proteasomes. Cut out caps from all proteasomes were merged and subjected to a further round of classification.
Figure S3: Subtomogram Averaging and Classification for TRiC/CCT and the 26S Proteasome, Related to Figure 2 and Figure 3. (A) Unsymmetrized average structure of TRiC/CCT at a resolution of 20.5 Å superimposed on its atomic model (purple; PDB 4V94). (B) Rotational correlation coefficient analysis of (A) indicating 8-fold symmetry. (C) Average structure of TRiC/CCT at a resolution of 17.1 Å upon application of D8 symmetry to (A) superimposed on its atomic model. Resolutions were estimated by gold-standard Fourier shell correlation using the 0.143 criterion. (D, F) Overall distribution of single-capped and double-capped 26S proteasomes within aggregates in (GA)175-GFP transduced neurons (D) and untransduced neurons (Asano et al., 2015) (F). (E, G) Distribution of the ground and substrate processing states for all regulatory particles within aggregates in neurons transduced with (GA)175-GFP (E) and untransduced (Asano et al., 2015) (G). (H) Gold-standard Fourier shell correlation curves of the four proteasome structures, yielding the following resolution estimations: 11.8 Å (GS1), 12.3 Å (GS2), 15.4 Å (SPS1) and 12.8 Å (SPS2).
Figure S4: Proteasome Abundance and Function at Poly-GA Aggregates, Related to Figure 2 and Figure 5. (A) Poly-GA-GFP aggregates (green) in primary neurons expressing a synthetic (GA)175-GFP construct with a start codon colocalize with proteasomes detected with a PSMC4/Rpt3 antibody (red). Proteasome staining is diffuse in GFP-expressing control cells. Nuclei are labeled with DAPI (blue). Scale bar: 50 μm. (B) Proteasome concentration for (GA)175-GFP aggregates (DIV 5 + 5), control neurons transduced with GFP only (DIV 5 + 5) or untransduced (DIV 10) (N = 9 tomograms, (GA)175-GFP; N = 17, GFP; N = 16, untransduced). For poly-GA aggregates, only tomograms with more than half volume occupied by aggregates were considered for the concentration calculation. The top and bottom boundaries of the boxes indicate ± 2x standard error, and whiskers extend to the maximum and minimum values. Proteasome concentration within poly-GA aggregates was significantly higher than in control conditions (***, p < 0.001 by Mann-Whitney test), whereas the difference between GFP-transfected and untransfected control neurons was not significant (n. s.). (C) Quantification of proteasome levels in the soluble and insoluble fractions as well as total, analyzed by immunoblotting. The data was normalized to the total proteasome levels in GFP-transduced control neurons. Error bars indicate standard error. * and ** indicate respectively p < 0.05 and p < 0.01 by paired t-test (N = 6 replicates from 4 independent experiments). Proteasomes are enriched in the insoluble fraction in the presence of poly-GA aggregates. (D) Poly-GA expression leads to the stabilization of a reporter for the ubiquitin-proteasome system. HEK293 cells stably expressing an unstable green fluorescent reporter for the ubiquitin-proteasome system (UbG76V-GFP) were transfected with the indicated constructs. After 72 h, UbG76V-GFP levels were analyzed by flow cytometry. The relationship between poly-GA-tagRFP levels on the x-axis and of UbG76V-GFP on the y-axis shows a concentration-dependent accumulation of UbG76V-GFP in the presence of poly-GA. The data shown are from a single representative experiment out of three independent repeats. (E) Poly-GA-GFP aggregates (green) in primary neurons expressing a non-ATG (G4C2)73 construct containing a downstream GFP in the poly-GA reading frame. RAN translation-derived poly-GA-GFP aggregates are stained by a poly-GA antibody (red) and colocalize with proteasomes detected with a PSMC4/Rpt3 antibody (white). Nuclei are labeled with DAPI (blue). Scale bar: 10 μm.
Figure S5: Macromolecule Mapping in Different Poly-GA Aggregates, Related to Figure 2. (A–C) Tomographic slices of aggregates within neurons transduced with (GA)175-GFP (main panels) and analyzed at DIV 5 + 5. Regions containing poly-GA ribbons are outlined in red. For the whole tomogram, proteasomes (green) and ribosomes (yellow) are mapped to their original positions and orientations by template matching and subtomogram averaging. For each tomogram, maximum intensity projection heat maps of poly-GA ribbons, proteasomes and ribosomes were generated (right column of each panel). (D) Numbers of proteasomes and ribosomes detected in the tomograms shown in (A–C) plotted versus the volume of the region containing poly-GA aggregates. The number of proteasomes detected shows a positive correlation with poly-GA aggregate volume. (E, F) Tomographic slices and mapped 26S proteasomes of aggregates within neurons transduced with (GA)175-GFP at an earlier time point (DIV 5 + 3) (E), or transfected with a RAN-translated (G4C2)73 construct with GFP in the poly-GA reading frame (F). Note that despite the smaller size compared to DIV 5 + 5 (GA)175-GFP aggregates, similar concentrations of proteasomes accumulated within these aggregates (~ 7 μM for (GA)175-GFP, DIV 5+3; ~ 5 μM for G4C2). Due to the lower thickness of the cryo-FIB lamella (70 nm vs. 100–200 nm in all other cases) fewer proteasomes are visible in (F). The inset in (F) shows a high magnification of a poly-GA ribbon (red arrowhead) decorated by GFP-associated additional densities (green arrowheads). Note that the morphology of RAN-translated poly-GA ribbons is indistinguishable from aggregates formed by the (GA)175-GFP construct. Tomographic slices are 7 nm thick. Scale bars: 200 nm in main panels, 20 nm in inset.
Table S1: Comparison of Atomic Models by RMSD Values, Related to Figure 3. Root mean square deviation (RMSD) values of RP atoms between the initial models (s1-s4 states from S. cerevisiae; Wehmer et al., 2017) and the atomic models fitted to the GS1, GS2, SPS1 and SPS2 classes. The Rpn1 and Rpn13 subunits were excluded due to their dramatically different conformation between yeast and mammalian proteasomes. The lowest RMSD values for GS1, GS2, SPS1 and SPS2 atomic models are for the s1, s1, s2 and s4 states respectively (underlined). Thus, GS1 and GS2 likely correspond to s1, SPS1 to s2 and SPS2 to the s4 state.
Movie S1: Mapping macromolecules to poly-GA inclusions, Related to Figure 2. Tomographic volume and 3D rendering of the neuronal (GA)175-GFP aggregate shown in Figure 2. Poly-GA ribbons (red), 26S proteasomes (green), ribosomes (yellow), TRiC/CCT chaperonins (purple). The macromolecules are mapped in their original locations and orientations, computationally determined by template matching and subtomogram averaging. Note that proteasomes accumulate in the aggregate interior and are often found direct proximity or even direct contact with poly-GA fibrils. In contrast, TRiC/CCT chaperonins and ribosomes are largely excluded from the aggregate and more abundant in its periphery.
Acknowledgments
We thank Radostin Danev, Günter Pfeifer, Jürgen Plitzko and Miroslava Schaffer for electron microscopy support, as well as Irina Dudanova and Eri Sakata for helpful discussions. The psPAX2 plasmid (Addgene plasmid # 12260) was a gift from Didier Trono. Q.G. is the recipient of postdoctoral fellowships from EMBO (EMBO ALTF 73-2015) and the Alexander von Humboldt Foundation. A. M.-S. is the recipient of a postdoctoral fellowship from the Séneca Foundation. This research has received funding from the European Commission (FP7 GA ERC-2012-SyG_318987–ToPAG, FP7 GA ERC-2013-CoG_617198 DPR-MODELS), the German Science Foundation [Excellence Cluster Center for Integrated Protein Science Munich (CIPSM), Munich Cluster for Systems Neurology (SyNergy) and SFB-1035/Project A01], the NOMIS Foundation, the Helmholtz Association and the National Institutes of Health (Center for Macromolecular Modeling and Bioinformatics, grant 9P41GM104601).
Footnotes
Author Contributions
Q. G. performed electron microscopy experiments, computationally analyzed the data and prepared figures. C. L., H. H. and M. S. H. performed cell culture and biochemistry experiments. A. M.-S. developed software procedures for data analysis. T. R. constructed structural models. F. B. contributed to the computational analysis of the data. M. P.-B. and F. F. contributed reagents. Q. G., M. S. H., F.-U. H., D. E., W. B. and R. F.-B. designed research. D. E. and R. F.-B. supervised experiments. Q. G., M. S. H., F.-U. H., D. E., W. B. and R. F.-B. wrote the manuscript.
Declaration of Interests
D. E. has a pending patent application on “Dipeptide-repeat proteins as therapeutic target in neurodegenerative diseases with hexanucleotide repeat expansion”.
References
- Asano S, Fukuda Y, Beck F, Aufderheide A, Forster F, Danev R, Baumeister W. A molecular census of 26S proteasomes in intact neurons. Science. 2015;347:439–442. doi: 10.1126/science.1261197. [DOI] [PubMed] [Google Scholar]
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, 3rd, Rademakers R, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufderheide A, Beck F, Stengel F, Hartwig M, Schweitzer A, Pfeifer G, Goldberg AL, Sakata E, Baumeister W, Forster F. Structural characterization of the interaction of Ubp6 with the 26S proteasome. Proc Natl Acad Sci U S A. 2015;112:8626–8631. doi: 10.1073/pnas.1510449112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashore C, Dambacher CM, Goodall EA, Matyskiela ME, Lander GC, Martin A. Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome. Nat Struct Mol Biol. 2015;22:712–719. doi: 10.1038/nsmb.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerlein FJB, Saha I, Mishra A, Kalemanov M, Martinez-Sanchez A, Klein R, Dudanova I, Hipp MS, Hartl FU, Baumeister W, et al. In Situ Architecture and Cellular Interactions of PolyQ Inclusions. Cell. 2017;171:179–187. e110. doi: 10.1016/j.cell.2017.08.009. [DOI] [PubMed] [Google Scholar]
- Beck M, Baumeister W. Cryo-Electron Tomography: Can it Reveal the Molecular Sociology of Cells in Atomic Detail? Trends Cell Biol. 2016;26:825–837. doi: 10.1016/j.tcb.2016.08.006. [DOI] [PubMed] [Google Scholar]
- Bennett EJ, Bence NF, Jayakumar R, Kopito RR. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol Cell. 2005;17:351–365. doi: 10.1016/j.molcel.2004.12.021. [DOI] [PubMed] [Google Scholar]
- Bharat TA, Scheres SH. Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION. Nat Protoc. 2016;11:2054–2065. doi: 10.1038/nprot.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YJ, Jeng US, Chiang YL, Hwang IS, Chen YR. The Glycine-Alanine Dipeptide Repeat from C9orf72 Hexanucleotide Expansions Forms Toxic Amyloids Possessing Cell-to-Cell Transmission Properties. J Biol Chem. 2016;291:4903–4911. doi: 10.1074/jbc.M115.694273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wu J, Lu Y, Ma YB, Lee BH, Yu Z, Ouyang Q, Finley DJ, Kirschner MW, Mao Y. Structural basis for dynamic regulation of the human 26S proteasome. Proc Natl Acad Sci U S A. 2016a;113:12991–12996. doi: 10.1073/pnas.1614614113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Randles L, Shi K, Tarasov SG, Aihara H, Walters KJ. Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome. Structure. 2016b;24:1257–1270. doi: 10.1016/j.str.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheroni C, Marino M, Tortarolo M, Veglianese P, De Biasi S, Fontana E, Zuccarello LV, Maynard CJ, Dantuma NP, Bendotti C. Functional alterations of the ubiquitin-proteasome system in motor neurons of a mouse model of familial amyotrophic lateral sclerosis. Hum Mol Genet. 2009;18:82–96. doi: 10.1093/hmg/ddn319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins GA, Goldberg AL. The Logic of the 26S Proteasome. Cell. 2017;169:792–806. doi: 10.1016/j.cell.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantuma NP, Bott LC. The ubiquitin-proteasome system in neurodegenerative diseases: precipitating factor, yet part of the solution. Front Mol Neurosci. 2014;7:70. doi: 10.3389/fnmol.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol. 2000;18:538–543. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- De Smet F, Saiz Rubio M, Hompes D, Naus E, De Baets G, Langenberg T, Hipp MS, Houben B, Claes F, Charbonneau S, et al. Nuclear inclusion bodies of mutant and wild-type p53 in cancer: a hallmark of p53 inactivation and proteostasis remodelling by p53 aggregation. J Pathol. 2017;242:24–38. doi: 10.1002/path.4872. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriziotis P, Andre R, Smith DM, Goold R, Kinghorn KJ, Kristiansen M, Nathan JA, Rosenzweig R, Krutauz D, Glickman MH, et al. Misfolded PrP impairs the UPS by interaction with the 20S proteasome and inhibition of substrate entry. EMBO J. 2011;30:3065–3077. doi: 10.1038/emboj.2011.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D, Haass C. An amyloid-like cascade hypothesis for C9orf72 ALS/FTD. Curr Opin Neurobiol. 2016;36:99–106. doi: 10.1016/j.conb.2015.10.009. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Muller B, Feng MT, Tubing F, Dittmar GA, Finley D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- Fernandez JJ, Laugks U, Schaffer M, Bauerlein FJ, Khoshouei M, Baumeister W, Lucic V. Removing contamination-induced reconstruction artifacts from cryo-electron tomograms. Biophys J. 2016;110:850–859. doi: 10.1016/j.bpj.2015.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D, Chen X, Walters KJ. Gates, Channels, and Switches: Elements of the Proteasome Machine. Trends Biochem Sci. 2016;41:77–93. doi: 10.1016/j.tibs.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Taylor JP. The Role of Dipeptide Repeats in C9ORF72-Related ALS-FTD. Front Mol Neurosci. 2017;10:35. doi: 10.3389/fnmol.2017.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126:829–844. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, Petrucelli L. Disease Mechanisms of C9ORF72 Repeat Expansions. Cold Spring Harb Perspect Med. 2017 doi: 10.1101/cshperspect.a024224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh BC, Hadden JA, Bernardi RC, Singharoy A, McGreevy R, Rudack T, Cassidy CK, Schulten K. Computational Methodologies for Real-Space Structural Refinement of Large Macromolecular Complexes. Annu Rev Biophys. 2016;45:253–278. doi: 10.1146/annurev-biophys-062215-011113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A, Varshavsky A. Basic Medical Research Award. The ubiquitin system. Nat Med. 2000;6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- Hipp MS, Bersuker K, Kopito RR. Live-cell imaging of ubiquitin-proteasome system function. Methods Mol Biol. 2012a;832:463–472. doi: 10.1007/978-1-61779-474-2_33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp MS, Park SH, Hartl FU. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 2014;24:506–514. doi: 10.1016/j.tcb.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Hipp MS, Patel CN, Bersuker K, Riley BE, Kaiser SE, Shaler TA, Brandeis M, Kopito RR. Indirect inhibition of 26S proteasome activity in a cellular model of Huntington’s disease. J Cell Biol. 2012b;196:573–587. doi: 10.1083/jcb.201110093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyama H, Yokoi M, Masutani C, Sugasawa K, Maekawa T, Tanaka K, Hoeijmakers JH, Hanaoka F. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J Biol Chem. 1999;274:28019–28025. doi: 10.1074/jbc.274.39.28019. [DOI] [PubMed] [Google Scholar]
- Hjerpe R, Bett JS, Keuss MJ, Solovyova A, McWilliams TG, Johnson C, Sahu I, Varghese J, Wood N, Wightman M, et al. UBQLN2 Mediates Autophagy-Independent Protein Aggregate Clearance by the Proteasome. Cell. 2016;166:935–949. doi: 10.1016/j.cell.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Zich J, Takeuchi J, Zhang M, Govaerts C, Coffino P. Glycine-alanine repeats impair proper substrate unfolding by the proteasome. EMBO J. 2006;25:1720–1729. doi: 10.1038/sj.emboj.7601058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabe T, Chen Y, Pfeffer S, Cuellar LK, Mangold AV, Forster F. PyTom: a python-based toolbox for localization of macromolecules in cryo-electron tomograms and subtomogram analysis. J Struct Biol. 2012;178:177–188. doi: 10.1016/j.jsb.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Huang X, Luan B, Wu J, Shi Y. An atomic structure of the human 26S proteasome. Nat Struct Mol Biol. 2016;23:778–785. doi: 10.1038/nsmb.3273. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW, 3rd, Sun S, Herdy JR, Bieri G, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci. 2015;18:1226–1229. doi: 10.1038/nn.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatter H, Myasnikov AG, Natchiar SK, Klaholz BP. Structure of the human 80S ribosome. Nature. 2015;520:640–645. doi: 10.1038/nature14427. [DOI] [PubMed] [Google Scholar]
- Kleijnen MF, Roelofs J, Park S, Hathaway NA, Glickman M, King RW, Finley D. Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites. Nat Struct Mol Biol. 2007;14:1180–1188. doi: 10.1038/nsmb1335. [DOI] [PubMed] [Google Scholar]
- Knowles TP, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- Kraut DA. Slippery substrates impair ATP-dependent protease function by slowing unfolding. J Biol Chem. 2013;288:34729–34735. doi: 10.1074/jbc.M113.512533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Kuhn PH, Wang H, Dislich B, Colombo A, Zeitschel U, Ellwart JW, Kremmer E, Rossner S, Lichtenthaler SF. ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. EMBO J. 2010;29:3020–3032. doi: 10.1038/emboj.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YB, Chen HJ, Peres JN, Gomez-Deza J, Attig J, Stalekar M, Troakes C, Nishimura AL, Scotter EL, Vance C, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5:1178–1186. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- Leitner A, Joachimiak LA, Bracher A, Monkemeyer L, Walzthoeni T, Chen B, Pechmann S, Holmes S, Cong Y, Ma B, et al. The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure. 2012;20:814–825. doi: 10.1016/j.str.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitskaya J, Sharipo A, Leonchiks A, Ciechanover A, Masucci MG. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein-Barr virus nuclear antigen 1. Proc Natl Acad Sci U S A. 1997;94:12616–12621. doi: 10.1073/pnas.94.23.12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Mao D, Bellen HJ. Amyotrophic Lateral Sclerosis Pathogenesis Converges on Defects in Protein Homeostasis Associated with TDP-43 Mislocalization and Proteasome-Mediated Degradation Overload. Curr Top Dev Biol. 2017;121:111–171. doi: 10.1016/bs.ctdb.2016.07.004. [DOI] [PubMed] [Google Scholar]
- Lu Y, Lee BH, King RW, Finley D, Kirschner MW. Substrate degradation by the proteasome: a single-molecule kinetic analysis. Science. 2015;348:1250834. doi: 10.1126/science.1250834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucic V, Yang T, Schweikert G, Forster F, Baumeister W. Morphological characterization of molecular complexes present in the synaptic cleft. Structure. 2005;13:423–434. doi: 10.1016/j.str.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Arzberger T, Kremmer E, Troost D, Lorenzl S, Mori K, Weng SM, Haass C, Kretzschmar HA, Edbauer D, et al. Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol. 2013;126:859–879. doi: 10.1007/s00401-013-1181-y. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Frick P, Grasser FA, Gendron TF, Petrucelli L, Cashman NR, Edbauer D, Kremmer E, Prudlo J, Troost D, et al. Quantitative analysis and clinico-pathological correlations of different dipeptide repeat protein pathologies in C9ORF72 mutation carriers. Acta Neuropathol. 2015;130:845–861. doi: 10.1007/s00401-015-1476-2. [DOI] [PubMed] [Google Scholar]
- Majounie E, Renton AE, Mok K, Dopper EG, Waite A, Rollinson S, Chio A, Restagno G, Nicolaou N, Simon-Sanchez J, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11:323–330. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Sanchez A, Garcia I, Asano S, Lucic V, Fernandez JJ. Robust membrane detection based on tensor voting for electron tomography. J Struct Biol. 2014;186:49–61. doi: 10.1016/j.jsb.2014.02.015. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- May S, Hornburg D, Schludi MH, Arzberger T, Rentzsch K, Schwenk BM, Grasser FA, Mori K, Kremmer E, Banzhaf-Strathmann J, et al. C9orf72 FTLD/ALS-associated Gly-Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol. 2014;128:485–503. doi: 10.1007/s00401-014-1329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Arzberger T, Grasser FA, Gijselinck I, May S, Rentzsch K, Weng SM, Schludi MH, van der Zee J, Cruts M, et al. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 2013a;126:881–893. doi: 10.1007/s00401-013-1189-3. [DOI] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013b;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- Mueller TD, Feigon J. Structural determinants for the binding of ubiquitin-like domains to the proteasome. EMBO J. 2003;22:4634–4645. doi: 10.1093/emboj/cdg467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myeku N, Clelland CL, Emrani S, Kukushkin NV, Yu WH, Goldberg AL, Duff KE. Tau-driven 26S proteasome impairment and cognitive dysfunction can be prevented early in disease by activating cAMP-PKA signaling. Nat Med. 2016;22:46–53. doi: 10.1038/nm.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickell S, Forster F, Linaroudis A, Net WD, Beck F, Hegerl R, Baumeister W, Plitzko JM. TOM software toolbox: acquisition and analysis for electron tomography. J Struct Biol. 2005;149:227–234. doi: 10.1016/j.jsb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144:67–78. doi: 10.1016/j.cell.2010.11.050. [DOI] [PubMed] [Google Scholar]
- Park SH, Kukushkin Y, Gupta R, Chen T, Konagai A, Hipp MS, Hayer-Hartl M, Hartl FU. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell. 2013;154:134–145. doi: 10.1016/j.cell.2013.06.003. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JV, Bernardi RC, Rudack T, Stone JE, Phillips JC, Freddolino PL, Schulten K. QwikMD - Integrative Molecular Dynamics Toolkit for Novices and Experts. Sci Rep. 2016;6:26536. doi: 10.1038/srep26536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigort A, Bauerlein FJ, Villa E, Eibauer M, Laugks T, Baumeister W, Plitzko JM. Focused ion beam micromachining of eukaryotic cells for cryoelectron tomography. Proc Natl Acad Sci U S A. 2012;109:4449–4454. doi: 10.1073/pnas.1201333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schludi MH, Becker L, Garrett L, Gendron TF, Zhou Q, Schreiber F, Popper B, Dimou L, Strom TM, Winkelmann J, et al. Spinal poly-GA inclusions in a C9orf72 mouse model trigger motor deficits and inflammation without neuron loss. Acta Neuropathol. 2017;134:241–254. doi: 10.1007/s00401-017-1711-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Finley D. Regulation of proteasome activity in health and disease. Biochim Biophys Acta. 2014;1843:13–25. doi: 10.1016/j.bbamcr.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder W, Martin KM, Lorensen WE. The visualization toolkit: an object-oriented approach to 3D graphics. New York: Kitware; 2006. [Google Scholar]
- Schweitzer A, Aufderheide A, Rudack T, Beck F, Pfeifer G, Plitzko JM, Sakata E, Schulten K, Forster F, Baumeister W. Structure of the human 26S proteasome at a resolution of 3.9 A. Proc Natl Acad Sci U S A. 2016;113:7816–7821. doi: 10.1073/pnas.1608050113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Chen X, Elsasser S, Stocks BB, Tian G, Lee BH, Shi Y, Zhang N, de Poot SA, Tuebing F, et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science. 2016:351. doi: 10.1126/science.aad9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai HC, Besche H, Goldberg AL, Schuman EM. Characterization of the brain 26S proteasome and its interacting proteins. Front Mol Neurosci. 2010;3:12. doi: 10.3389/fnmol.2010.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro Y, Urushitani M, Inoue H, Koike M, Uchiyama Y, Komatsu M, Tanaka K, Yamazaki M, Abe M, Misawa H, et al. Motor neuron-specific disruption of proteasomes, but not autophagy, replicates amyotrophic lateral sclerosis. J Biol Chem. 2012;287:42984–42994. doi: 10.1074/jbc.M112.417600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tivol WF, Briegel A, Jensen GJ. An improved cryogen for plunge freezing. Microsc Microanal. 2008;14:375–379. doi: 10.1017/S1431927608080781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49:174–180. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unverdorben P, Beck F, Sledz P, Schweitzer A, Pfeifer G, Plitzko JM, Baumeister W, Forster F. Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proc Natl Acad Sci U S A. 2014;111:5544–5549. doi: 10.1073/pnas.1403409111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zee J, Gijselinck I, Dillen L, Van Langenhove T, Theuns J, Engelborghs S, Philtjens S, Vandenbulcke M, Sleegers K, Sieben A, et al. A pan-European study of the C9orf72 repeat associated with FTLD: geographic prevalence, genomic instability, and intermediate repeats. Hum Mutat. 2013;34:363–373. doi: 10.1002/humu.22244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waelter S, Boeddrich A, Lurz R, Scherzinger E, Lueder G, Lehrach H, Wanker EE. Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol Biol Cell. 2001;12:1393–1407. doi: 10.1091/mbc.12.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters KJ, Kleijnen MF, Goh AM, Wagner G, Howley PM. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry. 2002;41:1767–1777. doi: 10.1021/bi011892y. [DOI] [PubMed] [Google Scholar]
- Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, Pestronk A, Whyte MP, Kimonis VE. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- Wehmer M, Rudack T, Beck F, Aufderheide A, Pfeifer G, Plitzko JM, Forster F, Schulten K, Baumeister W, Sakata E. Structural insights into the functional cycle of the ATPase module of the 26S proteasome. Proc Natl Acad Sci U S A. 2017;114:1305–1310. doi: 10.1073/pnas.1621129114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa M, Ito D, Honda T, Kubo K, Noda M, Nakajima K, Suzuki N. Characterization of the dipeptide repeat protein in the molecular pathogenesis of c9FTD/ALS. Hum Mol Genet. 2015;24:1630–1645. doi: 10.1093/hmg/ddu576. [DOI] [PubMed] [Google Scholar]
- Zang Y, Jin M, Wang H, Cui Z, Kong L, Liu C, Cong Y. Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM. Nat Struct Mol Biol. 2016;23:1083–1091. doi: 10.1038/nsmb.3309. [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Gendron TF, Grima JC, Sasaguri H, Jansen-West K, Xu YF, Katzman RB, Gass J, Murray ME, Shinohara M, et al. C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat Neurosci. 2016;19:668–677. doi: 10.1038/nn.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Jansen-West K, Xu YF, Gendron TF, Bieniek KF, Lin WL, Sasaguri H, Caulfield T, Hubbard J, Daughrity L, et al. Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol. 2014;128:505–524. doi: 10.1007/s00401-014-1336-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MA, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A. 2013;110:E4968–4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S2
Figure S1. Correlative Cryo-Light and Electron Microscopy Workflow, Related to Figure 1. (A) Rat cortical neurons were cultured on EM grids and transduced with (GA)175-GFP on day 5. Upon vitrification by plunge freezing 5 days after transfection, the grids were imaged by cryo-light microscopy. (A) shows an overlay of phase and GFP channels. (B) EM grids were transferred to a cryo-FIB/SEM microscope. To locate cells of interest (boxed), SEM images were aligned and superimposed with the GFP signal from the cryo-LM image. (C) FIB-induced secondary electron image of the cell shown in (B), superimposed with the GFP signal. Yellow boxes mark the regions of the cell removed by FIB to produce a lamella containing the GFP-positive region. (D) SEM image of the lamella resulting from FIB milling, with a final thickness of ~ 200 nm, superimposed with the GFP signal. (E) Cryo-TEM low magnification image of the lamella shown in (D). (F) 5 nm-thick tomographic slice of a tomogram recorded in the area marked by a white square in (E), containing the GFP signal. A dense network of poly-GA-GFP ribbons (red arrowheads) is visible. The inset shows a high magnification of a poly-GA-GFP ribbon decorated by GFP-associated additional densities (green arrowheads). Scale bar: 20 nm.
Figure S2: Subtomogram Averaging and Classification Workflow, Related to Figure 2 and Figure 3. (A) Unbiased template matching procedure. Several identical structures were first hand-picked from one tomogram, aligned and averaged. The resulting average was used as a template to search more tomograms for additional occurrences of the same structure. These additional particles were then visually inspected, aligned, classified and averaged again to produce a higher resolution average. This average was low-pass filtered and used as a template to search all the tomograms to produce the final subtomogram dataset. (B) All proteasome subtomograms were first classified into single-capped or double-capped. To further analyze the conformational status of the regulatory particles, all the subtomograms were cut in silico between the β-rings of the core particle (Asano et al., 2015), resulting in two independent particles for double-capped proteasomes. Cut out caps from all proteasomes were merged and subjected to a further round of classification.
Figure S3: Subtomogram Averaging and Classification for TRiC/CCT and the 26S Proteasome, Related to Figure 2 and Figure 3. (A) Unsymmetrized average structure of TRiC/CCT at a resolution of 20.5 Å superimposed on its atomic model (purple; PDB 4V94). (B) Rotational correlation coefficient analysis of (A) indicating 8-fold symmetry. (C) Average structure of TRiC/CCT at a resolution of 17.1 Å upon application of D8 symmetry to (A) superimposed on its atomic model. Resolutions were estimated by gold-standard Fourier shell correlation using the 0.143 criterion. (D, F) Overall distribution of single-capped and double-capped 26S proteasomes within aggregates in (GA)175-GFP transduced neurons (D) and untransduced neurons (Asano et al., 2015) (F). (E, G) Distribution of the ground and substrate processing states for all regulatory particles within aggregates in neurons transduced with (GA)175-GFP (E) and untransduced (Asano et al., 2015) (G). (H) Gold-standard Fourier shell correlation curves of the four proteasome structures, yielding the following resolution estimations: 11.8 Å (GS1), 12.3 Å (GS2), 15.4 Å (SPS1) and 12.8 Å (SPS2).
Figure S4: Proteasome Abundance and Function at Poly-GA Aggregates, Related to Figure 2 and Figure 5. (A) Poly-GA-GFP aggregates (green) in primary neurons expressing a synthetic (GA)175-GFP construct with a start codon colocalize with proteasomes detected with a PSMC4/Rpt3 antibody (red). Proteasome staining is diffuse in GFP-expressing control cells. Nuclei are labeled with DAPI (blue). Scale bar: 50 μm. (B) Proteasome concentration for (GA)175-GFP aggregates (DIV 5 + 5), control neurons transduced with GFP only (DIV 5 + 5) or untransduced (DIV 10) (N = 9 tomograms, (GA)175-GFP; N = 17, GFP; N = 16, untransduced). For poly-GA aggregates, only tomograms with more than half volume occupied by aggregates were considered for the concentration calculation. The top and bottom boundaries of the boxes indicate ± 2x standard error, and whiskers extend to the maximum and minimum values. Proteasome concentration within poly-GA aggregates was significantly higher than in control conditions (***, p < 0.001 by Mann-Whitney test), whereas the difference between GFP-transfected and untransfected control neurons was not significant (n. s.). (C) Quantification of proteasome levels in the soluble and insoluble fractions as well as total, analyzed by immunoblotting. The data was normalized to the total proteasome levels in GFP-transduced control neurons. Error bars indicate standard error. * and ** indicate respectively p < 0.05 and p < 0.01 by paired t-test (N = 6 replicates from 4 independent experiments). Proteasomes are enriched in the insoluble fraction in the presence of poly-GA aggregates. (D) Poly-GA expression leads to the stabilization of a reporter for the ubiquitin-proteasome system. HEK293 cells stably expressing an unstable green fluorescent reporter for the ubiquitin-proteasome system (UbG76V-GFP) were transfected with the indicated constructs. After 72 h, UbG76V-GFP levels were analyzed by flow cytometry. The relationship between poly-GA-tagRFP levels on the x-axis and of UbG76V-GFP on the y-axis shows a concentration-dependent accumulation of UbG76V-GFP in the presence of poly-GA. The data shown are from a single representative experiment out of three independent repeats. (E) Poly-GA-GFP aggregates (green) in primary neurons expressing a non-ATG (G4C2)73 construct containing a downstream GFP in the poly-GA reading frame. RAN translation-derived poly-GA-GFP aggregates are stained by a poly-GA antibody (red) and colocalize with proteasomes detected with a PSMC4/Rpt3 antibody (white). Nuclei are labeled with DAPI (blue). Scale bar: 10 μm.
Figure S5: Macromolecule Mapping in Different Poly-GA Aggregates, Related to Figure 2. (A–C) Tomographic slices of aggregates within neurons transduced with (GA)175-GFP (main panels) and analyzed at DIV 5 + 5. Regions containing poly-GA ribbons are outlined in red. For the whole tomogram, proteasomes (green) and ribosomes (yellow) are mapped to their original positions and orientations by template matching and subtomogram averaging. For each tomogram, maximum intensity projection heat maps of poly-GA ribbons, proteasomes and ribosomes were generated (right column of each panel). (D) Numbers of proteasomes and ribosomes detected in the tomograms shown in (A–C) plotted versus the volume of the region containing poly-GA aggregates. The number of proteasomes detected shows a positive correlation with poly-GA aggregate volume. (E, F) Tomographic slices and mapped 26S proteasomes of aggregates within neurons transduced with (GA)175-GFP at an earlier time point (DIV 5 + 3) (E), or transfected with a RAN-translated (G4C2)73 construct with GFP in the poly-GA reading frame (F). Note that despite the smaller size compared to DIV 5 + 5 (GA)175-GFP aggregates, similar concentrations of proteasomes accumulated within these aggregates (~ 7 μM for (GA)175-GFP, DIV 5+3; ~ 5 μM for G4C2). Due to the lower thickness of the cryo-FIB lamella (70 nm vs. 100–200 nm in all other cases) fewer proteasomes are visible in (F). The inset in (F) shows a high magnification of a poly-GA ribbon (red arrowhead) decorated by GFP-associated additional densities (green arrowheads). Note that the morphology of RAN-translated poly-GA ribbons is indistinguishable from aggregates formed by the (GA)175-GFP construct. Tomographic slices are 7 nm thick. Scale bars: 200 nm in main panels, 20 nm in inset.
Table S1: Comparison of Atomic Models by RMSD Values, Related to Figure 3. Root mean square deviation (RMSD) values of RP atoms between the initial models (s1-s4 states from S. cerevisiae; Wehmer et al., 2017) and the atomic models fitted to the GS1, GS2, SPS1 and SPS2 classes. The Rpn1 and Rpn13 subunits were excluded due to their dramatically different conformation between yeast and mammalian proteasomes. The lowest RMSD values for GS1, GS2, SPS1 and SPS2 atomic models are for the s1, s1, s2 and s4 states respectively (underlined). Thus, GS1 and GS2 likely correspond to s1, SPS1 to s2 and SPS2 to the s4 state.
Movie S1: Mapping macromolecules to poly-GA inclusions, Related to Figure 2. Tomographic volume and 3D rendering of the neuronal (GA)175-GFP aggregate shown in Figure 2. Poly-GA ribbons (red), 26S proteasomes (green), ribosomes (yellow), TRiC/CCT chaperonins (purple). The macromolecules are mapped in their original locations and orientations, computationally determined by template matching and subtomogram averaging. Note that proteasomes accumulate in the aggregate interior and are often found direct proximity or even direct contact with poly-GA fibrils. In contrast, TRiC/CCT chaperonins and ribosomes are largely excluded from the aggregate and more abundant in its periphery.