FAM64A positively regulates STAT3 activity to promote Th17 differentiation and colitis-associated carcinogenesis (original) (raw)
Significance
The transcription factor STAT3 plays pivotal roles in various physiological processes, including differentiation of Th cells. Its deregulation results in serious diseases, including inflammatory diseases and cancer. Understanding how STAT3 activity is regulated is important for deciphering the pathogenesis of such diseases. In this study, we identified a protein called FAM64A, which promotes STAT3 activity through modulating the DNA-binding activity of STAT3. Consequently, FAM64A also promotes Th17 differentiation and development of colitis and colitis-associated cancer. This study reveals a previously unreported function of FAM64A in the regulation of inflammation and tumorigenesis and provides a potential therapeutic target for inflammatory diseases and cancer.
Keywords: STAT3, FAM64A, Th17, colitis, CAC
Abstract
STAT3 is a transcription factor that plays central roles in various physiological processes, including differentiation of Th cells. Its deregulation results in serious diseases, including inflammatory diseases and cancer. The mechanisms related to how STAT3 activity is regulated remain enigmatic. Here we show that overexpression of FAM64A potentiates IL-6–induced activation of STAT3 and expression of downstream target genes, whereas deficiency of FAM64A has the opposite effects. FAM64A interacts with STAT3 in the nucleus and regulates binding of STAT3 to the promoters of its target genes. Deficiency of Fam64a significantly impairs differentiation of Th17 but not Th1 or induced regulatory T cells (iTreg). In addition, Fam64a deficiency attenuates experimental autoimmune encephalomyelitis (EAE) and dextran sulfate sodium (DSS)-induced colitis, which is correlated with decreased differentiation of Th17 cells and production of proinflammatory cytokines. Furthermore, Fam64a deficiency suppresses azoxymethane (AOM)/DSS-induced colitis-associated cancer (CAC) in mice. These findings suggest that FAM64A regulates Th17 differentiation and colitis and inflammation-associated cancer by modulating transcriptional activity of STAT3.
The transcription factor STAT3 plays crucial roles in many physiological processes, such as cell proliferation, survival, and differentiation. Various cytokines and growth factors, including IL-6, oncostatin M, IL-10, IL-21, IL-23, and EGF, can activate STAT3 (1). The binding of these ligands to their corresponding receptors leads to activation of JAKs. Activated JAKs then mediate phosphorylation of STAT3 at Y705. Phosphorylated STAT3 translocates into the nucleus and binds to consensus motifs in promoters of the downstream target genes to regulate their transcription (2).
STAT3 activity is delicately controlled. Aberrant activation of STAT3 results in diseases, including autoimmune disorders and tumors (3, 4). For example, STAT3 is persistently activated in the intestinal T cells of patients with inflammatory bowel disease (IBD), including Crohn’s disease and ulcerative colitis (5). It has been demonstrated that STAT3 promotes the expansion of T cells and regulates the balance of differentiation of Th17 and Treg cells during colitis development (6). STAT3 is also constitutively activated in many types of human malignancies and plays important roles in inflammation-associated tumorigenesis. Gene knockout studies in animal models have highlighted the NF-κB–IL-6–STAT3 axis in linking inflammation to tumorigenesis of various cancers, including colon, liver, and pancreatic cancers (4, 7–9).
Upon activation by antigen-presenting cells, naive CD4+ T cells differentiate into distinct subsets of Th cells such as Th1, Th2, Th17, T follicular helper (Tfh), and induced regulatory T (iTreg) cells. These distinct types of Th cells are characterized by different cytokine expression profiles and different biological functions. Th17 cells represent a population of cells that secrete signature cytokines such as IL-17A, IL-17F, IL-21, and IL-22. These cells contribute to host defenses against certain bacterial and fungal infections as well as to the pathogenesis of certain inflammatory diseases, such as psoriasis and IBD (10). The differentiation of Th17 cells requires TGF-β and IL-6, and recent studies have demonstrated an essential role of STAT3 in this process (11, 12). During differentiation of Th17 cells, activated STAT3 induces the expression of retinoic acid receptor-related orphan receptor gamma (RORγt) and RORα, which are the master transcription factors driving the lineage commitment to Th17 (13, 14). In addition, STAT3 can also inhibit TGF-β–induced expression of FOXP3, a transcription factor that binds and antagonizes the function of RORγt. Ablation of STAT3 in T cells impairs Th17 cell differentiation and leads to their skewing toward anti-inflammatory Treg cells (15, 16).
FAM64A (also called CATS and RSC1) was originally identified as a novel clathrin assembly lymphoid myeloid leukemia gene (CALM)-interacting protein expressed in the thymus and spleen (17). It has been shown that FAM64A is highly expressed in leukemia, lymphoma, and various tumor cell lines, and its protein levels strongly correlate with cell proliferation in both malignant and normal cells (18). FAM64A is a multifunctional protein that is involved in regulation of cell cycle progression, subcellular localization of the leukemogenic fusion protein CALM/AF10, and tumorigenesis (18–22). In a screen for proteins that regulate STAT3 activity, we identified FAM64A as a positive regulator of STAT3, which modulates binding of STAT3 to the promoters of its target genes. The deficiency of Fam64a significantly inhibited Th17 cell differentiation and suppressed experimental autoimmune encephalomyelitis (EAE), dextran sulfate sodium (DSS)-induced colitis, and azoxymethane (AOM)/DSS-induced CAC development in mice. These findings reveal a previously unreported function of FAM64A in regulation of STAT3 activity, Th17 differentiation, and inflammation-associated tumorigenesis.
Results
FAM64A Positively Regulates STAT3 Activity.
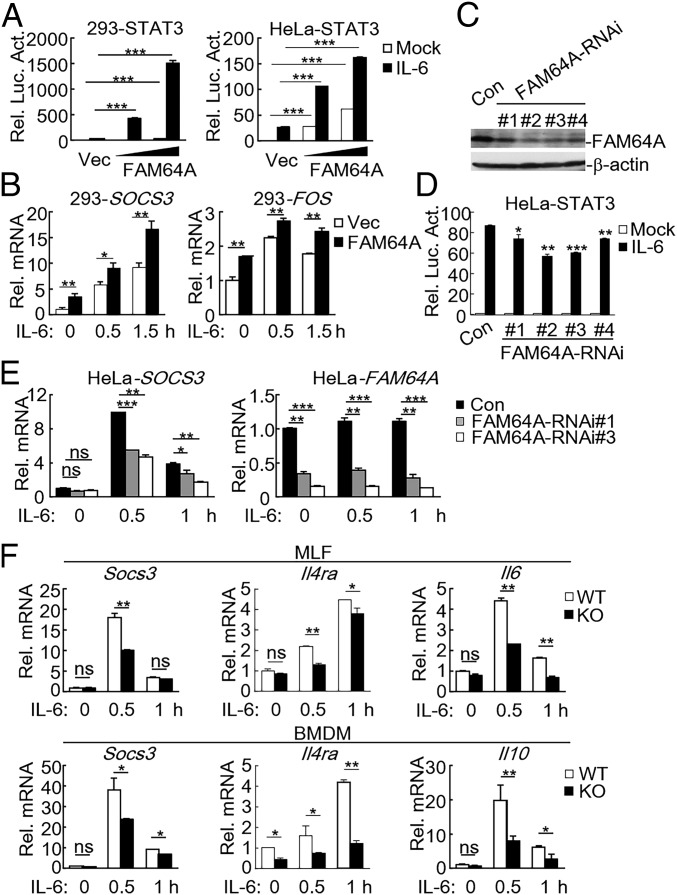
To identify candidate proteins that regulate the IL-6–STAT3 axis, we screened ∼13,000 independent human and murine cDNA expression plasmids by reporter assays (23). These screens led to identification of FAM64A as a positive regulator of STAT3 activation. As shown in Fig. 1_A_ and SI Appendix, Fig. S1_A_, overexpression of FAM64A and its murine homolog Fam64a potentiated IL-6–induced STAT3 activation in a dose-dependent manner. In similar experiments, FAM64A did not activate IFN-β–induced STAT1/2 activation (SI Appendix, Fig. S1_B_), suggesting that FAM64A specifically modulates STAT3 activity. Consistently, qPCR experiments showed that overexpression of FAM64A potentiated IL-6–induced transcription of downstream genes such as SOCS3 and FOS (Fig. 1_B_). To investigate whether endogenous FAM64A is involved in STAT3 activation, we constructed four RNAi plasmids which inhibited expression of FAM64A to different degrees (Fig. 1_C_). Reporter assays indicated that knockdown of FAM64A inhibited IL-6–induced STAT3 activation (Fig. 1_D_) but not IFN-β–induced STAT1/2 activation (SI Appendix, Fig. S1_C_) in HeLa cells. Consistently, knockdown of FAM64A also inhibited IL-6–induced transcription of the STAT3 target gene SOCS3 (Fig. 1_E_). These results suggest that FAM64A mediates IL-6–induced STAT3 activation.
Fig. 1.
FAM64A positively regulates STAT3 activity. (A) Effects of FAM64A on IL-6–induced STAT3 activation. HEK293 and HeLa cells (5 × 104) were transfected with STAT3 reporter (10 ng) and increased amounts of FAM64A expression plasmids. Twenty hours after transfection, cells were treated with IL-6 (20 ng/mL) or left untreated for 10 h in serum-free DMEM before relative luciferase activity (Rel. Luc. Act.) was determined with dual luciferase reporter assay system. (B) Effects of FAM64A on IL-6–induced transcription of SOCS3 and FOS genes. The experiments were performed as in A. Cells were stimulated with IL-6 (20 ng/mL) for the indicated times before qPCR experiments. (C) Effects of FAM64A-RNAi plasmids on expression of FAM64A. HeLa cells were transduced with a GFP control or FAM64A-RNAi plasmids by retroviral-mediated gene transfer. The expression of FAM64A in control (Con) and FAM64A-RNAi cell lines was analyzed by immunoblot. (D) Effects of FAM64A knockdown on IL-6–induced activation of STAT3 reporter. The experiments were performed as in A, except that control and FAM64A-RNAi HeLa cells were used. (E) Effects of FAM64A knockdown on IL-6–induced transcription of the SOCS3 gene. The control and FAM64A-RNAi HeLa cells (2 × 105) were stimulated with IL-6 (20 ng/mL) for the indicated times before qPCR experiments. (F) Effects of Fam64a deficiency on IL-6–induced transcription of Socs3, Il4ra, Il6, and Il10. WT and _Fam64a_−/− MLFs or BMDMs (2 × 105) were stimulated with IL-6 (20 ng/mL) for the indicated times before qPCR experiments. Data are representative of three experiments with similar results. Graphs show mean ± SD; n = 3. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
To further explore the functions of FAM64A in vivo, Fam64a-deficient mice were generated. Deficiency of Fam64a in the knockout mice was confirmed by genotyping and immunoblotting analysis (SI Appendix, Fig. S1_D_). We prepared primary murine lung fibroblasts (MLFs) and bone marrow-derived macrophages (BMDMs) from the knockout mice and found that deficiency of Fam64a significantly inhibited IL-6–induced transcription of STAT3 downstream genes, including Socs3, Il4ra, Il6, and Il10 in these cells (Fig. 1_F_). These results confirm that FAM64A is important for IL-6–mediated STAT3 activation.
FAM64A Facilitates Binding of STAT3 to the Promoters of Downstream Genes.
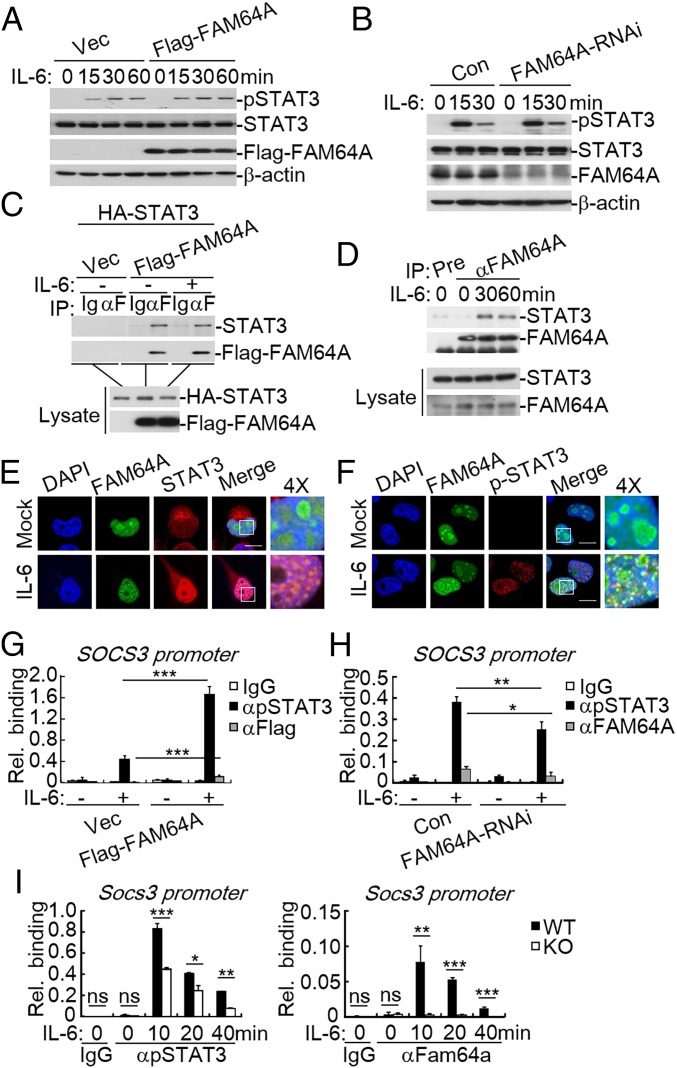
We next investigated the molecular mechanisms by which FAM64A regulates STAT3 activity. Since phosphorylation of STAT3 at Y705 is a hallmark of STAT3 activation, we examined whether FAM64A affects STAT3 phosphorylation. As shown in Fig. 2 A and B, neither overexpression nor knockdown of FAM64A had marked effects on STAT3 phosphorylation induced by IL-6. Next we investigated whether FAM64A interacts with STAT3. Coimmunoprecipitation experiments showed that FAM64A associated with STAT3 (Fig. 2 C and D and SI Appendix, Fig. S2_A_) and their association was enhanced upon IL-6 stimulation (Fig. 2_D_). Domain mapping experiments indicated that FAM64A interacted with multiple domains of STAT3 such as the N-terminal domain, DNA-binding domain, linker domain, SH2 domain, and transactivation domain (SI Appendix, Fig. S2_B_), while the N-terminal domain (1–129) of FAM64A was responsible for its interaction with STAT3 (SI Appendix, Fig. S2_C_). Since previous studies showed that FAM64A is exclusively located in the nucleus (17, 18), the interaction between FAM64A and STAT3 was further demonstrated by confocal microcopy. As shown in Fig. 2_E_, GFP-tagged FAM64A colocalized with endogenous STAT3 in the nucleus, and their colocalization was enhanced following IL-6 stimulation. In addition, phosphorylated STAT3 also colocalized with FAM64A in the nucleus following IL-6 stimulation (Fig. 2_F_). These results suggest FAM64A and STAT3 form complexes in the nucleus.
Fig. 2.
FAM64A regulates binding of STAT3 to the promoters of its target genes. (A) Effects of FAM64A on IL-6–induced STAT3 phosphorylation. HEK293 cells (2 × 105) were transfected with an empty vector or FAM64A expression plasmid (0.5 μg). Twenty hours after transfection, cells were starved with serum-free DMEM overnight, followed by IL-6 treatment (20 ng/mL) for the indicated times before immunoblot analysis. (B) Effects of FAM64A knockdown on IL-6–induced STAT3 phosphorylation. The control and FAM64A-RNAi HeLa cells (2 × 105) were stimulated with IL-6 (20 ng/mL) for the indicated times before immunoblot analysis. (C) FAM64A interacts with STAT3 in the mammalian overexpression system. HEK293 cells (2 × 106) were transfected with the indicated plasmids for 24 h. Coimmunoprecipitation and immunoblot analysis were performed with the indicated antibodies. (D) Endogenous FAM64A is associated with STAT3. HeLa cells (2 × 107) were starved overnight and then treated with IL-6 (50 ng/mL) or left untreated. Coimmunoprecipitation and immunoblot analysis were performed with the indicated antibodies. (E) Colocalization of FAM64A and STAT3. HeLa cells (1 × 105) were transfected with GFP-tagged FAM64A (0.2 μg). Twenty hours after transfection, cells were starved overnight followed by stimulation with IL-6 (50 ng/mL) for 30 min. Immunostaining was performed with anti-STAT3 antibody. (F) Colocalization of FAM64A and pY705-STAT3. The experiments were performed as in E, except that antibody against pY705-STAT3 was used. (G) Effects of FAM64A on STAT3 binding to the promoter of the Socs3 gene. HEK293 cells were transfected with an empty vector or FAM64A expression plasmid. Twenty hours after transfection, cells were starved with serum-free DMEM overnight, followed by IL-6 treatment (20 ng/mL), and were subjected to ChIP-qPCR assays with indicated antibodies. (H) Effects of FAM64A knockdown on STAT3 binding to the promoter of the Socs3 gene. The experiments were performed as in G, except that control and FAM64A-RNAi HeLa cells were used. (I) Effects of Fam64a deficiency on STAT3 binding to the promoter of the Socs3 gene. The experiments were performed as in G, except that WT and _Fam64a_−/− BMDMs were used. Data are representative of three experiments with similar results. Graphs show mean ± SD; n = 3. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant. Rel., relative.
We next investigated whether FAM64A affects the DNA-binding ability of STAT3. Chromatin immunoprecipitation (ChIP) assays indicated that overexpression of FAM64A significantly enhanced binding of STAT3 to the promoter of its target gene SOCS3 (Fig. 2_G_), while knockdown of FAM64A had the opposite effect (Fig. 2_H_). Consistently, deficiency of Fam64a dramatically inhibited binding of STAT3 to the promoter of Socs3 in BMDMs (Fig. 2_I_). Moreover, FAM64A was also recruited to the promoter of SOCS3, yet the intensity was relatively low in comparison with that of STAT3 (Fig. 2 G_–_I). Collectively, these results suggest that FAM64A facilitates binding of STAT3 to the promoters of its target genes.
Fam64a Deficiency Inhibits Th17 Differentiation.
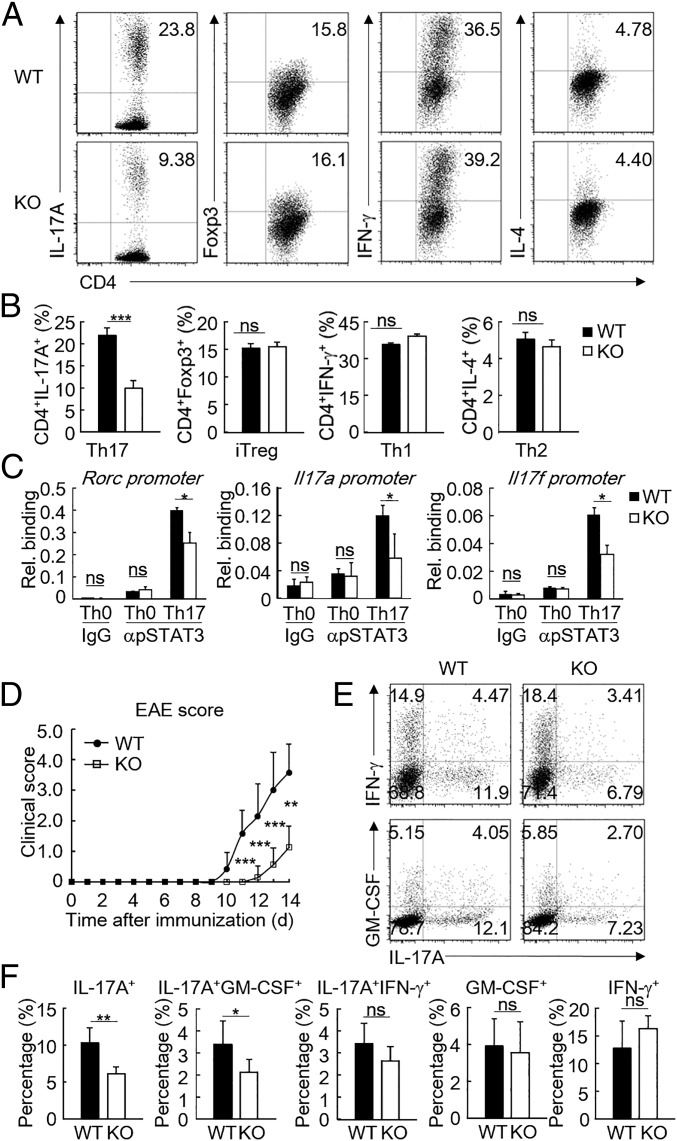
Since STAT3 is essential to the development of a subset of T cells, we next explored the function of FAM64A in T cell differentiation. We first analyzed the mRNA levels of Fam64a in thymic and splenic lymphoid populations and found that Fam64a was differentially expressed in various T cell subsets (SI Appendix, Fig. S3_A_). Fam64a-deficient mice showed no obvious difference in their T cell differentiation in the spleen, peripheral lymph nodes, and thymus compared with their wild-type littermates (SI Appendix, Fig. S3 B and C). Proliferation of wild-type and Fam64a −/− naive CD4+ T cells activated with anti-CD3 and anti-CD28 was also comparable (SI Appendix, Fig. S3_D_). However, compared with wild-type cells, Fam64a-deficient naive T cells activated by anti-CD3/anti-CD28 and cultured under various helper T cell-polarizing conditions exhibited decreased Th17 differentiation as characterized by reduced IL-17A expression (Fig. 3 A and B). In these experiments, Fam64a deficiency had no marked effects on differentiation of iTreg, Th1, or Th2 (Fig. 3 A and B). Consistently, Fam64a deficiency significantly suppressed transcription of the Th17 signature genes, such as Rora, Rorc, Il17a, and Il17f, but had no marked effects on transcription of Foxp3, Tbx21, and Gata3, the master transcription factors for iTreg, Th1, and Th2 cells, respectively (SI Appendix, Fig. S4_A_). Notably, the mRNA level of Fam64a was slightly up-regulated during Th17 differentiation (SI Appendix, Figs. S3_A_ and S4_A_). In addition, the amounts of IL-17A and IL-17F secreted by Fam64a-deficient Th17 cells were markedly lower than those of WT Th17 cells (SI Appendix, Fig. S4_B_). ChIP-qPCR assays indicated that Fam64a deficiency impaired binding of STAT3 to the promoters of Rorc, Il17a, Il17f, and Socs3 during Th17 differentiation (Fig. 3_C_ and SI Appendix, Fig. S5_A_). We then performed ChIP-sequencing to further evaluate the function of FAM64A in regulation of the DNA binding activity of STAT3 at the genome-wide level in Th17 cells. As shown in SI Appendix, Fig. S5_B_, the number of STAT3 binding peaks decreased in Fam64a-deficient Th17 cells. Moreover, the intensity of STAT3 also decreased at the Socs3 locus and other Th17 signature gene loci, including Rora, Rorc, and Il17a_–_Il17f (SI Appendix, Fig. S5_C_).
Fig. 3.
Fam64a deficiency inhibits Th17 differentiation. (A) Intracellular staining of IL-17A, Foxp3, IFN-γ, or IL-4 and surface staining of CD4 in WT and _Fam64a_−/− naive T cells activated with anti-CD3/anti-CD28 and cultured under various polarizing conditions for 72 h, then restimulated for 4 h with porbol-12-myristate-13-acetate and ionomycin. Numbers in the top right corners indicate the percentage of marker-positive CD4+ cells. (B) Frequency of IL-17A+, Foxp3+, IFN-γ+, or IL-4+ cells of WT and _Fam64a_−/− naive T cells cultured under various polarizing conditions. (C) ChIP assays of the binding of STAT3 to the promoters of Rorc, Il17a, and Il17f in WT and _Fam64a_−/− naive T cells cultured under the Th17-polarizing condition. Rel., relative. (D) The disease scores of female WT (n = 7) and _Fam64a_−/− mice (n = 7) in EAE. Disease severity was monitored and scored daily. (E) Intracellular staining of IL-17A, GM-CSF, and IFN-γ of infiltrated CD4+ T cells in the CNS of EAE mice. (F) Cellular populations and percentages of CNS-infiltrating cells. Student’s t test was used for the statistical test, n = 6. Results are represented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant. Data are representative of two experiments with similar results.
To determine the role of FAM64A in Th17 differentiation in vivo, EAE, a Th17 cell-mediated inflammatory disease, was induced in wild-type and Fam64a-deficient mice. As shown in Fig. 3_D_, _Fam64a_−/− mice exhibited significantly reduced clinical scores of EAE compared with their wild-type littermates. In addition, the frequencies of IL-17A+ and IL-17A+GM-CSF+ populations were significantly decreased in the central nervous system (CNS) of _Fam64a_−/− mice compared with the wild-type mice (Fig. 3 E and F). Notably, Fam64a mRNA level was increased in the spinal cords of mice that developed EAE (SI Appendix, Fig. S6_A_). Furthermore, to confirm whether Fam64a plays a T cell-intrinsic role in the development of EAE, we adoptively transferred isolated naive Fam64a+/+ or _Fam64a_−/− CD4+ T cells into Rag1−/− recipients and induced EAE 24 h later. Mice receiving _Fam64a_−/− CD4+ T cells had lower EAE score compared with mice receiving Fam64a+/+ CD4+ T cells (SI Appendix, Fig. S6_B_). In addition, reduced percentages of IL-17A+, IL-17A+IFN-γ+, and IL-17A+GM-CSF+ populations were observed in the CNS from mice that received _Fam64a_−/− CD4+ T cells compared with those receiving Fam64a+/+ CD4+ T cells (SI Appendix, Fig. S6 C and D). Collectively, these results indicate FAM64A is a positive regulator of Th17 differentiation both in vitro and in vivo.
Deficiency of FAM64A Attenuates DSS-Induced Colitis and Accumulation of Th17 in the Colon.
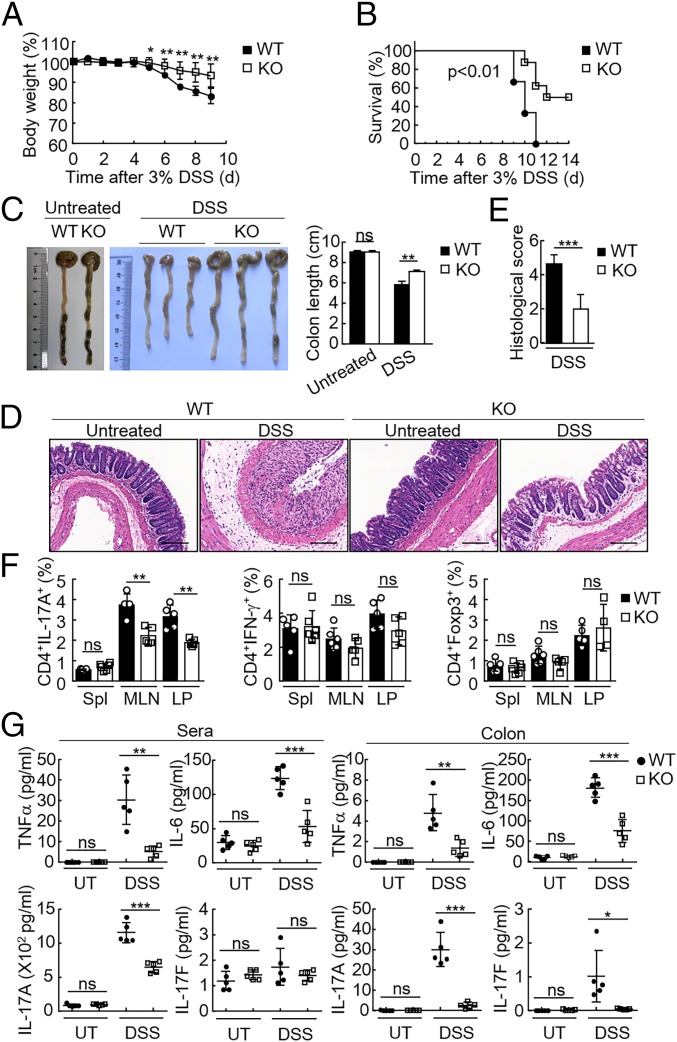
Since the DSS-induced mouse colitis model has been widely used to analyze the contribution of distinct T cell subsets in mucosal damage response, we next used this model to further examine the function of FAM64A in vivo. Mice were treated with 3% DSS in drinking water over a 10-d period to induce acute colitis. We found that _Fam64a_−/− mice displayed attenuated colitis with less weight loss (Fig. 4_A_), higher survival rate (Fig. 4_B_), and reduced colon shortening (Fig. 4_C_) compared with their wild-type littermates. Histopathological analysis revealed that the colonic mucosa of _Fam64a_−/− mice was more intact without apparent loss of crypt structures and mucosal ulceration. In addition, fewer inflammatory cells infiltrated in the colonic tissues of Fam64a −/− mice in comparison with their wild-type littermates after DSS challenge (Fig. 4_D_), which was also reflected in the pathological assessment of colitis severity scores (Fig. 4_E_). Notably, Fam64a mRNA level was increased in the colon tissues of mice after DSS exposure (SI Appendix, Fig. S7_A_). These results suggest that Fam64a deficiency attenuates the severity of DSS-induced colitis.
Fig. 4.
Deficiency of Fam64a attenuates DSS-induced colitis and inhibits the Th17 response during colitis. (A) WT and _Fam64a_−/− mice (female) were treated with 3% DSS over a 10-d period, and their body weights were monitored daily. Results are represented as mean ± SD. *P < 0.05; **P < 0.01. (B) Survival of mice described in A was monitored for 14 d. Results are represented as mean ± SD. **P < 0.01. (C) WT and _Fam64a_−/− mice were treated with 3% DSS or left untreated for 9 d before their colon lengths were measured. Results are represented as mean ± SD, n = 3. **P < 0.01; ns, not significant. (D) Representative images of hematoxylin and eosin staining of colon tissues of mice described in C. (Scale bar: 100 μM.) (E) Histological analysis of colon tissues described in D. The histological scores were determined in a double-blind manner. Results are represented as mean ± SD, n = 10. ***P < 0.001. (F) Frequency of IL-17A+, IFN-γ+, or Foxp3+ CD4+ T cells isolated from the spleen (Spl), MLNs, and LP of WT and Fam64a −/− mice treated for 9 d with 3% DSS in drinking water. Each symbol represents an individual mouse. (G) ELISA measurement of cytokine levels in the sera and colonic tissues of mice treated with DSS or left untreated (UT). Each symbol represents an individual mouse. Results in F and G are represented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant. Data are representative of two experiments with similar results.
To examine the correlation between alteration of T cell differentiation caused by Fam64a deficiency and the amelioration of colitis, the lymphocytes from spleens, mesenteric lymph nodes (MLNs), and lamina propria (LP) of wild-type and Fam64a −/− mice were isolated to perform intracellular cytokine staining. As shown in Fig. 4_F_ and SI Appendix, Fig. S7_B_, the proportion of CD4+IL-17A+ cells in the MLNs and LP of Fam64a −/− mice was decreased in comparison with that of their wild-type littermates. There was no significant difference between Fam64a-deficient mice and their wild-type littermates in the frequency of CD4+ Foxp3+ or CD4+ IFN-γ+ cells (Fig. 4_F_ and SI Appendix, Fig. S7_B_). In addition, levels of proinflammatory cytokines such as TNF-α, IL-6, IL-17A, and IL-17F in the sera and colonic mucosa were dramatically decreased in _Fam64a_−/− mice compared with their wild-type littermates (Fig. 4_G_), which was consistent with less severe colitis and the decreased proportion of Th17 cells in _Fam64a_−/− mice. Taken together, these results suggest that FAM64A facilitates Th17 differentiation to promote DSS-induced colitis.
Fam64a Deficiency Suppresses AOM/DSS-Induced CAC.
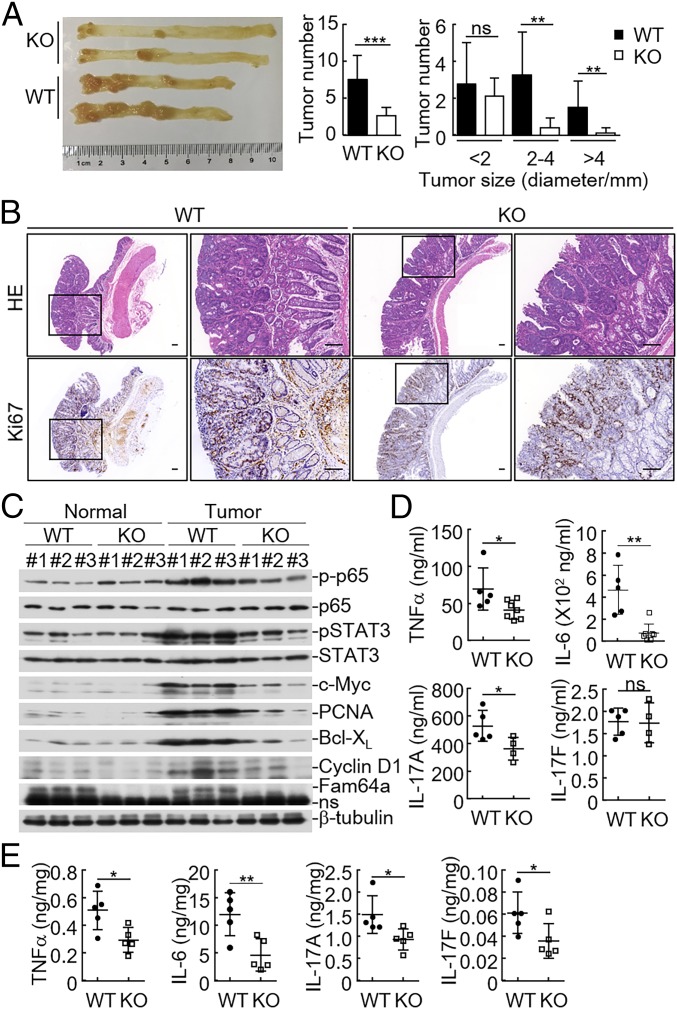
We next investigated the roles of FAM64A in inflammation-associated tumorigenesis with the AOM/DSS model (7). Mice were injected with AOM, followed by three rounds of 2% DSS exposure to induce CAC. The results indicated that _Fam64a_−/− mice developed fewer and smaller colon tumors compared with their wild-type littermates (Fig. 5_A_). Consistently, there was a significant reduction in proliferation rates of the colon cancers in Fam64a −/− mice as determined by Ki-67 nuclear staining (Fig. 5_B_). Immunoblot analysis indicated that the levels of phosphorylated p65 and STAT3 were markedly decreased in colon tumors in Fam64a-deficent mice, whereas levels of phosphorylated p65 and STAT3 showed no apparent differences in the tumor-adjacent normal colon tissues between wild-type and _Fam64a_−/− mice (Fig. 5_C_). Consistently, the protein levels of p65 and STAT3 downstream genes such as c-Myc, Pcna, Bcl-xl, and Ccnd1, which were responsible for the survival and proliferation of tumor cells, respectively, were decreased in colon tumors of _Fam64a_−/− mice (Fig. 5_C_). In addition, levels of inflammatory cytokines in the sera or colon tissues, such as TNF-α, IL-6, IL-17A, and IL-17F, decreased in _Fam64a_−/− mice (Fig. 5 D and E). Collectively, these results suggest that FAM64A plays important roles in CAC development.
Fig. 5.
Fam64a deficiency suppresses AOM/DSS-induced CAC. (A) The colons of WT and _Fam64a_−/− mice (female) were removed and photographed (Left). The tumor numbers (Middle) and tumor sizes (Right) were measured. Results are represented as mean ± SD, n = 10. **P < 0.01; ***P < 0.001; ns, not significant. (B) Representative images of immunohistochemical staining of colon tumors of WT and _Fam64a_−/−mice. (Scale bar: 100 μM.) (C) Immunoblotting analysis of colon tumors and adjacent normal tissues of WT and _Fam64a_−/− mice. Samples from three independent mice for each group were analyzed. (D) ELISA measurement of cytokine levels in the sera of AOM/DSS-treated WT and _Fam64a_−/− mice. (E) ELISA measurement of cytokine levels in the colonic tissues of AOM/DSS-treated WT and _Fam64a_−/− mice. Each symbol represents an individual mouse (D and E). Results in D and E are represented as mean ± SD. *P < 0.05; **P < 0.01; ns, not significant. Data are representative of two experiments with similar results.
Discussion
The activity of STAT3 is delicately regulated at different levels through distinct mechanisms to ensure proper biological functions. It has been reported that various tyrosine kinases, such as JAKs, Src, BTK, EGFR, and BMX, as well as other positive regulators, such as PASD1 and TRIM27, can mediate or potentiate STAT3 phosphorylation (23–26). In contrast, numerous protein phosphatases, such as LMW-PTP, LMW-DSP2, PTPRT, TCPTP, DUSP2, SHP1, SHP2, CD45, and PTP1B, as well as other negative regulators, such as SOCS3, GDX, and CUEDC2, can either directly or indirectly mediate dephosphorylation of STAT3 and JAKs (2, 27–29). In addition to phosphorylation, other posttranslational modifications of STAT3, such as methylation by EZH2, acetylation by CBP/p300, and deacetylation by HDACs and sirtuin 1, have also been reported to modulate its activity (30–33). Moreover, PIAS3 has been shown to inhibit STAT3 transcriptional activity by blocking its DNA-binding activity (34). In this study, we identified FAM64A as a positive modulator of STAT3. Overexpression of FAM64A potentiated IL-6–induced STAT3 activation and transcription of STAT3 downstream genes, whereas deficiency of FAM64A had opposite effects. We found that FAM64A facilitated binding of STAT3 to the promoters of its target genes and therefore enhanced STAT3 transcriptional activity.
IL-6–STAT3 signaling is essential for differentiation of Th17 cells. As a regulator of STAT3 activity, FAM64A plays important roles in Th17 cell differentiation. Deficiency of Fam64a inhibited Th17 cell differentiation but had no obvious effects on the differentiation of Th1, Th2, or iTreg cells. Some other proteins, such as SOCS3, DUSP2, TRIM28, and PKC-θ, have also been reported to regulate Th17 cell differentiation via modulating STAT3 activity (35–38). For examples, DUSP2 can mediate STAT3 dephosphorylation and suppress Th17 cell differentiation (35); TRIM28 is recruited to STAT3-occupied genes and mediates epigenetic activation during Th17 cell differentiation (36); PKC-θ can up-regulate STAT3 expression to promote Th17 cell differentiation (37).
Accumulating evidence suggests that Th17 cells and their related cytokines are crucial in the pathogenesis of IBD. In the model of DSS-induced colitis, _Fam64a_−/− mice displayed attenuated colitis in comparison with their wild-type littermates. Furthermore, Fam64a-deficient mice showed a decreased proportion of Th17 cells in colons and MLNs as well as decreased levels of inflammatory cytokines such as IL-6, TNF-α, IL-17Α, and IL-17F in the sera and/or colonic mucosa. Collectively, these results suggest an important role of FAM64A in differentiation of Th17 cells and development of colitis. Interestingly, a recent genome-wide association study of irritable bowel syndrome, a very common functional gastrointestinal disorder, identified 14 candidate risk loci and mapped a total of 93 genes, which includes FAM64A (39).
Chronic or unresolved inflammation promotes tumorigenesis. In the CAC model, Fam64a-deficient mice developed reduced Th17 differentiation and less severe inflammation. Consequently, activation of p65 and STAT3 in the colonic tissues, as well as the expression of their target genes, including c-Myc, Pcna, Bcl-xl, and Ccnd1, also decreased in Fam64a −/− mice, resulting in fewer and smaller colon tumors. These results suggest that FAM64A plays important roles in CAC development. Previously, it was shown that FAM64A is up-regulated in various tumor cells and is involved in regulation of cell proliferation, while knockdown of FAM64A inhibits growth of cancer cells in vitro (18, 21). In addition, FAM64A is significantly up-regulated in various types of tumor tissues, and high expression of FAM64A is associated with poor survival and clinical outcome, suggesting that FAM64A may be oncogenic in diverse types of cancers and it could be a potential prognostic marker and therapeutic target for cancer (40). In summary, our findings have identified a previously unreported function of FAM64A in the regulation of STAT3 activity, Th17 differentiation, colitis, and CAC development and have provided a potential therapeutic target for inflammatory disease and cancer.
Materials and Methods
All mouse studies were approved by the Animal Care Committees of the Wuhan University College of Life Sciences and the Wuhan Institute of Virology of the Chinese Academy of Sciences. The information on reagents, antibodies, constructs, PCR primers, and RNAi target sequences are described in SI Appendix, Materials and Methods. The methods for reporter assays, qPCR, establishment of stable cell lines, coimmunoprecipitation and immunoblot analysis, confocal microscopy, preparation of lymphocytes and flow cytometry, ChIP assays, EAE induction, colitis and CAC induction, and statistical analysis are presented in SI Appendix, Materials and Methods.
Supplementary Material
Supplementary File
Acknowledgments
We thank Juan Min (Core Facility Center, Wuhan Institute of Virology) for help with flow cytometry analysis. This work was supported by the National Science Fund for Distinguished Young Scholars (Grant 31425010), the Strategic Priority Research Program (XDB29010302) and Key Research Programs of Frontier Science funded by Chinese Academy of Sciences, the National Key R&D Program of China (Grants 2017YFA0505800, 2016YFA0502102), and the National Natural Science Foundation of China (Grants 91429304, 31630045, 31671465, 31521091, and 31700758).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Yu H, Pardoll D, Jove R (2009) STATs in cancer inflammation and immunity: A leading role for STAT3. Nat Rev Cancer 9:798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shuai K, Liu B (2003) Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol 3:900–911. [DOI] [PubMed] [Google Scholar]
- 3.Cho JH, Gregersen PK (2011) Genomics and the multifactorial nature of human autoimmune disease. N Engl J Med 365:1612–1623. [DOI] [PubMed] [Google Scholar]
- 4.Yu H, Lee H, Herrmann A, Buettner R, Jove R (2014) Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat Rev Cancer 14:736–746. [DOI] [PubMed] [Google Scholar]
- 5.Sugimoto K. (2008) Role of STAT3 in inflammatory bowel disease. World J Gastroenterol 14:5110–5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durant L, et al. (2010) Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity 32:605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greten FR, et al. (2004) IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell 118:285–296. [DOI] [PubMed] [Google Scholar]
- 8.Bromberg J, Wang TC (2009) Inflammation and cancer: IL-6 and STAT3 complete the link. Cancer Cell 15:79–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li N, Grivennikov SI, Karin M (2011) The unholy trinity: Inflammation, cytokines, and STAT3 shape the cancer microenvironment. Cancer Cell 19:429–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korn T, Bettelli E, Oukka M, Kuchroo VK (2009) IL-17 and Th17 cells. Annu Rev Immunol 27:485–517. [DOI] [PubMed] [Google Scholar]
- 11.Bettelli E, et al. (2006) Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441:235–238. [DOI] [PubMed] [Google Scholar]
- 12.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B (2006) TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 24:179–189. [DOI] [PubMed] [Google Scholar]
- 13.Yang XO, et al. (2008) T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR α and ROR γ. Immunity 28:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivanov II, et al. (2006) The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126:1121–1133. [DOI] [PubMed] [Google Scholar]
- 15.Harris TJ, et al. (2007) Cutting edge: An in vivo requirement for STAT3 signaling in TH17 development and TH17-dependent autoimmunity. J Immunol 179:4313–4317. [DOI] [PubMed] [Google Scholar]
- 16.Nurieva R, et al. (2007) Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature 448:480–483. [DOI] [PubMed] [Google Scholar]
- 17.Archangelo LF, Gläsner J, Krause A, Bohlander SK (2006) The novel CALM interactor CATS influences the subcellular localization of the leukemogenic fusion protein CALM/AF10. Oncogene 25:4099–4109. [DOI] [PubMed] [Google Scholar]
- 18.Archangelo LF, et al. (2008) The CALM and CALM/AF10 interactor CATS is a marker for proliferation. Mol Oncol 2:356–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao WM, et al. (2008) RCS1, a substrate of APC/C, controls the metaphase to anaphase transition. Proc Natl Acad Sci USA 105:13415–13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Archangelo LF, et al. (2013) The CATS (FAM64A) protein is a substrate of the kinase interacting stathmin (KIS). Biochim Biophys Acta 1833:1269–1279. [DOI] [PubMed] [Google Scholar]
- 21.Barbutti I, et al. (2016) CATS (FAM64A) abnormal expression reduces clonogenicity of hematopoietic cells. Oncotarget 7:68385–68396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hashimoto K, et al. (2017) Fam64a is a novel cell cycle promoter of hypoxic fetal cardiomyocytes in mice. Sci Rep 7:4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu ZS, et al. (2016) PASD1 promotes STAT3 activity and tumor growth by inhibiting TC45-mediated dephosphorylation of STAT3 in the nucleus. J Mol Cell Biol 8:221–231. [DOI] [PubMed] [Google Scholar]
- 24.Zhang HX, et al. (2018) TRIM27 mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis. Nat Commun 9:3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smithgall TE. (2002) Stat activation by Src, Fes and Btk tyrosine kinases. The Jak-Stat Pathway in Hematopoiesis and Disease (Landes Bioscience, Georgetown, TX: ), pp 51–77. [Google Scholar]
- 26.Guryanova OA, et al. (2011) Nonreceptor tyrosine kinase BMX maintains self-renewal and tumorigenic potential of glioblastoma stem cells by activating STAT3. Cancer Cell 19:498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekine Y, et al. (2006) Regulation of STAT3-mediated signaling by LMW-DSP2. Oncogene 25:5801–5806. [DOI] [PubMed] [Google Scholar]
- 28.Yoshimura A, Naka T, Kubo M (2007) SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol 7:454–465. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, et al. (2014) GdX/UBL4A specifically stabilizes the TC45/STAT3 association and promotes dephosphorylation of STAT3 to repress tumorigenesis. Mol Cell 53:752–765. [DOI] [PubMed] [Google Scholar]
- 30.Yuan ZL, Guan YJ, Chatterjee D, Chin YE (2005) Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science 307:269–273. [DOI] [PubMed] [Google Scholar]
- 31.Nie Y, et al. (2009) STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat Cell Biol 11:492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim E, et al. (2013) Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell 23:839–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dasgupta M, Dermawan JK, Willard B, Stark GR (2015) STAT3-driven transcription depends upon the dimethylation of K49 by EZH2. Proc Natl Acad Sci USA 112:3985–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung CD, et al. (1997) Specific inhibition of Stat3 signal transduction by PIAS3. Science 278:1803–1805. [DOI] [PubMed] [Google Scholar]
- 35.Lu D, et al. (2015) The phosphatase DUSP2 controls the activity of the transcription activator STAT3 and regulates TH17 differentiation. Nat Immunol 16:1263–1273. [DOI] [PubMed] [Google Scholar]
- 36.Jiang Y, et al. (2018) Epigenetic activation during T helper 17 cell differentiation is mediated by tripartite motif containing 28. Nat Commun 9:1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwon MJ, Ma J, Ding Y, Wang R, Sun Z (2012) Protein kinase C-θ promotes Th17 differentiation via upregulation of Stat3. J Immunol 188:5887–5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Z, et al. (2006) Selective regulatory function of Socs3 in the formation of IL-17-secreting T cells. Proc Natl Acad Sci USA 103:8137–8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonfiglio F, et al. (2018) Female-specific association between variants on chromosome 9 and self-reported diagnosis of irritable bowel syndrome. Gastroenterology 155:168–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu S, et al. (2017) Transcriptional response profiles of paired tumor-normal samples offer novel perspectives in pan-cancer analysis. Oncotarget 8:41334–41347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File