CDeep3M - Plug-and-Play cloud based deep learning for image segmentation (original) (raw)
. Author manuscript; available in PMC: 2019 Jun 4.
Published in final edited form as: Nat Methods. 2018 Aug 31;15(9):677–680. doi: 10.1038/s41592-018-0106-z
Abstract
As biomedical imaging datasets expand, deep learning will be vital for image processing. Yet, the need for complex computational environments and high performance compute resources still limits community access to these techniques. We address these bottlenecks with CDeep3M, a ready-to-use image segmentation solution that employs a cloud-based deep convolutional neural network. We benchmark CDeep3M on large and complex 2D and 3D imaging datasets from light, X-ray and electron microscopy.
Biomedical imaging prospers as technical advances provide enhanced temporal1 and spatial resolution with larger field of views2 with steadily decreasing acquisition times. Three-dimensional electron microscopy (EM) volumes, due to their extreme information content, anisotropy and increasing volume size2,3, are among the most challenging of segmentation problems. Substantial progress has been made with numerous deep neural networks4–7 to improve performance of computational image segmentation. However, generalized applicability of deep neural networks for biomedical image segmentation tasks is still limited and technical hurdles prevent the advances in speed and accuracy from reaching the mainstream of research applications. These limitations typically originate from the laborious steps required to recreate an environment that includes the numerous dependencies for each deep neural network. Further limitations arise from the scarcity of high-performance compute clusters and GPU nodes in individual laboratories, which are needed to train the network and process larger datasets within an acceptable timeframe.
With the goals of improving reproducibility and making deep learning algorithms available to the community, we built CDeep3M as a cloud based tool for image segmentation tasks. CDeep3M uses the underlying architecture of a state-of-the-art deep learning convolutional neural network (CNN), DeepEM3D7, which was integrated in the Caffe deep learning framework8. While there are a growing number of deep-learning algorithms, we were attracted to the features offered by the CNN built in DeepEM3D7, since it was designed for anisotropic data - which is the product of many microscopy techniques - by using a 2D-3D hybrid network. The deep neural network of DeepEM3D uses inception and residual modules as well as multiple dilated convolutions and an ensemble of three models integrating one, three and five consecutive image frames7.
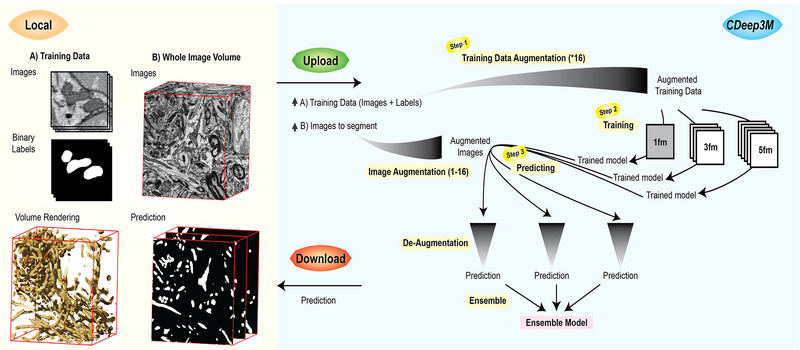
For CDeep3M, we modified all required components to make the CNN applicable for a wide range of segmentation tasks, permit processing of very large image volumes, and automated data processing. In brief, large image volumes are automatically split into sub-volumes with overlap, augmented, processed on GPU(s), de-augmented and both training and processing are parallelized on multi-GPU nodes. We also implemented a modular structure and created batch processing pipelines, for ease of use and to minimize idle time on the cloud instance. To reflect the broad applicability of our implementation to data of multiple microscopy modalities (e.g., X-ray microscopy (XRM), light microscopy (LM) and EM), we named this toolkit Deep3M. To give users easy access and to eliminate configuration issues and hardware requisites, we implemented steps to facilitate launching the cloud-based version CDeep3M on Amazon Web Services (AWS), which can be readily used for training the deep neural network on 2D or 3D image segmentation tasks. We minimized the number of required steps (Fig. 1), while still allowing for a flexible use of the code and enabling advanced settings (e.g. hyper-parameter tuning). Complete processing instructions are provided (see Supplementary Note), from generating training images to performing segmentation with CDeep3M and applying transfer learning.
Figure 1: Image segmentation workflow with CDeep3M.
In Steps 1-2 a new trained model is generated, based on training images and labels. For 3D segmentation tasks CDeep3M trains three different models seeing 1 frame (1fm), seeing 3 frames (3fm) and seeing 5 frames (5fm) that are applied in Step 3 to provide three predictions. Those are merged into a single ensemble model at the post-processing step (Step 3).
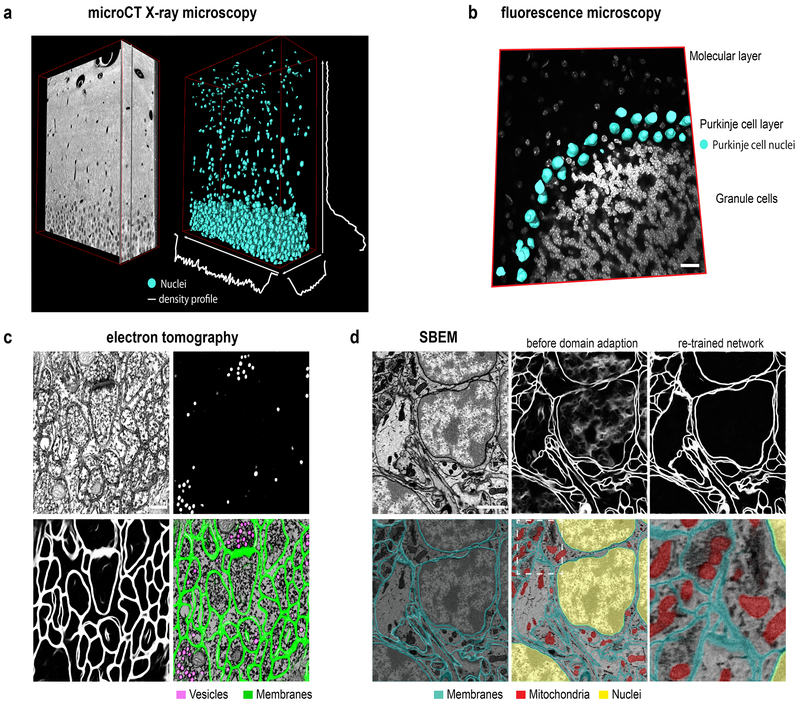
We applied and benchmarked CDeep3M on numerous image segmentation tasks in 2D and 3D, such as cellular organelle segmentation (nuclei, mitochondria, synaptic vesicles) and cell counting and classification. We established a cell density profile by training CDeep3M for the segmentation of nuclei in XRM data acquired from a hippocampal brain section prepared for EM. The density profile of segmented nuclei was extracted by applying a 20μm sliding volume across one axis at a time in x, y and z directions (Fig. 2a). We found an average cell density of 5.8×105 cells/mm3 in a volume (Volume size: 291μmx50μmx33μm) restricted to the suprapyramidal blade of the dentate gyrus (DG), which is consistent with stereology results reported previously 9,10. We find a gradual decrease towards the molecular layer (Supplementary Fig. 1), where the average cell density was at 3×104 cells/mm3. Training on fluorescence microscopy images of DAPI stained brain sections enabled us to distinguish neurons, solely based on their chromatin pattern and segment one individual cell-type (Fig. 2b). The object classification accuracy was compared to Ilastik11, which is often used for easier cell counting tasks on LM datasets (F1 values CDeep3M: 0.9114 and Ilastik: 0.425). To further test the performance of CDeep3M on diverse imaging data volumes, we analysed data acquired through multi-tilt electron tomography (ET), a form of transmission EM used to achieve high-resolution 3D volumes of biological specimens. More specifically, we applied CDeep3M to serial section ET of high-pressure frozen brain tissue and were able to automatically annotate synaptic vesicles (CDeep3M: Precision: 0.9789; Recall: 0.9769; F1 value: 0.9779) and membranes with high accuracy (Fig. 2c), which facilitates the 3D segmentation of synapses and neuronal processes and which was not possible using conventional machine learning tools (F1 value for vesicles using CHM12: 0.9071; see Supplementary Fig. 2 for membrane segmentations).
Figure 2: Multimodal image segmentation using CDeep3M.
(a) Segmentation of nuclei in XRM volume of a 50μm mouse brain slice containing the hippocampal DG area used for cell counting and establishing a cell density profile across x-y-z. (b) Segmentation of cell type specific DNA profile allows identification of Purkinje cells. Overlay of 3D surface mesh of nuclei on light microscopic image of DAPI-stained mouse cerebellar brain section. Scale bar: 20μm**.** (c) Segmentation of vesicles and membranes on multi-tilt electron tomography of high-pressure frozen mouse brain section. Scale bar: 200nm**.** (d) Upper row: SBEM micrograph (left) Scale bar: 1μm, segmentation using pre-trained model before (middle) and after domain adaption (right). Lower row: segmentation of membranes, mitochondria and nuclei overlaid on SBEM data
We further used CDeep3M for the segmentation of intracellular constituents (nuclei, membranes and mitochondria) of cells in a serial block-face scanning electron microscopy (SBEM) dataset (Fig. 2d). To improve our understanding of the role of intracellular organelles and alterations in diseases, parameters - such as the precise volume, the distribution, and fine details like contact points between organelles - is of utmost importance. Therefore, we evaluated the performance of CDeep3M based on predictions per pixel, which is more representative (compared to object detection) to determine accurate segmentations and distribution of intracellular organelles. To compensate for inaccuracies of human annotations along the borders of objects an exclusion zone around the ground truth objects was used, as described previously 13. We compared CDeep3M to the three-class Conditional Random Field (3C-CFR) mitochondria specific machine learning (ML) segmentation method13, using the same published training and validation datasets. We found CDeep3M outperformed 3C-CFR with (e.g. 2 voxel exclusion zone CDeep3M: 0,9266 3C-CFR: 0.85 Jaccard index) as well as without (0.8361 versus 0.741 Jaccard index) exclusion zone (Supplementary Fig. 3a, b). We further found the segmentation accuracy of CDeep3M equals the one of human expert annotators compared to the consensus ground truth (CDeep3M_:_ Precision: 0,9871 Recall: 0,9656 F1: 0,9762 Jaccard:0,9536 Humans: Precision: 0,9727, Recall: 0,9843, F1: 0,9782, Jaccard: 0,9574, one voxel exclusion zone; Supplementary Fig. 3c, d; for training and validation loss and accuracy see Supplementary Fig. 4).
Since training is time consuming, both to generate manual ground truth labeling (particularly for membrane annotation) and to train a completely naïve CNN, the re-use and refinement of previously trained neural networks (transfer learning) is of eminent interest. To test our ability to re-use a pre-trained model for a new dataset, we performed a type of transfer learning, domain adaption14. We therefore trained a model on the recognition of membranes using a published training dataset of a serial section SEM volume15 (similar to 7) with a voxel size of 6×6×30 nm. As expected, applying the network without further refinement on a SBEM dataset with similar voxel size (5.9×5.9×40 nm) but with staining differences and new staining features (cellular nuclei) led to unsatisfactory results (Fig. 2d, upper middle). Histogram matching was insufficient to remove the ambiguity caused by new features (nucleus) and staining differences. However, we found that using only 1/5th of the training data (20 instead of 100 training images) and 1/10th of the original training time (2000 and fewer iterations; from 22000 to 24000 iterations for 5fm; Fig. 2d; 14454 to 15757 for 3fm; and 16000 to 18000 for 1fm, Supplementary Fig. 5) was sufficient to fully adapt the network. This shows that adaption of pre-trained models can reduce the effort and time required for training by up to 90%. We additionally enabled a feature on CDeep3M to reduce the number of variations used for predictions and hence increase processing speed. We noticed in several cases that the accuracy remained sufficiently high when performing predictions on eight, four or even no additional variation of the same dataset, which can be tested on each dataset specifically. Lastly, to distinguish the nuclear membranes from the other membranes a separate training and prediction was performed for cellular nuclei (Fig. 2d).
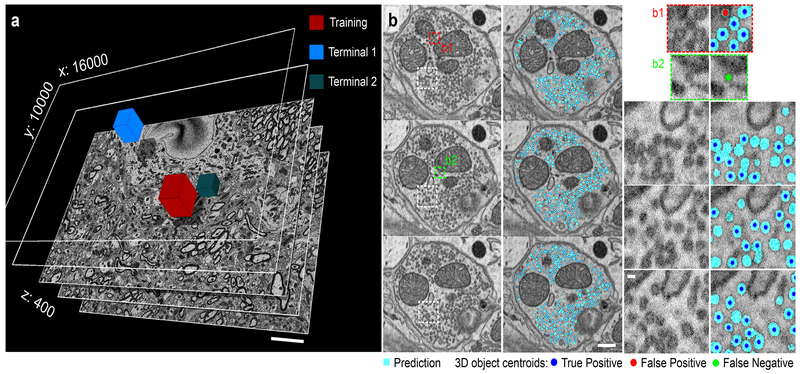
Encouraged by the performance of CDeep3M on complex SBEM datasets, we wondered about the feasibility of one of the most challenging tasks on those data, to identify and accurately count synaptic vesicles within presynaptic terminals. These are hundreds to thousands of small objects in each presynaptic terminal (sometimes referred to as ‘vesicles clouds’), some with sharply delineated membranes and others with much blurrier edges, spanning between one to a few sections. This combination makes it a particular difficult task for human and computer segmentation to identify individual objects, which would tremendously benefit from automation. We performed training on a small area (512×512×100 voxel) of one terminal and applied to a dataset of 16000×12000×400 voxel. To verify the segmentation accuracy of CDeep3M and how well the model generalizes, two terminals at random locations within the volume were manually annotated by human experts for comparison (Fig. 3). CDeep3M with a 3D analysis of circular objects was used to separate densely packed vesicles (and place 3D centroids, see Fig. 3) and compare vesicle counts and calculate error rates (Terminal 1: Vesicle counts CDeep3M: 3487; Humans: 3258, 3435, 3146, 2895; mean F1 value: 0.934; Terminal 2: Vesicle counts CDeep3M: 1105; Humans: 1121, 938, 942; mean F1 value: 0.937).
Figure 3: Synaptic vesicle counts on SBEM data using CDeep3M.
(a) SBEM volume acquired at 2.4nm × 24nm voxelsize (16000×10000×400 voxel). Performance tests were done on two terminals (Terminal 1: 3183 vesicles and Terminal 2: 1000 vesicles) comparing to several independent human counts. Scale: 5μm (b) Three consecutive sections of Terminal 1 in overview (left panels, Scale: 200nm) alongside the CDeep3M predictions and zoomed in (right panels, Scale: 40nm) show comparisons to human counts. Centroids of 3D objects occurring across sections are marked on most prevalent plane.
Altogether, CDeep3M takes advantage of cloud resources, which is scalable in times of high demand within the same laboratory and free of cost and maintenance when unused. Our cloud-based solution to provide an open AWS image is efficient for end-users, minimizing time spent for software / hardware configuration and updating numerous software packages, while alleviating the burden on algorithm developers to support a community with a multitude of underlying systems and platforms. Altogether this should facilitate the analysis of large and complex imaging data and render CDeep3M a widely applicable tool for the biomedical community.
Online Methods
Animals were used in accordance with a protocol approved by the Institutional Animal Care and Use Committee at the University of California, San Diego.
Tissue preparation and imaging for serial block-face scanning electron microscopy (SBEM) and X-ray microscopy (XRM)
C57BL/6NHsd mice (Envigo) at age 4-6 weeks were anesthetized using ketamine / xylazine and transcardially perfused with Ringers solution followed by a fixative mix composed of 2.5% glutaraldehyde, 2% formaldehyde, 2 mM CaCl2, in 150mM cacodylate buffer. Fixation was started at 37ºC and cooled to 4ºC during the 15 minute perfusion. The brain was removed, post-fixed in the same fixative mix for 2 hours at 4ºC. Free floating brain sections of 100 μm thickness were prepared in 150mM cacodylate buffer with 2 mM CaCl2 using a vibratome (Leica), sagittal from the cerebellum and coronal from the lateral habenula for SBEM and coronal from the hippocampus for XRM. The tissue was processed as described in Deerinck et al. 201016. Briefly, sequential staining consisted of 2% OsO4 / 1.5% potassium ferrocyanide, 0.5% thiocarbohydrazide and OsO4, followed by en-bloc uranyl acetate (2%). Sections were dehydrated by a sequence of increasing concentrations of ethanol, followed by dry acetone and then placed into 50:50 Durcupan ACM:acetone overnight. Slices were immersed in 100% Durcupan resin overnight in vacuum, then flat embedded between glass slides and left to harden at 60 °C for 48 hours. SBEM was performed using a Merlin SEM (Zeiss, Oberkochen, Germany) with a Gatan 3View system at high vacuum. The XRM tilt series was collected using a Zeiss Xradia 510 Versa (Zeiss X-Ray Microscopy, Pleasanton, CA, USA) operated at 40 kV (76 μA current) with a 40× magnification and 0.416μm pixel size. XRM volumes were generated from a tilt series of 3201 projections using XMReconstructor (Xradia), resulting in a final reconstructed volume of 391μm × 405μm × 395μm in x/y/z.
Electron tomograms
High Pressure Freezing and Freeze Substitution
For high-pressure freezing the brain was removed and post-fixed in the fixative mix for 1 hour at 4ºC. Vibratome sections of 100 μm thickness were transferred into 0.15M cacodylate buffer with 2 mM CaCl2 before high-pressure freezing. A small portion of the tissue was punched out and placed into a 100 μm deep membrane carrier and surrounded with 20% BSA in 0.15 M cacodylate buffer. The specimens were high pressure frozen with a Leica EM PACT2. Freeze substitution was carried out in extra dry acetone (Acros) as follows: 0.1% tannic acid at −90ºC for 24 hours, wash 3× 20 min in acetone at −90ºC, transferred to 2% osmium tetroxide / 0.1% uranyl acetate and kept at −90ºC for 48 hours, warmed to −60ºC over 15 hours, kept at −60ºC for 10 hours, warmed to 0ºC for 16 hours. The specimens were then washed with ice-cold acetone and allowed to come to room temperature and washed twice more with acetone. The specimens were infiltrated with 1:3 Durcupan ACM resin:acetone for several hours, 1:1Durcupan:acetone for 24 hours, 3:1 Durcupan:acetone for several hours, 100% Durcupan:acetone overnight, and then fresh Durcupan for several hours. The 100% Durcupan steps were done under vacuum. The specimens were then placed in Durcupan in 60ºC oven for 48 hours. The epoxy blocks were cut with a Leica UCT ultramicrotome into 300 nm thick sections. No on-grid staining was performed. Ribbons were collected on slot grids with a 50 nm thick support film (Luxel Corp, Friday Harbor, WA). The grids were glow discharged for 10 seconds on both sides and then coated with 10 nm colloidal gold diluted 1:2 with 0.05M bovine serum albumin solution (Ted Pella, Redding, CA).
Electron tomogram acquisition
Electron tomography was used to digitally reconstruct a small portion (about 1.5μm×1.5μm×1.5μm) of a plastic embedded high-pressure frozen mouse cerebellum specimen at a voxelsize of 1.6nm. The final volume was assembled from 7 consecutive tomograms (serial sections), each generated after a 4-tilt series scheme in which the sample is tilted every 0.5° from −60° to 60° at four distinct azimuthal angles (0°, 90°, 45° and 135°) in an electron beam. The micrographs were acquired on a FEI Titan operating at 300kV with a Gatan Ultrascan 4k×4k CCD camera. An iterative scheme was used during the tomographic processing to reduce the influence of reconstruction artifacts17.
Fluorescence microscopy
Tissue was collected at 6 weeks from the cerebellum of C57BL/6NHsd mice (Envigo). Mice were anesthetized using ketamine / xylazine and transcardially perfused with Ringers solution followed by a fixative mix composed of 4% formaldehyde in 1× Phosphate buffered saline (PBS). Fixation was started at 37ºC and cooled to 4ºC during the 30-minute perfusion. The brain was removed, post-fixed in the same fixative mix for 2 hours at 4ºC. Free floating brain sections of 50 μm thickness were prepared using a vibratome (Leica) and stained for 1 hour in 2.5 μg/mL 4’,6-diamidino-2-phenylindole (DAPI) in 1× PBS. Sections were mounted using ProLong Gold Antifade Reagent (Molecular Probes) and imaged with the Olympus Fluoview FV1000 confocal laser scanning microscope using 60× magnification lens at a pixel size of 0.21μm in x/y and a step size of 0.3μm in z.
Evaluation of CDeep3M performance
To define the performance of CDeep3M compared to ground truth segmentations, established by human expert annotators, following formulas where used. Pixels or objects are classified in one of four categories, TP: True positive TN: True negative FP: False positive FN: False negative. Precision = TP / (TP+FP); Recall = TP / (TP+FN); F1 value = 2*TP /(2*TP+FP+FN); Jaccard index = TP / (FP + TP + FN)
Areas used to evaluate performance did not include areas used for training.
Size of training data volumes were as follows in x/y/z: LM: 512×1024×92 voxel; XRM nuclei: 382×974×101 voxel; ET vesicles: 514×514×100 voxel; ET membranes: 1024×1024×20 voxel; ssTEM mitochondria: 1024×768×165 voxel; SBEM nuclei: 1024×1024×15 voxel; SBEM mitochondria: 1024×1024×80 voxel; ssSEM membranes: 1024×1024×100 voxel; SBEM membranes for transfer learning: 1024×1024×20 voxel; SBEM vesicles: 512×512×100 voxel
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data and software availability
CDeep3M source code and documentation are available for download on GitHub (https://github.com/CRBS/cdeep3m) and is free for non-profit use. Amazon AWS CloudFormation templates are available with each release enabling easy customization and deployment of CDeep3M for AWS cloud compute infrastructure. For the end user ~10 minutes after creating the CloudFormation stack, a p2x or p3x instance with a fully installed version of CDeep3M will be available to process data. Example data are included in the release. Further data will be made available from the corresponding authors upon reasonable request.
Supplementary Material
Supplemental
Supplemental Note
Acknowledgments
We thank the DIVE lab for making DeepEM3D publicly available and T. Zeng for initial discussion. We thank S. Viana da Silva for critical feedback on the manuscript. We thank S. Yeon, N. Allaway and C. Nava-Gonzales for help with ground truth segmentations for the membrane training data and mitochondria segmentations and C. Li, J. Shergill, I. Tang, M.M. and R.A. for synaptic vesicle annotations. M.G.H. and R.A. proof edited and performed other ground truth segmentations. Research published in this manuscript leveraged multiple NIH grants (5P41GM103412, 5P41GM103426 and 5R01GM082949) supporting the National Center for Microscopy and Imaging Research (NCMIR), the National Biomedical Computation Resource (NBCR), and the Cell Image Library (CIL), respectively. M.G.H. was supported by a postdoctoral fellowship from an interdisciplinary seed program at UCSD to build multiscale 3D maps of whole cells, called the Visible Molecular Cell Consortium. This work benefitted from the use of compute cycles on the Comet cluster, a resource of the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number ACI-1548562. This research benefitted from the use of credits from the National Institutes of Health (NIH) Cloud Credits Model Pilot, a component of the NIH Big Data to Knowledge (BD2K) program.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Chen BC et al. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science (80-. ). 346, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bock DD et al. Network anatomy and in vivo physiology of visual cortical neurons. Nature 471, 177–184 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briggman KL, Helmstaedter M & Denk W Wiring specificity in the direction-selectivity circuit of the retina. Nature 471, 183–190 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T & Ronneberger O 3D U-net: Learning dense volumetric segmentation from sparse annotation. Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics) 9901 LNCS, 424–432 (2016). [Google Scholar]
- 5.Quan TM, Hildebrand DGC & Jeong W-K FusionNet: A deep fully residual convolutional neural network for image segmentation in connectomics. (2016).
- 6.Badrinarayanan V, Kendall A & Cipolla R SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39, 2481–2495 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Zeng T, Wu B & Ji S DeepEM3D: approaching human-level performance on 3D anisotropic EM image segmentation. Bioinformatics d, 2555–2562 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia Y et al. Caffe: Convolutional Architecture for Fast Feature Embedding. (2014). doi: 10.1145/2647868.2654889 [DOI]
- 9.Jinno S & Kosaka T Stereological estimation of numerical densities of glutamatergic principal neurons in the mouse hippocampus. Hippocampus 20, 829–840 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Abusaad I et al. Stereological estimation of the total number of neurons in the murine hippocampus using the optical disector. J. Comp. Neurol 408, 560–566 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Sommer C, Straehle C, Kothe U & Hamprecht FA Ilastik: Interactive learning and segmentation toolkit. in Proceedings - International Symposium on Biomedical Imaging 230–233 (2011). doi: 10.1109/ISBI.2011.5872394 [DOI] [Google Scholar]
- 12.Perez AJ et al. A workflow for the automatic segmentation of organelles in electron microscopy image stacks. Front. Neuroanat 8, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lucchi A, Becker C, Márquez Neila P & Fua P Exploiting Enclosing Membranes and Contextual Cues for Mitochondria Segmentation in Medical Image Computing and Computer-Assisted Intervention -- MICCAI 2014 (eds. Golland P, Hata N, Barillot C, Hornegger J & Howe R) 65–72 (Springer International Publishing, 2014). [DOI] [PubMed] [Google Scholar]
- 14.Pan SJ & Yang Q A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 22, 1345–1359 (2010). [Google Scholar]
- 15.Kasthuri N et al. Saturated Reconstruction of a Volume of Neocortex. Cell 162, 648–661 (2015). [DOI] [PubMed] [Google Scholar]
Methods-only References
- 16.Deerinck T et al. Enhancing Serial Block-Face Scanning Electron Microscopy to Enable High Resolution 3-D Nanohistology of Cells and Tissues. Microsc. Microanal 16, 1138–1139 (2010). [Google Scholar]
- 17.Phan S et al. 3D reconstruction of biological structures: automated procedures for alignment and reconstruction of multiple tilt series in electron tomography. Adv. Struct. Chem. Imaging 2, 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental
Supplemental Note
Data Availability Statement
CDeep3M source code and documentation are available for download on GitHub (https://github.com/CRBS/cdeep3m) and is free for non-profit use. Amazon AWS CloudFormation templates are available with each release enabling easy customization and deployment of CDeep3M for AWS cloud compute infrastructure. For the end user ~10 minutes after creating the CloudFormation stack, a p2x or p3x instance with a fully installed version of CDeep3M will be available to process data. Example data are included in the release. Further data will be made available from the corresponding authors upon reasonable request.