Cell cycle progression in confining microenvironments is regulated by a growth-responsive TRPV4-PI3K/Akt-p27Kip1 signaling axis (original) (raw)
A growth-responsive TRPV4-PI3K-p27 pathway regulates cell cycle progression for cancer cells in confining 3D microenvironments.
Abstract
In tissues, cells reside in confining microenvironments, which may mechanically restrict the ability of a cell to double in size as it prepares to divide. How confinement affects cell cycle progression remains unclear. We show that cells progressed through the cell cycle and proliferated when cultured in hydrogels exhibiting fast stress relaxation but were mostly arrested in the G0/G1 phase of the cell cycle when cultured in hydrogels that exhibit slow stress relaxation. In fast-relaxing gels, activity of stretch-activated channels (SACs), including TRPV4, promotes activation of the phosphatidylinositol 3-kinase (PI3K)/Akt pathway, which in turn drives cytoplasmic localization of the cell cycle inhibitor p27Kip1, thereby allowing S phase entry and proliferation. Cell growth during G1 activated the TRPV4-PI3K/Akt-p27Kip1 signaling axis, but growth is inhibited in the confining slow-relaxing hydrogels. Thus, in confining microenvironments, cells sense when growth is sufficient for division to proceed through a growth-responsive signaling axis mediated by SACs.
INTRODUCTION
In tissues, cells are often spatially confined by the surrounding microenvironment, which includes adjacent cells and extracellular matrix (ECM) (1, 2). Mechanical interactions of cells with their microenvironments play a key role in development, homeostasis, regeneration, and disease, and it is known that the elastic properties of ECM regulate various cell behaviors in both two-dimensional (2D) and 3D cultures (2–5). However, many soft tissues and biological ECMs are viscoelastic and exhibit stress relaxation, with their resistance to a deformation relaxing over time (Table 1) (1, 2, 6–16). The viscoelastic properties of ECM have been found to influence cell behaviors such as spreading and differentiation (1, 16–18). However, it remains unclear how viscoelastic properties of ECM influence cell cycle progression.
Table 1. Stress relaxation properties of selected soft tissues that exhibit substantial stress relaxation.
Tabulated values from stress relaxation tests of selected tissues. _t_i and _t_f, respectively, indicate the time when stress relaxation tests started and ended. _E_i is the initial modulus measured at _t_i in units of kilopascal. The stress relaxation time, τ1/2, indicates the time at which the initial stress relaxes to half its original value. _E_f /_E_i is the normalized relaxation modulus measured at _t_f. Times and stresses reported here are approximate. Note that, in some stress relaxation tests, the stress reached an equilibrium stress by the end of the tests, while in other tests, the test may not have been sufficiently long for the stress to reach an equilibrium value or zero. For testing method, comp. and indent. represent compression and indentation, respectively. (–) indicates not reported.
| Tissue type | Animal | Strain (%) | Initial modulus | Stress relaxation time | Final relaxation modulus | Testing method | References | ||
|---|---|---|---|---|---|---|---|---|---|
| _t_i (s) | _E_i (kPa) | τ1/2 (s) | _t_f (s) | _E_f/_E_i (norm) | |||||
| Bone marrow | Rat | 15 | 0.1 | – | 10 | 500 | 0.25 | Comp. | (1) |
| Breast cancer | Human | 40 | 0.01 | 1.8 | 10 | 3500 | 0.02 | Indent. | fig. S1 |
| Murine | 2–10 | 0 | 2 | 20–50 | 180 | 0.2–0.4 | Indent. | (11) | |
| Adipose | Porcine | 0.1 | 0.01 | 35 | 0.1 | 50 | 0.06 | Shear | (9) |
| Rat | 15 | 0.1 | – | 100 | 1000 | 0.5 | Comp. | (1) | |
| Brain | Bovine | 10.6 | 0.01 | 0.7 | 1 | 500 | 0.21 | Shear | (10) |
| Rat | 10 | 0.01 | 0.55 | 1 | 40 | 0.18 | Shear | (2) | |
| 15 | 0.1 | – | 100 | 1000 | 0.25 | Comp. | (1) | ||
| Liver | Bovine | 0.13 | 0.01 | 5 | 0.1 | 200 | 0.08 | Shear | (14) |
| Rat | 25 | 7 | 0.06–0.08 | 50 | 1000 | 0.25 | Shear | (15) | |
| 15 | 0.1 | – | 100 | 1000 | 0.3 | Comp. | (1) | ||
| Embryonic tissues | Chicken | – | 0 | – | 15–70 | 170 | 0.2–0.5 | Comp. | (12) |
| Muscle | Murine | 10 | 10 | 1.25 | 70 | 600 | 0.3 | Shear | (16) |
| Hematoma | Human | 15 | 0 | – | 200 | 1000 | 0.3 | Comp. | (13) |
| Skin | Rat | – | 1 | – | 650 | 1500 | 0.47 | Tension | (7) |
| Swine | 5–15 | 0 | – | 200–1200 | 1200 | 0.3–0.5 | Tension | (8) | |
| Human | 30 | 0 | – | 1200 | 9000 | 0 | Radial tension | (6) |
Cell division is a mechanical process that is responsive to mechanical cues. It is known that tension in epithelial monolayers promotes cell cycle entry through signaling from cadherins and induces rapid division in cells paused in the G2 phase through the activation of the Piezo1 channel (19–21). Conversely, increased osmotic stress or compressional stress inhibits cell proliferation (22–24). During the early phase of the cell cycle, G1 phase, cells must typically double in size to prepare to divide (25–27). Therefore, confining microenvironments would be expected to mechanically restrict this growth with compressive stresses (28–30). While increased ECM stiffness is known to promote cell cycle progression in 2D culture (31), where cells can grow and divide unrestricted, less is known about how confinement may regulate cell cycle progression in 3D culture. Here, we investigate the impact of confinement on cell cycle progression by studying cells cultured within confining viscoelastic hydrogels with different levels of stress relaxation. Stress relaxation has been previously found to correspond to mechanical confinement, with faster stress relaxation translating to a reduced mechanical confinement (1, 29). We find that hydrogel stress relaxation, or confinement, regulates cell cycle progression through a growth-responsive phosphatidylinositol 3-kinase (PI3K)/Akt-p27Kip1 signaling axis mediated by stretch-activated channels (SACs).
RESULTS
Hydrogel relaxation regulates the growth of tumor spheroids
To elucidate the impact of confinement on cell cycle progression, we first studied cells cultured in 3D in viscoelastic alginate hydrogels. Alginate is an inert polymer derived from algae that exhibits minimum susceptibility to degradation by mammalian enzymes and forms a nanoporous hydrogel when cross-linked with calcium, thereby providing a confining microenvironment to cells (29, 32). The stress relaxation properties of alginate gels were modulated by using different molecular weights (MWs) of the alginate and tuning ionic cross-link density. Decreasing the MW while increasing the density of ionic cross-links lead to faster stress relaxation, because of the reduced network connectivity of the shorter alginate chains, while maintaining the same initial elastic modulus (1). In this study, alginate hydrogels with an initial elastic modulus of ~3 and ~16 kPa and that exhibit slow, medium, and fast stress relaxation in response to a constant strain were used (Fig. 1, A to D, and fig. S1, A to D). Initial elastic moduli and stress relaxation responses were within the range of those measured in soft tissues generally, including those of breast cancer tissues (fig. S1, E and F, and Table 1). As stress relaxation, or simply, relaxation, corresponds to a decrease in resistance to deformation over time, hydrogels with faster relaxation provide a microenvironment where mechanical confinement is lower (29). Faster relaxation corresponds to higher viscosity and greater creep in the alginate hydrogels (fig. S1, G to J) (29). Initially, to remove any ligand-dependent effects, no cell adhesion ligands were coupled to the inert alginate hydrogels and cancer cell lines were encapsulated in the gels, as these cells exhibit anchorage-independent growth and proliferation.
Fig. 1. Faster stress relaxation in alginate hydrogels promotes tumor spheroid growth.
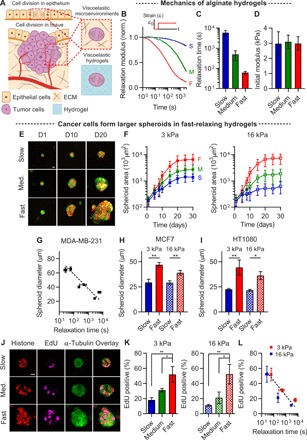
(A) Schematics of cells dividing in physiological tissues such as an epithelial monolayer and a growing tumor. Physiological tissues provide a viscoelastic confining microenvironment to cells. Viscoelastic hydrogels can be used to mimic these microenvironments. (B) Stress relaxation tests on alginate hydrogels exhibiting slow (S), medium (M), and fast (F) relaxation. Relaxation modulus was normalized by the initial modulus in response to 10% compressional strain. (C) Time scale at which the relaxation modulus is relaxed to half of its initial value from stress relaxation tests. (D) The initial modulus of gels in (B). (E) Fluorescence images of spheroids formed by MDA-MB-231 cancer cells in slow-, medium-, and fast-relaxing gels at the indicated days. Here and in all other figures, red and green fluorescence indicate red fluorescent protein (RFP) histones and green fluorescent protein (GFP) microtubules, respectively. (F) The growth curves of MDA-MB-231 spheroids cultured in gels with an initial modulus of 3 or 16 kPa and varying relaxation, over 30 days (n = 10 to 77 spheroids). (G) The diameter of spheroids formed by MDA-MB-231 as a function of relaxation time at 15 days. The diameter of spheroids for (H) MCF7 at 15 days (n = 11 to 47 spheroids) and (I) HT1080 at 14 days (n = 20 to 43 spheroids). (J) Fluorescence images of spheroids formed by MDA-MB-231 for EdU staining at day 10. (K) The fraction of EdU-positive MDA-MB-231 cells cultured in gels with an initial modulus of 3 or 16 kPa and varying relaxation [soft and stiff, n = 3, measured in 16 to 38 cells; one-way analysis of variance (ANOVA) tests; *P < 0.05 and **P < 0.01]. (L) The fraction of EdU-positive MDA-MB-231 cells as a function of relaxation time. Data are shown as means ± SD, except for (G and I), where data are shown as means ± SEM. Scale bars, 10 μm (for all figures).
With this system, we assessed the impact of confinement on cancer cell proliferation. In fast-relaxing hydrogels, MDA-MB-231 cancer cells formed large spheroids in hydrogels with an initial elastic modulus of 3 and 16 kPa, while spheroid growth was much lower in slow-relaxing hydrogels (Fig. 1, E to G, and fig. S2, A to E). The concentration of calcium cross-linker did not determine spheroid diameter (fig. S2, D and E). We found similar results for both MCF7 and HT1080 cells (Fig. 1, H and I, and fig. S2, F and G). The proliferation of MDA-MB-231 cells, indicated by nuclear staining of EdU (5-ethynyl-2′-deoxyuridine), was enhanced in the hydrogels with faster relaxation but was reduced in the gels with slower relaxation (Fig. 1, J to L). By contrast, levels of apoptosis were not affected by changes in stress relaxation (fig. S2, H and I). These findings demonstrate that hydrogel stress relaxation mediates the proliferation of cancer cells.
Next, we used flow cytometry to quantify DNA content and assess the fraction of cells in G0/G1, S, and G2/M phases of the cell cycle as a function of stress relaxation. Most cells were arrested at the G0/G1 phase in hydrogels with slow relaxation and an initial modulus of both 3 and 16 kPa, while a substantially higher fraction of cells was found in S and G2/M phases in hydrogels with fast relaxation (Fig. 2, A to C, and fig. S3, A to C). Measurements of nuclear staining of Ki-67, a marker of cell cycle entry (20), were used to estimate the numbers of cells at the G0 and G1 phases (fig. S3, D to G). This analysis indicates that faster stress relaxation promotes cell cycle progression beyond the G1 phase, although cell cycle entry was also increased by faster relaxation (fig. S3H). Together, these results establish that hydrogel stress relaxation regulates cell cycle progression beyond the G1 phase (Fig. 2D).
Fig. 2. Hydrogel stress relaxation regulates cell cycle progression from the G1 phase to the S phase.
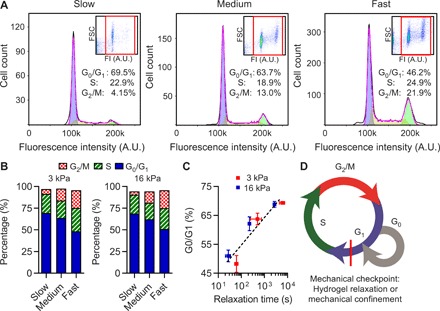
(A) Cell cycle analysis of cells cultured in slow-, medium-, and fast-relaxing gels for 10 days, measured by flow cytometry. Inset, fluorescence intensity (FI) versus forward scatter (FSC). Red rectangle indicates a gate of cell population. A.U., arbitrary units. (B) Population of cells in G0/G1, S, and G2/M phases in soft (3 kPa) and stiff (16 kPa) gels with varying relaxation (n = 2 to 4 per each condition). (C) Population of cells in the G0/G1 phase as a function of relaxation time. (D) A schematic of cell cycle progression including a mechanical checkpoint associated with hydrogel relaxation or mechanical confinement identified by the studies. Data are shown as means ± SD.
SACs, PI3K/Akt, and p27Kip1 localization regulate cell cycle progression
We then investigated the molecular mechanism regulating the S phase entry of cells cultured in the hydrogels. p27Kip1, a cyclin-dependent kinase inhibitor regulating progression from the G1 phase to the S phase, halts progression in response to increased osmotic pressure, making it a logical candidate to study (22). The localization of p27Kip1 in hydrogel-cultured cells was assessed because the cell cycle arrest activity of p27Kip1 depends on its nuclear localization (33–35). p27Kip1 localized to nucleus in slow-relaxing hydrogels, while cells in fast-relaxing hydrogels exhibited cytoplasmic p27Kip1, corresponding to low and high levels of proliferation, respectively (Fig. 3, A to C). Thus, hydrogel stress relaxation regulates p27Kip1 localization, and cytoplasmic p27Kip1 is correlated with the S phase entry of cells in the hydrogels.
Fig. 3. TRPV4 activation regulates PI3K/Akt-p27Kip1 signaling activation and proliferation.
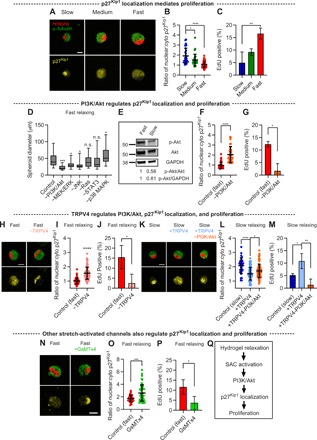
(A) Fluorescence images of p27Kip1 in single cells cultured in slow-, medium-, and fast-relaxing gels. Here and in all other figures, yellow fluorescence indicates p27Kip1, unless otherwise specified. (B) The ratio of the nuclear p27Kip1 to the cytoplasmic p27Kip1 in cells in soft (3 kPa) gels with varying relaxation (n = 30 to 88 cells). (C) The fraction of EdU-positive cells (n = 3, measured in 10 to 48 cells). (D) The diameter of spheroids cultured in soft and fast-relaxing gels for 10 days in the presence of the indicated small-molecule inhibitors (n = 22 to 34 spheroids). n.s., not significant. (E) Immunoblot of Akt and phosphorylated Akt (p-Akt) for cells in soft gels with slow and fast relaxation. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (F) The ratio of the nuclear p27Kip1 to the cytoplasmic p27Kip1 in cells in soft and fast-relaxing gels with and without PI3K/Akt inhibitor (n = 44 to 57 cells). (G) The fraction of EdU-positive cells in soft and fast-relaxing gels with and without PI3K/Akt inhibitor (n = 3, measured in 15 to 28 cells). (H) Fluorescence images of p27Kip1 in cells cultured in fast-relaxing gels with and without TRPV4 antagonist. (I) The ratio of the nuclear p27Kip1 to the cytoplasmic p27Kip1 in cells treated with TRPV4 antagonist (n = 51 to 123 cells). (J) The fraction of EdU-positive cells in fast-relaxing gels with and without TRPV4 antagonist (n = 3, measured in 12 to 30 cells). (K) Fluorescence images of p27Kip1 in cells cultured in slow-relaxing gels with and without TRPV4 agonist and PI3K/Akt inhibitor. (L) The ratio of the nuclear p27Kip1 to the cytoplasmic p27Kip1 in cells in the indicated conditions (n = 34 to 92 cells). (M) The fraction of EdU-positive cells (n = 3, measured in 11 to 36 cells). (N) Fluorescence images of p27Kip1 in cells cultured in fast-relaxing gels with and without GsMTx4. (O) The ratio of the nuclear p27Kip1 to the cytoplasmic p27Kip1 in cells treated with GsMTx4 (n = 35 to 51 cells). (P) The fraction of EdU-positive cells in fast-relaxing gels with GsMTx4 (n = 3, measured in 10 to 19 cells). (Q) A proposed mechanism underlying cell cycle progression in confining hydrogels. In (B) to (D), (L), and (M), one-way ANOVA tests were used, and in (F), (G), (I), (J), (O), and (P), Student’s t tests were used; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. The box plots show 25th to 75th percentiles, and whiskers show minimum and maximum. Data are shown as means ± SD. Scale bars, 10 μm [for (A), (H), (K), and (N)].
Next, we examined the molecular pathways regulating p27Kip1 localization and cell cycle progression. A screening of inhibitors of molecules or pathways known to regulate proliferation was conducted for cells in hydrogels with fast relaxation. Pathways or molecules inhibited included mitogen-activated protein kinase (MAPK) kinase (MEK)/extracellular signal–regulated kinase (ERK), p38 MAPK, PI3K/Akt, c-Jun N-terminal kinase (JNK), signal transducer and activator of transcription 3 (STAT3), and Ras (36, 37). After 10 days of culture, cells treated with PI3K/Akt, MEK/ERK, and JNK pathway inhibitors formed significantly smaller spheroids than did the untreated cells, with the inhibition of PI3K/Akt pathway having the strongest effect (Fig. 3D and fig. S4). Immunoblot analysis for the phosphorylation of Akt confirmed that the PI3K/Akt pathway exhibited higher levels of activation in fast-relaxing hydrogels relative to slow-relaxing hydrogels (Fig. 3E). The inhibition of the PI3K/Akt pathway in cells in fast-relaxing hydrogels led to higher levels of nuclear p27Kip1 and lower levels of proliferation than untreated cells (Fig. 3, F and G). These results point toward the role of PI3K/Akt activation in mediating p27Kip1 localization and thereby coordinating cell cycle progression.
We then explored the molecular mechanism by which hydrogel relaxation regulates the activation of the PI3K/Akt pathway. While the hydrogels did not themselves present cell adhesion ligands to the cells, cells could potentially secrete and deposit ECM molecules to bind to. However, immunohistochemical staining revealed that cells did not secrete type 1 collagen or laminin-5 during single-cell experiments (fig. S5, A to D). In addition, the inhibition of actomyosin-based contractility with blebbistatin did not decrease the S phase entry (fig. S5E). Therefore, cells may be using an adhesion-independent mechanism to sense hydrogel stress relaxation. SACs are activated by membrane stretching and function independently of adhesions, suggesting them as a logical class of molecules to investigate. Among SACs, a previous study linked the activation of the PI3K/Akt pathway to the opening of TRPV4 ion channels, providing one possible target (38). Conveniently, both agonist and antagonist specific to TRPV4 are commercially available. Therefore, we studied the role of TRPV4 in mediating cell cycle progression in our system. The inhibition of TRPV4 with a TRPV4 antagonist (GSK205) or RNA silencing led to a reduction in phosphorylated Akt in single cells in hydrogels with fast relaxation (fig. S6, A and B). Further, TRPV4 inhibition induced the nuclear localization of p27Kip1 and hindered the S phase entry (Fig. 3, H to J, and fig. S6C). Conversely, the activation of TRPV4 in slow-relaxing hydrogels, using TRPV4 agonists GSK1016790A or 4α-Phorbol 12,13-didecanoate (4α-PDD), increased phosphorylated Akt, led to cytoplasmic p27Kip1, and raised the fraction of cells entering the S phase (Fig. 3, K to M, and fig. S6, A, D, and E). The inhibition of the PI3K/Akt pathway in cells with activated TRPV4 resulted in the nuclear localization of p27Kip1 and reduced the S phase entry, placing PI3K/Akt directly downstream of TRPV4 (Fig. 3, K to M, and fig. S6A). As with TRPV4 inhibition of MDA-MB-231 cells, the inhibition of TRPV4 in MCF7 and HT1080 cells in fast-relaxing hydrogels resulted in increased nuclear levels of p27Kip1 and reduced the S phase entry (fig. S6, F to I). The inhibition of TRPV4 inhibited the S phase entry of cells in multicellular spheroids in fast-relaxing hydrogels, while the activation of TRPV4 promoted cell cycle progression in multicellular spheroids in slow-relaxing hydrogels (fig. S6, J to P). Together, these results demonstrate that TRPV4 activity regulates the PI3K/Akt pathway in response to hydrogel relaxation, which, in turn, controls p27Kip1 localization.
We next investigated whether other SACs could be involved in regulating cell cycle progression. Cells were encapsulated in fast-relaxing gels and exposed to GsMTx4, a nonspecific inhibitor of SACs including Piezo1, TRPC1, and TRPC6 (39). Treatment with GsMTx4 reduced the level of phosphorylated Akt, induced nuclear p27Kip1, and reduced the S phase entry (Fig. 3, N to P). These results indicate that hydrogel relaxation regulates cell cycle progression through the activation of SACs and the PI3K/Akt-p27Kip1 signaling axis (Fig. 3Q).
Hydrogel relaxation regulates the growth of single cells
After identifying that the PI3K/Akt-p27Kip1 signaling axis mediated by SACs promoted cell cycle progression in fast-relaxing hydrogels, we sought to identify the biophysical link between hydrogel relaxation and activation of this signaling axis. It is known that SACs are activated by physical stimuli such as membrane stretching generated by cell swelling (40). During the G1 phase of the cell cycle, cells typically grow to roughly double their size in preparation for cell division (25, 26). We conducted finite element modeling to predict the impact of confinement on cell growth, with the simple assumption that cells exert a constant outward mechanical stress to drive growth. The simulations indicated that cell growth should be regulated by relaxation of the surrounding microenvironment, with faster relaxation allowing cells to grow and slower relaxation blocking growth (Fig. 4, A to C, and fig. S7). To experimentally test the prediction, we encapsulated single cells in gels of varying relaxation and cultured them in the presence of mimosine, a drug blocking cell cycle at late G1 phase, to isolate cell growth during the G1 phase. Consistent with the result from the computational simulations, the diameter of single cells in the hydrogels increased as the hydrogel relaxation was enhanced (Fig. 4, D to F, and fig. S8, A to D). In fast-relaxing hydrogels, cells approximately doubled their size during the G1 phase. Live-cell imaging revealed that cells expand their diameter at a rate of ~0.025 μm/min in fast-relaxing gels and at a rate of ~0.007 μm/min in slow-relaxing gels (fig. S8, E to G). Because the hydrogels are nanoporous, cell growth must be accommodated by hydrogel deformation. In the gels with fast relaxation, we observed hydrogel densification around cells because of deformation resulting from cell growth (fig. S8, H to J). Together, these results support the hypothesis that the fast relaxation of hydrogels allows single-cell growth during the G1 phase and implicate the role of cell growth in activating the signaling axis mediated by SACs to drive proliferation.
Fig. 4. Single-cell growth, regulated by hydrogel stress relaxation or osmotic pressure, is mediated by the Na+/H+ exchanger and controls p27Kip1 localization.
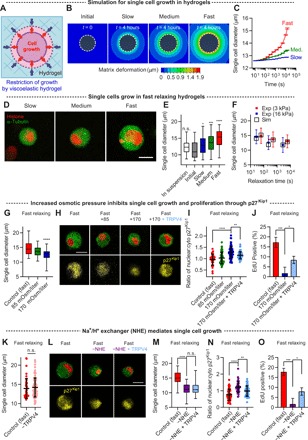
(A) A schematic of a cell growing in a hydrogel. Cells must exert mechanical stress to allow cell growth in the confining hydrogels. (B) 2D planar view of 3D computational simulations of hydrogel deformation due to cells exerting a constant outward stress for a time scale of 4 hours. Note that the viscoelastic parameters used for the simulations were derived from creep tests, which more closely approximate the case of a cell applying a constant stress to the hydrogel. White dashed lines indicated cell boundaries. (C) The diameter of the simulation models in response to a constant stress for a time scale of 4 hours. (D) Fluorescence images of single cells growing during the G1 phase in soft gels with varying relaxation for 2 days. (E) Quantification of the diameter of single cells. The diameter of single cells before encapsulation is indicated as “in suspension” and right after encapsulation is denoted as “initial” (n = 43 to 65 cells, one-way ANOVA tests and Tukey’s comparisons with respect to the initial condition). (F) Comparison of the experimentally and computationally obtained diameter of single cells as a function of relaxation time. The time scale for the simulations was 2 days for all cases, except for the case of soft gels with fast relaxation, where the time scale was 4 hours. (G) The diameter of single cells cultured in soft and fast-relaxing hydrogels with varying osmotic pressure (n = 31 to 52 cells). (H) Fluorescence images of p27Kip1 in cells under the different osmotic conditions. TRPV4 agonist was additionally added to cells under an osmotic pressure of 170 mOsm/liter. (I) The ratio of the nuclear p27Kip1 to the cytoplasmic p27Kip1 in cells under conditions corresponding to (H) (n = 31 to 58 cells). (J) The fraction of EdU-positive cells with and without an osmotic pressure of 170 mOsm/liter and TRPV4 agonist (n = 3, measured in 12 to 28 cells). (K) The diameter of single cells in fast-relaxing gels with and without TRPV4 antagonist (n = 46 to 97 cells). (L) Fluorescence images of p27Kip1 in cells in soft and fast-relaxing gels with an NHE inhibitor. TRPV4 agonist was additionally added to cells with an NHE inhibitor. (M) The diameter of single cells (n = 44 to 50 cells). (N) The ratio of the nuclear p27Kip1 to the cytoplasmic p27Kip1 in cells under the conditions corresponding to (L) (n = 44 to 50 cells). (O) The fraction of EdU-positive cells (n = 3, measured in 13 to 33 cells). In (E), (G), (I), (J), and (M) to (O), one-way ANOVA tests were used, and in (K), Student’s t tests were used; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Data are shown as means ± SD. The box plots show 25th to 75th percentiles, and whiskers show minimum and maximum. Scale bars, 10 μm [for (D), (H), and (L)].
Thus, we tested whether cell growth controlled the activation of the SAC-mediated signaling axis to regulate proliferation. Cell growth in fast-relaxing hydrogels was suppressed by increasing osmotic pressure with the addition of polyethylene glycol to the cell culture medium (29, 41). Increased osmotic pressure decreased cell size (Fig. 4G). Decreased cell size reduced the level of phosphorylated Akt, led to nuclear localization of p27Kip1, and diminished the S phase entry (Fig. 4, H to J, and fig. S9, A and B). TRPV4 activation by its agonist in cells under high osmotic pressure led to a recovery in phosphorylated Akt, the cytoplasmic localization of p27Kip1, and progression through the S phase (Fig. 4, H to J, and fig. S9, A and B). The inhibition of TRPV4 in cells in fast-relaxing hydrogels did not affect cell growth, indicating that TRPV4 activation is downstream of cell growth (Fig. 4K). A previous study found the sodium-hydrogen ion exchanger (NHE) to be involved in cell volume increase (25). The inhibition of NHE with 5-(_N_-ethyl-_N_-isopropyl)amiloride (EIPA) decreased the diameter of single cells, reduced the phosphorylation of Akt, induced the nuclear transport of p27Kip1, and impeded the S phase entry (Fig. 4, L to O, and fig. S9, C and D). The activation of TRPV4 with the agonist in the NHE-inhibited cells led to a recovery of phosphorylated Akt, the retention of p27Kip1 in the cytoplasm, and enhanced progression through the S phase (Fig. 4, L to O, and fig. S9, C and D). This further confirmed SAC activity to be downstream of cell growth. Whether modulated by the inhibition of NHE, osmotic pressure, or stress relaxation, cell size was strongly correlated with the S phase progression (fig. S9, E to G). Together, these results establish that cell growth, involving NHE, regulates the activation of the SAC-mediated PI3K/Akt pathway to control p27Kip1 localization and progression to the S phase.
Relevance to other 3D microenvironments
Last, we investigated whether these findings were relevant to 3D culture models of ligand-rich microenvironments and to human breast cancer. TRPV4 was first inhibited in MCF10A mammary epithelial cells cultured in viscoelastic interpenetrating networks of alginate and reconstituted basement matrix (rBM), in which they normally form organotypic acini (5). In addition, TRPV4 inhibition was also applied to 3T3 mouse fibroblasts cultured in gels of type 1 collagen, which models the microenvironment of collagen-rich stromal tissue that fibroblasts reside in. The collagen gels exhibit strain-enhanced stress relaxation (42). Notably, the inhibition of TRPV4 led to the nuclear localization of p27Kip1 and reduced the S phase progression (Fig. 5, A to D). In tissue from patients with breast cancer, the increased average cell size in tumors exhibited a significant correlation with the decreased average ratio of nuclear p27Kip1 to cytoplasmic p27Kip1, and cells with cytoplasmic and nuclear p27Kip1 were significantly larger on average than cells with nuclear p27Kip1 (fig. S10). Both of these findings are consistent with the expectation from our results, although the image analysis of slices of tumors from patient tissue cannot be directly compared to our 3D in vitro image analysis of single cells. Together, these findings indicate that the SAC-PI3K/Akt-p27Kip1 signaling axis represents a broadly relevant mechanism connecting cell growth to the S phase progression in 3D culture.
Fig. 5. A growth-responsive TRPV4-PI3K/Akt-p27Kip1 signaling axis controls the S phase progression in confining 3D microenvironments.
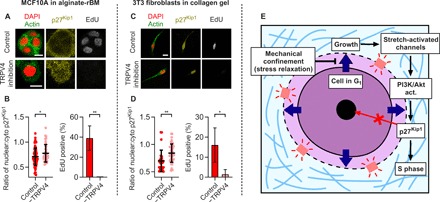
(A) Florescence images of p27Kip1 and EdU staining of MCF10A cells cultured in alginate-rBM gels with and without TRPV4 antagonist. (B) The ratio of nuclear p27Kip1 to cytoplasmic p27Kip1 (n = 40 to 97 cells) and quantification of EdU-positive cells for MCF10A cells (n = 3, measured in 15 to 36 cells). (C) Florescence images of p27Kip1 and EdU staining of 3T3 fibroblasts cultured in collagen gels with and without TRPV4 antagonist. (D) The ratio of nuclear p27Kip1 to cytoplasmic p27Kip1 (n = 26 to 44 cells) and quantification of EdU-positive cells for 3T3 fibroblasts (n = 3, measured in 21 to 48 cells). In (B) and (D), Student’s t tests were used; *P < 0.05 and **P < 0.01. (E) When mechanical confinement is low, for example, in hydrogels with fast stress relaxation, cells grow through NHE during the G1 phase. Cell growth promotes the activity of SACs, which activate the PI3K/Akt pathway, which, in turn, drives cytoplasmic localization of p27Kip1, thereby promoting the S phase entry and proliferation. In microenvironments with higher confinement, such as hydrogels with slow stress relaxation, cell growth is inhibited so that SACs are not activated, and therefore, nuclear localization of p27Kip1 blocks the S phase entry and proliferation. Data are shown as means ± SD. Scale bars, 10 μm [for (A) and (C)].
DISCUSSION
Together, our findings reveal how confinement regulates cell cycle progression and mechanistically links cell growth to the promotion of cell cycle progression through the activation of a SAC-PI3K/Akt-p27Kip1 signaling axis (Fig. 5E). This signaling axis provides a long-sought mechanism that connects cell size and cell growth to cell cycle progression (43–46). Doubling in size is required for cells to maintain a stable size through successive generations of cell division. Therefore, this mechanism allows cells to sense when growth is sufficient for mitosis to progress normally in confining microenvironments.
Our findings identify stress relaxation as a key mechanical parameter controlling the proliferation of cancer cells in confining microenvironments. Many soft tissues and cells are viscoelastic and exhibit stress relaxation, indicating the physiological relevance of this investigation (Table 1). Stress relaxation can arise from unbinding of weak bonds that link ECM proteins together (42). Cancer cells proliferated in hydrogels with stress relaxation times on the order of 60 s, while proliferation was diminished in hydrogels with stress relaxation times on the order of 5000 s. While both time scales are far below the time scale of the full cell cycle, growth during the G1 phase of the cell cycle occurs continuously so that the different time scales of stress relaxation are expected to lead to the differential accumulation of stress. While cells did not secrete type 1 collagen or laminin during single-cell experiments, they could deposit significant ECM as they form spheroids over a longer culture period. Thus, the growth of single cells in spheroids would be expected to be affected by not only hydrogel relaxation but also the viscoelastic properties of the surrounding cells, cell-cell signaling, and matrix deposited within the spheroid. Therefore, the mechanisms underlying cell cycle progression in tumor spheroids are likely to be more complex. Nonetheless, our findings that TRPV4 inhibition reduced proliferation in multicellular spheroids in fast-relaxing gels while TRPV4 activation promoted proliferation in spheroids in gels with slow relaxation indicate that the SAC-PI3K/Akt-p27Kip1 signaling axis found still plays a key role in regulating proliferation in spheroids.
Mechanistically, our results indicate that cell growth, involving NHE, regulates the activation of the SAC-PI3K/Akt pathway to control p27Kip1 localization and progression to the S phase (Fig. 5E). There is evidence that the pathway is relevant to tumors. Previous studies investigating the localization of p27Kip1 on breast cancer tissues revealed that p27Kip1 localization was mediated by PI3K/Akt and that increased cytoplasmic localization led to the decrease in survival rate of patients, both consistent with our findings (33–35). However, none of these studies examined cell size. Here, we find that larger cell sizes in human breast cancer tissue are associated with the increased cytoplasmic localization of p27Kip1 (fig. S10). However, we note that, in tumors, cells are confined by neighboring cells and the surrounding adhesion ligand–rich ECM. Such a microenvironment is distinct from that of our 3D culture model where cells are encapsulated as single cells in hydrogels that do not contain any adhesion ligands. Thus, our model system does not capture the impact of cell-cell signaling or signaling from cell-matrix adhesions, which could affect the mechanisms occurring in vivo. In addition, a recent study showing that the high expression of TRPV4 is found to be associated with the reduced survival in patients with human breast cancer suggests the pathological relevance of TRPV4 to human cancer (47). While our study focused on TRPV4, we also demonstrate that other SACs contribute. This suggests that other SACs provide redundancy and could potentially be substituted for TRPV4 in regulating the growth-responsive pathway.
Broadly, these findings reveal confinement, or stress relaxation, as a key parameter that regulates cell growth and cell cycle progression, which could be relevant to various physiological contexts. In growing tumors, cancer cells are surrounded by other cancer cells and the surrounding tissue, which would be expected to resist tumor growth. A recent study found that tumors in the breast, pancreas, and brain are under mechanical compression by the surrounding tissues (48), indicating the mechanical restriction of growth by individual cancer cells within the tumor. In addition, dense collagen-rich stromal tissue has been found to fully confine individual cells (49). Alternatively, in epithelial monolayers, cells must divide in a monolayer crowded by adjacent cells, a process that occurs during development, in which monolayers undergo expansion (50), and homeostasis, where cell division is required to maintain cell numbers as cells are extruded and undergo apoptosis (51). While confinement in monolayers is not fully 3D, cells do divide within the plane of the monolayer and increased cell crowding inhibits proliferation (19, 27). However, in other microenvironments where there is asymmetric confinement, cells can grow and divide along the path of least confinement (52–54). During development, confinement may also be relevant to mesenchymal condensations (50). Thus, while the focus of this study has been on elucidating the role of confinement on cell cycle progression in cancer, growth-responsive cell cycle progression could be a universally relevant mechanism, relevant to these other various physiological contexts.
MATERIALS AND METHODS
Study design
The aim of this study was to investigate the impact of confinement on cell cycle progression by studying cells cultured within confining viscoelastic hydrogels with different levels of stress relaxation in 3D matrices. The number of samples was determined on the basis of experimental approach, availability, and feasibility required to obtain definitive results. All samples were allocated randomly into experimental groups. No data were excluded from the analyses. The numbers of replicates are specified in the figure legends. The researchers were not blinded during data collection or analysis.
Cell culture
MDA-MB-231 human cells transfected with green fluorescent protein (GFP)–labeled α-tubulin and red fluorescent protein (RFP)–labeled histone were gifts from B. Weaver (University of Wisconsin–Madison). MDA-MB-231, MCF7, and HT1080 were cultured in standard Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) containing 10% fetal bovine serum (HyClone) and 1% penicillin/streptomycin (Gibco/Thermo Fisher Scientific, Waltham, MA). MCF10A cells (American Type Culture Collection) were cultured in DMEM/F12 50/50 medium (Thermo Fisher Scientific) containing 5% horse serum (Thermo Fisher Scientific), epidermal growth factor (20 ng/ml; Peprotech Inc.), hydrocortisone (0.5 μg/ml; Sigma-Aldrich), cholera toxin (100 ng/ml; Sigma-Aldrich), insulin (10 μg/ml; Sigma-Aldrich), and penicillin/streptomycin (100 U/ml; Thermo Fisher Scientific).
Encapsulation of cells in hydrogels and 3D cell culture
For cell encapsulation in alginate hydrogels, cells were first trypsinized, washed in serum-free DMEM, and resuspended as a single-cell suspension in serum-free DMEM at 10× the final concentration of 3 to 4 million/ml measured using a Vi-CELL (Beckman Coulter). Cells were then mixed with alginates reconstituted in serum-free DMEM in one Luer lock syringe (Cole-Parmer). The cell alginate solution in the syringe was then homogeneously mixed with DMEM containing various concentrations of calcium sulfate in another syringe, using a female-female Luer lock coupler (Value Plastics). The final concentrations of calcium sulfate and corresponding initial elastic moduli are listed in table S1. The mixture of the cell alginate–calcium sulfate solution was deposited on a hydrophobic surface of a glass plate and then covered with another glass plate with 1 to 2 mm spacing between the plates. The cell alginate mixture was fully gelled for 35 to 45 min. Hydrogels were punched out using a biopsy punch (a diameter of 6 mm) and immersed in the cell culture medium supplemented with 1 mM calcium chloride (Sigma-Aldrich) to the growth medium to help maintain the initial elastic modulus of the hydrogels during the long culture period, as was done in a recent study (29). The medium was changed every 2 to 3 days during cell culture. For single-cell experiments, cells were cultured in serum-free DMEM 1 day before hydrogel encapsulation to synchronize cell cycle at the G0/G1 phase. For matrix densification experiments, alginate coupled with fluorescein and wild-type MDA-MB-231 cells were used.
Matrigel was purchased from Corning (catalog no. 354230) and used as the rBM matrix at a final concentration of 4.4 mg/ml. To encapsulate MCF10A cells in rBM-rich matrix, rBM-alginate hydrogels were formed, as described previously (5). Briefly, high-MW alginate was mixed with rBM, cells, and DMEM/F12 and loaded into one Luer lock syringe on ice. The cell alginate–rBM solution was mixed with the other syringe containing CaSO4. The mixture was then rapidly deposited into wells precoated with 50 μl of gelled rBM.
Alginate preparation
Sodium alginate with an MW of 280 kDa (high-MW; LF20/40) was purchased from FMC BioPolymer (Philadelphia, PA) and was prepared as has been described previously (1). Mid- and low-MW alginates were produced by irradiating high-MW alginate with 3 or 8 Mrad from a cobalt source. The average MW of mid- and low-MW alginates was 70 and 35 kDa, respectively. The alginates were dialyzed against deionized water for 2 to 3 days (MW cutoff of 3.5 kDa), filtered with activated charcoal, and then reconstituted at 3 weight % in serum-free DMEM (Life Technologies). High-, mid-, and low-MW alginates were used to form slow-, medium-, and fast-relaxing hydrogels. The initial modulus of hydrogels was modulated by adjusting the concentration of calcium sulfate, as has been described previously (29) and is listed in table S1.
Mechanical characterization of alginate hydrogels
The initial elastic modulus of hydrogels was characterized with unconfined compression tests using a mechanical tester (5848 MicroTester, Instron). Stress relaxation tests were performed using both unconfined and confined compression tests. Alginate gel disks (a diameter of 6 mm, a thickness of 2 mm) were formed and equilibrated in DMEM for 24 hours. The alginate gel disks were placed on the machine and compressed to a strain of 10% at a deformation rate of 1 mm mm−1. The slope of the stress-strain curve up to a strain of 5 to 10% was measured as the initial elastic modulus (obtained on the time scales of ~0.2 s). Subsequently, stress relaxation tests were conducted. A 10% strain was held constant while measuring stress over time. The stress relaxation time was quantified as the time at which the initial modulus of the gels was relaxed to half of its initial value. To remove the noise and select an appropriate value for the initial and half moduli, a Savitzky-Golay filter was used with a polynomial order of 20 and a window length of 421 points. We note that the values of relaxation time from unconfined and confined compression tests were similar. While the time scales from unconfined compression tests were used to describe the hydrogels to be consistent with previous literatures, confined compression tests may be more relevant to the physiological context of cells expanding within a hydrogel. Creep tests of alginate gels in shear were conducted using an AR-G2 stress-controlled rheometer (TA Instruments) equipped with the 25-mm-diameter top and bottom plates (42, 55). Alginate solution was directly deposited on the bottom plate of the rheometer immediately after mixing with calcium sulfate, and the top plate was lowered into contact with the solution immediately before gelation. Mineral oil (Sigma-Aldrich) was deposited around the exposed gel surface between the rheometer plates to avoid dehydration of the alginate. The storage modulus was monitored at a strain of 0.01 and a frequency of 1 rad/s during the gelation of alginate. Creep tests were conducted once the storage modulus of alginates reached an equilibrium value, typically around 45 to 60 min. In creep tests, a constant shear stress of 100 Pa was applied for 2 hours, while strain in response to the stress was recorded over time.
Experiments with pharmacological and small-molecule inhibitors
For the inhibition experiments, the following inhibitors were added to the medium at the indicated concentrations, based on concentrations used in similar studies: GsMTx4 (10 μM), TRPV4 agonists GSK1016790A (G101; 100 nM) and 4α-PDD (20 μM) and TRPV4 antagonist GSK205 (G205, 10 to 30 μM), NHE inhibitor EIPA (50 μM), MEK/ERK inhibitor PD98059 (10 μM), JNK inhibitor SP600125 (10 μM), p38 MAPK inhibitor SB203580 (10 μM), PI3K/Akt inhibitor LY294002 (10 μM), Stat3 inhibitor 5,15-diphenylporphyrin (10 μM), Ras inhibitor Salirasib (10 μM), myosin inhibitor blebbistatin (10 μM), and cell cycle inhibitor at late G1 phase mimosine (400 μM).
RNA interference experiments
To knock down TRPV4, cells were transiently transfected with human TRPV4-targeting small interfering RNA (siRNA; Dharmacon) or nontargeting control siRNA at a final concentration of 100 to 200 nM using the transfection reagent, according to the manufacturer’s instructions 48 to 72 hours before encapsulation.
Proliferation assay (EdU)
For the single-cell EdU assay, cells were first encapsulated in hydrogels, and EdU (Thermo Fisher Scientific) was added to the culture medium at a final concentration of 10 μM. After 2 to 3 days of culture, cells were fixed and EdU was stained according to the manufacturer’s instructions. For the tumor spheroid proliferation assay, cells were encapsulated in hydrogels and cultured for 10 days and EdU was added to the culture medium for 24 hours before fixation.
Apoptosis assay (TUNEL)
For the apoptosis assay, cells were encapsulated in hydrogels and fixed after 1 week. The TUNEL (terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling) Assay Kit (Thermo Fisher Scientific) was used according to the manufacturer’s instructions.
Osmotic pressure studies
Osmotic pressure was modulated by adding 400-Da polyethylene glycol (PEG 400, TCI America) in the culture medium (29). Briefly, cells were encapsulated in soft (3 kPa) and fast-relaxing hydrogels, and sterilized PEG 400 was added to the medium at the concentration of 0% (wt/vol; control), 3%, and 6%, respectively. Osmotic pressure was reported as osmolarity.
Immunohistochemistry
Gels containing cells were first fixed with 4% paraformaldehyde in serum-free DMEM at 37°C for 30 to 45 min. The gels were then washed in phosphate-buffered saline (PBS) containing calcium (cPBS; GE Healthcare), incubated in 30% sucrose (Thermo Fisher Scientific) at 4°C overnight, and placed in a mixture of 50% of a 30% sucrose in cPBS solution and 50% O.C.T. (Tissue-Tek) for 4 hours. The mix solution was removed, and the gels were then embedded in O.C.T., frozen, and sectioned with a thickness of 30 to 80 μm using a cryostat (CM1950, Leica). The sectioned samples were stained using standard immunohistochemistry protocols. The samples were permeabilized with Dulbecco’s PBS (DPBS) containing 0.5% Triton X-100 (Sigma-Aldrich), blocked with DPBS containing 1% bovine serum albumin (Sigma-Aldrich), 10% goat serum (Invitrogen), 0.3 M glycine (Thermo Fisher Scientific), and 0.1% Triton X-100. The following antibodies and reagents were used for immunohistochemistry: p27Kip1 antibody (catalog no. ab32034, Abcam), Ki-67 antibody (catalog no. RM-9106S, Thermo Fisher Scientific), collagen antibody (catalog no. ab34710, Abcam), fibronectin (catalog no. ab2413, Abcam), laminin-5 (catalog no. ab78286, Abcam), α-tubulin (catalog no. 3873, Cell Signaling Technology), and Prolong Gold antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen) and Alexa Fluor 488 phalloidin to stain actin (Invitrogen). The Click-IT EdU cell proliferation assay (Invitrogen) and TUNEL assay (Thermo Fisher Scientific) were used to assess the proliferation and apoptosis of cells, respectively.
Immunoblot
To harvest cells in hydrogels, gels containing cells were incubated in cold PBS containing 50 mM EDTA (Sigma-Aldrich) to chelate calcium for 5 min while pipetting to break up the gels. Cells were centrifuged at 500_g_ for 10 min, and the supernatant was removed. For SDS–polyacrylamide gel electrophoresis of whole-cell lysates, cells were lysed in Pierce radioimmunoprecipitation assay buffer (catalog no. 89900, Thermo Fisher Scientific) supplemented with protease inhibitor cocktail tablets (catalog no. 11836170001, Roche) and PhosSTOP phosphatase inhibitor cocktail tablets (catalog no. 04906845001, Roche) following the manufacturer’s instructions. The concentration of harvested protein from the cell lysates was quantified using the Pierce Bicinchoninic Acid Protein Assay Kit (catalog no. 23227, Thermo Fisher Scientific). Laemmli sample buffer (catalog no. 1610747, Bio-Rad) was added to lysates, and samples were boiled for 10 min before loading 25 μg of protein in each lane of a polyacrylamide gradient gel (catalog no. 4561086, Bio-Rad). Proteins in the polyacrylamide gels were transferred to nitrocellulose at 100 V for 1 hour, blocked with 5% milk in TBS-T [137 mM NaCl, 2.7 mM KCl, 19 mM tris base, 0.1% Tween (pH 7.4)], incubated in primary antibodies overnight, and then incubated in IRDye 680– or IRDye 800–conjugated secondary antibodies for 1 hour. Blots were visualized using a Li-COR Odyssey imaging system (Li-COR Biotechnology). The following antibodies were used for immunoblotting: AkT antibody (catalog no. 2920S, Cell Signaling Technology), phosphorylated Akt antibody (catalog no. 4060S, Cell Signaling Technology), TRPV4 (catalog no. ab191580, Abcam), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (catalog no. ab181602, Abcam).
Gene expression analysis
To harvest RNA in cells encapsulated in hydrogels, gels containing cells were frozen in liquid nitrogen, ground, and treated with ice-cold PBS containing 50 mM EDTA to break up the hydrogels. RNA was extracted using a commercially available RNA extraction kit (Epoch) with TRIzol (Invitrogen) following the manufacturer’s instructions. The RNA harvested from the hydrogels was then transcribed to complementary DNA (cDNA) using a polymerase chain reaction (PCR) with the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Gene expression was measured using real-time PCR with a Fast SYBR green master mix (Applied Biosystems). The relative gene expression was calculated using the 2−ΔΔ_Ct_ method, normalized to β-actin and compared to cells cultured in soft (3 kPa), fast-relaxing hydrogels. The following primers were used: CCNE1 (forward, 5′-ACAGATTGCAGAGCTGTTGG-3′; reverse, 5′-AAATGATACAAGGCCGAAGC-3′), CCNA2 (forward, 5′-ACAAAGCTGGCCTGAATCAT-3′; reverse, 5′-GTGGTGCTTTGAGGTAGGTCT-3′), and β-actin (forward, 5′-GCTCGTCGTCGACAACGGCTC-3′; reverse, 5′-CAAACATCATCTGGGTCATCTTCTC-3′).
Flow cytometry
Cells were harvested from alginate hydrogels by dissolving the gels using 50 mM EDTA. Cell cycle progression was then analyzed using flow cytometry (Stanford Shared FACS Facility, Scanford). Briefly, all cells harvested from the alginate gels were washed with DPBS and then fixed with cold 70% (v/v) ethanol overnight at 4°C. Cells were then resuspended in DPBS containing RNaseA (a concentration of 20 μg/ml) and propidium iodide (a concentration of 20 μg/ml) to stain DNA for 20 min. The histogram of DNA content was obtained by gating cell population on a fluorescence intensity–versus–forward scatter dot plot. Once viable cells were gated, the fraction of cells at each cell cycle was quantified by fitting the experimentally obtained histogram with the Watson model in FlowJo version 10.2 software.
Live-cell imaging
Live-cell imaging of cells was conducted using bright-field microscopy (SP8, Leica) under standard cell culture conditions (37°C, 5% CO2). Images were taken using 20×/0.75–numerical aperture (NA) objective.
Mechanical testing of human tumors
Human breast carcinoma specimens were obtained from the Stanford Tissue Bank. An International Review Board waiver was obtained for these studies, as the specimens were excess tissue not collected for the current research and were de-identified before use. Tissue samples were cut into 6-mm-diameter plugs and stored in serum-free RPMI 1640 at 4°C before testing. For mechanical testing, stress relaxation tests were performed using an Instron 5848 material testing system with a 4-mm indenter. The indenter was brought down to ~40% of strain, with a ramp of 0.5 mm mm−1 min−1 and held for 1 hour. The measurements were conducted with fully hydrated samples.
Analysis of growth rate using the Gompertz model
To estimate the growth rate of spheroids, we used the Gompertz model, an empirical relationship for tumor growth lnV/V0=lnVmaxV0[1−exp(−αt)], where V is a measure of spheroid size, _V_0 is the initial size, and _V_max is the final size. The growth rate is interpreted as parameter α. Spheroid growth curves were replotted as f(V)=ln[1−ln(VV0)/ln(VmaxV0)] versus time, and the single linear fit of the curves was used to calculate the values of parameter α, the growth rate. The growth rate was then reported as the absolute value of α.
Analysis for p27Kip1 localization
To quantify p27Kip1 localization between the nucleus and cytoplasm, cells in hydrogels were fixed and stained for p27Kip1. For MDA-MB-231 human cells transfected with GFP-labeled α-tubulin and RFP-labeled histone, the nucleus and cytoplasm were not further stained. For the other cell lines, DAPI and phalloidin were used to stain the nucleus and cytoplasm. The ratio of nuclear p27Kip1 to cytoplasmic p27Kip1 was quantified using the Cell Profiler (Broad Institute) software. Briefly, images were thresholded on each color channel to determine the nuclear and cytoplasmic areas outside of the nucleus. The nuclear p27Kip1 was then determined as the summed intensity of the p27Kip1 signal within the nucleus normalized by the nuclear area. The cytoplasmic p27Kip1 was measured as the summed intensity of the p27Kip1 signal outside of the nucleus normalized by the non-nuclear cytoplasmic area. Last, the ratio of nuclear p27Kip1 to cytoplasmic p27Kip1 was calculated as the averaged intensity of nuclear p27Kip1 divided by the averaged intensity of cytoplasmic p27Kip1.
Tissue staining and analysis
Immunofluorescence (IF) staining was performed on paraffin-embedded tissue microarray (TMA) sections (Stanford TA390). Tissues were from patients with ductal carcinoma in situ. An International Review Board waiver was obtained for these studies, as the specimens were excess tissue not collected for the current research and were de-identified before use. The TMAs were cut into 4-μm-thick sections. Four-micrometer TMA sections were deparaffinized in three changes of xylene for 10 min each, hydrated in gradient series of ethyl alcohol, subjected to antigen retrieval, and then stained with p27Kip1 (1:2400 dilution) and α-tubulin (1:8000 dilution) for 45 mins at room temperature and counterstained with DAPI. IF stains were imaged using Ariol 3.4v (Leica Biosystems) at 40×. Five different fields of view of each human patient sample were randomly selected, and the Cell Profiler was used to quantify p27Kip1 localization and cell size. Note that images of 2D slices may not be directly comparable with 3D in vitro images, as the 2D slices may not capture the maximum cross section of any given cell and therefore underestimate the actual cell size. Because it was difficult to distinguish cell-cell boundaries in a human patient sample and to measure single-cell size, averaged measurements were performed instead of single-cell measurements. The averaged ratio of nuclear p27Kip1 to cytoplasmic p27Kip1 was calculated as the total intensity of nuclear p27Kip1 measured in 21 to 203 cells divided by the total intensity of cytoplasmic p27Kip1. The averaged cell size was calculated as the total area of cells divided by the total number of cells.
Estimation of fraction of cells in G0 and G1
The fraction of cells in G0 and G1 phases of the cell cycle was estimated by analyzing the results from Ki-67 staining and flow cytometry. First, the fraction of cells at the G0 phase was quantified as the fraction of cells that did not stain positive for Ki-67. Then, the fraction of cells at the G1 phase was determined by subtracting the estimated fraction of cells at the G0 phase from the fraction of cells determined to be at G0/G1 from flow cytometry (cells at G0/G1 from flow cytometry − cells at G0 from Ki-67 staining).
Image analyses for human samples from The Human Protein Atlas
Images of human patient breast tissues were obtained from The Human Protein Atlas (www.proteinatlas.org) (56). Images classified as strong p27Kip1 intensity were used. Images were already designated as having nuclear or cytoplasmic and nuclear p27Kip1 in The Human Protein Atlas. Single cells with identifiable cell boundary were selected for manual measurement of cell size.
Image analysis
All images were taken using a confocal microscope (SP8, Leica) with a 25×/0.95-NA water immersion or 20×/0.75-NA objective. The diameter and area of spheroids and the diameter of single cells were measured using Image J.
Simulations
Computational simulations with a 3D finite element model were performed using Abaqus 6.14 (Dassault Systèmes) to study the growth of single cells in hydrogels with varying relaxation. It was assumed that cells exert a constant stress on the hydrogels during growth. The simulation model was designed to include two parts: a rectangular hexahedron with depth, width, and height of 200, 200, and 100 [in arbitrary units (A.U.)] and a hemispherical hole at the center with a diameter of 30 (A.U.). The hemispherical hole and the rectangular hexahedron represented a cell and a hydrogel surrounding the cell, respectively. The mechanical properties of the hydrogels in the simulation were determined by the experimentally measured values. The initial modulus of hydrogels was ~3 and ~16 kPa. Because the cells exerting a constant stress on the hydrogels over time more closely resemble to a creep test, in which a constant stress is applied and the strain is measured over time (as opposed to a stress relaxation test in which a constant strain is applied and the stress is measured), the shear viscoelastic parameters used in the simulation models were taken from hydrogel creep measurements. To incorporate the creep properties of hydrogels in the simulation, linear viscoelastic models such as Burger and generalized standard linear solid (GSLS) models were used. These linear viscoelastic models consist of a combination of springs and dashpots. The governing equation for the Burger model can be described as σ+(η2μ1+η1+η2μ1)σ˙+(η1η2μ1μ2)σ¨=η2(ε˙+η1μ1ε¨), whereas the GSLS model was written as σ(1μ1+1μ2+1μ3)+σ˙(η1μ1(1μ2+1μ3)+η2μ2(1μ1+1μ3))+σ¨(η1η2μ1μ2μ3)=ε+ε˙(η1μ1+η2μ2)+ε¨ η1η2μ1μ2, where σ and ε represent stress and strain, respectively, and η1, η2, μ1, μ2, and μ3 represent viscosities of dashpots and spring constants of springs, respectively. After establishing the viscoelastic models with the experimentally obtained creep results, the viscoelastic properties were implemented in the simulation. Slow- and medium-relaxing gels were found to be well modeled with standard linear solid, while fast-relaxing gels were captured with the Burger model (fig. S7). With the assumption that the hydrogels are an isotropic material, bulk viscoelastic parameters can be estimated from the shear parameters and input into the model. The Poisson ratio used for the simulation model was 0.49. The stress of ~100 Pa was uniformly applied in the direction to the hydrogels for a time scale of 2 days, while displacement in response to the stress was measured. Displacement was scaled by comparing the diameter of the hemispherical hole to the actual cell sizes. While the assumption that hydrogels are isotropic may lead to inaccuracies, these simulations provide general insight into how matrix viscoelasticity would affect cell growth.
Statistical analysis
Statistical analyses were performed using Student’s t tests to compare two groups or one-way analysis of variance (ANOVA) with a post hoc Tukey’s multiple comparisons to compare more than two groups through GraphPad Prism. Statistical analysis for correlation was performed using two-tailed Pearson’s correlation tests with an assumption of Gaussian distribution through GraphPad Prism. Statistical significance was determined on the basis of P values evaluated by Pearson’s correlation test, and Pearson’s correlation coefficients and _R_2 are specified in the figure legends.
Supplementary Material
http://advances.sciencemag.org/cgi/content/full/5/8/eaaw6171/DC1
Download PDF
Acknowledgments
We thank Chaudhuri laboratory members and J. Nelson for helpful discussions and M. Levenston for use of the rheometer and Instron. Funding: This work was supported by a Samsung scholarship to S.N. and by a Hellman Fellows Award, a grant from the National Science Foundation (CMMI-1536736), and a National Institutes of Health National Cancer Institute award (R37 CA214136) to O.C. Author contributions: S.N. and O.C. designed the experiments, analyzed the data, and wrote the manuscript. H.-p.L. suggested the idea of cell growth–mediated TRPV4 activation in viscoelastic environments. S.N., V.K.G., J.Y.L., K.M.W., E.M.F., and C.D. conducted the experiments, and S.N. ran the simulations. S.V. and R.B.W. conducted the tissue IF staining. Competing interests: All authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/8/eaaw6171/DC1
Fig. S1. Mechanical characterization of alginate hydrogels and human breast cancer tissue.
Fig. S2. Tumor growth in hydrogels with varying stress relaxation.
Fig. S3. Cell cycle progression and cell cycle entry are enhanced in hydrogels with faster relaxation.
Fig. S4. The inhibition of the PI3K/Akt pathway diminishes the diameter of spheroids formed by HT1080 and MCF7 cells.
Fig. S5. Cells did not secrete and deposit collagen and laminin-5, and the inhibition of actomyosin contractility did not affect cell cycle progression.
Fig. S6. Impact of TRPV4 inhibition on proliferation.
Fig. S7. 3D computational simulations of hydrogel deformation due to cell exerting a constant outward stress in hydrogels with varying relaxation.
Fig. S8. The growth of single cells in hydrogels with varying stress relaxation.
Fig. S9. Increasing osmotic pressure and NHE inhibition regulate cell cycle progression.
Fig. S10. Human patient samples show that cancer cells with cytoplasmic and nuclear p27Kip1 exhibit larger cell size than cells with nuclear p27Kip1.
Table S1. List of calcium concentrations and corresponding viscoelastic properties.
REFERENCES AND NOTES
- 1.Chaudhuri O., Gu L., Klumpers D., Darnell M., Bencherif S. A., Weaver J. C., Huebsch N., Lee H. P., Lippens E., Duda G. N., Mooney D. J., Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat. Mater. 15, 326–334 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levental I., Georges P. C., Janmey P. A., Soft biological materials and their impact on cell function. Soft Matter 3, 299–306 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Discher D. E., Janmey P., Wang Y.-L., Tissue cells feel and respond to the stiffness of their substrate. Science 310, 1139–1143 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Liu A. P., Chaudhuri O., Parekh S. H., New advances in probing cell–extracellular matrix interactions. Integr. Biol. 9, 383–405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaudhuri O., Koshy S. T., Branco da Cunha C., Shin J. W., Verbeke C. S., Allison K. H., Mooney D. J., Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat. Mater. 13, 970–978 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Reihsner R., Menzel E. J., Two-dimensional stress-relaxation behavior of human skin as influenced by non-enzymatic glycation and the inhibitory agent aminoguanidine. J. Biomech. 31, 985–993 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Purslow P. P., Wess T. J., Hukins D. W., Collagen orientation and molecular spacing during creep and stress-relaxation in soft connective tissues. J. Exp. Biol. 201, 135–142 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Liu Z., Yeung K., Liu Z., The preconditioning and stress relaxation of skin tissue. J. Biomed. Pharm. Eng. 21, 22–28 (2008). [Google Scholar]
- 9.Geerligs M., Peters G. W. M., Ackermans P. A. J., Oomens C. W. J., Baaijens F. P. T., Linear viscoelastic behavior of subcutaneous adipose tissue. Biorheology 45, 677–688 (2008). [PubMed] [Google Scholar]
- 10.Bilston L. E., Liu Z., Phan-Thien N., Large strain behaviour of brain tissue in shear: Some experimental data and differential constitutive model. Biorheology 38, 335–345 (2001). [PubMed] [Google Scholar]
- 11.Qiu S., Zhao X., Chen J., Zeng J., Chen S., Chen L., Meng Y., Liu B., Shan H., Gao M., Feng Y., Characterizing viscoelastic properties of breast cancer tissue in a mouse model using indentation. J. Biomech. 69, 81–89 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Forgacs G., Foty R. A., Shafrir Y., Steinberg M. S., Viscoelastic properties of living embryonic tissues: A quantitative study. Biophys. J. 74, 2227–2234 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darnell M., Young S., Gu L., Shah N., Lippens E., Weaver J., Duda G., Mooney D., Substrate stress-relaxation regulates scaffold remodeling and bone formation in vivo. Adv. Healthc. Mater. 6, 10.1002/adhm.201601185 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z., Bilston L., On the viscoelastic character of liver tissue: Experiments and modelling of the linear behaviour. Biorheology 37, 191–201 (2000). [PubMed] [Google Scholar]
- 15.Perepelyuk M., Chin L., Cao X., van Oosten A., Shenoy V. B., Janmey P. A., Wells R. G., Normal and fibrotic rat livers demonstrate shear strain softening and compression stiffening: A model for soft tissue mechanics. PLOS ONE 11, e0146588 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKinnon D. D., Domaille D. W., Cha J. N., Anseth K. S., Biophysically defined and cytocompatible covalently adaptable networks as viscoelastic 3D cell culture systems. Adv. Mater. 26, 865–872 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaudhuri O., Gu L., Darnell M., Klumpers D., Bencherif S. A., Weaver J. C., Huebsch N., Mooney D. J., Substrate stress relaxation regulates cell spreading. Nat. Commun. 6, 6364 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nam S., Chaudhuri O., Stowers R., Lou J., Xia Y., Varying PEG density to control stress relaxation in alginate-PEG hydrogels for 3D cell culture studies. Biomaterials 200, 15–24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gudipaty S. A., Lindblom J., Loftus P. D., Redd M. J., Edes K., Davey C. F., Krishnegowda V., Rosenblatt J., Mechanical stretch triggers rapid epithelial cell division through Piezo1. Nature 543, 118–121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benham-Pyle B. W., Pruitt B. L., Nelson W. J., Mechanical strain induces E-cadherin-dependent Yap1 and β-catenin activation to drive cell cycle entry. Science 348, 1024–1027 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uroz M., Wistorf S., Serra-Picamal X., Conte V., Sales-Pardo M., Roca-Cusachs P., Guimerà R., Trepat X., Regulation of cell cycle progression by cell-cell and cell-matrix forces. Nat. Cell Biol. 20, 646–654 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Delarue M., Montel F., Vignjevic D., Prost J., Joanny J. F., Cappello G., Compressive stress inhibits proliferation in tumor spheroids through a volume limitation. Biophys. J. 107, 1821–1828 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng G., Tse J., Jain R. K., Munn L. L., Micro-environmental mechanical stress controls tumor spheroid size and morphology by suppressing proliferation and inducing apoptosis in cancer cells. PLOS ONE 4, e4632 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desmaison A., Frongia C., Grenier K., Ducommun B., Lobjois V., Mechanical stress impairs mitosis progression in multi-cellular tumor spheroids. PLOS ONE 8, e80477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Son S., Kang J. H., Oh S., Kirschner M. W., Mitchison T. J., Manalis S., Resonant microchannel volume and mass measurements show that suspended cells swell during mitosis. J. Cell Biol. 211, 757–763 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zlotek-Zlotkiewicz E., Monnier S., Cappello G., Le Berre M., Piel M., Optical volume and mass measurements show that mammalian cells swell during mitosis. J. Cell Biol. 211, 765–774 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramkumar N., Baum B., Coupling changes in cell shape to chromosome segregation. Nat. Rev. Mol. Cell Biol. 17, 511–521 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Helmlinger G., Netti P. A., Lichtenbeld H. C., Melder R. J., Jain R. K., Solid stress inhibits the growth of multicellular tumor spheroids. Nat. Biotechnol. 15, 778–783 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Lee H. P., Gu L., Mooney D. J., Levenston M. E., Chaudhuri O., Mechanical confinement regulates cartilage matrix formation by chondrocytes. Nat. Mater. 16, 1243–1251 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nam S., Chaudhuri O., Mitotic cells generate protrusive extracellular forces to divide in three-dimensional microenvironments. Nat. Phys. 14, 621–628 (2018). [Google Scholar]
- 31.Klein E. A., Yin L., Kothapalli D., Castagnino P., Byfield F. J., Xu T., Levental I., Hawthorne E., Janmey P. A., Assoian R. K., Cell-cycle control by physiological matrix elasticity and in vivo tissue stiffening. Curr. Biol. 19, 1511–1518 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee K. Y., Mooney D. J., Alginate: Properties and biomedical applications. Prog. Polym. Sci. 37, 106–126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang J., Zubovitz J., Petrocelli T., Kotchetkov R., Connor M. K., Han K., Lee J. H., Ciarallo S., Catzavelos C., Beniston R., Franssen E., Slingerland J. M., PKB/Akt phosphorylates p27, impairs nuclear import of p27 and opposes p27-mediated G1 arrest. Nat. Med. 8, 1153–1160 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Shin I., Yakes F. M., Rojo F., Shin N. Y., Bakin A. V., Baselga J., Arteaga C. L., PKB/Akt mediates cell-cycle progression by phosphorylation of p27Kip1 at threonine 157 and modulation of its cellular localization. Nat. Med. 8, 1145–1152 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Viglietto G., Motti M. L., Bruni P., Melillo R. M., D’Alessio A., Califano D., Vinci F., Chiappetta G., Tsichlis P., Bellacosa A., Fusco A., Santoro M., Cytoplasmic relocalization and inhibition of the cyclin-dependent kinase inhibitor p27Kip1 by PKB/Akt-mediated phosphorylation in breast cancer. Nat. Med. 8, 1136–1144 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Zhang W., Liu H. T., Liu, MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 12, 9–18 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Liang J., Slingerland J. M., Multiple roles of the PI3K/PKB (Akt) pathway in cell cycle progression. Cell Cycle 2, 336–342 (2014). [PubMed] [Google Scholar]
- 38.Thodeti C. K., Matthews B., Ravi A., Mammoto A., Ghosh K., Bracha A. L., Ingber D. E., TRPV4 channels mediate cyclic strain-induced endothelial cell reorientation through integrin-to-integrin signaling. Circ. Res. 104, 1123–1130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blumenthal N. R., Hermanson O., Heimrich B., Shastri V. P., Stochastic nanoroughness modulates neuron–astrocyte interactions and function via mechanosensing cation channels. Proc. Natl. Acad. Sci. U.S.A. 111, 16124–16129 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vriens J., Watanabe H., Janssens A., Droogmans G., Voets T., Nilius B., Cell swelling, heat, and chemical agonists use distinct pathways for the activation of the cation channel TRPV4. Proc. Natl. Acad. Sci. U.S.A. 101, 396–401 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo M., Pegoraro A. F., Mao A., Zhou E. H., Arany P. R., Han Y., Burnette D. T., Jensen M. H., Kasza K. E., Moore J. R., Mackintosh F. C., Fredberg J. J., Mooney D. J., Lippincott-Schwartz J., Weitz D. A., Cell volume change through water efflux impacts cell stiffness and stem cell fate. Proc. Natl. Acad. Sci. U.S.A. 114, E8618–E8627 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nam S., Hu K. H., Butte M. J., Chaudhuri O., Strain-enhanced stress relaxation impacts nonlinear elasticity in collagen gels. Proc. Natl. Acad. Sci. U.S.A. 113, 5492–5497 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tzur A., Kafri R., Lebleu V. S., Lahav G., Kirschner M. W., Cell growth and size homeostasis in proliferating animal cells. Science 325, 167–171 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varsano G., Wang Y., Wu M., Probing mammalian cell size homeostasis by channel-assisted cell reshaping. Cell Rep. 20, 397–410 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Wells W. A., Does size matter? J. Cell Biol. 158, 1156–1159 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ginzberg M. B., Chang N., D’Souza H., Patel N., Kafri R., Kirschner M. W., Cell size sensing in animal cells coordinates anabolic growth rates and cell cycle progression to maintain cell size uniformity. eLife 7, e26957 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee W. H., Choong L. Y., Mon N. N., Lu S. Y., Lin Q., Pang B., Yan B., Krishna V. S. R., Singh H., Tan T. Z., Thiery J. P., Lim C. T., Tan P. B. O., Johansson M., Harteneck C., Lim Y. P., TRPV4 regulates breast cancer cell extravasation, stiffness and actin cortex. Sci. Rep. 6, 27903 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nia H. T., Liu H., Seano G., Datta M., Jones D., Rahbari N., Incio J., Chauhan V. P., Jung K., Martin J. D., Askoxylakis V., Padera T. P., Fukumura D., Boucher Y., Hornicek F. J., Grodzinsky A. J., Baish J. W., Munn L. L., Jain R. K., Solid stress and elastic energy as measures of tumour mechanopathology. Nat. Biomed. Eng. 1, 0004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raviraj V., Zhang H., Chien H. Y., Cole L., Thompson E. W., Soon L., Dormant but migratory tumour cells in desmoplastic stroma of invasive ductal carcinomas. Clin. Exp. Metastasis 29, 273–292 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Mammoto A., Mammoto T., Ingber D. E., Mechanosensitive mechanisms in transcriptional regulation. J. Cell Sci. 125, 3061–3073 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eisenhoffer G. T., Loftus P. D., Yoshigi M., Otsuna H., Chien C. B., Morcos P. A., Rosenblatt J., Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature 484, 546–549 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He L., Chen W., Wu P. H., Jimenez A., Wong B. S., San A., Konstantopoulos K., Wirtz D., Local 3D matrix confinement determines division axis through cell shape. Oncotarget 7, 6994–7011 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lesman A., Notbohm J., Tirrell D. A., Ravichandran G., Contractile forces regulate cell division in three-dimensional environments. J. Cell Biol. 205, 155–162 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tse H. T. K., Weaver W. M., Carlo D., Increased asymmetric and multi-daughter cell division in mechanically confined microenvironments. PLOS ONE 7, 1–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nam S., Lee J., Brownfield D. G., Chaudhuri O., Viscoplasticity enables mechanical remodeling of matrix by cells. Biophys. J. 111, 2296–2308 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uhlen M., Zhang C., Lee S., Sjöstedt E., Fagerberg L., Bidkhori G., Benfeitas R., Arif M., Liu Z., Edfors F., Sanli K., von Feilitzen K., Oksvold P., Lundberg E., Hober S., Nilsson P., Mattsson J., Schwenk J. M., Brunnström H., Glimelius B., Sjöblom T., Edqvist P. H., Djureinovic D., Micke P., Lindskog C., Mardinoglu A., Ponten F., A pathology atlas of the human cancer transcriptome. Science 357, eaan2507 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
http://advances.sciencemag.org/cgi/content/full/5/8/eaaw6171/DC1
Download PDF
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/8/eaaw6171/DC1
Fig. S1. Mechanical characterization of alginate hydrogels and human breast cancer tissue.
Fig. S2. Tumor growth in hydrogels with varying stress relaxation.
Fig. S3. Cell cycle progression and cell cycle entry are enhanced in hydrogels with faster relaxation.
Fig. S4. The inhibition of the PI3K/Akt pathway diminishes the diameter of spheroids formed by HT1080 and MCF7 cells.
Fig. S5. Cells did not secrete and deposit collagen and laminin-5, and the inhibition of actomyosin contractility did not affect cell cycle progression.
Fig. S6. Impact of TRPV4 inhibition on proliferation.
Fig. S7. 3D computational simulations of hydrogel deformation due to cell exerting a constant outward stress in hydrogels with varying relaxation.
Fig. S8. The growth of single cells in hydrogels with varying stress relaxation.
Fig. S9. Increasing osmotic pressure and NHE inhibition regulate cell cycle progression.
Fig. S10. Human patient samples show that cancer cells with cytoplasmic and nuclear p27Kip1 exhibit larger cell size than cells with nuclear p27Kip1.
Table S1. List of calcium concentrations and corresponding viscoelastic properties.