Genetic Spectrum of Arrhythmogenic Cardiomyopathy (original) (raw)
. Author manuscript; available in PMC: 2020 Mar 1.
The morphologically distinct forms of cardiomyopathy, dilated, hypertrophic, right ventricular arrhythmogenic and restrictive (DCM, HCM, ARVC and RCM, respectively), each have high heritable component, and the precise genes implicated help predict arrhythmia complications and progression.1 When evaluating the patient with cardiomyopathy, the probability of an informative genetic test ranges from 20–60%, depending on the presence of family history and type of cardiomyopathy. HCM is more genetically restricted, dividing HCM into sarcomere-HCM and non-sarcomere-HCM, with clear differences in outcome.2 DCM is more genetically heterogeneous with truncations of the TTN gene accounting for 20% of DCM.3 The next most frequently implicated genes in DCM each account for 2–8% of cases and include LMNA, SCN5A, and RBM20.4 The precise frequency of each gene’s contribution evolves as datasets enlarge and with increased use of genetic testing in clinical practice. Mutations in the RBM20 gene encoding RNA Binding Motif Protein 20 were initially described in two large families with DCM.5 RBM20 is an RNA splicing regulator and among its targets is the TTN gene encoding titin.6 RBM20 mutations were suggested to indicate a rapid progression to heart failure and the presence of life-threatening arrhythmias.7 Early genetic reports may overestimate clinical findings due to ascertainment bias in which the most severely affected families are overrepresented in genetic research. Over time, especially if genotype status leads the investigation, genotype-phenotype correlations reveal a broader phenotype with milder presentations. The gnomAD dataset indicates that RBM20 is particularly intolerant of loss of function variation (http://gnomad.broadinstitute.or/gene/ENSG00000203867),8 and that some RBM20 exons are not well covered with exome sequencing possibly underestimating RBM20 in DCM. RBM20 was included on targeted cardiomyopathy gene sequencing panels in 2012–2014, and with this inclusion, more data has become available on RBM20’s role DCM.
Parikh et al. now describe a larger cohort of RBM20 gene mutation carriers ascertained from two clinical US testing laboratories.9 The study captured 171 unique RBM20 variants from 403 clinical genetic tests. They observed significant enrichment of cardiomyopathy-associated RBM20 variants in exons 9 and 11, similar to earlier studies.5, 10 Exon 9 encodes an RNA binding domain of the RBM20 protein, while no function is ascribed to the exon 11 encoded region, and these regions were not enriched with variation in gnomAD. ClinVar, a public database of clinically relevant genetic variants, reports 12 RBM20 missense gene variants classified as likely pathogenic or pathogenic with 11 falling within the pathogenic regions (https://www.ncbi.nlm.nih.gov/clinvar/?term=RBM20%5Bgene%5D). An additional seven variants in these regions are classified as variants uncertain significance (VUS). Three exon 9 variants have conflicting reports of pathogenicity, while four exon 11 variants are uncertain significance. These inconsistencies highlight the difficulty of defining pathogenicity in single probands.
Parikh et al. found RBM20 variants in exons 9 and 11 associated with more severe clinical phenotypes. In particular, these patients were more likely to have a non-sustained ventricular tachycardia (NSVT), atrial fibrillation, a shorter PR interval, and a family history of cardiac death. Although variation in exons 9 and 11 may be easier to interpret as pathogenic, this does not exclude variants in other regions of RBM20 as causing disease. Interpreting RBM20 variation outside these regions is aided by family segregation analysis. In family-based studies, arrhythmia phenotypes like increased NSVT may serve as milder presentations of cardiac findings in gene-positive family members. In a related study, 80 RBM20 gene carriers from 15 families highlighted male sex as a risk factor for faster progression.11
In addition to rare pathogenic variants, there are GWAS signals for RBM20 in cardiac dysrhythmias. Specifically, two intronic RBM20 variants associate with atrial fibrillation and QRS complex measures.12 One of these variants, rs10749053, falls within intron 9, which is close to the regions identified by Parikh et al. The finding that RBM20 variants associate with atrial fibrillation and QRS interval further underscores the importance of RBM20 function for cardiac conduction system and arrhythmia risk.
RMB20 is in the subset of cardiomyopathy genes associated with increased risk for arrhythmias. This increased arrhythmia risk is best described for LMNA in which the risk for arrhythmias occurs in the absence of cardiomyopathy LMNA gene mutations are associated with atrioventricular nodal heart block and atrial fibrillation, as well as increased risk for ventricular arrhythmias.1, 4 Parikh et al. observed that RBM20 cardiomyopathy patients have comparable ventricular arrhythmia risk to LMNA cardiomyopathy patients and a trend towards reduced atrial fibrillation. However, the GWAS link between RBM20 intronic SNPs and atrial fibrillation may suggest the absence of association could relate to the small sample size.12
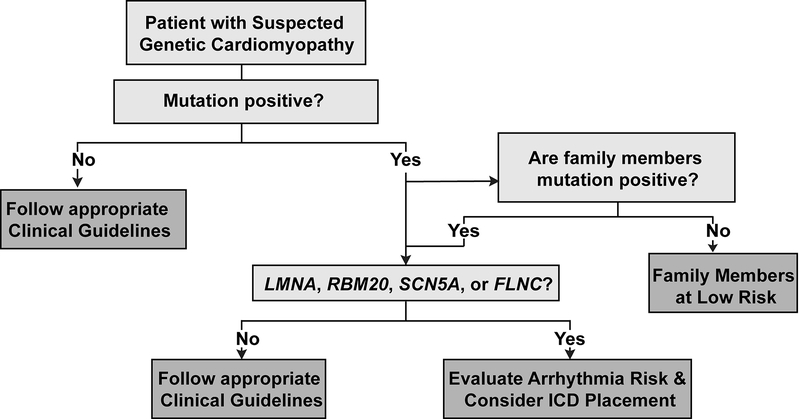
The AHA/ACC/HRS guidelines for managing ventricular arrhythmias recommend placement of an implantable cardiac defibrillator (ICD) in patients with LMNA cardiomyopathy and ≥ 2 risk factors (NSVT, EF < 45%, nonsense mutations, or male sex).13 Cardiomyopathy patients with pathogenic mutations in RBM20 appear to be at similar ventricular arrhythmia risk as LMNA cardiomyopathy patients. As such, these gene carriers should be monitored closely, and standard guidelines for primary prevention ICD placement may be inadequate. Due to the high penetrance of RBM20 mutations, family members of affected individuals should have site-specific genetic testing and managed based on genotype (Figure 1).
Figure 1:
Basic Workflow for Evaluating Patients with Genetic Cardiomyopathy.
Mutations in the DCM genes, SCN5A and FLNC, are also associated with higher rates of arrhythmias.14, 15 These specific arrhythmia associations with pathogenic mutations in LMNA, RBM20, SCN5A, or FLNC are of urgent importance, and genetic evaluation should be standard of care for DCM patients to better delineate this risk. The growing use of genome sequencing in medicine allows the use of personalized genetic information to inform clinical care of patients and family members. While it can be difficult to find clinically actionable associations, RBM20 mutations and arrhythmia risk is an excellent example. Genetic testing for DCM is at the forefront of personalized genomic medicine in predicting medically manageable risk.
Footnotes
Conflict of Interest Statement
EMM serves as a consultant to Invitae Inc, Tenaya Therapeutics, Exonics, AstraZeneca, and Cytokinetics. AMG has no conflicts of interest to report.
REFERENCES
- 1.Hershberger RE, Hedges DJ and Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nature reviews Cardiology. 2013;10:531–547. [DOI] [PubMed] [Google Scholar]
- 2.Ho CY, Day SM, Ashley EA, Michels M, Pereira AC, Jacoby D, Cirino AL, Fox JC, Lakdawala NK, Ware JS, Caleshu CA, Helms AS, Colan SD, Girolami F, Cecchi F, Seidman CE, Sajeev G, Signorovitch J, Green EM and Olivotto I. Genotype and Lifetime Burden of Disease in Hypertrophic Cardiomyopathy: Insights from the Sarcomeric Human Cardiomyopathy Registry (SHaRe). Circulation. 2018;138:1387–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herman DS, Lam L, Taylor MR, Wang L, Teekakirikul P, Christodoulou D, Conner L, DePalma SR, McDonough B, Sparks E, Teodorescu DL, Cirino AL, Banner NR, Pennell DJ, Graw S, Merlo M, Di Lenarda A, Sinagra G, Bos JM, Ackerman MJ, Mitchell RN, Murry CE, Lakdawala NK, Ho CY, Barton PJ, Cook SA, Mestroni L, Seidman JG and Seidman CE. Truncations of titin causing dilated cardiomyopathy. The New England journal of medicine. 2012;366:619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McNally EM and Mestroni L. Dilated cardiomyopathy: genetic determinants and mechanisms. Circulation research. 2017;121:731–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brauch KM, Karst ML, Herron KJ, de Andrade M, Pellikka PA, Rodeheffer RJ, Michels VV and Olson TM. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J Am Coll Cardiol. 2009;54:930–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo W, Schafer S, Greaser ML, Radke MH, Liss M, Govindarajan T, Maatz H, Schulz H, Li S, Parrish AM, Dauksaite V, Vakeel P, Klaassen S, Gerull B, Thierfelder L, Regitz-Zagrosek V, Hacker TA, Saupe KW, Dec GW, Ellinor PT, MacRae CA, Spallek B, Fischer R, Perrot A, Ozcelik C, Saar K, Hubner N and Gotthardt M. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nature medicine. 2012;18:766–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Refaat MM, Lubitz SA, Makino S, Islam Z, Frangiskakis JM, Mehdi H, Gutmann R, Zhang ML, Bloom HL, MacRae CA, Dudley SC, Shalaby AA, Weiss R, McNamara DM, London B and Ellinor PT. Genetic variation in the alternative splicing regulator RBM20 is associated with dilated cardiomyopathy. Heart Rhythm. 2012;9:390–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ and MacArthur DG. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parikh V, Caleshu C, Reuter C, Lazzeroni L, Ingles J, Garcia J, McCaleb K, Adesiyun T, Sedaghat-Hemdani F, Kumar S, Graw S, Gigli M, Stolfo D, Dal Ferro M, Nussbaum R, Funke B, Wheeler M, Hershberger RE, Cook SA, Steinmetz LM, Lakdawala NK, Taylor MR, Mestroni L, Merlo M, Sinagra G, Semsarian C, Meder B, Judge D and Ashley EA. Regional variation in RBM20 causes a highly penetrant arrhythmogenic cardiomyopathy. Circulation:Heart Failure. 2019;12:e005371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beqqali A, Bollen IA, Rasmussen TB, van den Hoogenhof MM, van Deutekom HW, Schafer S, Haas J, Meder B, Sorensen KE, van Oort RJ, Mogensen J, Hubner N, Creemers EE, van der Velden J and Pinto YM. A mutation in the glutamate-rich region of RNA-binding motif protein 20 causes dilated cardiomyopathy through missplicing of titin and impaired Frank-Starling mechanism. Cardiovasc Res. 2016;112:452–463. [DOI] [PubMed] [Google Scholar]
- 11.Hey T, Rasmussen TB, Madsen T, Aagard MM, Harbo M, Molgaard H, Moller JE, Eiskjaer H and Mogensen J. Pathogenic RBM20 variants are associated wtih a severe disease expression in male patients with dilated cardiomyopathy. Circulation Heart failure. 2019;12:e005700. [DOI] [PubMed] [Google Scholar]
- 12.Nielsen JB, Thorolfsdottir RB, Fritsche LG, Zhou W, Skov MW, Graham SE, Herron TJ, McCarthy S, Schmidt EM, Sveinbjornsson G, Surakka I, Mathis MR, Yamazaki M, Crawford RD, Gabrielsen ME, Skogholt AH, Holmen OL, Lin M, Wolford BN, Dey R, Dalen H, Sulem P, Chung JH, Backman JD, Arnar DO, Thorsteinsdottir U, Baras A, O’Dushlaine C, Holst AG, Wen X, Hornsby W, Dewey FE, Boehnke M, Kheterpal S, Mukherjee B, Lee S, Kang HM, Holm H, Kitzman J, Shavit JA, Jalife J, Brummett CM, Teslovich TM, Carey DJ, Gudbjartsson DF, Stefansson K, Abecasis GR, Hveem K and Willer CJ. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat Genet. 2018;50:1234–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Khatib SM, Stevenson WG, Ackerman MJ, Bryant WJ, Callans DJ, Curtis AB, Deal BJ, Dickfeld T, Field ME, Fonarow GC, Gillis AM, Granger CB, Hammill SC, Hlatky MA, Joglar JA, Kay GN, Matlock DD, Myerburg RJ and Page RL. 2017 AHA/ACC/HRS guideline for management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: Executive summary: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Rhythm Society. Heart Rhythm. 2018;15:e190–e252. [DOI] [PubMed] [Google Scholar]
- 14.McNair WP, Sinagra G, Taylor MR, Di Lenarda A, Ferguson DA, Salcedo EE, Slavov D, Zhu X, Caldwell JH and Mestroni L. SCN5A mutations associate with arrhythmic dilated cardiomyopathy and commonly localize to the voltage-sensing mechanism. Journal of the American College of Cardiology. 2011;57:2160–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ortiz-Genga MF, Cuenca S, Dal Ferro M, Zorio E, Salgado-Aranda R, Climent V, Padron-Barthe L, Duro-Aguado I, Jimenez-Jaimez J, Hidalgo-Olivares VM, Garcia-Campo E, Lanzillo C, Suarez-Mier MP, Yonath H, Marcos-Alonso S, Ochoa JP, Santome JL, Garcia-Giustiniani D, Rodriguez-Garrido JL, Dominguez F, Merlo M, Palomino J, Pena ML, Trujillo JP, Martin-Vila A, Stolfo D, Molina P, Lara-Pezzi E, Calvo-Iglesias FE, Nof E, Calo L, Barriales-Villa R, Gimeno-Blanes JR, Arad M, Garcia-Pavia P and Monserrat L. Truncating FLNC Mutations Are Associated With High-Risk Dilated and Arrhythmogenic Cardiomyopathies. J Am Coll Cardiol. 2016;68:2440–2451. [DOI] [PubMed] [Google Scholar]