A Single Injection of an Adeno-Associated Virus Vector into Nuclei with Divergent Connections Results in Widespread Vector Distribution in the Brain and Global Correction of a Neurogenetic Disease (original) (raw)
Abstract
Neurogenetic disorders typically affect cells throughout the brain. Adeno-associated virus (AAV) vector-mediated transfer of a normal cDNA can correct the metabolic defects at the site of injection, but treatment of the entire brain requires widespread delivery of the normal gene and/or protein. Current methods require multiple injections for widespread distribution. However, some AAV vectors can be transported along neuronal pathways associated with the injected region. Thus, targeting widely dispersed systems in the CNS might be a pathway for gene dispersal from a limited number of sites. We tested this hypothesis in the ventral tegmental area (VTA), a region with numerous efferent and afferent projections. A single 1 μl injection resulted in transport of the vector genome to projection sites in distal parts of the brain. When compared with injections into the striatum, the VTA injection resulted in higher enzyme levels in more regions of the brain. The AAV-9 serotype vector was the most widely disseminated, but AAV-Rh.10 and AAV-1 were also transported after VTA injection. The effect on global lesions of a neurogenetic disease was tested in the mouse model of MPS VII (mucopolysaccharidosis VII), a lysosomal storage disorder. Widespread distribution of the vector genome after AAV-9 VTA injection resulted in even further distribution of the enzyme product, by secretion and uptake by surrounding cells, and complete correction of the storage lesions throughout the entire brain. This unprecedented level of correction from a single injection into the developed brain provides a potential strategy to correct a large volume of brain while minimizing the number of injections.
Keywords: adeno-associated virus, vector transport, axonal transport, gene therapy, MPS VII, ventral tegmental area, hippocampus, striatum, AAV-9, AAV-Rh.10, AAV-1, enzyme transport
Introduction
Many neurogenetic diseases affect cells throughout the brain. The use of viral vectors to treat these diseases has been limited because of the large number of injections required for widespread dispersal of the therapeutic gene. A method that allows the vector and recombinant protein to reach a maximum volume of brain from a limited number of injections will be important to advance gene therapy to the treatment of neurogenetic diseases clinically.
There are many adeno-associated virus (AAV) serotypes available, each incorporating a different viral capsid protein and each mediating different transduction characteristics within the brain (Davidson et al., 2000; Passini et al., 2003; Vite et al., 2003; Burger et al., 2004; Cearley and Wolfe, 2006). Some AAV serotypes are transported along neuronal projections of the injected nucleus (Kaspar et al., 2003; Passini et al., 2005; Provost et al., 2005; Cearley and Wolfe, 2006). We hypothesized that an injection of a transportable vector into a region with divergent connections would result in widespread transport of the therapeutic gene, thereby producing a large area of therapeutic correction with only a single injection.
To test this, we injected AAV into the ventral tegmental area (VTA), a group of nuclei with divergent efferent and afferent projections. Injections of AAV-1, AAV-9, or AAV-Rh.10, expressing the cDNA for the lysosomal enzyme β-glucuronidase (GUSB), resulted in genome transport to distal projection sites of the VTA. In addition, some lysosomal enzymes themselves can be transported along some neuronal pathways (Passini et al., 2002; Hennig et al., 2003; Dodge et al., 2005; Luca et al., 2005; Cearley and Wolfe, 2006), and this resulted in enzyme delivery to even further distances, showing that the two transport mechanisms are able to work synergistically to maximize potential therapeutic correction with a single injection.
AAV-9 resulted in more genome-positive cells in many of the VTA projection sites and in higher levels of enzyme expression throughout most of the brain, so it was used in the remainder of the studies. Projections from the striatum are less extensive than the VTA, and injections of the same volume of AAV-9 into the striatum resulted in mRNA transport only to the striatal connections; far less enzyme expression was seen with a striatum injection compared with an injection into the VTA.
To test the extent to which injection of AAV into the VTA could result in correction of disease pathology, we used a mouse model of a lysosomal storage disease, mucopolysaccharidosis VII (MPS VII), which is caused by mutations in the GUSB gene. A single injection of AAV-9 GUSB into the VTA of MPS VII animals resulted in lysosomal storage correction in all regions of the brain examined at 2 months after injection. This is an unprecedented level of correction with a single small injection of AAV into the brain.
Materials and Methods
Experimental animals.
Normal and MPS VII-affected C3H/HeOuJ mice were produced in our breeding colony. Identification of MPS VII-affected mice, which contain a single base pair deletion in exon 10 of the GUSB gene, was verified by PCR (Wolfe and Sands, 1996). All animals receiving injections were older than 2 months of age at the time of injection. All procedures were approved by the Institutional Care and Use Committee at the Children's Hospital of Philadelphia.
AAV vector construction and packaging.
The detailed construction of the recombinant AAV genome used in the present study was reported previously (Passini and Wolfe, 2001). Briefly, the cross-packaging of the AAV-9, AAV-1, or AAV-Rh.10 vector, all expressing the human GUSB (hGUSB) promoter and transgene in the AAV-2 recombinant genome, used a chimeric AAV-2Rep/(AAV-9, AAV-1, or AAV-Rh.10) plasmid during the triple-transfection procedure. The packaging, purification, and determination of vector titers were performed by the University of Pennsylvania Vector Core. Recombinant vectors were purified using the CsCl sedimentation method, and genome copy (GC) titers were determined as described previously (Gao et al., 2000). Injection titers were between 1.2 and 1.3 × 1013 GC/ml for AAV-9, AAV-1, and AAV-Rh.10.
Cyanine 3 vector labeling.
The covalent linkage of cyanine 3 (Cy3) to the protein coat of the AAV-9 virions was done as described previously (Passini and Wolfe, 2001). Briefly, Cy3 was covalently linked to AAV-9 using a CyDye reactive dye pack (Amersham Biosciences, Piscataway, NJ). Unconjugated dye molecules were separated from labeled viral particles using a slide-a-lyzer (7000 molecular weight) dialysis chamber (Pierce, Rockford, IL). Purified Cy3-labeled viral particles were brought up to 20% glycerol and stored at −80°C until use. To localize Cy-3 particles, a subset of sections was stained with mouse anti-NeuN (Chemicon, Temecula, CA) and a goat anti-mouse Alexa Fluor 488-conjugated secondary antibody (Invitrogen, Eugene, OR) and coverslipped with Vectashield hardmount with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA).
Stereotaxic injections into adult mice.
Under sterile conditions, the animals were anesthetized with isofluorane and secured in a stereotaxic frame (David Kopf Instruments, Tujunga, CA), holes the size of the injection needle were drilled into the skull, and injections were done unilaterally with 1 μl of vector per brain region. The injection syringe (Hamilton, Reno, NV) delivered vector at a constant volume of 0.5 μl/min using a syringe pump (KD Scientific, Holliston, MA). The needle was left in place for 3 min after each injection to minimize upward flow of viral solution after raising the needle. For normal C3H/HeOuJ mice, coordinates [in millimeters; rostral (=) or caudal (−) to bregma, left of midline, ventral to pial surface] for the VTA were −3.28, 0.3, and 4.1; for the hippocampus were −2.06, 1.26, and 1.70; and for the striatum were = 0.5, 2.0, and 2.5. The MPS VII mouse brain is altered because of the abnormal shape of the skull (Skorupa et al., 1999; Passini et al., 2002), so the coordinates were slightly different for mutant animals. Coordinates for the MPS VII mice were determined using dye injections. For MPS VII animals, the coordinates for the VTA were −2.8, 0.35, and 4.1.
Tissue collection and preparation.
Tissue collection and preparation of unaffected mouse brains was done as described previously (Cearley and Wolfe, 2006). Brains from MPS VII-affected mice were put in 4% paraformaldehyde overnight after perfusion, after which they were sliced into 500 μm sections using a McIlwaine tissue chopper (Brinkman, Westbury, NY), stained with X-Gluc (Research Organics, Cleveland, OH), embedded in JB4 resin (Polysciences, Warrington, PA), and sectioned at a thickness of 1 μm.
Enzyme histochemistry.
Frozen tissue sections (20 μm) from unaffected mice were assayed for GUSB enzymatic activity using a naphthol AS-BI β-d-glucuronide substrate (Wolfe and Sands, 1996). The low levels of endogenous GUSB in the brains of C3H/HeOuJ mice were heat inactivated as reported previously (Passini and Wolfe, 2001). Heat-inactivated and non-heat-inactivated control slides of cells from normal C3H mice, cells from rat transfected with a retroviral vector expressing hGUSB, and cells from MPS VII heterozygous mice were used for comparison. For sections that were to be further processed using toluidine blue, tissue sections (500 μm) were assayed for GUSB enzymatic activity using an X-Gluc histochemical reaction. Sections were washed in PBS and sodium acetate, stained with X-Gluc for 48 h, and fixed with 2% glutaraldehyde for 24 h (Skorupa et al., 1999). X-Gluc-stained sections were then dehydrated and embedded in JB4 resin (Polysciences) for further processing.
Enzyme analysis.
Coronal sections (20 μm), which underwent enzyme histochemistry, from unaffected mice were scanned into the computer using a Microtek (Carson, CA) Artixscan scanner that had been manipulated to allow scanning of tissues on glass slides. Three sections, at a distance of 100 μm between each section, were taken at every 1 mm distance from bregma. The three sections were averaged to give a value at that 1 mm distance. The average and SE for all mice in a group (n = 3 per group) were calculated and graphed using Excel (Microsoft, Redmond, WA).
Colorometric
in situ hybridization with hGUSB mRNA riboprobe. The in situ hybridization procedure was performed as reported previously (Cearley and Wolfe, 2006). Control slides included GUSB transgenic and normal uninjected C3H/HeOuJ mouse sections stained with sense and antisense riboprobe. In situ positive cell counting was performed using a Zeiss (Thornwood, NY) microscope. Three sections, at a distance of 100 μm between each section, from each region were manually counted, and the average was used for the number of positive cells for that region for each mouse. All regions were counted in three mice, and the average and SE for all three mice were calculated using Excel.
Fluorescent in situ hybridization and tyrosine hydroxylase detection.
Fluorescent in situ hybridization was performed using the same digoxigenin (DIG)-labeled hGUSB mRNA riboprobes as in the colorometric in situ hybridization. After incubation in hybridization solution containing antisense riboprobe, sections were incubated in horseradish peroxidase (POD)-conjugated anti-DIG–POD antibody (Roche, Indianapolis, IN) and Cy3 labeled using the Tyramide Signal Amplification System (PerkinElmer Life Sciences, Boston, MA). After fluorescent in situ hybridization, sections were colabeled using a rabbit anti-tyrosine hydroxylase (TH) antibody (Pel-Freez, Rogers, AK) and a goat anti-rabbit Alexa Fluor 488-conjugated secondary antibody (Invitrogen) and coverslipped with Vectashield hardmount with DAPI (Vector Laboratories).
Histology.
Uninjected X-Gluc-stained MPS VII and normal control brains, as well as AAV-9-injected, X-Gluc-stained MPS VII brains, were mounted in JB4 resin, sectioned at a thickness of 1 μm, and analyzed for lysosomal storage vacuoles using a toluidine blue stain, as reported previously (Skorupa et al., 1999).
Results
Vector and enzyme distribution after a VTA injection
To determine whether an AAV vector could be transported along neuronal projections of the VTA, adult mice were given injections of 1 μl AAV-9 hGUSB unilaterally into the VTA. The injection coordinates were near the midline, allowing vector to transduce the VTA on either side of the brain, further increasing the level of potential genome transport to bilateral distal projection sites of the VTA. Normal C3H mice were used for the initial vector and enzyme distribution study, because these animals have naturally very low levels of endogenous GUSB enzyme expression (Casal and Wolfe, 1998) and any levels of mouse GUSB they do have can be heat inactivated, whereas most hGUSB activity remains (Herrup and Mullen, 1977; Moullier et al., 1993; Casal and Wolfe, 2001). The brains were analyzed for hGUSB enzymatic activity by a histochemical staining reaction (Wolfe and Sands, 1996) and for vector genome expression using in situ hybridization with an antisense DIG-labeled riboprobe against the hGUSB mRNA sequence.
After a single injection of AAV-9 into the VTA, hGUSB mRNA-positive cells were found in widely distributed areas throughout most of the brain and GUSB enzyme was found in every brain section examined (Fig. 1). The extensive distribution of GUSB enzyme is the result of two properties of lysosomal enzymes: cross-correction and enzyme transport. Cross-correction allows enzyme produced by genetically corrected cells and released into the extracellular space to be taken up by receptor-mediated endocytosis on neighboring cells. Because the neighboring cells cannot make enzyme, cross-correction results in a sphere of correction around each genetically corrected cell (Taylor and Wolfe, 1997; Heuer et al., 2002). GUSB enzyme has also been shown to be transported along neuronal projections (Passini et al., 2002; Hennig et al., 2003; Cearley and Wolfe, 2006), providing a second method for distribution.
Figure 1.
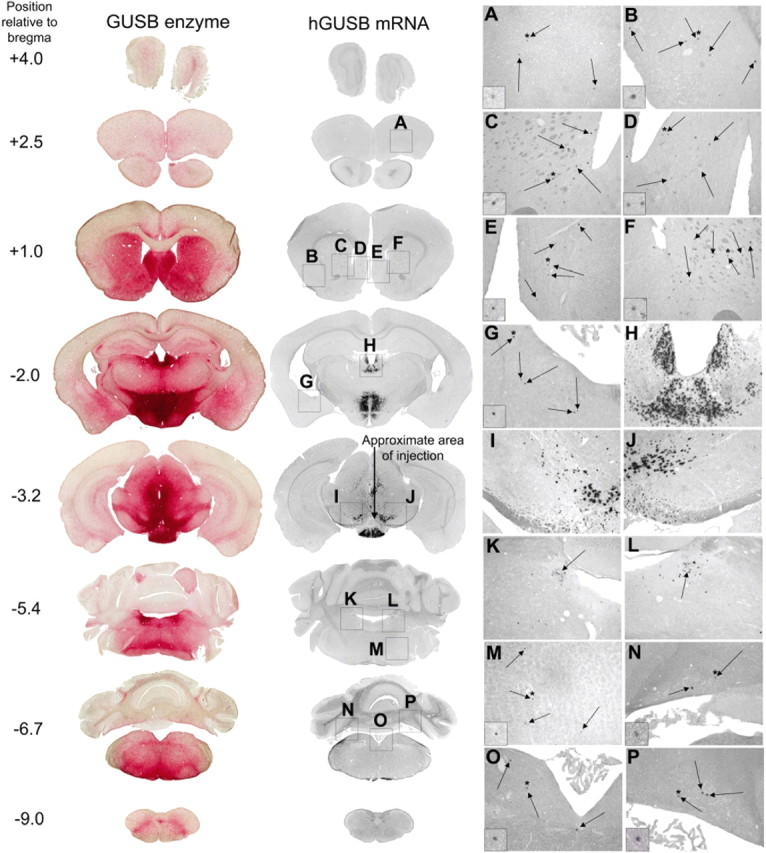
A single injection of AAV-9 into the VTA resulted in vector-positive cells in many regions of the brain known to be projection sites of the VTA and enzyme-positive cells throughout the brain. Normal animals (n = 4) were given injections of 1 μl of AAV-9 hGUSB into the unilateral VTA and analyzed 2 months after injection. Shown are the enzyme histochemistry and corresponding in situ hybridization. The approximate position of the section relative to bregma is given on the left. The arrow in the in situ hybridization section at −3.2 mm shows the approximate injection tract, with the injection at the arrowhead. Boxes A–P (not drawn to scale) on the in situ hybridization sections correspond to the magnified images on the right from the prefrontal cortex (A), piriform cortex (B), striatum (C, F), septal nuclei (D, E), amygdala (G), habenula and mediodorsal thalamic nucleus (H), substantia nigra (I, J), locus ceruleus (K, L), reticular nuclei (M), medial vestibular/prepositus nucleus and tegmental nuclei (O), and cerebellum (N, P). All of these regions have known projections with the VTA. Arrows within A–G and M–P show in situ positive cells. Arrows in K and L show the location of the locus ceruleus. Insets in A–G and M–P are further magnified images showing representative in situ positive cells. Asterisks are shown above the cells that have been magnified.
Areas surrounding the VTA and injection tract, such as the deep mesencephalic nucleus, red nucleus, and medial lemniscus, also received vector, either by direct transduction or diffusion from the injection site, resulting in some vector genome transport to nonprojection sites of the VTA. However, the VTA has far more projection sites than surrounding nuclei, and the extensive vector transport seen after injection is accounted for by the widespread projections of the VTA. Most regions distal from the injection site in which hGUSB mRNA-positive cells were found are known to be connected with the VTA and surrounding regions, although there are some differences between studies in the literature (Beckstead et al., 1979; Phillipson, 1979; Simon et al., 1979; Swanson, 1982; Geisler and Zahm, 2005). Of the regions with mRNA-positive cells shown, those thought to have reciprocal projections with the VTA include the prefrontal cortex (Fig. 1A), septal nuclei (Fig. 1D,E), habenula (Fig. 1H), thalamus (Fig. 1H), locus ceruleus (Fig. 1K,L), tegmental nuclei (Fig. 1O), and cerebellar nuclei (Fig. 1N,P). In situ positive areas thought to receive projections from the VTA include the piriform cortex (Fig. 1B), striatum (Fig. 1C,F), and amygdala (Fig. 1G). In situ positive areas thought to send projections to the VTA include the medial vestibular/prepositus nucleus (Fig. 1O) and the reticular nuclei (Fig. 1M). The substantia nigra (Fig. 1I,J) and zona incerta also had numerous in situ positive nuclei and sent projections to the VTA. However, these regions lie in close proximity to the VTA, so we could not distinguish between whether these regions received GUSB genome via vector transport or diffusion from the injection site.
Vector transport characteristics
Covalent linkage of Cy3 to the capsid of the AAV vector shell allows the localization of AAV virions to be visualized at short time points after injection (Bartlett and Samulski, 1998; Passini and Wolfe, 2001). AAV-9 vector particles were labeled with Cy3, and the distribution of the labeled particles was examined at 1 and 24 h after injection into the hippocampus or VTA (Fig. 2). At 1 h after injection, Cy3-labeled particles were mostly found along the injection tracts. There was evidence of diffusion at 1 h but no evidence of vector transport. With the hippocampal injection, the concentration of vector appears to be greater in the figure, although we attribute this to the limited area available for the vector to diffuse into, because of the enclosed hippocampal anatomy. Both the hippocampus and VTA were injected with the same volume of vector (1 μl). At 24 h after injection (Fig. 2 A–P), vector transport was evident, in that the Cy3-labeled particles were found in projection sites of the injected regions. With the hippocampus injection, labeled particles were found in many of the layers of the hippocampus on the injected side, such as the dentate gyrus (Fig. 2F), pyramidal cell layer (Fig. 2C), and oriens cell layer (Fig. 2G). Cy3-labeled particles were also found in the retrosplenial cortex (Fig. 2B) and entorhinal cortex (Fig. 2D,H), both known projection sites of the hippocampus (Witter et al., 2000; Van Groen and Wyss, 2003). On the contralateral side, Cy3-labeled particles were found in select layers of the hippocampus, such as the dentate gyrus caudal to the injection (Fig. 2E). One region that did not receive vector particles was the CA1 region at the rostrocaudal level of injection, on the contralateral side (Fig. 2A).
Figure 2.
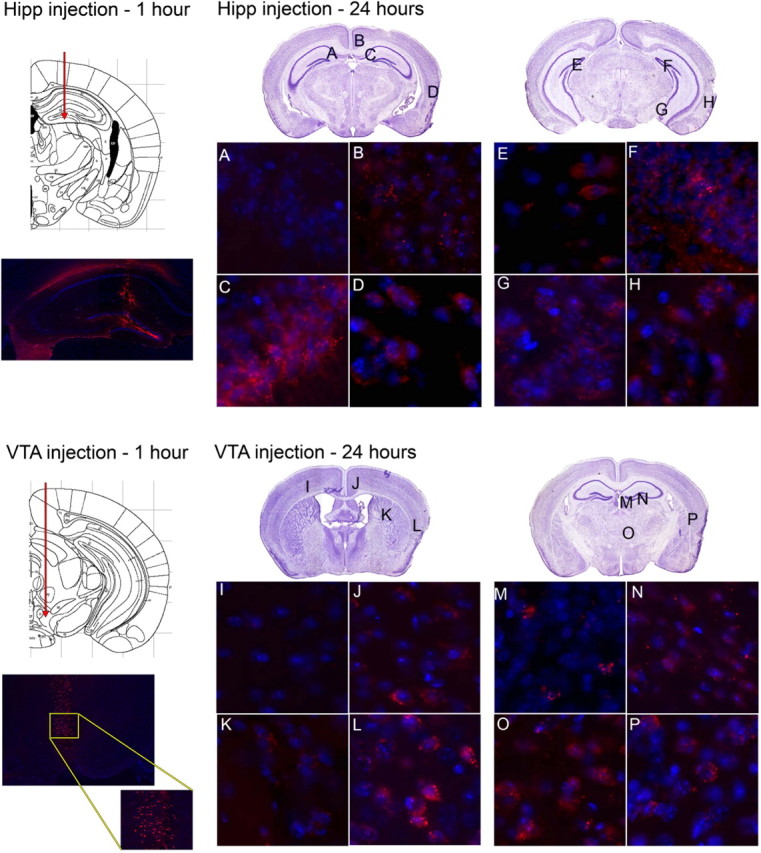
Cy3-labeled AAV-9 vector distribution after injection into the hippocampus or VTA. Animals were given injections of Cy3-labeled AAV-9 virions into the hippocampus (Hipp; top) or VTA (bottom) and killed at 1 h (left) or 24 h (right) after injection. Slides were costained with DAPI. The 1 h injection time-point panels show the hippocampus or VTA injection tracts (above) [images adapted from Paxinos and Franklin (2001)] and the presence of fluorescent vector (below), demonstrating the desired location of the injection. The inset in the 1 h VTA injection panel is a higher magnification of the selected region. The 24 h panels show sites analyzed by fluorescent microscopy. Locations designated by letters on Nissl-stained sections (from Paxinos and Franklin, 2001) are shown at high magnification at the bottom half of each panel. Images are from areas that did not receive Cy3-labeled particles (A, I) or did receive Cy3-labeled particles (B–H, J–P).
With the VTA injection, Cy3-labeled particles were found in many areas of the retrosplenial cortex (Fig. 2J), striatum (Fig. 2K), habenula (Fig. 2M), thalamus (Fig. 2N,O), and endopiriform nuclei (Fig. 2L,P), all regions that are known projection sites of the VTA (Beckstead et al., 1979; Simon et al., 1979). No Cy3 particles were found in the contralateral cortex (Fig. 2I). We did not detect labeled particles in all projection sites of the VTA or hippocampus that were found to have mRNA-positive cells at 2 months after injection. This could be attributable to limitations in detection of the Cy3 particles, or to the time course of transport to some regions being longer than to others. The finding of Cy3 particles in the projection sites of the VTA suggests that it is the vector itself that is being transported, not the hGUSB mRNA.
We have previously shown that AAV-9 preferentially transduces neurons (Cearley and Wolfe, 2006). To determine whether the Cy3-labeled particles in distal sites were also in neurons, we colabeled sections from animals given injections of Cy3-labeled AAV-9 with an antibody against NeuN. We found that Cy3 particles were specifically localized in neurons in all regions distal to the injection site. Within the neurons, Cy3 particles did not appear to be nuclear in the distal neurons (Fig. 3A,B), which could be attributable to the delay it takes for AAV to enter the nucleus (Bartlett et al., 2000) or could be attributable to the covalently linked Cy3 inhibiting the AAV vector from entering the nucleus in vivo.
Figure 3.
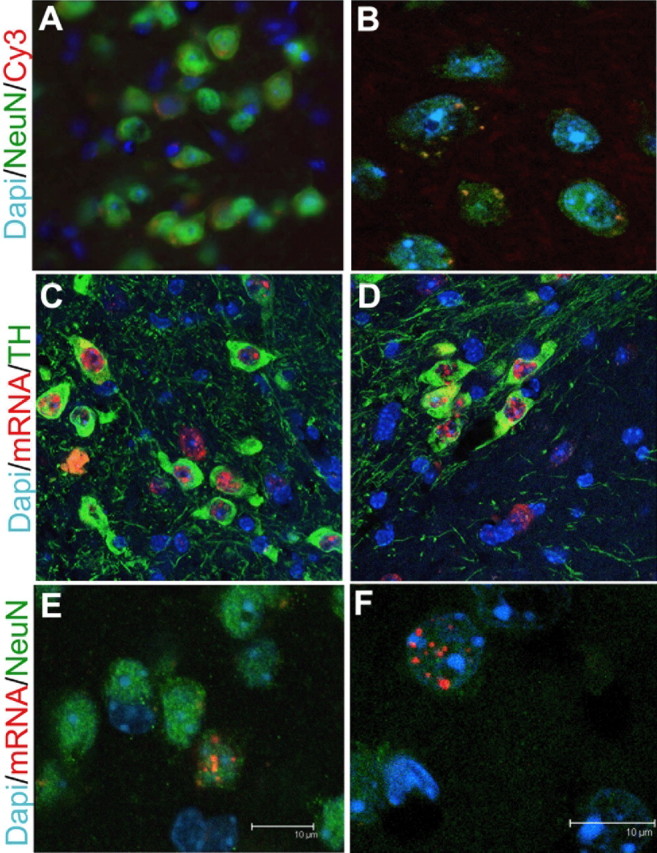
Localization of vector after an injection of AAV-9 into the VTA. A, B, To demonstrate the location of Cy3-labeled particles at 24 h after injection, sections from animals injected with Cy-3-labeled AAV-9 vector (red) into the VTA were costained with DAPI (blue) to label nuclei and NeuN (green) to label neurons. Pictures were taken using nonconfocal (40×, 2× zoom) (A) and confocal (100×, 2× zoom) (B) microscopy and are of the cortex (A) and striatum (B). C, D, Fluorescent in situ hybridization (FISH) using a DIG-labeled riboprobe against the hGUSB mRNA sequence was used to label hGUSB mRNA-positive cells (red) on sections from animals given injections of AAV-9 into the VTA and killed 1 month after injection. Sections were then costained with an anti-TH antibody, and slides were coverslipped using a mounting medium with DAPI. Pictures were taken using 100× confocal magnification and are of the VTA (C) or substantia nigra (D). E, F, To determine whether the hGUSB mRNA was located within the nucleus of striatal neurons, anterograde targets of the VTA, FISH was performed on sections that were then costained with anti-NeuN and coverslipped with DAPI. E is a compressed _z_-stack, and F represents a single plane of another cell to demonstrate nuclear localization. Scale bars: E, F, 10 μm.
Although it is known that AAV-9 transduces neurons, tropism for specific types of neurons is not known. The VTA contains many dopaminergic neurons (Swanson, 1982). To determine whether AAV-9 was transducing the dopaminergic neurons within the injected VTA and surrounding regions, we used an anti-TH antibody to label dopaminergic cells and looked for coexpression of hGUSB mRNA using fluorescent in situ hybridization (Fig. 3C,D). Animals that received injections 1 month before they were killed were used to ensure that vector uncoating and second-strand synthesis had occurred. Within the VTA, hGUSB mRNA could be found in every cell labeled with anti-TH and could also be found in cells that were not labeled with anti-TH (Fig. 3C). In the substantia nigra, a region that lies near the VTA and contains many dopaminergic cells, the TH-labeled cells also contained hGUSB mRNA (Fig. 3D). The substantia nigra is a retrograde target of the VTA but also lies near enough to the VTA that vector may have reached this region via diffusion. However, vector was mainly localized within the dopaminergic cells of the substantia nigra pars compacta (Fig. 3B), which is the specific part of the substantia nigra that sends projections to the VTA (Simon et al., 1979). Very few vector genome-positive cells were seen in the substantia nigra pars reticulata, which does not send projections to the VTA, suggesting that transport of the AAV vector genome occurred rather than just diffusion from the injection site.
Comparison of AAV serotypes in the VTA
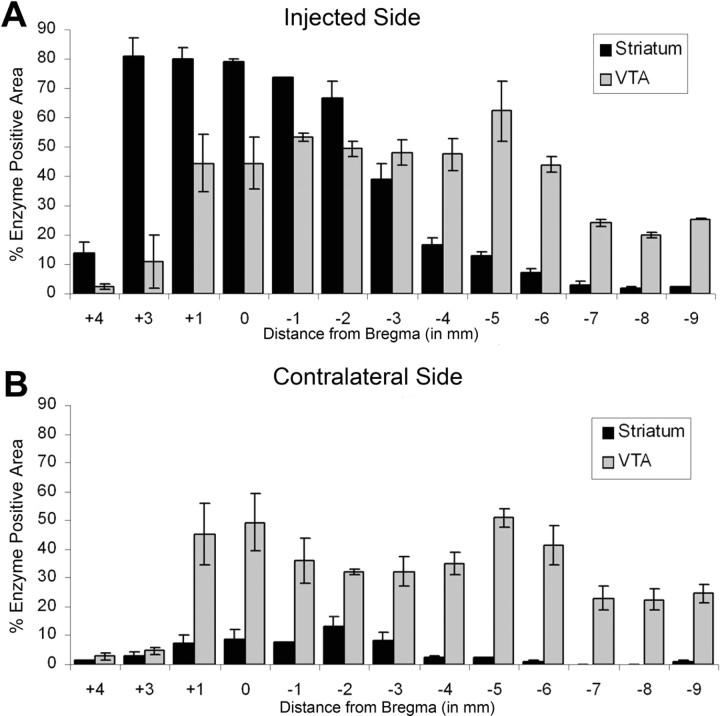
AAV-1 and AAV-Rh.10 are two other AAV serotypes currently considered to work well at transducing cells of the adult mouse CNS. Both serotypes have been shown to result in extremely high transduction levels at the site of injection with vector distribution far from the injected region (Wang et al., 2003; Burger et al., 2004; Dodge et al., 2005; Cearley and Wolfe, 2006; Sondhi et al., 2007). We compared the level of enzyme production, enzyme transport, and potential vector transport of AAV serotypes 1, 9, and Rh.10 after an injection into the VTA. For a visual comparison of the vectors, see supplemental Figure S1 (available at www.jneurosci.org as supplemental material). To quantitatively compare enzyme levels, coronal sections from animals given injections of the three serotypes were stained for GUSB enzyme using a histochemical reaction and analyzed using methods described and validated previously (Skorupa et al., 1999; Cearley and Wolfe, 2006). Sections were analyzed at each 1 mm distance from bregma based on known coordinates (Paxinos and Franklin, 2001), from 4 mm rostral (olfactory bulb) to 9 mm caudal (spinal cord) from bregma (Fig. 4). The single unilateral injection into the VTA resulted in substantial enzyme expression for all three vectors, on both the injected (Fig. 4B) and contralateral (Fig. 4C) sides. On the injected side (Fig. 4B), AAV-9 and AAV-1 resulted in higher enzyme levels than AAV-Rh.10 rostral to the injection site, but all three vectors resulted in similar levels at the level of injection and caudal to the injection site. On the contralateral side of the brain (Fig. 4C), AAV-9 resulted in higher levels of enzyme far rostral to the injection site, but all vectors resulted in similar levels of enzyme in the rest of the brain. A decrease in the total percentage of enzyme positive area for all three vectors at 6 and 7 mm caudal to bregma was attributable to the lower amount of enzyme expression in the cerebellum.
Figure 4.
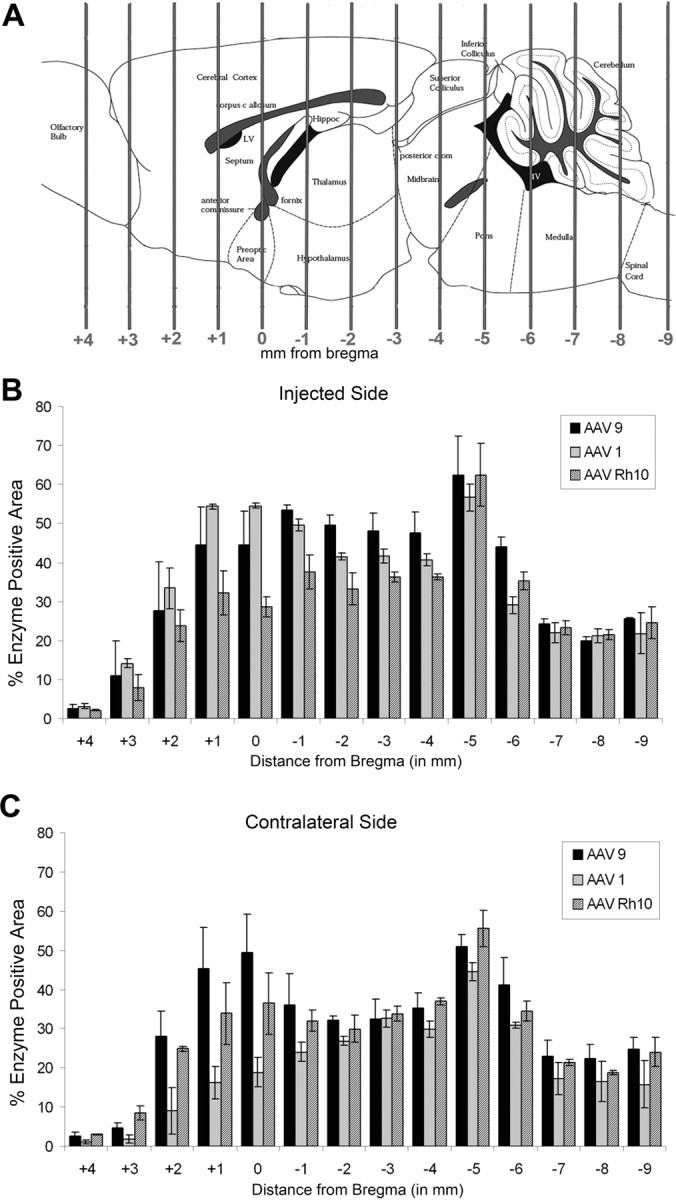
GUSB enzyme expression resulting from a single injection of AAV-9, AAV-1, or AAV-Rh.10 hGUSB into the VTA. Animals were given injections of 1 μl of AAV-9, AAV-1, or AAV-Rh.10 hGUSB into the unilateral VTA and analyzed 2 months after injection. A, The location of the VTA is represented on the brain illustration (adapted from Paxinos and Franklin, 2001). B, C, Tissue sections that underwent enzyme histochemistry were scanned into the computer and analyzed using a custom-designed Image-Pro program, which calculated the percentage of red area in each section. Three 20 μm sections at 100 μm apart for every 1 mm region were analyzed for each animal (n = 3 per group), the values were averaged, and the SEMs were calculated to give the values shown on the chart. The _x_-axis shows the distance from bregma. Hippoc, Hippocampus; LV, lateral ventricle; Pons, pontine nuclei. The injected side (B) was analyzed separately from the noninjected side (C). Enzyme expression from AAV-9-injected animals is shown by black bars, from AAV-1-injected animals is shown by light gray bars, and from AAV-Rh.10-injected animals is shown by striped bars.
To determine the extent of vector transport, hGUSB mRNA-positive cells were counted in the projection sites of the VTA (Table 1). There was some variability between animals, which is likely caused by slight differences in the exact region of the VTA injected, because different parts of the VTA have projections with different parts of the brain. Distal to the site of injection, most of the projection sites of the VTA are reciprocal. Of the nonreciprocal retrograde projection sites, the superior colliculus falls near the injection tract, so although many positive cells were found in this region for all three vectors, this could be attributable to diffusion from the injection tract. For the anterograde projection targets, the nucleus reunions lies close enough to the VTA that it is possible that many of the vector genome-positive cells in this region were also caused by diffusion from the injected region.
Table 1.
hGUSB in situ hybridization positive cells in projection sites of the VTA 1 month after AAV–GUSB injection
| Direction of projection | Structure | In situ positive cells (±SEM) | |||||
|---|---|---|---|---|---|---|---|
| AAV-9 | AAV-1 | AAV-Rh.10 | |||||
| Inj | Cont | Inj | Cont | Inj | Cont | ||
| Retrograde | Superior colliculus | 239.8 ± 41.7a | 182.4 ± 77.5 | 271.3 ± 27.6 | 221.9 ± 20.9 | 95.9 ± 38.4 | 81.0 ± 46.2 |
| Reticular nuclei | 1.7 ± 1.1 | 0.4 ± 0.3 | 3.8 ± 1.6 | 2.0 ± 1.0 | 0.8 ± 0.4 | 0.4 ± 0.3 | |
| Anterograde | Bed nucleus of stria terminalis | 9.6 ± 5.6 | 4.6 ± 3.6 | 4.1 ± 2.1 | 0.6 ± 0.4 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Amygdala | 2.3 ± 2.2 | 0.8 ± 0.6 | 3.9 ± 1.8 | 2.1 ± 2.0 | 0.4 ± 0.4 | 1.1 ± 1.0 | |
| Striatum | 78.3 ± 42.9 | 47.9 ± 47.2 | 85.8 ± 8.5 | 0.2 ± 0.2 | 0.8 ± 0.6 | 0.8 ± 0.6 | |
| Nucleus reunions of thalamus | 54.6 ± 7.0 | 48.2 ± 6.1 | 24.1 ± 11.6 | 21.1 ± 8.0 | 25.3 ± 6.4 | 23.0 ± 5.3 | |
| Reciprocal | Locus ceruleus | 28.4 ± 8.2 | 10.2 ± 5.3 | 21.6 ± 5.2 | 3.0 ± 1.2 | 22.0 ± 6.4 | 8.7 ± 0.8 |
| Dorsal raphe | 195.1 ± 23.9 | 173.3 ± 27.8 | 218.4 ± 29.8 | 183.2 ± 27.4 | 194.0 ± 7.1 | 167.0 ± 7.7 | |
| Medial raphe | 65.2 ± 20.1 | 39.8 ± 13.3 | 34.6 ± 2.5 | 23.9 ± 5.4 | 37.0 ± 24.5 | 24.1 ± 16.8 | |
| Deep cerebellar nucleus | 0.9 ± 0.3 | 1.1 ± 1.1 | 0.9 ± 0.2 | 0.9 ± 0.6 | 0.2 ± 0.2 | 0.3 ± 0.2 | |
| Medial forebrain bundle | 1.8 ± 1.0 | 0.4 ± 0.8 | 5.4 ± 0.8 | 0.8 ± 0.1 | 0.4 ± 0.4 | 0.2 ± 0.1 | |
| Medial habenula | 263.4 ± 44.2 | 214.7 ± 49.3 | 169.4 ± 34.3 | 38.9 ± 20.1 | 235.1 ± 55.2 | 205.9 ± 38.6 | |
| Lateral habenula | 93.0 ± 20.7 | 70.0 ± 11.2 | 54.7 ± 12.2 | 21.4 ± 7.9 | 123.9 ± 42.3 | 75.9 ± 21.5 | |
| Nucleus accumbens | 6.9 ± 6.1 | 4.6 ± 4.2 | 12.7 ± 3.6 | 0.6 ± 0.4 | 0.4 ± 0.1 | 0.4 ± 0.4 | |
| Mediodorsal nucleus of thalamus | 110.1 ± 30.0 | 94.7 ± 12.78 | 69.6 ± 26.5 | 43.8 ± 19.8 | 102.6 ± 22.6 | 102.2 ± 20.5 | |
| Lateral septal nucleus | 2.6 ± 1.4 | 4.8 ± 3.4 | 0.8 ± 0.4 | 1.6 ± 1.2 | 2.8 ± 2.5 | 0.3 ± 0.3 | |
| Prefrontal cortex | 1.7 ± 1.1 | 0.9 ± 0.9 | 4.1 ± 1.4 | 0.1 ± 0.1 | 0.6 ± 0.3 | 0.1 ± 0.1 | |
| Tegmental nuclei | 20.9 ± 15.0 | 16.0 ± 11.6 | 12.3 ± 3.5 | 4.8 ± 1.8 | 12.0 ± 5.8 | 8.1 ± 4.1 |
The substantia nigra pars compacta, which was found to contain many vector-positive cells, lies very close to the injected region, and mRNA likely reached this region via both diffusion and retrograde transport of vector. Both the substantia nigra pars compacta and the VTA send projections to the striatum. The striatum is very distal from the injected region, and sections between the two regions were not found to have hGUSB mRNA-positive cells, so any positive cells in the striatum were not caused by diffusion. The striatum was found to have numerous vector-positive cells after an AAV-9 or AAV-1 injection, suggesting that both of these vectors are able to undergo anterograde transport after an injection into the VTA. Interestingly, AAV-Rh.10 injection resulted in high expression of GUSB enzyme at the level of the striatum (Fig. 4B), but the striatum did not have hGUSB mRNA-positive cells after the AAV-Rh.10 injection, suggesting that the enzyme seen in this region was anterogradely transported from the AAV-Rh.10-injected VTA. Other anterograde targets of the VTA, such as the amygdala and bed nucleus of stria terminalis, were found to have a few hGUSB mRNA-positive cells with the AAV-9 and AAV-1 injections but not with the AAV-Rh.10 injection. Vector genome presence in cell bodies of anterograde targets suggests that the vector is able to be released by cells transporting the vector anterogradely and then picked up by other cells in the anterograde projection region. To determine whether in situ hybridization was labeling mRNA at axon terminals in the striatum or whether cell bodies were being labeled, we ran fluorescent in situ hybridization on striatum sections and colocalized vector genome with the neuronal antibody NeuN and the nuclear stain DAPI. We found that the fluorescent in situ hybridization-labeled mRNA was localized within neuronal cell bodies (Fig. 3E) and appeared to be nuclear (Fig. 3F). It is possible that cells within the substantia nigra pars reticulata were able to take up vector and transport it retrogradely to their cell bodies in the striatum. However, very little transduction of the substantia nigra pars reticulata was noted, so this is not likely.
Most of the regions that have reciprocal projections with the VTA had numerous hGUSB mRNA-positive cells after injections with any of the three vectors. Several regions that have known connections with the VTA rarely had positive cells, including the reticular nuclei, medial forebrain bundle, prefrontal cortex, and deep cerebellar nuclei. Most of these regions are located very distal to the injected region, and not as many projection fibers go to these regions, suggesting that AAV transport may be limited by distance or by numbers of projection fibers.
Regions distal to the VTA were found to have numerous positive cells on both the injected and contralateral side of the brain; this is most likely attributable to the fact that the VTA has many bilateral projections and because of the injection being so close to the midline. This was especially true with the AAV-9 injection in which the contralateral side of the brain had vector-positive cells in almost all of the projection sites. The injection of AAV-1 or AAV-Rh.10 resulted in vector genome-positive cells on the opposite side, but not in as many of the contralateral projection sites as seen with AAV-9. An injection of AAV-9 into the VTA resulted in higher levels of enzyme throughout most of the brain (Fig. 4), in transport of vector genome to most of the projection sites of the VTA to a slightly higher extent than seen with AAV-1 or AAV-Rh.10 on the ipsilateral side, and to a more consistent vector transport on the contralateral side (Table 1). For this reason, the remaining studies were performed using AAV-9.
Comparison of structures for vector transport
We determined whether the degree of enzyme and vector distribution was specific to the VTA by comparing injections of AAV-9 into the VTA, the striatum, or the hippocampus. The striatum has extensive retrograde connections with many other midbrain nuclei, such as the substantia nigra. It does not have connections with very caudal structures, does not have many anterograde projections, and does not have many projections reaching the contralateral side of the brain. The hippocampus has extensive connections within the structure itself but more limited connections to other areas of the brain compared with the VTA. The extent of enzyme spread after a unilateral injection of AAV-9 into the hippocampus has been shown previously (Cearley and Wolfe, 2006) and is much more limited than that with a VTA injection. To compare the VTA injection with a striatum injection, we quantitated the enzyme levels 1 month after a unilateral VTA or striatum injection (Fig. 5) using the same enzyme quantitation methods used with the serotype comparison. The distances on the _x_-axis can be compared with those shown in the illustration in Figure 4A.
Figure 5.
GUSB enzyme expression resulting from a single injection of AAV-9 into the striatum or VTA. The analysis was the same as that for Figure 4. Animals were given injections of AAV-9 into the unilateral striatum (Str) or VTA and analyzed at 1 month after injection (n = 3 per group). The injected side (A) was analyzed separately than the contralateral side (B). The enzyme expression levels after VTA injection are represented by light gray bars, whereas the enzyme expression levels after striatum injection are represented by black bars. The distances from bregma can be mapped to the illustration in Figure 4A. Because of the mounting process, sections around 2 mm rostral to bregma from striatum-injected brains were lost, so are not shown on the graph. Error bars indicate SEMs.
At 1 month after injection of AAV-9 into the striatum, areas near the injected region had almost 80% enzyme expression (Fig. 5A) which was higher than enzyme expression from an injection into the VTA. However, enzyme levels resulting from the striatum injection decreased as the distance from the striatum increased, whereas enzyme levels after an injection into the VTA remained high far distal to the injection site. Much lower enzyme amounts were present on the contralateral side of the brain with the striatal injection, whereas with a VTA injection, enzyme levels were high on both sides of the brain (Fig. 5B). For coronal sections showing the extent of enzyme distribution after striatal injection, see Figure 7.
Figure 7.
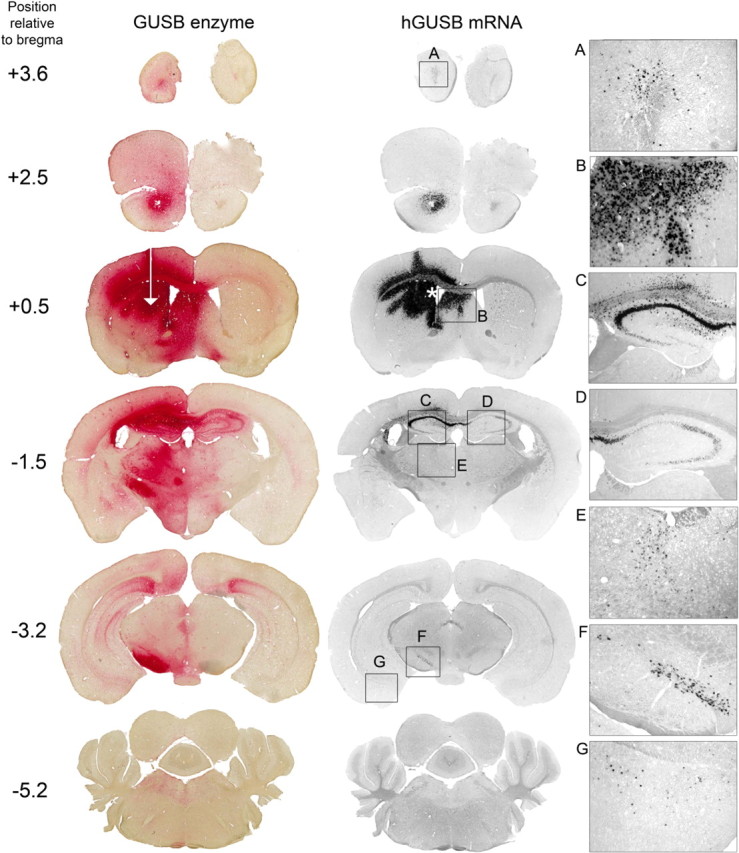
An injection of AAV-9 into the striatum results in vector transport along known projections. Animals (n = 3) were injected into the unilateral striatum and analyzed 1 month after injection. The injection site is represented by the white arrow. Resultant enzyme and vector genome (hGUSB mRNA) are shown. Boxes A–G (not drawn to scale) on the in situ hybridization sections correspond to panels A–G on the right. An injection into the striatum also resulted in transduction of the SVZ (asterisk) and septal nuclei (B) by diffusion. Vector genome was transported to the olfactory bulb (A), ipsilateral hippocampus (C), contralateral hippocampus (D), thalamus (E), substantia nigra pars compacta (F), and amygdala (G). Images in panels A–G were taken using 10× magnification.
Injection of AAV-9 into the hippocampus or striatum resulted in vector transport to the projection sites of these regions. Vector genome transport after a hippocampal injection has been shown (Cearley and Wolfe, 2006), but we repeated this experiment to provide a more detailed analysis of the extent of vector transport after an injection into this region. Vector genome transport was examined 2 months after injection into the hippocampus and was found to be variable between animals (Fig. 6). Variability appeared to be caused by where the exact injection site was within the hippocampus (Fig. 6C,D). An injection into the CA2/3 region (Fig. 6C) resulted in vector genome expression in the CA1 region near the injection and in the CA1 region of the contralateral hippocampus (Fig. 6E). Vector genome was also found in the lateral and medial septal nuclei on both sides of the brain (Fig. 6G) but not in the entorhinal cortex (Fig. 6I). The areas that received hGUSB mRNA correspond with regions known to have projections with the CA2/3 region (Fig. 6A). The CA3 region of the hippocampus sends many fibers to the contralateral CA1 region (Laurberg, 1979; Laurberg and Sorensen, 1981), so it is possible that vector mRNA got to this region by anterograde transport. The medial septal nuclei send projections to CA2/3 (Meibach and Siegel, 1977; Swanson and Cowan, 1979; Oka and Yoshida, 1985), so vector genome transport in this region is most likely caused by retrograde transport. The lateral septal nuclei on both sides of the brain receive projections from the ipsilateral CA3 region (Swanson, 1977; Swanson and Cowan, 1977), so vector genome presence in the bilateral septal nuclei suggests anterograde transport from the CA3 injection. Although the entorhinal cortex sends some fibers to the ipsilateral CA3 region, this projection is weak, which may explain why the entorhinal cortex was not found to have vector-positive cells (Wyss, 1981).
Figure 6.
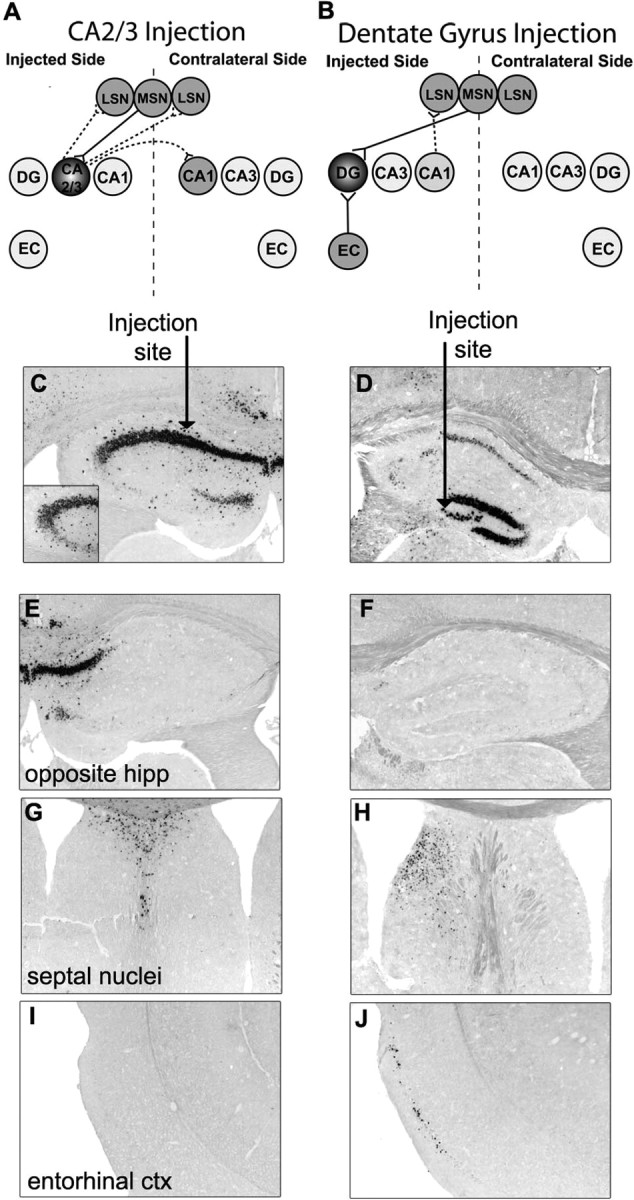
An injection of AAV-9 into the hippocampus results in vector transport along known projections. Animals were given injections of 1 μl of AAV-9 GUSB into the unilateral hippocampus and analyzed 2 months after injection. C, D, Injections into the hippocampus were variable with some animals receiving injections into the CA2/3 region (C) and some animals receiving injections into the dentate gyrus (DG) region (D). The arrow represents the site of injection. The inset in C shows transduction of the CA3 region. A, B, E–J, Connections of the CA2/3 region (A) and DG region (B) are known and correspond with areas where hGUSB mRNA expression was or was not found, including the opposite hippocampus (hipp) (E, F), medial (MSN) and lateral (LSN) septal nuclei (G, H), and entorhinal cortex (EC; ctx) (I, J). Within the hippocampus connection diagrams (A, B), solid lines represent retrograde targets and dotted lines represent anterograde targets of the injected region. The regions injected are darkly shaded, whereas the distal sites that were found to have hGUSB mRNA are shaded lighter.
An injection into the dentate gyrus (Fig. 6D) resulted in vector genome in the entorhinal cortex (Fig. 6J) and the lateral and medial septal nuclei on the injected side (Fig. 6H) but no genome expression in the contralateral lateral septal nuclei (Fig. 6H) or contralateral hippocampus (Fig. 6F). The areas that received hGUSB mRNA-positive cells correspond to regions with known projections to and/or from the dentate gyrus or CA1, which lies along the injection tract (Fig. 6B). The dentate gyrus receives a large number of projections from the entorhinal cortex (Steward, 1976; Wyss, 1981), and the hilar region receives projections from the medial septal nuclei (Oka and Yoshida, 1985), so vector genome expression in these regions is most likely caused by retrograde transport. The expression in the lateral septal nuclei is most likely attributable to the transduction of cells along the injection tract, including cells in the CA1 region, which send projections to the lateral septal nuclei on the ipsilateral side (Swanson, 1977; Swanson and Cowan, 1977), suggesting anterograde transport.
Vector genome distribution after striatal injection was quantitated at 1 month after injection of AAV-9 and was found in many of the known projection sites of the injected region (Table 2, Fig. 7 A–G). The striatum receives projections from the cortex, substantia nigra pars compacta, amygdala, VTA, raphe nuclei, subiculum, and the thalamus (Kelley et al., 1982; Berendse and Groenewegen, 1990). The striatum sends projections to the substantia nigra pars reticulata and medial globus pallidus (Tulloch et al., 1978; Gandia and Gimenez-Amaya, 1991). Some projection sites of the striatum, such as the cortex, were not counted because they fall within the injection tract. A single AAV-9 injection resulted in many vector genome-positive cells in the retrograde projection targets of the striatum, with a few positive cells found in anterograde targets (Table 2).
Table 2.
hGUSB in situ hybridization positive cells in projection sites of the striatum 1 month after AAV-9 injection
| Direction of projection | Structure | In situ positive cells (±SEM) |
|---|---|---|
| Retrograde | SNpc | 75.4 ± 13.8 |
| Dorsal raphe | 2.3 ± 0.9 | |
| Hippocampus | 79.2 ± 28.4 | |
| VTA | 10.3 ± 5.2 | |
| Amygdala | 163.8 ± 35.4 | |
| Thalamus (parafascicular and medial nuclei) | 49.9 ± 10.7 | |
| Subiculum | 3.7 ± 2.1 | |
| Anterograde | Medial GP | 6.6 ± 1.6 |
| SNpr | 6.0 ± 3.0 |
The CA3 region of the dorsal hippocampus, which is not a direct projection site of the striatum, had many in situ hybridization-positive cells, on both the ipsilateral and contralateral side (Fig. 7C,D). This can be explained by the transduction of the septal nuclei via diffusion from the striatal injection site (Fig. 7B). The medial septal nuclei send projections to the CA1–CA3 region, which suggests that anterograde transport is responsible for the vector genome presence in these regions on the injected side of the brain. The lateral septal nuclei receive projections from the bilateral CA1 and CA3 nuclei, suggesting retrograde transport is responsible for the positive cells found in the CA1 and CA3 regions on both sides of the brain. Interestingly, many vector genome-positive cells were also found in the olfactory bulb (Fig. 7A). This is most likely because of transduction of the neural progenitor cells in the subventricular zone (SVZ), which lies near the injection site and is the source of the rostral migratory stream, a pathway for neural stem cells that ends in the olfactory bulb (Altman, 1969). The transport of GUSB enzyme to the olfactory bulb after transduction of cells in the SVZ with AAV-2 serotype has been documented (Passini et al., 2002), but this is the first evidence of the AAV vector genome being transported to the olfactory bulb from the SVZ.
Effect of vector and enzyme distribution from a VTA injection on disease lesions
To determine whether the widespread distribution of vector would result in reversal of lysosomal storage in MPS VII animals, adult affected mice were given injections of AAV-9 hGUSB and analyzed 2 months after injection for enzyme expression and resolution of the characteristic lysosomal distension (Fig. 8). For this analysis, the X-Gluc substrate was used to stain for enzyme expression (Fig. 8A,E). Unlike the naphthol AS-BI β-d-glucuronide stain used on frozen sections and shown in Figure 1, the X-Gluc stain allows brain slices to be reacted without freezing so they can be further processed in plastic embedding (Fig. 8 B–D), which is necessary for visualization of lysosomal storage. Although the X-Gluc stain is not as sensitive as the naphthol stain (Skorupa et al., 1999), there was substantial enzyme staining widely distributed throughout the brains of the treated animals (Fig. 8E) compared with nontreated animals (Fig. 8A), which demonstrated the presence of active GUSB enzyme in the AAV-9-treated brain.
Figure 8.
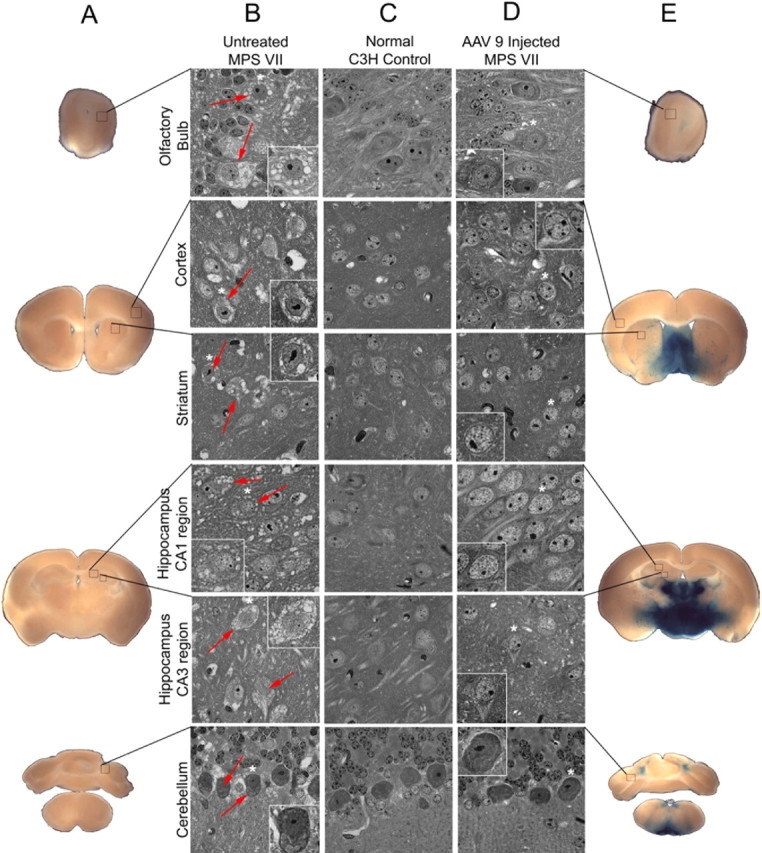
Enzyme expression and correction of lysosomal storage 2 months after AAV-9 hGUSB injection into the VTA of MPS VII animals. A, E, Brains from AAV-9 hGUSB-injected MPS VII mice (n = 3), noninjected MPS VII mice (n = 3), and normal mice (n = 3) were divided into 500-μm-thick slabs and stained for enzyme expression using an X-Gluc reaction to demonstrate enzyme expression in the AAV-treated brain (E) compared with the untreated MPS VII brain (A). B–D, Slabs were then embedded in JB4 resin, sectioned into 1-μm-thick sections, and stained with toluidine blue to visualize storage vacuoles. Boxes (not drawn to scale) on the X-Gluc-stained sections (A, E) show the area that the magnified images (B–D) represent. There were numerous storage vacuoles present in many regions of the untreated MPS VII brain (B). Examples of cells with storage vacuoles are shown with red arrows. Within each magnified image from untreated (B) or treated (D) MPS VII mice, higher-magnified images (outlined in white) show single cells. The cells represented by these higher-magnification images have asterisks above them in the lower-magnification images. The extensive storage found in the untreated MPS VII animals is shown in the olfactory bulb, cortex, striatum, hippocampus, and cerebellum (B). No storage was found in any of these representative regions in either the normal control brains (C) or in the AAV-9-injected MPS VII brains (D).
To analyze the extent of lysosomal storage reversal after an injection of AAV-9 into the VTA, semithin sections (1 μm) from the brains of AAV-9-injected MPS VII, noninjected MPS VII, and noninjected normal animals were stained with toluidine blue to highlight the presence of storage vacuoles (Fig. 8 B–D). The untreated MPS VII mouse brain (Fig. 8B) had extensive storage in areas such as the olfactory bulb, cortex, striatum, hippocampus, and cerebellum compared with the normal brain (Fig. 8C). Storage was reversed in all these regions after a single injection of AAV-9 hGUSB into the VTA (Fig. 8D). Sections from the treated MPS VII animals did not have lysosomal storage in any of the brain regions examined and were indistinguishable from the normal mouse brain sections.
Discussion
It is currently estimated that treatment using AAV vectors would require between 50 and 350 injection tracts to achieve widespread dispersal of a lysosomal enzyme in an infant brain (Vite et al., 2003, 2005). To be useful in treating metabolic disorders, gene therapy strategies need to be developed in which the therapeutic gene or protein can reach the entire brain from a more limited number of injections.
Lysosomal enzymes are transported in some axonal pathways, resulting in correction of disease pathology at distal sites from the injection (Heuer et al., 2002; Passini et al., 2002; Hennig et al., 2003; Luca et al., 2005). However, areas between the injected site and the distal site receiving enzyme via transport still have significant lysosomal storage (Passini et al., 2002), suggesting that only a limited volume of brain can be corrected using enzyme transport alone.
Some AAV vector serotypes mediate genome transport in the retrograde direction (Kaspar et al., 2003; Passini et al., 2005; Provost et al., 2005; Cearley and Wolfe, 2006). The vector transport of AAV-9 results in increased GUSB enzyme distribution (Cearley and Wolfe, 2006) compared with studies in which the vector was not transported (Heuer et al., 2002; Passini et al., 2002).
We sought to use the transport capabilities of AAV to achieve more widespread dispersal of vector and enzyme. We hypothesized that an injection of a transportable vector into an area with multiple dispersed projections, may allow transport along those projections to distribute the therapeutic gene to a larger volume of brain. The VTA was chosen because it has projections from, or projects to, most regions of the brain. Data on the retrograde (Phillipson, 1979; Simon et al., 1979; Geisler and Zahm, 2005) and anterograde (Beckstead et al., 1979; Simon et al., 1979; Swanson, 1982) targets of the VTA suggest that the majority of the projections are reciprocal. Because of this, it was not possible to conclusively determine which direction transport occurred. However, the injection of AAV-1 or AAV-9 resulted in vector-positive cells in the anterograde targets of the VTA to a greater extent than the injections of AAV-Rh.10. In addition, the pattern of expression of vector-positive cells after AAV-9 or AAV-1 injection appeared to match patterns seen in anterograde tract tracing studies (Simon et al., 1979), suggesting that anterograde transport may be contributing to the distribution observed. The finding of vector mRNA in cell bodies within the anterograde projection site, the striatum, is interesting because it suggests that the vector is transported anterogradely, released, and taken up by local cells in the anterograde projection target region. Additional studies need to be done to determine the mechanism by which this could occur. Overall, the injection of AAV-9 resulted in hGUSB mRNA-positive cells in the highest numbers of projection sites of the VTA, in the highest numbers of positive cells in many of the projection sites, and in the highest levels of enzyme throughout most of the brain compared with AAV serotypes 1 and Rh.10.
The differences in transport between AAV serotypes 1, 9, and Rh.10 suggest that it is the vector virus itself that is being transported. If it were the GUSB mRNA only that was being transported, it would be expected that transport would occur equally with the three vectors because they share the same genome. The mechanism(s) the vector uses for transport of vector genome to distal neurons has not been identified. Knowing the speed at which transport occurs can provide insights into the possible mechanisms a vector uses. When an AAV vector enters a cell, it must uncoat to expose the AAV genome, and then it must undergo second-strand synthesis before mRNA is detectable within the transduced cells, a process that takes from days to weeks depending on the serotype used (Ferrari et al., 1996; Hauck et al., 2004). Covalent Cy3 labeling of AAV vectors provides a way to track the location of AAV virions at early time points after injection (Bartlett et al., 1998; Sanlioglu et al., 2000; Passini and Wolfe, 2001; Ding et al., 2006). The Cy3-labeled vector was transported to projection sites of the injected regions between 1 and 24 h after injection. The Cy3-labeled particles appeared to be outside the nucleus in distal cell bodies, which may be attributable to the delay it takes for nuclear entry (Bartlett et al., 2000).
Although the Cy3 data and the 1 month mRNA data were consistent in most regions, the Cy3 signal was not detected in all projection sites of the VTA that were later found to have GUSB mRNA, such as the amygdala. The lack of signal in some projection sites could reflect a threshold of detection of the particles. Alternatively, because the mechanism of transport for this vector is unknown, it could take longer for vector to be transported to some regions than others. Cy3-labeled particles were detected in regions that were not mRNA positive at 1 month after injection, such as the cortex. The presence of Cy3-labeled vector particles in distal sites supports the idea that the vector itself is being transported, not the GUSB mRNA. The finding that transport occurs by 24 h after injection suggests that fast axonal transport is involved, which would imply that microtubules may play a role in the transport. AAV vectors, including AAV-Rh.10, have been shown to bind to microtubules, an interaction mediated by dynein, to facilitate trafficking to the nucleus (Kelkar et al., 2006). It is possible that the retrograde and anterograde axonal transport of AAV vectors also rely on microtubule-dependent processes, perhaps using the molecular motors kinesin, which moves in the anterograde direction (Vale et al., 1985), or MAP-1C (microtubule-associated protein 1C), which moves in the retrograde direction (Paschal and Vallee, 1987).
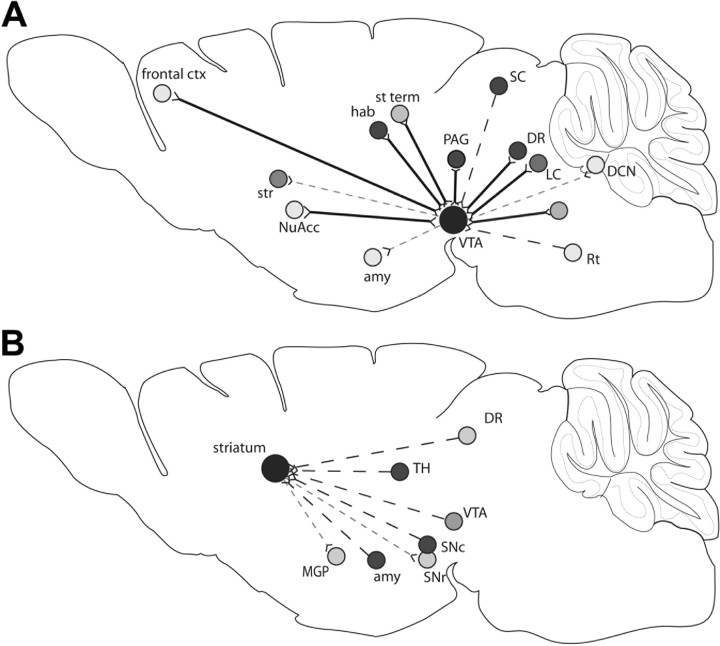
The AAV-9 vector was transported to projection sites of the VTA (Fig. 9A) and striatum (Fig. 9B) in different amounts. This could be attributable to different regions of the VTA or striatum projecting to different parts of the brain or could be attributable to a limit on the transport, in that the injected region may need to have a minimum number of projection fibers in order for the vector transport to be detectable. Two of the regions that received few cells from the VTA injection include the frontal cortex and deep cerebellar nuclei, and these regions appear to have fewer projection fibers than other regions such as the habenula or dorsal raphe (Beckstead et al., 1979; Simon et al., 1979).
Figure 9.
Projection sites of the VTA or striatum and the presence or absence of vector genome within the regions after injection of AAV-9. Diagrams illustrate the retrograde (dashed black line) anterograde (dashed gray line), and reciprocal (solid black line) projection sites of the VTA (A) or striatum (B). Projection sites are represented by shaded circles, and the shade represents the relative level of vector genome-positive cells in that region, with the darker the circle the more cells found to have vector genome. ctx, Cortex; str, striatum; NuAcc, nucleus accumbens; amy, amygdale; Rt, reticular nuclei; DCN, deep cerebellar nuclei; LC, locus ceruleus; DR, dorsal raphe; SC, superior colliculus; PAG, periaqueductal gray; st term, bed nucleus of the stria terminalis; hab, habenula; LGP, lateral globus pallidus; MGP, medial globus pallidus; Snr, substantia nigra pars reticulate; Snc, substantia nigra pars compacta; TH, thalamus.
The ability of the GUSB enzyme to be transported along neuronal projections increases the potential distance of enzymatic correction from the injection site. However, there are still limits to the extent of correction because the GUSB enzyme appears to only be transported to direct projections of the enzyme-producing cells. The limits to enzyme dispersal are evident after injection into the striatum, which does not have extensive distal or contralateral projections. An injection into this region resulted in vector genome transport to the projection sites, but because the projections are not widespread, resultant enzyme levels were only high near the site of injection. An injection into a region with widespread projections, such as the VTA, allowed the AAV vector to be transported to distal projection sites of the VTA where cells were then able to make GUSB enzyme, which was further transported to projection sites of these cells, with the final result being GUSB enzyme distribution throughout the entire brain after a single small volume injection.
The MPS VII mouse provides an ideal model system in which to test gene therapy vectors. The phenotype of the mouse is essentially the same as in the human disease (Birkenmeier et al., 1989). When MPS VII animals were given injections into the VTA with AAV-9 GUSB and analyzed 2 months after injection, the levels of storage were indistinguishable from normal mice in all regions of the brain. The mouse brain, which weighs about 400 mg, was injected with 1 μl of vector. A 1-year-old human brain weighs about 800 g, which would require an injection of 2 ml if the distribution scales up proportionally. It is also possible that an injection of <1 μl would be enough to allow the extent of vector and enzyme transport that we saw in our experiment.
Treating genetic disorders of the CNS has been difficult because of the invasive nature of the treatment methods required. Targeting of the injection to areas of the brain that have widespread connections may be a way to increase the dispersal of a transportable AAV vector from a limited number of injections. Here we present a strategy that allows a maximum area of brain to be treated with only a single injection.
Footnotes
This work was supported by National Institutes of Health Grants NS-38690, DK-47757, and DK-63973. C.N.C. was supported by NeuroVirology Training Grant NS-07180. We thank A. Polesky, T. Clarke, E. Cabacungan, A. Radu, and the Vector Core at the University of Pennsylvania for expert technical assistance and R. Valentino and M. Passini for helpful discussions.
References
- Altman J. Autoradiographic and histological studies of postnatal neurogenesis. IV. Cell proliferation and migration in the anterior forebrain, with special reference to persisting neurogenesis in the olfactory bulb. J Comp Neurol. 1969;137:433–457. doi: 10.1002/cne.901370404. [DOI] [PubMed] [Google Scholar]
- Bartlett JS, Samulski RJ. Fluorescent viral vectors: a new technique for the pharmacological analysis of gene therapy. Nat Med. 1998;4:635–637. doi: 10.1038/nm0598-635. [DOI] [PubMed] [Google Scholar]
- Bartlett JS, Samulski RJ, McCown TJ. Selective and rapid uptake of adeno-associated virus type 2 in brain. Hum Gene Ther. 1998;9:1181–1186. doi: 10.1089/hum.1998.9.8-1181. [DOI] [PubMed] [Google Scholar]
- Bartlett JS, Wilcher R, Samulski RJ. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J Virol. 2000;74:2777–2785. doi: 10.1128/jvi.74.6.2777-2785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckstead RM, Domesick VB, Nauta WJ. Efferent connections of the substantia nigra and ventral tegmental area in the rat. Brain Res. 1979;175:191–217. doi: 10.1016/0006-8993(79)91001-1. [DOI] [PubMed] [Google Scholar]
- Berendse HW, Groenewegen HJ. Organization of the thalamostriatal projections in the rat, with special emphasis on the ventral striatum. J Comp Neurol. 1990;299:187–228. doi: 10.1002/cne.902990206. [DOI] [PubMed] [Google Scholar]
- Birkenmeier EH, Davisson MT, Beamer WG, Ganschow RE, Vogler CA, Gwynn B, Lyford KA, Maltais LM, Wawrzyniak CJ. Murine mucopolysaccharidosis type VII. Characterization of a mouse with beta-glucuronidase deficiency. J Clin Invest. 1989;83:1258–1266. doi: 10.1172/JCI114010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger C, Gorbatyuk OS, Velardo MJ, Peden CS, Williams P, Zolotukhin S, Reier PJ, Mandel RJ, Muzyczka N. Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Mol Ther. 2004;10:302–317. doi: 10.1016/j.ymthe.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Casal ML, Wolfe JH. Variant clinical course of mucopolysaccharidosis type VII in two groups of mice carrying the same mutation. Lab Invest. 1998;78:1575–1581. [PubMed] [Google Scholar]
- Casal ML, Wolfe JH. In utero transplantation of fetal liver cells in the mucopolysaccharidosis type VII mouse results in low-level chimerism, but overexpression of beta-glucuronidase can delay onset of clinical signs. Blood. 2001;97:1625–1634. doi: 10.1182/blood.v97.6.1625. [DOI] [PubMed] [Google Scholar]
- Cearley CN, Wolfe JH. Transduction characteristics of adeno-associated virus vectors expressing cap serotypes 7, 8, 9, and Rh10 in the mouse brain. Mol Ther. 2006;13:528–537. doi: 10.1016/j.ymthe.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Davidson BL, Stein CS, Heth JA, Martins I, Kotin RM, Derksen TA, Zabner J, Ghodsi A, Chiorini JA. Recombinant adeno-associated virus type 2, 4, and 5 vectors: transduction of variant cell types and regions in the mammalian central nervous system. Proc Natl Acad Sci USA. 2000;97:3428–3432. doi: 10.1073/pnas.050581197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W, Zhang LN, Yeaman C, Engelhardt JF. rAAV2 traffics through both the late and the recycling endosomes in a dose-dependent fashion. Mol Ther. 2006;13:671–682. doi: 10.1016/j.ymthe.2005.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge JC, Clarke J, Song A, Bu J, Yang W, Taksir TV, Griffiths D, Zhao MA, Schuchman EH, Cheng SH, O'Riordan CR, Shihabuddin LS, Passini MA, Stewart GR. Gene transfer of human acid sphingomyelinase corrects neuropathology and motor deficits in a mouse model of Niemann-Pick type A disease. Proc Natl Acad Sci USA. 2005;102:17822–17827. doi: 10.1073/pnas.0509062102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari FK, Samulski T, Shenk T, Samulski RJ. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J Virol. 1996;70:3227–3234. doi: 10.1128/jvi.70.5.3227-3234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandia JA, Gimenez-Amaya JM. A neuroanatomical analysis of the rostral striatopallidal pathway in the rat. J Hirnforsch. 1991;32:79–88. [PubMed] [Google Scholar]
- Gao G, Qu G, Burnham MS, Huang J, Chirmule N, Joshi B, Yu QC, Marsh JA, Conceicao CM, Wilson JM. Purification of recombinant adeno-associated virus vectors by column chromatography and its performance in vivo. Hum Gene Ther. 2000;11:2079–2091. doi: 10.1089/104303400750001390. [DOI] [PubMed] [Google Scholar]
- Geisler S, Zahm DS. Afferents of the ventral tegmental area in the rat-anatomical substratum for integrative functions. J Comp Neurol. 2005;490:270–294. doi: 10.1002/cne.20668. [DOI] [PubMed] [Google Scholar]
- Hauck B, Zhao W, High K, Xiao W. Intracellular viral processing, not single-stranded DNA accumulation, is crucial for recombinant adeno-associated virus transduction. J Virol. 2004;78:13678–13686. doi: 10.1128/JVI.78.24.13678-13686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig AK, Levy B, Ogilvie JM, Vogler CA, Galvin N, Bassnett S, Sands MS. Intravitreal gene therapy reduces lysosomal storage in specific areas of the CNS in mucopolysaccharidosis VII mice. J Neurosci. 2003;23:3302–3307. doi: 10.1523/JNEUROSCI.23-08-03302.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrup K, Mullen RJ. Biochemical and genetic factors in the heat inactivation of murine beta-glucuronidase. Biochem Genet. 1977;15:641–653. doi: 10.1007/BF00484095. [DOI] [PubMed] [Google Scholar]
- Heuer GG, Passini MA, Jiang K, Parente MK, Lee VM, Trojanowski JQ, Wolfe JH. Selective neurodegeneration in murine mucopolysaccharidosis VII is progressive and reversible. Ann Neurol. 2002;52:762–770. doi: 10.1002/ana.10373. [DOI] [PubMed] [Google Scholar]
- Kaspar BK, Llado J, Sherkat N, Rothstein JD, Gage FH. Retrograde viral delivery of IGF-1 prolongs survival in a mouse ALS model. Science. 2003;301:839–842. doi: 10.1126/science.1086137. [DOI] [PubMed] [Google Scholar]
- Kelkar S, De BP, Gao G, Wilson JM, Crystal RG, Leopold PL. A common mechanism for cytoplasmic dynein-dependent microtubule binding shared among adeno-associated virus and adenovirus serotypes. J Virol. 2006;80:7781–7785. doi: 10.1128/JVI.00481-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley AE, Domesick VB, Nauta WJ. The amygdalostriatal projection in the rat–an anatomical study by anterograde and retrograde tracing methods. Neuroscience. 1982;7:615–630. doi: 10.1016/0306-4522(82)90067-7. [DOI] [PubMed] [Google Scholar]
- Laurberg S. Commissural and intrinsic connections of the rat hippocampus. J Comp Neurol. 1979;184:685–708. doi: 10.1002/cne.901840405. [DOI] [PubMed] [Google Scholar]
- Laurberg S, Sorensen KE. Associational and commissural collaterals of neurons in the hippocampal formation (hilus fasciae dentatae and subfield CA3) Brain Res. 1981;212:287–300. doi: 10.1016/0006-8993(81)90463-7. [DOI] [PubMed] [Google Scholar]
- Luca T, Givogri MI, Perani L, Galbiati F, Follenzi A, Naldini L, Bongarzone ER. Axons mediate the distribution of arylsulfatase A within the mouse hippocampus upon gene delivery. Mol Ther. 2005;12:669–679. doi: 10.1016/j.ymthe.2005.06.438. [DOI] [PubMed] [Google Scholar]
- Meibach RC, Siegel A. Efferent connections of the septal area in the rat: an analysis utilizing retrograde and anterograde transport methods. Brain Res. 1977;119:1–20. doi: 10.1016/0006-8993(77)90088-9. [DOI] [PubMed] [Google Scholar]
- Moullier P, Bohl D, Heard JM, Danos O. Correction of lysosomal storage in the liver and spleen of MPS VII mice by implantation of genetically modified skin fibroblasts. Nat Genet. 1993;4:154–159. doi: 10.1038/ng0693-154. [DOI] [PubMed] [Google Scholar]
- Oka H, Yoshida K. Septohippocampal connections to field CA1 of the rat identified with field potential analysis and retrograde labeling by horseradish peroxidase. Neurosci Lett. 1985;58:19–24. doi: 10.1016/0304-3940(85)90322-2. [DOI] [PubMed] [Google Scholar]
- Paschal BM, Vallee RB. Retrograde transport by the microtubule-associated protein MAP 1C. Nature. 1987;330:181–183. doi: 10.1038/330181a0. [DOI] [PubMed] [Google Scholar]
- Passini MA, Wolfe JH. Widespread gene delivery and structure-specific patterns of expression in the brain after intraventricular injections of neonatal mice with an adeno-associated virus vector. J Virol. 2001;75:12382–12392. doi: 10.1128/JVI.75.24.12382-12392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passini MA, Lee EB, Heuer GG, Wolfe JH. Distribution of a lysosomal enzyme in the adult brain by axonal transport and by cells of the rostral migratory stream. J Neurosci. 2002;22:6437–6446. doi: 10.1523/JNEUROSCI.22-15-06437.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passini MA, Watson DJ, Vite CH, Landsburg DJ, Feigenbaum AL, Wolfe JH. Intraventricular brain injection of adeno-associated virus type 1 (AAV1) in neonatal mice results in complementary patterns of neuronal transduction to AAV2 and total long-term correction of storage lesions in the brains of beta-glucuronidase-deficient mice. J Virol. 2003;77:7034–7040. doi: 10.1128/JVI.77.12.7034-7040.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passini MA, Macauley SL, Huff MR, Taksir TV, Bu J, Wu IH, Piepenhagen PA, Dodge JC, Shihabuddin LS, O'Riordan CR, Schuchman EH, Stewart GR. AAV vector-mediated correction of brain pathology in a mouse model of Niemann-Pick A disease. Mol Ther. 2005;11:754–762. doi: 10.1016/j.ymthe.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. The mouse brain in stereotaxic coordinates. Ed 2. San Diego: Academic; 2001. [Google Scholar]
- Phillipson OT. Afferent projections to the ventral tegmental area of Tsai and interfascicular nucleus: a horseradish peroxidase study in the rat. J Comp Neurol. 1979;187:117–143. doi: 10.1002/cne.901870108. [DOI] [PubMed] [Google Scholar]
- Provost N, Le Meur G, Weber M, Mendes-Madeira A, Podevin G, Cherel Y, Colle MA, Deschamps JY, Moullier P, Rolling F. Biodistribution of rAAV vectors following intraocular administration: evidence for the presence and persistence of vector DNA in the optic nerve and in the brain. Mol Ther. 2005;11:275–283. doi: 10.1016/j.ymthe.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Sanlioglu S, Benson PK, Yang J, Atkinson EM, Reynolds T, Engelhardt JF. Endocytosis and nuclear trafficking of adeno-associated virus type 2 are controlled by rac1 and phosphatidylinositol-3 kinase activation. J Virol. 2000;74:9184–9196. doi: 10.1128/jvi.74.19.9184-9196.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon H, Le Moal M, Calas A. Efferents and afferents of the ventral tegmental-A10 region studied after local injection of [3H]leucine and horseradish peroxidase. Brain Res. 1979;178:17–40. doi: 10.1016/0006-8993(79)90085-4. [DOI] [PubMed] [Google Scholar]
- Skorupa AF, Fisher KJ, Wilson JM, Parente MK, Wolfe JH. Sustained production of beta-glucuronidase from localized sites after AAV vector gene transfer results in widespread distribution of enzyme and reversal of lysosomal storage lesions in a large volume of brain in mucopolysaccharidosis VII mice. Exp Neurol. 1999;160:17–27. doi: 10.1006/exnr.1999.7176. [DOI] [PubMed] [Google Scholar]
- Sondhi D, Hackett NR, Peterson DA, Stratton J, Baad M, Travis KM, Wilson JM, Crystal RG. Enhanced survival of the LINCL mouse following CLN2 gene transfer using the rh. 10 rhesus macaque-derived adeno-associated virus vector. Mol Ther. 2007;15:481–491. doi: 10.1038/sj.mt.6300049. [DOI] [PubMed] [Google Scholar]
- Steward O. Topographic organization of the projections from the entorhinal area to the hippocampal formation of the rat. J Comp Neurol. 1976;167:285–314. doi: 10.1002/cne.901670303. [DOI] [PubMed] [Google Scholar]
- Swanson LW. The anatomical organization of septo-hippocampal projections. Ciba Found Symp. 1977;58:25–48. doi: 10.1002/9780470720394.ch4. [DOI] [PubMed] [Google Scholar]
- Swanson LW. The projections of the ventral tegmental area and adjacent regions: a combined fluorescent retrograde tracer and immunofluorescence study in the rat. Brain Res Bull. 1982;9:321–353. doi: 10.1016/0361-9230(82)90145-9. [DOI] [PubMed] [Google Scholar]
- Swanson LW, Cowan WM. An autoradiographic study of the organization of the efferent connections of the hippocampal formation in the rat. J Comp Neurol. 1977;172:49–84. doi: 10.1002/cne.901720104. [DOI] [PubMed] [Google Scholar]
- Swanson LW, Cowan WM. The connections of the septal region in the rat. J Comp Neurol. 1979;186:621–655. doi: 10.1002/cne.901860408. [DOI] [PubMed] [Google Scholar]
- Taylor RM, Wolfe JH. Decreased lysosomal storage in the adult MPS VII mouse brain in the vicinity of grafts of retroviral vector-corrected fibroblasts secreting high levels of beta-glucuronidase. Nat Med. 1997;3:771–774. doi: 10.1038/nm0797-771. [DOI] [PubMed] [Google Scholar]
- Tulloch IF, Arbuthnott GW, Wright AK. Topographical organization of the striatonigral pathway revealed by anterograde and retrograde neuroanatomical tracing techniques. J Anat. 1978;127:425–441. [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Schnapp BJ, Mitchison T, Steuer E, Reese TS, Sheetz MP. Different axoplasmic proteins generate movement in opposite directions along microtubules in vitro. Cell. 1985;43:623–632. doi: 10.1016/0092-8674(85)90234-x. [DOI] [PubMed] [Google Scholar]
- Van Groen T, Wyss JM. Connections of the retrosplenial granular b cortex in the rat. J Comp Neurol. 2003;463:249–263. doi: 10.1002/cne.10757. [DOI] [PubMed] [Google Scholar]
- Vite CH, Passini MA, Haskins ME, Wolfe JH. Adeno-associated virus vector-mediated transduction in the cat brain. Gene Ther. 2003;10:1874–1881. doi: 10.1038/sj.gt.3302087. [DOI] [PubMed] [Google Scholar]
- Vite CH, McGowan JC, Niogi SN, Passini MA, Drobatz KJ, Haskins ME, Wolfe JH. Effective gene therapy for an inherited CNS disease in a large animal model. Ann Neurol. 2005;57:355–364. doi: 10.1002/ana.20392. [DOI] [PubMed] [Google Scholar]
- Wang C, Wang CM, Clark KR, Sferra TJ. Recombinant AAV serotype 1 transduction efficiency and tropism in the murine brain. Gene Ther. 2003;10:1528–1534. doi: 10.1038/sj.gt.3302011. [DOI] [PubMed] [Google Scholar]
- Witter MP, Wouterlood FG, Naber PA, Van Haeften T. Anatomical organization of the parahippocampal-hippocampal network. Ann NY Acad Sci. 2000;911:1–24. doi: 10.1111/j.1749-6632.2000.tb06716.x. [DOI] [PubMed] [Google Scholar]
- Wolfe JH, Sands MS. Murine mucopolysaccharidosis type VII: a model system for somatic gene therapy of the central nervous system. In: Lowenstein PR, Enquist LW, editors. Protocols for gene transfer in neuroscience: towards gene therapy of neurological disorders. London: Wiley; 1996. pp. 263–274. [Google Scholar]
- Wyss JM. An autoradiographic study of the efferent connections of the entorhinal cortex in the rat. J Comp Neurol. 1981;199:495–512. doi: 10.1002/cne.901990405. [DOI] [PubMed] [Google Scholar]