Tumor Mutational Burden and Response Rate to PD-1 Inhibition (original) (raw)
. Author manuscript; available in PMC: 2019 Jun 5.
Published in final edited form as: N Engl J Med. 2017 Dec 21;377(25):2500–2501. doi: 10.1056/NEJMc1713444
TO THE EDITOR:
Inhibitors of programmed death 1 (PD-1) protein or its ligand (PD-L1) have shown remarkable clinical benefit in many cancers.1 One emerging biomarker of response to anti–PD-1 therapy is the tumor mutational burden (i.e., the total number of mutations per coding area of a tumor genome). This finding is supported by the clinical activity of anti–PD-1 therapy in colorectal cancer with mismatch repair deficiency, a tumor subtype with a high tumor mutational burden, as compared with the colorectal cancer subtype with mismatch repair proficiency, which has a significantly lower tumor mutational burden and a poor response to these agents.2,3
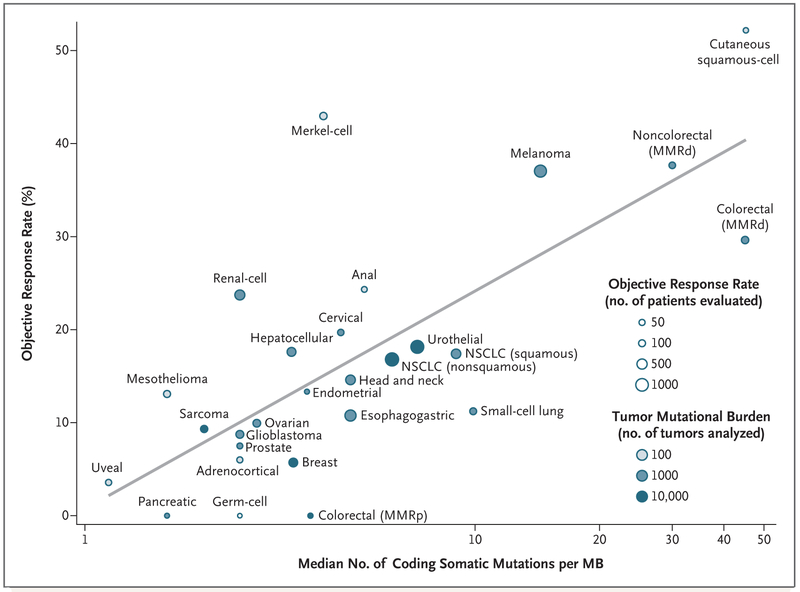
To evaluate the relationship between the tumor mutational burden and the objective response rate, we plotted the objective response rate for anti–PD-1 or anti–PD-L1 therapy against the corresponding median tumor mutational burden across multiple cancer types (Fig. 1). Through an extensive literature search, we identified 27 tumor types or subtypes for which data regarding the objective response rate are available. For each tumor type, we pooled the response data from the largest published studies that evaluated the objective response rate. We included only studies of anti–PD-1 or anti–PD-L1 monotherapy that enrolled at least 10 patients who were not selected for PD-L1 tumor expression. (Details about the methods are provided in the Supplementary Appendix, available with the full text of this letter at NEJM.org.) The median tumor mutational burden for each tumor type was obtained from a validated comprehensive genomic profiling assay performed and provided by Foundation Medicine.4
Figure 1. Correlation between Tumor Mutational Burden and Objective Response Rate with Anti–PD-1 or Anti-PD–L1 Therapy in 27 Tumor Types.
Shown is the median number of coding somatic mutations per megabase (MB) of DNA in 27 tumor types or subtypes among patients who received inhibitors of programmed death 1 (PD-1) protein or its ligand (PD-L1), as described in published studies for which data regarding the objective response rate are available. The number of patients who were evaluated for the objective response rate is shown for each tumor type (size of the circle), along with the number of tumor samples that were analyzed to calculate the tumor mutational burden (degree of shading of the circle). Data on the x axis are shown on a logarithmic scale. MMRd denotes mismatch repair-deficient, MMRp mismatch repair-proficient, and NSCLC non–small-cell lung cancer.
We observed a significant correlation between the tumor mutational burden and the objective response rate (P<0.001). The correlation coefficient of 0.74 suggests that 55% of the differences in the objective response rate across cancer types may be explained by the tumor mutational burden. Some cancer subtypes have a response to therapy that is better than would be predicted by the tumor mutational burden (e.g., Merkel-cell carcinoma), and some have a response that is worse than would be predicted (e.g., colorectal cancer with mismatch repair proficiency). The higher-than-anticipated objective response rates for Merkel-cell carcinoma and some other cancers that have been associated with viruses suggest that the presentation of viral antigens on certain tumor types may confer an increased response rate to anti–PD-1 therapy.5
Our linear correlation formula — objective response rate = 10.8 × loge(X) – 0.7, where “X” is the number of coding somatic mutations per megabase of DNA — can be used to make hypotheses with respect to the objective response rate in tumor types for which anti–PD-1 therapy has not been explored. For example, we anticipate a clinically meaningful objective response rate of 40.1% (95% confidence interval [CI], 31.2 to 50.6) for basal-cell carcinoma of the skin and of 20.6% (95% CI, 16.7 to 24.5) for sarcomatoid carcinoma of the lung on the basis of a median tumor mutational burden of 47.3 and 7.2, respectively.4 We anticipate a low objective response rate (<5%) for several other cancers (e.g., pilocytic astrocytoma and small-intestine carcinoid).4 A limitation of our analysis is that the sequenced tumor specimens were probably not the same ones for which clinical responses were assessed. Many different factors modulate the clinical response to an immune checkpoint inhibitor, but our findings highlight the strong relationship between the tumor mutational burden and the activity of anti–PD-1 therapies across multiple cancers.
Supplementary Material
Appendix
Disclosures
| THIS WEEK’S LETTERS | |
|---|---|
| 2500 | Tumor Mutational Burden and Response Rate to PD-1 Inhibition |
| 2502 | Once-Weekly Exenatide and Cardiovascular Outcomes in Type 2 Diabetes |
| 2503 | Nivolumab and Ipilimumab in Advanced Melanoma |
| 2504 | Hospital-Readmission Risk — Isolating Hospital Effects |
Footnotes
Disclosure forms provided by the authors are available with the full text of this letter at NEJM.org.
References
- 1.Brahmer JR, Tykodi SS, Chow LQM, et al. Safety and activity of anti–PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012;366:2455–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le DT, Uram JN, Wang H, et al. PD-1 Blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015;372:2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nghiem PT, Bhatia S, Lipson EJ, et al. PD-1 blockade with pembrolizumab in advanced Merkel-cell carcinoma. N Engl J Med 2016;374:2542–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Disclosures