Switching Mature Retinal Ganglion Cells to a Robust Growth State In Vivo: Gene Expression and Synergy with RhoA Inactivation (original) (raw)
Abstract
The inability of mature CNS neurons to regenerate injured axons has been attributed to a loss of inherent growth potential of cells and to inhibitory signals associated with myelin and the glial scar. The present study investigated two complementary issues: (1) whether mature CNS neurons can be stimulated to alter their gene expression profile and switch into a strong growth state; and (2) whether inactivating RhoA, a convergence point for multiple inhibitory signals, is sufficient to produce strong regeneration even without activating the growth state of neurons. In the mature rat, retinal ganglion cells (RGCs) normally fail to regenerate axons through the injured optic nerve but can be stimulated to do so by activating macrophages in the eye (e.g., by lens injury). To investigate underlying changes in gene expression, we retrogradely labeled RGCs with a fluorescent dye, performed optic nerve surgery with or without lens injury, and 4 d later, dissociated retinas, isolated RGCs by fluorescence-activated cell sorting, and examined their profiles of gene expression using microarrays. To investigate the effects of inactivating RhoA, we transfected RGCs with adeno-associated viruses carrying a gene for C3 ribosyltransferase. Our results show that, with appropriate stimulation, mature CNS neurons can undergo dramatic changes in gene expression comparable with those seen in regenerating neurons of the PNS, and that RhoA inactivation by itself results in moderate regeneration, and strongly potentiates axon regeneration through the mature optic nerve when the growth state of neurons is activated.
Keywords: retina, optic nerve, axon, regeneration, retinal ganglion cells, gene therapy, microarrays, explants, RhoA
Introduction
In the primary visual pathway, as in other parts of the mature CNS, projection neurons cannot regenerate axons beyond the site of an injury. Mature retinal ganglion cells (RGCs) can extend injured axons through a peripheral nerve graft, but the number that do so is small (Aguayo et al., 1991; Ramon y Cajal, 1991), suggesting that most RGCs may irreversibly lose their capacity for robust axon growth postnatally (Goldberg et al., 2002b). However, if an inflammatory reaction is induced in the eye, trophic factors secreted by macrophages strongly enhance the number of RGCs that regenerate axons through a peripheral nerve graft and even enable RGCs to regenerate lengthy axons through the optic nerve itself (Leon et al., 2000; Fischer et al., 2000, 2001; Yin et al., 2003). Axon regeneration in the mature optic nerve is marked by an induction of GAP-43 (Berry et al., 1996; Leon et al., 2000; Yin et al., 2003; Fischer et al., 2004), but, in general, it is unknown whether RGCs, or any type of mature CNS neuron, can reactivate a growth program comparable with that seen in regenerating peripheral neurons (Fernandes and Tetzlaff, 2001; Costigan et al., 2002; Xiao et al., 2002; Tanabe et al., 2003). We have investigated this question by using fluorescent-activated cell sorting to isolate RGCs undergoing either regeneration or degeneration in vivo, then using microarrays to characterize their gene expression profiles.
A complementary question addressed here is whether counteracting extrinsic inhibitory signals can promote extensive optic nerve regeneration even when the growth program of RGCs is not fully activated. We previously showed that expression of a dominant-negative form of the Nogo receptor (NgR) does not promote optic nerve regeneration unless RGCs are exposed to macrophage-derived factors (Fischer et al., 2004). NgR mediates the inhibitory effects of the myelin proteins OMgp and MAG and the C-terminal region of Nogo (Fournier et al., 2001; Domeniconi et al., 2002; Wang et al., 2002a). However, additional inhibitors are still present in the injured optic nerve after NgR is neutralized, and a more comprehensive strategy to overcome these might be sufficient to produce extensive regeneration (Lehmann et al., 1999; Niederost et al., 2002; Dubreuil et al., 2003). The small GTPase RhoA lies in the final common pathway of signals mediated through NgR and of other inhibitors, including the N terminus of NogoA, chondroitin-sulfate proteoglycans, semaphorins, and ephrins (Hall et al., 2001; Shamah et al., 2001; Niederost et al., 2002; Monnier et al., 2003; Yamashita and Tohyama, 2003). Here, we have used a gene therapy approach to transfect mature RGCs with a gene encoding ADP ribosyl transferase C3 to inactivate RhoA.
Our results show (1) that mature CNS neurons can be induced to undergo changes in gene expression comparable with those seen in regenerating peripheral neurons, and (2) that irreversibly blocking RhoA by itself leads to modest regeneration in vivo but greatly enhances the effects of activating the growth program of neurons.
Materials and Methods
Induction of axon regeneration
Adult female Sprague Dawley rats, 220-250 gm, were anesthetized by intraperitoneal injection of ketamine (60-80 mg/kg) and xylazine (10-15 mg/kg), and a 1-1.5 cm incision was made in the skin above the right orbit. The optic nerve was surgically exposed under an operating microscope, the epineurium was opened longitudinally, and the nerve was crushed 0.5 mm behind the eye for 10 sec using jeweler's forceps, avoiding injury to the ophthalmic artery. Nerve injury was verified by the appearance of a clearing at the crush site; the vascular integrity of the retina was verified by fundoscopic examination. Lens injury was induced through a retrolenticular approach, puncturing the lens capsule with the narrow tip of a microcapillary tube; inflammation was enhanced by injecting 10 μl of PBS intravitreally after retrieving the same volume from the anterior chamber of the eye (Fischer et al., 2000). Controls received PBS injections only. All surgical procedures were approved by the Institutional Animal Care and Use Committee of Children's Hospital.
Retinal explants
Rats were killed, and their retinas were dissected 0-7 d after crushing the optic nerve and either injuring the lens or performing sham intraocular surgery (n = 5 animals per group). Additional controls received no treatment (n = 5) or lens injury without nerve crush (n = 5). Retinas were cut into eight radial pieces, which were cultured in astrocyte-microglia growth medium (PromoCell, Heidelberg, Germany) in laminin-poly-d-lysine-coated dishes (Bahr et al., 1988). In some cases, we coated culture plates with myelin (courtesy of Dr. Zhigang He, Children's Hospital, Boston, MA), as described (Wang et al., 2002a). The number of axons extending ≥50 μm from each explant was counted after 24 and 48 hr using inverted phase-contrast optics (200×; Axiovert; Zeiss, Thornwood, NY) and a calibrated ocular micrometer. In cases with strong regeneration, some fiber fasciculation was observed, and these were counted as one axon. Results from individual explants were averaged within each experimental group, and intergroup differences were evaluated by Student's t test. Growth velocities were estimated after at least five axons had extended from the edge of the explant. The lengths of these five axons were measured at 4, 6, 12, 18, 24, 36, and 48 hr.
Retrograde labeling of RGCs in vivo
Rats were anesthetized as above, a midline incision was made on the scalp, a bone flap was opened above the right occipital cortex, and the underlying cortex was vacuum-aspirated to expose the superior colliculus. For cell-sorting studies, small crystals of the carbocyanine dye _N_-4-[4-didecylaminostryryl]-_N_-methyl-pyridinium iodide (4Di-10ASP; Molecular Probes, Eugene, OR) were placed into the right superior colliculus, small pieces of Gelfoam (Upjohn, Kalamazoo, MI) were inserted, the incision was closed, and animals were kept for 7 d to allow the dye to be transported back to RGC somata before performing additional surgery. For some studies, RGCs were visualized by injecting Fluorogold (Fluorochrome, Englewood, CO) into the superior colliculus.
Isolation of RGCs by fluorescence-activated cell sorting
Four days after optic nerve surgery, 4Di-10ASP-labeled animals were killed with an overdose of anesthesia. Retinas were rapidly dissected, incubated in a digestion solution containing papain (0.7 U/ml; Worthington, Lakewood, NJ) and l-cysteine (0.3 mg/ml; Sigma, St. Louis, MO) in DMEM (37°C for 40 min; CO2 incubator; Sigma) for 30 min, rinsed twice in 5 ml of DMEM, and triturated in 4 ml of DMEM containing 1:50 B27 supplement (Invitrogen, Carlsbad, CA). Dissociated cells were passed through a cell strainer (40 μm nylon net; BD-Falcon, Bedford, MA), cooled to 4°C, and sorted immediately using a FACSVantage SE cell sorter (Becton-Dickinson, San Jose, CA) controlled by Cell-Quest software. A 530/30 filter was used to detect 4Di-10ASP-labeled RGCs. Cells were sorted at a speed of 12,000 objects/sec directly into 50 μl of RNA-later (Ambion, Austin, TX), frozen immediately, and stored at -80°C until further use. Optimal digestion and sorting conditions were determined empirically by varying papain concentration, digestion time, and the size and fluorescence cutoff levels used for cell sorting. After each experiment, sorted cells were plated in serum-free media without growth factors, and the yield, purity, and viability of RGCs were determined after 3 hr in culture.
RNA extraction and amplification
Supplemental material (available at www.jneurosci.org) summarizes the sample preparation and results of these studies. In all, five independent sets of samples were prepared, each containing ∼50,000 RGCs (pooled from four to six individually sorted retinas) for each of three experimental conditions: RGCs subjected to axotomy alone, RGCs subjected to axotomy and exposed to the effects of lens injury, or normal controls. We did not investigate changes in gene expression associated with lens injury alone. After cell sorting, sample volumes were adjusted to 300 μl with 3.5% β-mercaptoethanol, and 1050 μl of RNeasy lysis buffer (RNeasy kit; Qiagen, Valencia, CA) plus 750 μl of ethanol were added. Samples were mixed gently (Vortex), and 700 μl was applied three consecutive times to the same RNeasy mini-column and centrifuged. Columns were washed and eluted following the manufacturer's protocol. Extracted RNA was concentrated by centrifugation (SpeedVac; Savant, New York, NY) to 4 μl.
T7 antisense RNA amplification. A modified version of the T7 antisense RNA amplification method was used (Van Gelder et al., 1990). A double-stranded cDNA library containing a T7 RNA polymerase promoter site in the 5′ end was made from input mRNA and transcribed using T7 polymerase. This process was repeated twice.
First round. For reverse transcription (RT), 0.1 μg of (dT)-T7 primer [5′-GCATTAGCGGCCGCGAAATTAATACGACTCACTATAGGGAGA-(T)21V-3′] was added to total RNA and denatured at 70°C for 4 min. The RT reaction was performed in a 10 μl volume with 100 U of Superscript II (Invitrogen), 20 U of Rnasin (Promega, Madison, WI), 5% T4gp32 (8 mg/ml; United States Biochemicals, Cleveland, OH), and 500 μm deoxy NTPs in first-strand buffer (Invitrogen) at 42°C for 60 min. The reaction was terminated by incubation at 65°C for 15 min. For second-strand synthesis, 65 μl containing second-strand buffer, 20 U of DNA polymeraseI, 1 U of Escherichia coli Rnase H, and 5 U of E. coli DNA ligase were added. The mix was incubated at 15°C for 2 hr. DNA was polished by adding 10 U of T4-polymerase (5 U/μl; Invitrogen), and incubation was continued at 15°C for 15 min. The reaction was terminated by incubation at 70°C for 10 min. cDNA was purified by phenol-chloroform extraction, followed by chromatography on a BioGel p-6 column (Bio-Rad, Richmond, CA) and ethanol precipitation with 5 μg of linear polyacrylamide (Ambion). Transcription reactions using the T7 Megascript kit (Ambion) were performed in a volume of 20 μl at 37°C for 6 hr according to the manufacturer's instructions. cRNA was isolated with the RNeasy kit and quantified by absorbance at 260 and 280 nm.
Second round. Random primers (0.5 μg; Invitrogen) were added to 100 ng of cRNA from the first round, denatured at 70°C for 10 min, and cooled on ice. Nine microliters of first-strand mixture were added and incubated at 37°C for 20 min, 42°C for 20 min, 50°C for 10 min, 55°C for 10 min, and 65°C for 15 min. One unit of RnaseH was added, and incubation was continued at 37°C for 30 min, followed by 95°C for 2 min. After a brief centrifugation, 100 ng of (dT)-T7 primer was added, and the mixture was incubated at 42°C for 10 min, then put on ice. Second-strand synthesis mixture without E. coli ligase (65 μl) was added, and the sample was incubated at 15°C for 2 hr. The double-stranded cDNA was polished by adding T4-DNA polymerase, and incubation continued at 15°C for 15 min. The enzymes were heat inactivated at 65°C for 10 min, and the cDNA was extracted with phenol-chloroform, passed through the BioGel p-6 column, and precipitated with ethanol. For in vitro transcription, the BioArray HighYield RNA Transcript Labeling kit (T7; Enzo; Affymetrix, Santa Clara, CA) was used according to the manufacturer's protocol at 42°C for 4 hr. cRNA was extracted using the RNeasy kit (Qiagen).
Array hybridization and scanning
Labeled cRNA samples were analyzed on Affymetrix Rat Genome RAE 230 A chips, on which 15,866 named genes and expressed-sequence tag (EST) sequences are represented. Sample fragmentation, hybridization to the array, and array scanning were performed according to standard Affymetrix protocols (Thibault et al., 2000). In brief, aliquots of fragmented cRNA (10 μg in a 200 μl master mix) were hybridized to RAE 230 A chips at 45°C for 16 hr in a rotisserie oven (60 rpm). Arrays were then washed and stained with streptavidin-phycoerythrin (Molecular Probes) using an Affymetrix Fluidics Station. Hybridization signals were amplified by incubating arrays at 25°C with 3 μg/ml biotinylated goat anti-streptavidin antibody (Vector Laboratories, Burlingame, CA) in the presence of 0.1 mg/ml normal goat IgG (Sigma) in staining buffer. Arrays were restained with streptavidin-phycoerythrin, washed with nonstringent buffer, and scanned using a dedicated confocal scanner (Hewlett-Packard, Palo Alto, CA).
Initial processing of microarray data, including calculation of “average difference” expression intensity levels, was performed using Microarray Suite software (MAS version 4; Affymetrix). Arrays were normalized by correction to the set value for median total hybridization intensity. Scaling factors for all arrays were between 2 and 2.5. Quality of array hybridization was further assessed by ensuring that the ratio of 3′ to 5′ end probes of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) did not exceed 1.2-1.4. Pairwise scattergram comparisons of all arrays showed a highly linear behavior across all intensity classes (see below).
Additional analyses, including clustering analysis, was done using DNA-Chip software (Li and Wong, 2001). All arrays were normalized, and samples from each series were compared independently with each other, with significance based on all three of the following criteria: (1) ratio of the hybridization signals ≥1.8 or ≤1/1.8; (2) a p value for a paired t test <0.025; and (3) a difference between signals ≥500. The “resulting fold change values” were averaged for the values obtained in three independent experiments. Genes showing a significant change between either group were clustered using the 1-correlation method based on a p value of 0.1 for calling a cluster significant.
Quantitative real-time PCR
Total RNA (100 ng) or amplified cRNA (10 ng) was reverse transcribed and amplified using TaqMan One-Step RT-PCR Master Mix (Applied Biosystems, Foster City, CA). Primers and probes were synthesized by BioSource International (Camarillo, CA): gap-43: forward TGCAGAAAGCAGCCAAGCT, reverse CCTGTCGGGCACTTTCCTTA, TaqMan probe FAM-AGGAGGAGAAAGAAGCTGTAGATGAAGCCAAAC-TAMRA; sprr1A: forward GTCCATAGCCAAGCCTGAAGA, reverse GGCAATGGGACTCATAAGCAG, TaqMan probe FAM-CTGATCACCAGATGCTGAGGCTGCTTTC-TAMRA; l1: forward CTGCCATGTCGCATGAGATC, reverse CAGTCTCCTTCGGCCATTTG, TaqMan probe FAM-AGCTCGTGGCTGAGGGTGCCC-TAMRA; c/ebp-δ: forward AGAACGAGAAGCTGCATCAGC, reverse TTGAGAACTGCCGGAGGC, TaqMan probe FAM-CAGCTCACCCGGGACCTGGC-TAMRA; C/ebp-β: forward AAGATGCGCAACCTGGAGAC, reverse CCTTCTTCTGCAGCCGCTC, TaqMan probe FAMCACAAGGTGCTGGAGCTGACGGC-TAMRA; MafK: forward CCATCGTCAAATCCGCAGA, reverse CAGACACTAGGAGGCGGCTG, TaqMan probe FAM-CCCTCCTCCACCTCTGTGCCCTTCTAMRA; NgR: forward TGCACTCAAGGGACGTGTGCCTC, reverse AGTCATTGATGTGCCGTGGG, TaqMan probe FAM-TGACACTCCACCAGGCAATGGCTC-TAMRA; Narp: forward TCCCCCACAAAAGAGGAAC, reverse CACAGTGGGCTGAGCATCAA, TaqMan probe FAM-CCATTAGAAGCCAGCCTCCCCTCCT-TAMRA.
Primers and VIC/TAMRA-labeled TaqMan probe for the rodent GAPDH internal control were purchased from Applied Biosystems. The samples were analyzed on an ABI PRISM 7700 Sequence Detector, and the primer sets were designed by Primer Express 1.5 software. Changes in gene expression relative to naive RGCs were evaluated using cRNA samples generated from three to five independent cell sorts. Each sample was run in duplicate within each run.
Immunohistochemistry
Animals were killed with a lethal overdose of anesthesia and perfused through the heart with cold saline plus heparin, followed by 4% paraformaldehyde. Eyes with optic nerves segments attached were dissected free from connective tissue, postfixed overnight, transferred to 30% sucrose overnight (4°C), and frozen. Frozen sections were cut longitudinally on a cryostat, thaw-mounted onto coated glass slides (Superfrost plus; Fisher Scientific, Pittsburgh, PA), and stored at -20°C until additional use. As primary antibodies, we used rabbit anti-narp (1:1400; generously provided by Dr. Paul Worley, Johns Hopkins University, Baltimore, MD), rabbit anti-Mn-SOD (1:100; Upstate Biotechnology, Lake Placid, NY), rabbit anti-MafK (1:500; Santa Cruz Biotechnology, Santa Cruz, CA), sheep anti-GAP-43 (Benowitz et al., 1988), and rabbit anti-SPRR1A (1: 300; kindly provided by Dr. Steven Strittmatter, Yale University, New Haven, CT). To visualize RGCs in double-labeling experiments, we used the monoclonal mouse TUJ1 antibody (Babco, Richmond, CA) at a dilution of 1:500. Secondary antibodies included a cyanine 3-conjugated anti-rabbit IgG antibody (1:600; Jackson ImmunoResearch, West Grove, PA) and anti-mouse IgG conjugated to Alexa Fluor 488 (1:500; Molecular Probes). Fluorescent sections were covered using Vectashield mounting medium (Vector Laboratories) and analyzed under a fluorescent microscope.
Visualization of RhoA activation by Rho-binding domain-glutathione S_-transferase staining_
The Rho-binding domain (RBD) of the protein rhotekin binds selectively to the active (GTP-bound) form of RhoA and can be used as a reagent to visualize RhoA-GTP in cell homegenates or in situ (Dubreuil et al., 2002). Bacteria expressing a glutathione _S_-transferase (GST)-RBD fusion protein in a pGEX vector (a gift from John Collard, Division of Cell Biology, Netherlands Cancer Institute, Amsterdam, The Netherlands) were grown in L-broth with 100 μl/ml ampicillin. Overnight cultures were diluted 1:10 into 1000 ml of L-broth and incubated in a shaking bacterial incubator at 37°C for 1 hr. Isopropyl-β-d-thiogalactopyranoside was then added to the incubating cultures for 2 hr, resulting in a final concentration of 0.1 mm. Bacteria were collected by centrifugation at 6000 × g for 20 min. The pellets were resuspended in 10 ml of lysis buffer (50 mm Tris, pH 7.5, 1% Triton-X, 150 mm NaCl, 5 mm MgCl2, 1 mm DTT, 10 μg/ml leupeptin, 10 μg/ml aprotinin, and 1 mm PMSF), sonicated, and lysates were spun at 14,000 rpm for 30 min at 4°C. The clarified bacterial lysate was diluted 1:100 and used for in situ binding studies. Paraformaldehyde-fixed retinal cryostat sections were incubated with diluted lysate overnight at 4°C, washed three times in TBS, blocked in 5% BSA in TBS with 0.05% Tween 20 for 1 hr at room temperature, and incubated with an anti-GST antibody (Immunology Consultants Laboratory, Newberg, OR) and with the TUJ1 antibody (Babco) overnight at 4°C as described (Dubreuil et al., 2002). Sections were washed in TBS and incubated for 2 hr at room temperature with Alexa Fluor 488 and 594-conjugated secondary antibodies (1:500; Molecular Probes).
Viral construction
cDNA encoding a modified form of the ADP ribosyl transferase C3 was generated by PCR from the pET-3a-C3 plasmid, generously provided by Dr. S. Narumiya (Kyoto University, Kyoto, Japan) (Kumagai et al., 1993), using the following primers: forward, 5′-TATGGCTAGCTATGC AGATACTTTCACAGAATT-3′; reverse, 5′-CTATTTAAATATCATTGCTGTAAT CATAATTTGTC-3′. The encoded form (Fournier et al., 2001) and the dipeptide Met-Ala is attached to Ser1. The cDNA was inserted into the AAV-MCS2-IGFP plasmid, developed by the Harvard Gene Therapy Initiative (HGTI). In addition, we ligated in-frame a sequence encoding the first 10 amino acids of GAP-43 to target the protein to the cell membrane (Zuber et al., 1989; Liu et al., 1994). Gene expression was driven by a cytomegalovirus promoter; constructs also expressed enhanced green fluorescent protein (GFP) from an internal ribosome entry site (IRES). Controls were transfected with viruses expressing GFP alone. Virus production was performed at the HGTI Core Facility.
Viral transfections
To transfect RGCs, female Sprague Dawley rats (160-180 gm) were anesthetized with ketamine-xylazine, and the back of the eye was exposed intraorbitally. After withdrawing 10 μl of fluid from the eye, ∼1011 AAV particles in 10 μl of PBS were injected into the vitreous body using a micropipette, with care taken to avoid injury to the lens. Injections were done 2 weeks before optic nerve surgery to obtain high levels of transgene expression during the course of regeneration (Cheng et al., 2002).
Results
Activating a strong growth state in mature RGCs: explant studies
Initial studies were performed to determine when injured RGCs go into a regenerative state. Explants of normal retinas showed no outgrowth after 1-2 d in culture (Fig. 1_a_), and retinas from animals subjected to optic nerve crush without lens injury showed very little (Fig. 1_b_). In contrast, retinas explanted 3 d after nerve crush plus lens injury showed strong outgrowth, which reached maximal levels in retinas explanted after 4 d in vivo or longer (Fig. 1_c,d_). Axons in these cultures arise exclusively from RGCs, as demonstrated by βIII tubulin immunostaining and by expression of a transgene expressed only in RGCs (see Fig. 9) (Fischer et al., 2004). Retinas subjected to axotomy and lens injury in vivo exhibited 10-14 times more axon regeneration than retinas exposed to axotomy alone (p < 0.001) (Fig. 1_d_). Lens injury also increased the rate of neurite outgrowth. When evaluated 4-7 d after surgery, axons from retinas exposed to axotomy alone grew at a rate of 13.9 μm/hr, whereas those from retinas exposed to axotomy plus lens injury grew at 32.2 μm/hr (difference significant at p < 0.001; data not shown). Explants from animals that sustained lens injury without nerve crush 3 or 5 d earlier showed little outgrowth (data not shown). Thus, extensive regeneration requires both axotomy and lens injury and becomes fully developed after 4 d in vivo.
Figure 1.

RGCs go into a regenerative state 4 d after axotomy accompanied by lens injury. a-c, Retinal explants from mature rats after 48 hr in culture. Retinas were subjected to no previous treatment (a), axotomy alone 4 d before explanting (b), or axotomy plus lens injury 4 d beforehand (c). d, Quantitation of results: axons per explant after 24 or 48 hr in culture. Data represent results from normal retinas (0) or from retinas subjected to axotomy (Axot), with or without lens injury, 1-7 d beforehand, as indicated.
Figure 9.
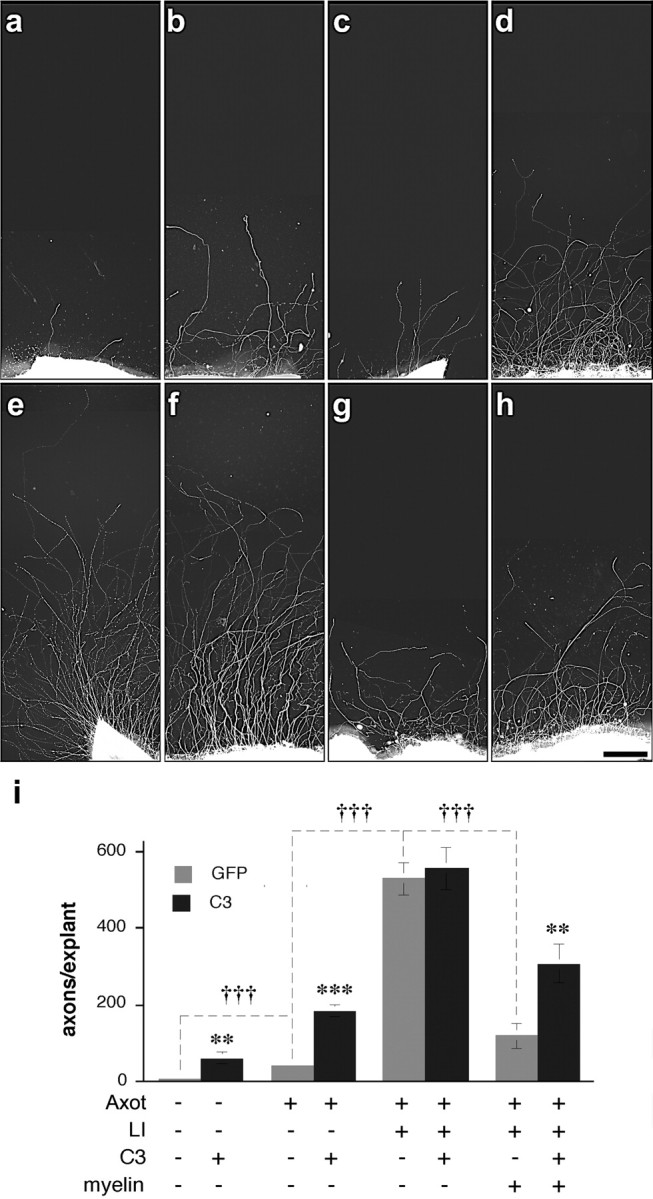
The effect RhoA inactivation on axon regeneration depends on growth state and substrate: in vitro studies. Retinal explants were grown on poly-l-lysine-laminin substrate without (a-f) or with (g, h) myelin proteins 2 weeks after transfecting RGCs in vivo with genes expressing GFP alone (a, c, e, g) or C3 plus GFP (b, d, f, h). Controls not exposed to previous axotomy show almost no spontaneous outgrowth (a), and C3 expression has a small stimulatory effect under these conditions (b). Optic nerve injury 4 d before explanting increases outgrowth slightly relative to controls (c), and C3 expression enhances this growth considerably (d). Exposure of axotomized RGCs to the effects of lens injury increases outgrowth greatly (e), but C3 expression has no additional effect (f). Myelin proteins diminish outgrowth from growth-activated RGCs (g), and C3 expression partially reverses this inhibition (h). i, Quantitation of results. Significance of C3 expression: **p < 0.02; ***p < 0.001; †††differences between experimental treatments significant at p < 0.001. Scale bar, 250 μm.
To investigate changes in RGC viability, we retrogradely labeled RGCs with 4Di-10ASP and then either performed optic nerve surgery 1 week later or left the nerve intact. Retinas were dissected and flat mounted 4-7 d later, and the number of surviving RGCs was quantified. Even in the absence of lens injury, RGC survival remained unaltered 4 or 5 d after axotomy (data not shown), and cell morphology (size, position of nucleus) was normal (Fig. 2_a_). However, by 6 d, RGC survival had decreased by ∼20% and surviving RGCs showed eccentric nuclei (Fig. 2_b_); labeled microglia were first detected on day 7 (Fig. 2_b_, inset), reflecting phagocytosis of labeled RGCs (Berkelaar et al., 1994; Koeberle and Ball, 1998). From these results, we can conclude that cells sorted 4 d after axotomy are unlikely to include microglia or dying neurons.
Figure 2.
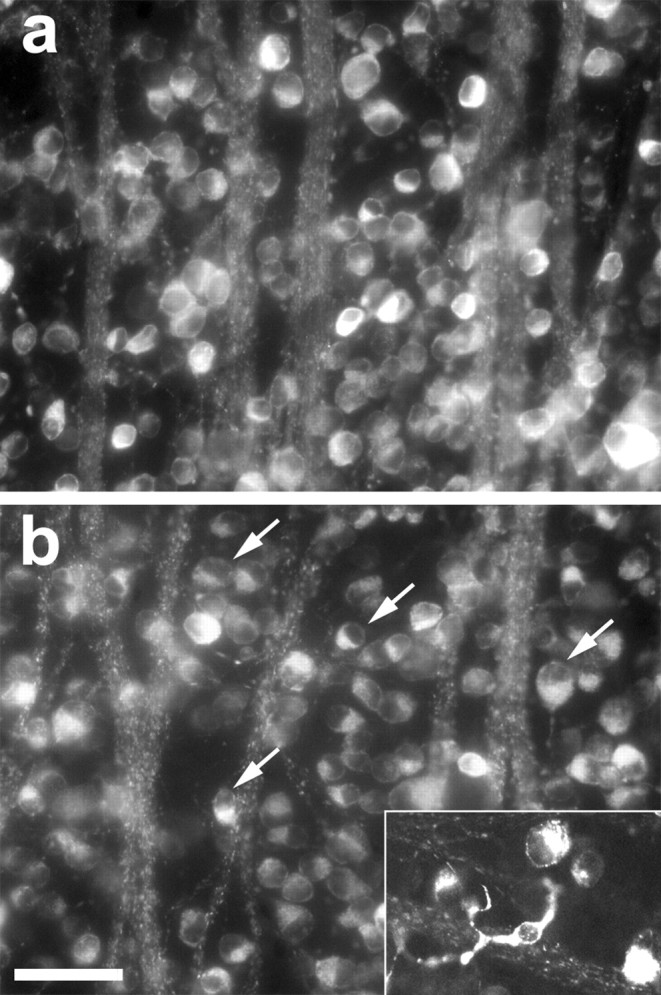
RGC survival after optic nerve injury. RGC morphology, survival, and phagocytosis were visualized in flat-mounted retinas dissected 4-7 d after axotomy; RGCs were retrogradely labeled with 4Di-10ASP 1 week before axotomy. RGCs appear normal 4 d after axotomy (a) but show eccentric nuclei after 6 d (b; arrows). Labeled microglia appear by day 7 (inset). Scale bar, 50 μm.
Axotomy combined with lens injury leads to striking changes in gene expression
Isolation of RGCs by fluorescence-activated cell sorting
Because RGCs represent only a small percentage of the total cell population in the retina (Fig. 3_a,b_), it was necessary to isolate these cells to investigate changes in gene expression associated with regeneration. Near-purification was achieved using fluorescence-activated cell sorting (FACS), taking advantage of the fact that RGCs would be the only cells in the retina that become labeled after injecting 4Di-10ASP into the superior colliculus. The above results suggest that day 4 may be the optimal time point for investigating changes in gene expression associated with regeneration: whereas our explant studies show that RGCs are not yet in a fully developed regenerative state before day 4, the survival studies show that axotomized RGCs not exposed to the effects of lens injury begin to die within 2 d of this time point, diminishing the validity of comparing axotomized RGCs in different growth states. In addition, important immediate-early genes might be missed at later time points, and labeled microglia could contaminate the sorted cell population. Because lens injury alone did not promote axon outgrowth from retinal explants, we did not investigate changes in gene expression associated with this condition; thus, we cannot exclude the possibility that some of the changes seen after axotomy plus lens injury are attributable to the effects of lens injury per se.
Figure 3.
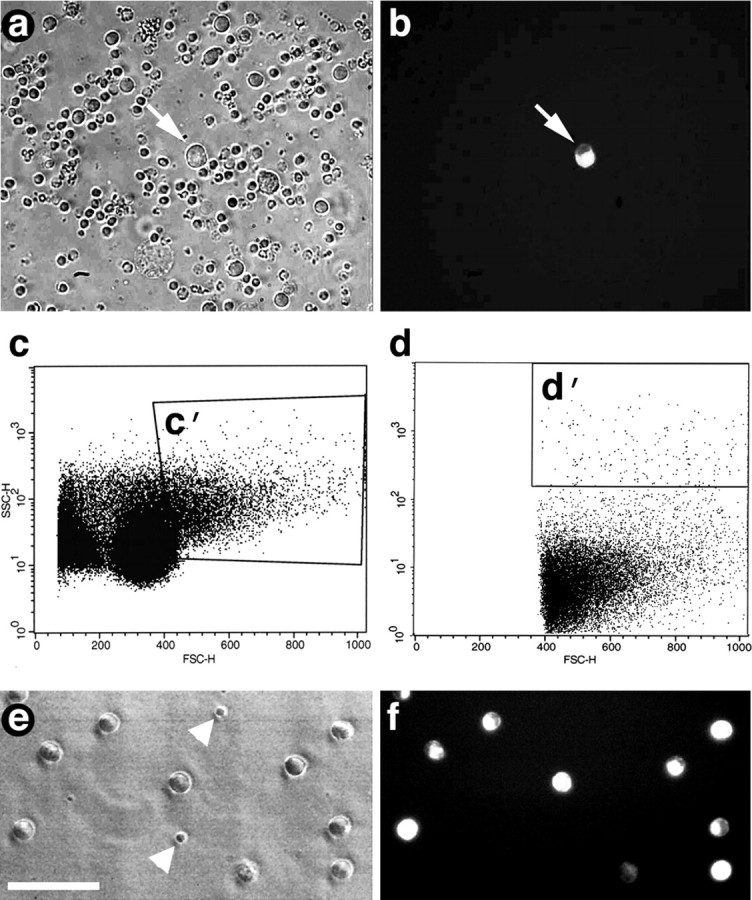
Isolation of RGCs by FACS. a, Dissociated retinal cells viewed under inverted phase-contrast optics before sorting. b, Field includes one fluorescent RGC (arrow) among many unlabeled cells. c, d, Analysis of retinal cells by FACS. _c, x_- and _y_-axes represent two different parameters of cell size. _c_′, Population of large cells analyzed further in d. d, The _x_-axis represents cell size; the _y_-axis represents intensity of fluorescence (logarithmic scale). _d_′, Large cells with high fluorescence selected for sorting. e, f, Cells enclosed with in d ′, viewed by phase-contrast (e) and fluorescence (f) microscopy, consist mostly of RGCs; impurities, mostly small cells, are shown with arrowheads. Scale bar, 50 μm.
FACS analysis demonstrates that most cells of the dissociated retina fall in the small and intermediate size ranges and exhibit only low levels of fluorescence (Fig. 3_c,d_). A relatively small population contains larger, highly fluorescent cells (Fig. 3_d_′), and when these were evaluated in culture, 85-93% appeared to be RGCs by virtue of their size and labeling (Fig. 3_e,f_). After being cultured for 3 hr in serum-free media without growth factors, 50-60% of these RGCs appeared to be viable, as visualized by coincident labeling of 4Di-10ASP and calcein (Molecular Probes). However, this number is likely to underestimate the viability of the RGCs that were used to study gene expression, because the latter were preserved immediately after FACS. Most of the impurities in these cultures were small cells, perhaps photoreceptors (Fig. 3_e_), or cell fragments. Our yield was 7500-16,000 viable RGCs per retina.
In all, five sets of samples were sorted (supplemental material, available at www.jneurosci.org), each of which contained ∼50,000 each of intact RGCs, RGCs subjected to axotomy plus lens injury, and RGCs subjected to axotomy alone. RGCs within each sample, pooled from four to six individually sorted retinas, yielded 500-800 ng of total RNA. The first round of amplification started with ∼200 ng of total RNA per sample and yielded 7.7-9.7 μg of cRNA. The second round started with 200 ng of cRNA from the first round and yielded 54-75 μg of biotinylated cRNA for each sample. RNA from three of the five experiments was used to probe Affymetrix rat RAE 230A nucleotide arrays, and RNA from all five experiments was used to confirm microarray results by quantitative real-time PCR (QRT-PCR).
Microarray analyses: overview
Microarray results were highly reproducible across experiments. A comparison of gene expression profiles for RGCs subjected to axotomy alone in two independent studies showed signal intensities to have a correlation coefficient of 0.99 (Fig. 4_a_); only 0.02% of all genes on the arrays met our criteria for being differentially expressed [i.e., ratio of signals ≥1.8 (or ≤1/1.8) and an absolute difference in signal intensity ≥500]. The false-positive rate would be far lower when comparing two different experimental conditions, because (1) signal intensities for each condition are averaged across three independent samples, (2) normalized averaged signal ratio across all three sets must be ≥1.8 (or ≤1/1.8), and (3) p < 0.025 for paired t tests to be considered significant.
Figure 4.
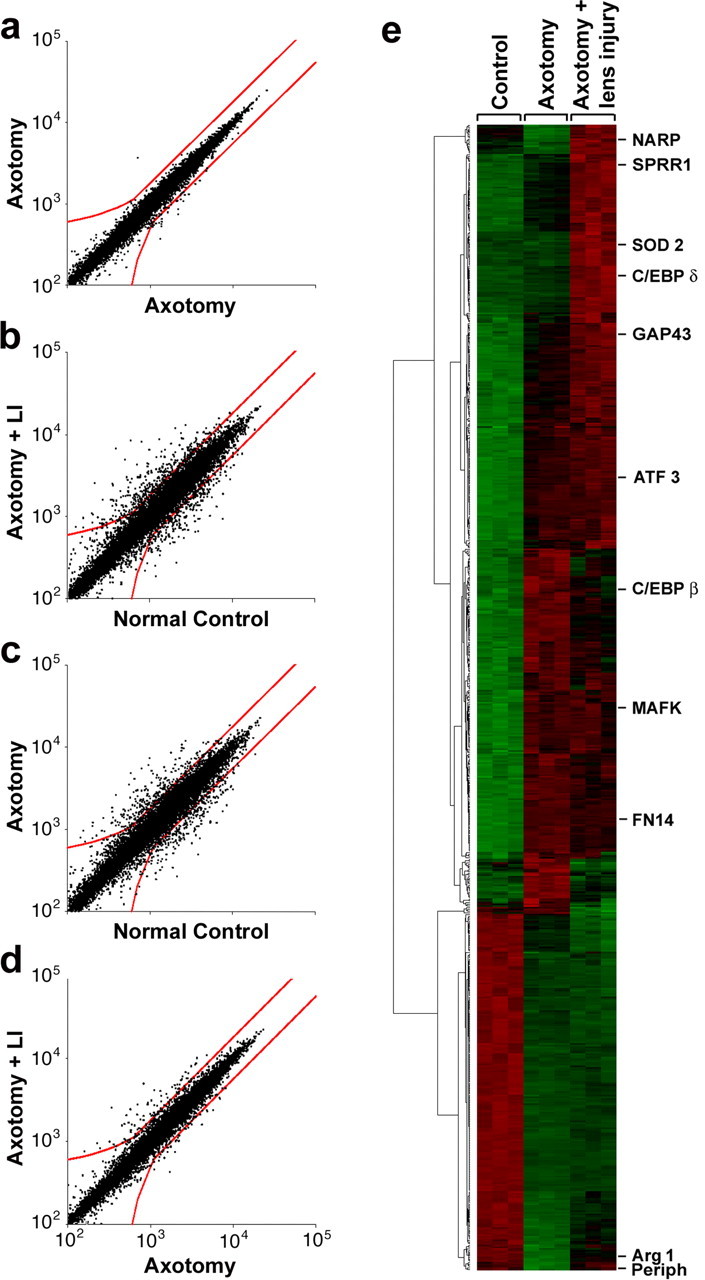
Microarray analysis of RGC gene expression. a-c, Cross-correlation of hybridization signals. Red lines bound points that fall below the criteria for significance. a, Control for reproducibility: hybridization signals for RGCs subjected to axotomy alone in two independent studies (correlation coefficient, 0.99). b, c, Hybridization signals for RGCs subjected to axotomy plus lens injury (b) or axotomy alone (c) compared with normal controls (correlation coefficient, 0.94 for both). d, Hybridization signals for RGCs subjected to axotomy plus lens injury versus axotomy alone (correlation coefficient, 0.98). e, Cluster diagram of gene expression changes from three independent experiments, each of which profiled mRNA levels for RGCs in the three conditions shown on top. Diagram includes 730 genes that change ≥1.8-fold between any two experimental conditions. Representative genes showing different patterns of changes are shown on far right.
Axotomy combined with lens injury strongly altered the program of gene expression of RGCs. The correlation coefficient of the hybridization signals declined to 0.94 (Fig. 4_b_), with 550 genes, or 3.5% of all genes represented on the arrays, changing relative to normal controls; most of these changes (60%) represented increases in gene expression. Axotomy alone altered the expression of 514 genes, only half of which represented increases in expression relative to normal controls (Fig. 4_c_). A comparison between RGCs subjected to axotomy with lens injury versus RGCs subjected to axotomy alone (Fig. 4_d_) showed 94 genes to be differentially expressed between the two conditions, with the vast majority representing increases in the former state. The cluster diagram shown in Figure 4_e_ includes all 730 identified genes or ESTs that showed significant differences between any two experimental conditions, clustered using standard algorithms of the dChip program (Li and Wong, 2001). This diagram reveals a high reproducibility within any one experimental condition across the three data sets, contrasted with strong differences in gene expression among the three experimental conditions. Note that clustering is based on the pattern, rather than magnitude, of the changes. Examples of genes showing different patterns of changes are indicated to the right of Fig. 4_e_.
Microarray analyses: quantitation of gene expression changes
Table 1 lists identified genes, the expression of which changed threefold or greater in RGCs as a result of axotomy plus lens injury (relative to normal control RGCs). In a number of instances, more quantitative results were obtained using QRT-PCR (see below), and these results were substituted for the microarray data in Table 1 (superscript a). Some of the strongest changes in gene expression were seen for the well established growth-associated proteins SPRR-1A and GAP-43. Several transcription factors (C/EBP-δ, C/EBP-β, MafK, fos-related antigen, CREM, ATF3, cysteine-rich protein 3/LIM) were upregulated threefold or greater compared with normal RGCs, as were genes encoding proteins related to cell survival (TIMP-1, SOCS-3, HSP 27, sphingosine kinase I, and GADD 45-α and -γ). Genes that were downregulated in response to axotomy plus lens injury include ones that encode ion channels and acetylcholine receptor subunits. Many of the changes noted here have been observed in neurons undergoing regeneration in the PNS, or are associated with CNS development, long-term potentiation, or altered cell survival (Table 1).
Table 1.
Changes in RGC gene expression during axon regeneration
| Name/function | Axot plus LI | Axot | Ratio | Other observations | Probe set |
|---|---|---|---|---|---|
| SPRR1 (axon growth, cytoskeleton)a | 6297 | 1912 | 3.3 | PNS (Bonilla et al., 2002) | 1371248_at |
| C/EBP-σ (transcription)a | 26.5 | 5.5 | 4.8 | LTP (Taubenfeld et al., 2001) | 1387343_at |
| Cysteine-rich protein 3/LIM | 14.3 | 8.3 | 1.8 | PNS (Tanabe et al., 2003) | 1398243_at |
| SOCS-3 (cytokine signaling) | 11.5 | 1.6 | 6.9 | Survival (Raghavendra Rao et al., 2002) | 1377092_at |
| Lipocalin 2 | 10.4 | 7.0 | 1.4 | 1387011_at | |
| Sphingosine kinase 1 | 9.4 | 1.6 | 5.8 | Survival (Edsall et al., 1997) | 1368254_a_at |
| GAP-43 (axon growth, plasticity)a | 9.3 | 2.9 | 3.2 | PNS (Benowitz and Routtenberg, 1997) | 1367930_at |
| 4-Hydroxyphenylpyruvic acid dioxygenase | 7.3 | 1.8 | 4.2 | 1368188_at | |
| Galanin (neuropeptide) | 7.1 | 2.9 | 2.6 | PNS (Zigmond and Sun, 1997) | 1387088_at |
| HSP 27 | 6.7 | 7.5 | −1.2 | PNS (Costigan et al., 2002) | 1367577_at |
| Retinoic acid-binding protein 2 | 6.6 | 4.9 | 1.3 | PNS (Costigan et al., 2002) | 1370391_at |
| GADD45-γ | 6.0 | 3.7 | 1.7 | PNS (Tanabe et al., 2003) | 1388792_at |
| iGb3 synthase | 5.6 | 1.8 | 3.1 | PNS (Tanabe et al., 2003) | 1370561_at |
| MAFK | 5.3 | 6.9 | −1.3 | PC12 (Torocsik et al., 2002) | 1372211_at |
| GADD 45-α | 5.3 | 4.8 | 1.1 | PNS (Tanabe et al., 2003) | 1368947_at |
| Best5 | 4.9 | 1.3 | 3.8 | 1370913_at | |
| Metallothionein | 4.7 | 2.9 | 1.7 | PNS (Chung et al., 2003) | 1371237_a_at |
| ATF 3 (transcription) | 4.6 | 4.1 | 1.1 | PNS (Tsujino et al., 2000) | 1369268_at |
| Retinol-binding protein 1 | 4.6 | 2.0 | 2.2 | PNS (Zhelyaznik et al., 2003) | 1367939_at |
| GFAP | 4.5 | 1.7 | 2.6 | PNS (Aldskogius and Kozlova, 1998) | 1368353_at |
| Fn14 (FGF-regulated protein 2) | 4.5 | 5.0 | −1.2 | PNS (Tanabe et al., 2003) | 1371785_at |
| ATP-binding cassette subfamily B (MDR/TAP)-1A | 4.4 | 5.7 | −1.3 | PNS (Tanabe et al., 2003) | 1370464_at |
| Serine (or cysteine) proteinase inhibitor, G-1 | 4.4 | 1.7 | 2.5 | 1372254_at | |
| Myxovirus (influenza virus) resistance 3 | 4.3 | 1.1 | 3.9 | 1387283_at | |
| ASCT2 (Na+ -dependent amino acid transporter) | 4.3 | 2.4 | 1.8 | 1371040_at | |
| Tumor-associated glycoprotein pE4 | 4.3 | 3.2 | 1.3 | PNS (Costigan et al., 2002) | 1370177_at |
| Importin β-3 (karyopherin β-3 subunit) | 4.3 | 2.5 | 1.8 | 1373955_at | |
| Lysozyme | 4.2 | 2.8 | 1.5 | PNS (Tanabe et al., 2003) | 1370154_at |
| Anthrax toxin receptor 2 | 4.1 | 3.3 | 1.2 | 1389017_at | |
| Tissue inhibitor of metalloproteinase 1 | 4.0 | 1.4 | 2.9 | Survival (La Fleur et al., 1996) | 1367712_at |
| Interferon regulatory factor 7 (IRF-7) | 4.0 | 1.3 | 3.2 | 1383564_at | |
| Cytochrome c oxidase, subunit Vla, polypeptide 2 | 3.9 | 5.0 | −1.3 | PNS (Tanabe et al., 2003) | 1367782_at |
| Endothelin converting enzyme-like 1 | 3.9 | 4.3 | −1.1 | PNS (Nakagomi et al., 2000) | 1368923_at |
| Glycoprotein (transmembrane) nmb | 3.8 | 4.8 | −1.3 | 1368187_at | |
| Small cell adhesion glycoprotein (smagp) | 3.7 | 1.6 | 2.3 | 1372734_at | |
| Galectin 3 | 3.7 | 6.4 | −1.8 | PNS (Costigan et al., 2002) | 1386879_at |
| Heme oxygenase | 3.7 | 3.3 | 1.1 | PNS (Magnusson et al., 2000) | 1370080_at |
| Moesin | 3.6 | 3.9 | −1.1 | 1371575_at | |
| Syntenin | 3.6 | 3.9 | −1.1 | 1376973_at | |
| Kinesin family member 22 | 3.6 | 2.6 | 1.4 | 1372516_at | |
| C/EBP-β (transcription)a | 3.5 | 4.4 | −1.3 | LTP(Taubenfeld et al., 2001) | 1387087_at |
| CD24 antigen | 3.5 | 3.9 | −1.1 | PNS (Shewan et al., 1995) | 1369953_a_at |
| Protein tyrosine phosphatase, non-receptor, type 5 | 3.4 | 1.6 | 2.1 | PNS (Tanabe et al., 2003) | 1368421_at |
| Endothelial monocyte-activating polypeptide | 3.3 | 2.2 | 1.6 | 1375170_at | |
| Kidney predominant protein NCU-G1 | 3.3 | 3.4 | −1.0 | 1371946_at | |
| Serine protease inhibitor | 3.2 | 1.0 | 3.1 | 1368224_at | |
| CREM | 3.2 | 1.8 | 1.7 | PNS (Costigan et al., 2002) | 1369737_at |
| Cell death activator CIDE-A | 3.1 | 2.4 | 1.3 | 1389179_at | |
| 3-Phosphoglycerate dehydrogenase | 3.0 | 3.0 | −1.0 | 1367811_at | |
| fos-related antigen | 3.0 | 3.4 | −1.2 | 1373035_at | |
| CTL target antigen | 3.0 | 3.4 | −1.2 | 1367838_at | |
| Neurolysin (metallopeptidase M3 family) | 3.0 | 3.1 | −1.1 | PNS (Tanabe et al., 2003) | 1369669_at |
| LPS-induced TNF-α factor | 3.0 | 2.2 | 1.3 | 1370928_at | |
| Cholinergic receptor, nicotinic, α6 | −3.0 | −2.8 | −1.0 | 1369845_at | |
| Netrin-G1a | −3.1 | −2.5 | −1.3 | 1376311_at | |
| Inwardly-rectifying K+ -channel, K4 | −3.2 | −3.2 | 1.0 | PNS (Xiao et al., 2002) | 1370110_at |
| Proline transporter | −3.4 | −2.0 | −1.7 | 1398457_at | |
| Phosphofructokinase | −3.4 | −3.6 | 1.0 | PNS (Xiao et al., 2002) | 1386961_at |
| Thyroid hormone-response protein-1 | −3.5 | −2.9 | −1.1 | 1370302_at | |
| Glutamate receptor, ionotropic, 2 | −3.7 | −2.9 | −1.3 | PNS (Costigan et al., 2002) | 1387171_at |
| Rab 3B protein | −3.7 | −3.4 | −1.1 | 1370061_at | |
| Monoglyceride lipase | −3.7 | −4.3 | 1.1 | 1388644_at | |
| Leucine-rich repeat protein 3, neuronal | −3.8 | −3.7 | −1.1 | 1368964_at | |
| Thyroid hormone-responsive protein (spot14) | −3.9 | −3.3 | −1.2 | 1371400_at | |
| FXYD domain-containing ion transport reg.7 | −4.2 | −7.4 | 1.8 | 1368696_at | |
| Insulin-like growth factor I | −4.3 | −3.3 | −1.3 | 1388469_at | |
| Rabphilin 3A | −4.3 | −5.0 | 1.2 | 1390477_at | |
| Calcyon; D1 receptor-interacting protein | −5.1 | −5.5 | 1.1 | 1376345_at | |
| Opsin 4 (melanopsin) | −5.4 | −4.4 | −1.3 | 1374302_at | |
| Na+ -channel, voltage-gated, type 1 α polypeptide | −5.7 | −4.5 | −1.4 | PNS (Xiao et al., 2002) | 1369210_at |
| Visinin-like 1 | −5.8 | −3.5 | −1.7 | PNS (Xiao et al., 2002) | 1368853_at |
| Cholinergic receptor, nicotinic, α3 | −6.3 | −6.5 | 1.1 | 1369001_at | |
| Neuritin | −9.2 | −5.0 | −1.8 | 1386969_at |
Axotomy by itself altered the expression of many of the same genes that changed after axotomy combined with lens injury, although the magnitude of the changes was frequently smaller after axotomy alone. Bold entries in Table 1 indicate genes with an expression in axotomized RGCs after lens injury that was ≥1.8-fold higher than in RGCs subjected to axotomy alone. Some of the genes that are upregulated after axotomy alone (e.g., caspase 3) may be associated with the degeneration that occurs if RGCs are not exposed to macrophage-derived growth factors. Supplemental material (available at www.jneurosci.org) lists genes that are differentially expressed after axotomy with versus without lens injury.
Verifying microarray results by quantitative RT-PCR and immunohistochemistry
Validation of the method
Because of the limited amount of RNA available from purified RGCs, we performed a pilot study to determine whether the relative abundance of various mRNA species in the original samples would be preserved after RT and mRNA amplification. We extracted total RNA from PC12 cells that were either exposed to NGF for 24 hr or left untreated and amplified a portion of this to obtain cRNA. QRT-PCR reactions were performed using either 100 ng of total, unamplified RNA (of which 1-2% is expected to be mRNA) or 10 ng of amplified cRNA per reaction. NGF induction of gap-43 was found to be 5.44 ± 0.87-fold in the original RNA samples and 6.24 ± 0.81-fold in the amplified cRNA samples (mean induction ± SEM); for l1, NGF induction was 2.50 ± 0.12 based on the original RNA and 2.9 ± 0.01 from the cRNA. NGF treatment diminished ngr expression in PC12 cells, as evaluated from the original RNA (ratio, 0.62 ± 0.00) or the cRNA (ratio, 0.46 ± 0.11) but did not alter levels of mafk or _c/ebp-_β (ratios, 1 ± 0.07%); narp was undetectable in all samples (data not shown). From these studies, we conclude that RNA amplification does not bias QRT-PCR results significantly.
We next used QRT-PCR to verify changes in the expression of representative genes that show different patterns of change (Fig. 4_e_) or that are associated with different cellular functions. In addition, in some instances, we used immunohistochemistry to investigate (1) whether observed changes in gene expression are translated into changes at the protein level at 4 d, (2) whether these changes occur in most RGCs, and (3) whether they are restricted to RGCs.
Growth-associated proteins
QRT-PCR showed that the increases in GAP-43 and SPRR1A expression that occur in axotomized RGCs as a result of lens injury are considerably higher than estimated on the microarrays. By QRT-PCR, the increase in GAP-43 mRNA levels in axotomized RGCs after lens injury was approximately threefold higher than found by microarrays and approximately twofold higher than found on the microarrays after axotomy alone (n = 5 experiments) (Fig. 5). This discrepancy might be explained by the fact that, after axotomy, the gap-43 signal becomes one of the highest on the arrays, perhaps saturating binding sites and attenuating the apparent change. Increased GAP-43 expression is apparent at the protein level during optic nerve regeneration (Leon et al., 2000; Yin et al., 2003).
Figure 5.
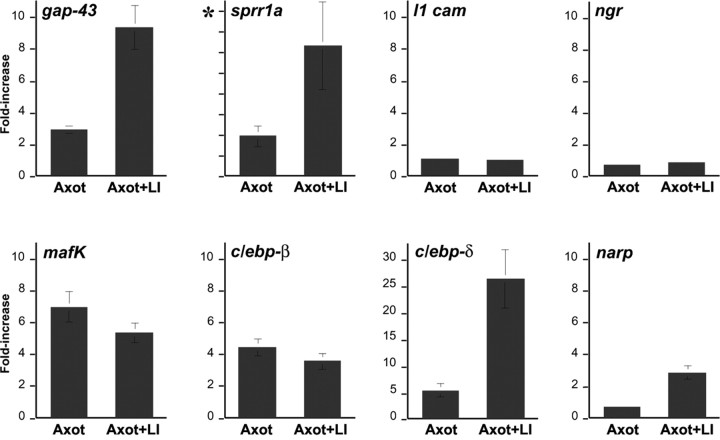
Changes in RGC gene expression: QRT-PCR. Changes in gene expression were examined in fluorescence-activated cell-sorted RGCs 4 d after axotomy (Axot) or 4 d after axotomy and simultaneous lens injury (Axot + LI). Results are normalized to expression levels in control RGCs. Representative genes encode growth-associated proteins (gap-43, sprr1a, l1-cam), Nogo receptor (ngr), transcription factors (_mafk, c/ebp-_β, and c/ebp-d), and the immediate-early gene narp. All ordinate scales are 0-10, except for _c/ebp-_δ (0-30) and sprr1a, in which the changes are >1000-fold because of almost undetectable levels in control RGCs (asterisk).
The cytoskeleton-associated protein SPRR1A is not expressed in developing peripheral neurons but is strongly expressed during axon regeneration (Bonilla et al., 2002). QRT-PCR showed SPRR1A mRNA to be extremely low in control RGCs and to increase >6000-fold in axotomized RGCs after lens injury (Fig. 5); microarray analysis estimated this increase to be ∼16-fold, presumably because of nonspecific hybridization in the control condition. At the protein level, we observe only trace SPRR1A immunoreactivity in control RGCs (Fig. 6_a_) but high levels in nearly all RGCs after axotomy without (Fig. 6_b_) and with (Fig. 6_c_) lens injury.
Figure 6.
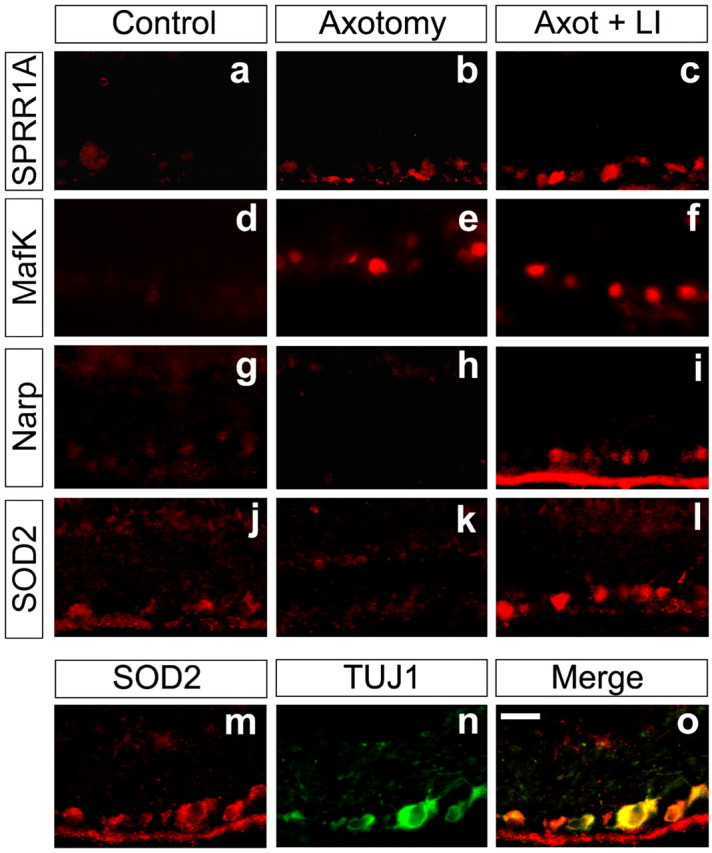
Changes in protein expression resulting from axotomy and lens injury. The proteins SPRR1A (a-c), MafK (d-f), Narp (g-i), and SOD-2 (j-l) were detected by immunofluorescence in normal rat RGCs (a, d, g, j) or in RGCs 4 d after axotomy without (b, e, h, k) or with lens injury (c, f, i, l). m-o, Representative colocalization study: cells were stained for SOD2 (n) and βIII-tubulin (m). The merged image (o) demonstrates that changes in SOD2 expression occur specifically in RGCs. Scale bar, 20 μm.
The cell adhesion molecule L1 is upregulated in goldfish RGCs during axon regeneration (Vielmetter et al., 1991; Petrausch et al., 2000), and L1 immunostaining is detected in rat RGC axons regenerating through a peripheral nerve graft (Jung et al., 1997). However, both microarray analysis and QRT-PCR showed no upregulation of L1 mRNA 4 d after axotomy, with or without lens injury (Fig. 5).
The NgR signaling pathway
Counteracting NgR enhances optic nerve regeneration in axotomized RGCs after lens injury (Fischer et al., 2004). This finding implies that NgR remains present and dampens the amount of regeneration that occurs under these conditions. In conformity with this, QRT-PCR showed that NgR mRNA levels decline only slightly after axotomy (ratio relative to controls, 0.72 ± 0.13) or after axotomy combined with lens injury (ratio, 0.83 ± 0.07) (Fig. 4). p75, a coreceptor of NgR (Wang et al., 2002b), showed a low signal strength in our microarrays under all conditions (data not shown). RhoA and Rho kinase (ROCK) are downstream mediators of NgR and other growth inhibitors, and levels of their mRNAs did not change appreciably after axotomy, regardless of lens injury (ratios for RhoA and Rho kinase mRNA levels between experimental conditions and controls were between 1.0 and 1.3; data not shown). The continued expression of these inhibitory mediators in regenerating RGCs suggests that axon regeneration through the optic nerve might be enhanced significantly if RhoA were inactivated (see below).
Transcription factors
Because transcription factors that are upregulated early in the regenerative process could be important for the expression of multiple downstream genes, we investigated the expression of several of these in further detail. By QRT-PCR, expression of c/ebp-δ was found to increase 26.5-fold above baseline in axotomized RGCs after lens injury and 5.5-fold after axotomy alone (Fig. 5) (n = 4). The related transcriptional activator c/ebp-β increased approximately fourfold in axotomized RGCs, regardless of lens injury (Fig. 5). The gene encoding MafK showed a similar pattern of expression (Fig. 5), and by immunohistochemistry, MafK protein increased dramatically within the nuclei of almost all RGCs after optic nerve damage, regardless of lens injury (Fig. 6_d-f_).
Other changes
mRNA encoding the immediate-early gene product Narp declined after axotomy alone but increased above baseline after axotomy combined with lens injury (Fig. 5). Narp protein was evident in naive RGCs (Fig. 6_g_), undetectable 4 d after axotomy (Fig. 6_h_), and strongly elevated in most RGCs after axotomy plus lens injury (Fig. 6_i_).
Superoxide dismutase 2 (SOD2) is upregulated in spinal motorneurons after sciatic nerve crush (Rosenfeld et al., 1997) and is protective for several types of neurons when overexpressed (Gonzalez-Zulueta et al., 1998; Keller et al., 1998). Axotomized RGCs showed elevated SOD2 mRNA levels by microarray analysis, and the majority of RGCs showed increases at the protein level (Fig. 6_l_).
In summary, for every gene studied, QRT-PCR showed the changes in mRNA levels to be as great or greater than seen on the microarrays; in addition, immunohistochemistry demonstrated that, within most RGCs, these changes were translated into changes at the protein level by day 4.
Transfection of RGCs with AAV expressing C3 ADP-ribosyltransferase
As shown above, RhoA and Rho kinase continue to be expressed in axotomized RGCs after lens injury. Thus, if RhoA signaling were inactivated, it is possible that many more axons might regenerate beyond the lesion site when RGCs are in an active growth state. Another possibility is that, because RhoA is a convergence point of many inhibitory signals, its inactivation might be sufficient to cause strong regeneration even when the growth state of RGC is unaltered. To investigate these questions, we injected mature rats intravitreally with AAV expressing either GFP alone (AAV-GFP) or clostridium botulinum C3 ADP-ribosyltransferase (and GFP after an IRES: AAV-C3-IGFP) to inactivate RhoA. By virtue of AAV2 being neuron specific, and by virtue of RGC somata and axons being superficial in the retina, this method results in the transfection of ∼75% of RGCs but little transfection of other cell types (Di Polo et al., 1998; Martin et al., 2002; Fischer et al., 2004). RT-PCR demonstrated a strong C3 signal in retinas transfected with AAV-C3-IGFP but none in controls transfected with AAV-GFP (Fig. 7_a_). The high efficiency and specificity of transfection was verified by double-labeling studies showing the GFP reporter to be expressed in the same cells that express the RGC-specific tubulin isoform βIII tubulin (Fig. 7_b-f_). Using RBD-GST for in situ_“pull-down assays” to detect RhoA in the active (GTP-bound) state (Dubreuil et al., 2003), we observed considerable binding in normal RGCs (Fig. 7_i_) but much less in RGCs transfected with AAV-C3-IGFP (7_j). Thus, AAV transfection leads to strong transgene expression in RGCs, and in the case of C3 expression, this inactivates RhoA.
Figure 7.
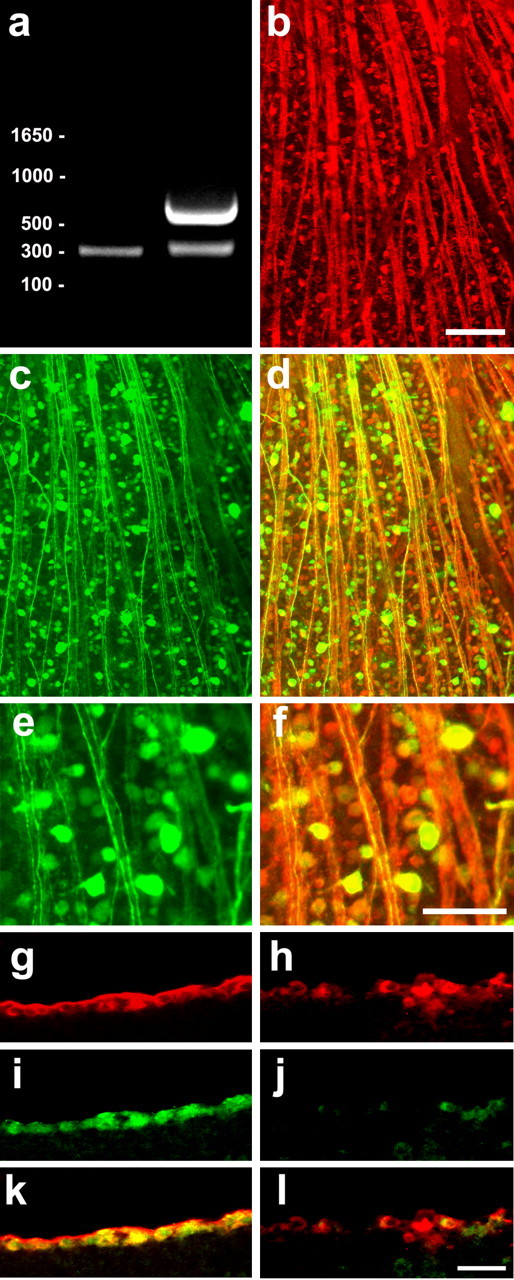
Expression of transgenic proteins and inactivation of RhoA in RGCs. RGCs were transfected in vivo with AAV carrying a gene encoding C3 and GFP after an IRES. a, The C3 transcript is detected by RT-PCR in transfected retinas (right) but not in normal retinas. RT-PCR reactions include primers for GAPDH as a control (C3 band at 615 bp; GAPDH at 300 bp). b-d, Low-power photomicrographs of flat-mounted whole retina stained for βIII tubulin (b) and the GFP reporter (c). d, The merged image shows that most RGCs express the transgene. e, f, Higher-magnification of areas from c and d, respectively. Note intense staining in RGC somata and their axons, which coalesce to form the optic nerve. g-l, C3 transfection decreases RhoA activation. Sections through the retina were prepared 2 weeks after transfection with either AAV-GFP (g, i, k) or AAV-C3-IGFP (h, j, l) and were stained with TUJ1 to visualize all RGCs (g, h) and RBD-GST, which binds to active (GTP-bound) RhoA, followed by antibodies to detect GST (i, j). Note low levels of active RhoA in C3-transfected RGCs (j) compared with control RGCs (i). k, l, Merged images showing colocalization of RBD binding in TUJ1-positive RGCs. Scale bars: b-d, 50 μm; e, f, 25 μm.
RhoA inactivation and macrophage activation have synergistic effects in vivo
After allowing 2 weeks for transgenic C3 protein levels to become sufficiently high in RGCs, rats were re-anesthetized, the left optic nerve was crushed, and the lens was either injured or was left intact. Regeneration was evaluated 2 weeks later by GAP-43 immunostaining (Berry et al., 1996; Leon et al., 2000). As expected, AAV-GFP-transfected animals subjected to nerve crush alone showed no axons growing ≥500 μm beyond the lesion site 2 weeks after surgery (Fig. 8_a_), whereas similarly transfected animals with lens injury had, on average, ∼400 axons extending ≥500 μm beyond the lesion site (Fig. 8_b,e_) (cf. Leon et al., 2000; Yin et al., 2003; Fischer et al., 2004). Even in the absence of lens injury, rats expressing C3 showed a modest number of axons passing through the lesion site; a higher percentage of these continued to extend ≥500 μm than was seen in GFP-expressing cases with lens injury, although the total number of axons reaching that criterion was lower (Fig. 8_c,e_). Combining C3 expression with lens injury resulted in unprecedented levels of axon regeneration. In every animal in this group, axon growth was so high as to obscure the discontinuity in GAP-43 immunostaining that is otherwise seen at the injury site (Fig. 8_d_). The number of axons extending ≥500 μm beyond the injury site was 4.5 times greater than after lens injury or C3 expression alone (Fig. 8_e_) (n = 9; p < 0.001) and higher than the effects of two added together. Thus, inactivation of RhoA and activation of the growth state of RGCs have synergistic effects in vivo.
Figure 8.
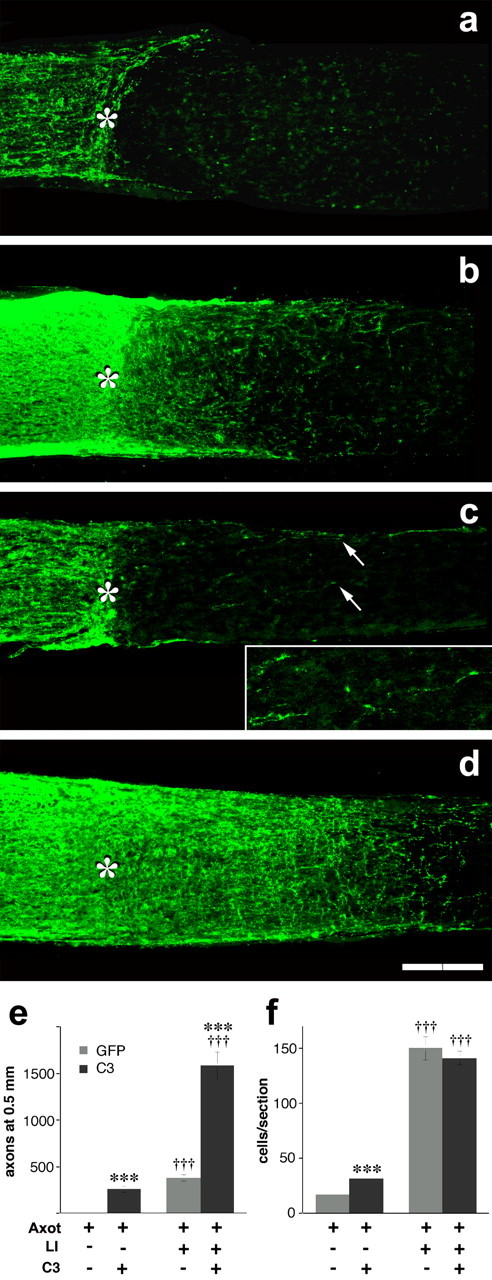
Activation of the growth program of RGCs and inactivation of RhoA have synergistic effects in vivo. a-d, GAP-43-positive axons visualized in longitudinal sections through the adult rat optic nerve 2 weeks after axotomy with (b, d) or without (a, c) lens injury (asterisk indicates crush site). RGCs were transfected with AAV expressing GFP alone (a, b) or C3 plus GFP (c, d). a, Absence of regeneration after axotomy alone. b, Lens injury increases the number of GAP-43+ axons proximal to the injury site (to the left of the asterisk) and the number that regenerate beyond this point. c, C3 expression stimulates some regeneration even in the absence of lens injury. The inset shows regenerating axons at increased magnification and illumination. d, C3 expression greatly enhances axon regeneration when axotomized RGCs are exposed to the effects of lens injury. e, Quantitation of outgrowth (number of axons growing ≥500 μm beyond the injury site per optic nerve). f, RGC survival (TUJ1+ RGCs per retinal cross section). Axot, Axotomy; LI, lens injury. ***Effect of C3 expression significant at p < 0.001; †††effect of intravitreal macrophage activation significant at p < 0.001. Scale bar, 200 μm.
C3 expression enhances RGC survival
RhoA inactivation by C3 has been reported to protect neurons and other cells from apoptotic cell death (Dubreuil et al., 2003). To investigate whether C3 affects RGC survival in vivo, we counted the number of TUJ1-positive cells from four to six cross sections through each retina (near the level of the optic nerve head) 2 weeks after nerve crush and lens injury. C3 expression increased RGC survival after nerve crush approximately twofold relative to controls expressing GFP alone but did not enhance the strong neuroprotective effects of lens injury any further (Fig. 8_f_).
The effects of C3 expression depend on growth state and substrate
To investigate the effects of C3 expression in more detail, we examined the growth of retinal explants expressing C3 or GFP in culture. On a permissive laminin-poly-L-lysine substrate, control RGCs transfected with GFP showed almost no outgrowth, and C3 expression increased growth only slightly (Fig. 9_b,i_) (p < 0.01). Subjecting GFP-transfected RGCs to axotomy alone 4 d beforehand caused a moderate increase in regeneration compared with control RGCs (Fig. 9_c,i_) (p < 0.001) (compare Fig. 1), and C3 transfection increased growth 4.6-fold when RGCs were in this state (p < 0.001) (Fig. 9_d,i_). Axotomy combined with lens injury increased growth 14-fold relative to RGCs subjected to axotomy alone, and this growth was not enhanced further by C3 transfection (Fig. 9_e,f,i_). Thus, when extrinsic inhibitors are absent, RhoA inactivation has only a small effect when the growth program of RGCs is not activated, a strong effect when the growth program is weakly activated by axotomy alone, but no additional effect when the growth program of RGCs is strongly activated.
When plated on a substrate containing myelin proteins, RGCs subjected to axotomy and lens injury showed far less growth than on poly-L-lysine-laminin (Fig. 9_g,i_) (p < 0.001) (cf. Fischer et al., 2004). Under these conditions, C3 expression increased the number of axons regenerating ≥50 μm 2.6-fold (Fig. 9_h,i_) (p < 0.02) and increased the number of axons growing ≥0.5 mm 3.8-fold (p = 0.001; data not shown). Thus, when RGCs are in an active growth state, RhoA inactivation (by C3 expression) helps overcome the inhibitory effects of myelin.
Discussion
Although mature RGCs cannot normally regenerate axons through the injured optic nerve, this situation can be at least partially reversed by injuring the lens, injecting the inflammatory agent Zymosan into the eye, or implanting a fragment of peripheral nerve into the vitreous (Berry et al., 1996; Leon et al., 2000; Fischer et al., 2001, 2004; Yin et al., 2003). All of these treatments increase the number of activated macrophages in the eye (Leon et al., 2000), and macrophages have been shown to secrete proteins that enable dissociated RGCs to extend lengthy axons in the presence of mannose, which is abundant intravitreally, and elevated [cAMPi] (Li et al., 2003; Yin et al., 2003).
In vivo, axotomy combined with lens injury leads to striking changes in the growth state of RGCs and in the underlying program of gene expression. RGCs from normal controls or animals with lens injury alone showed little regenerative capacity when explanted, and RGCs from rats subjected to axotomy alone showed only weak outgrowth and subsequent cell death. In contrast, when explanted 3-4 d after axotomy combined with lens injury, RGCs showed extensive outgrowth, and, as shown here, this state is associated with a marked shift in gene expression.
Most of the known genes that are strongly upregulated in RGCs undergoing regeneration in vivo have also been reported to change in regenerating neurons of the PNS (Table 1); in the case of several prominent growth-associated proteins (e.g., GAP-43, SPRR1A), the magnitude of the changes seen here are even greater than have been reported in the PNS (Costigan et al., 2002; Xiao et al., 2002; Tanabe et al., 2003). However, direct comparisons between the present studies and those performed in the PNS are complicated by the fact that (1) changes in the expression of a particular gene are reported relative to baseline levels of expression, which are likely to differ considerably between cell types; (2) peripheral ganglia comprise several types of neurons that may not be synchronized in their rates of regeneration; (3) the cell type analyzed in this study was nearly homogeneous, whereas studies in the PNS have not separated projection neurons from other cells, the gene expression patterns of which may or may not be changing; and (4) most published PNS studies have either used different microarrays from those used here or have reported a select subset of results. Nonetheless, to a first approximation, the resemblance between the gene expression patterns that characterize the regenerative states of RGCs and peripheral neurons is striking.
Although only a fraction of all RGC axons grow into the distal optic nerve after lens injury, with C3 expression, the number that does so increases to the point that the discontinuity in GAP-43 immunostaining normally seen at the lesion site is obscured (Fig. 8_d_). Likewise, Fischer et al. (2000) showed that up to 40% of RGCs exposed to the effects of lens injury can extend axons through the permissive environment of a peripheral nerve graft. After axotomy and lens injury, nearly all RGCs showed increased expression of proteins encoded by genes associated with the regenerative state. Nonetheless, it remains uncertain whether all RGCs or just a subpopulation go into a strong regenerative state under these circumstances. A related issue is whether our methods selected for a subset of RGCs that show higher survival. In vivo, no cell death was apparent at the time that we dissected retinas, although it is conceivable that certain RGCs are prone to die after dissociation and cell sorting. After 3 hr in serum-free media, the viability of FACS-sorted RGCs diminishes significantly, but this is unlikely to reflect the viability of RGCs preserved for gene expression analysis immediately after FACS.
RGCs exposed to axotomy alone showed many of the same changes in gene expression observed in the regenerative state, although a number of these changes were smaller in magnitude (Table 1) (supplemental material, available at www.jneurosci.org). Some of the differences between the two conditions may help account for the greater outgrowth and survival that occur when axotomized RGCs are exposed to macrophage-derived factors. For example, GAP-43 and SPRR1A, which are differentially expressed in the regenerating condition, contribute directly to axon outgrowth (Benowitz and Routtenberg, 1997; Bomze et al., 2001; Bonilla et al., 2002), whereas proteins such as SOCS-3, sphingosine kinase 1, and tissue inhibitor of metalloproteinase-1 (TIMP-1) are likely to enhance cell survival (La Fleur et al., 1996; Edsall et al., 1997; Raghavendra Rao et al., 2002). In contrast, some of the genes that are upregulated or downregulated to a similar extent in the two states (e.g., transcription factors such as MafK and ATF3) may lay the groundwork for a strong regenerative response, but they are clearly not sufficient. Because lens injury in the absence of axotomy did not stimulate axon outgrowth in retinal explants, we did not investigate changes in gene expression associated with lens injury alone. Thus, it remains possible that some of the changes associated with axotomy plus lens injury result, in part, from the effects of lens injury alone, despite the inadequacy of these changes to cause frank regeneration. Finally, whether axotomized RGCs exposed to appropriate growth factors revert to the same growth state seen during the initial development of retinofugal projections, or whether certain features of the latter state are irreversibly lost (Goldberg et al., 2002b), remains to be determined.
A great deal of attention has been focused on improving regeneration by overcoming inhibitory molecules associated with myelin and the glial scar. We previously showed that transfecting RGCs with a gene encoding a dominant-negative form of NgR was insufficient to cause significant axon regeneration through the mature optic nerve unless RGCs were exposed to the effects of lens injury (Fischer et al., 2004). Along with the inhibitory proteins that signal through NgR (e.g., MAG, OMgp, and the C-terminal region of NogoA), regrowing RGC axons face additional growth inhibitors, including the C-terminal region of NogoA (Spillmann et al., 1998; Fournier et al., 2001), chondroitin-sulfate proteoglycans (Snow et al., 1991; Selles-Navarro et al., 2001), and semaphorin 5A (Oster et al., 2003; Goldberg et al., 2004). RhoA activation is part of the final common pathway activated by all of these (Hall et al., 2001; Hu et al., 2001; Shamah et al., 2001; Dergham et al., 2002; Niederost et al., 2002; Perrot et al., 2002; Monnier et al., 2003; Yamashita and Tohyama, 2003; Gallo and Letourneau, 2004). A second issue addressed in this study was whether RhoA inactivation would be sufficient to produce significant regeneration even when RGCs were not in an active growth state. RhoA was inactivated using a gene therapy approach to express clostridium C3 toxin constitutively in the majority of RGCs. When RGCs were subjected to axotomy alone, C3 expression stimulated a modest amount of regeneration in vivo; these results are consistent with those of a previous study in which C3 protein was introduced into RGC terminals at the time of nerve injury (Lehmann et al., 1999) and with studies showing that RhoA inactivation stimulates modest regeneration of corticospinal tract axons in the injured spinal cord (Dergham et al., 2002; Fournier et al., 2003). However, as shown here, when C3 expression was combined with exposure to appropriate growth factors, the two acted synergistically, resulting in stronger regeneration than has ever been observed in the mature rat optic nerve (cf. Fischer et al., 2004).
Although lens injury, and to a lesser extent C3 expression, increase the survival of RGCs after axotomy, increased survival alone is not likely to be sufficient to account for the increased growth seen under these conditions: complete prevention of cell death by expressing the anti-apoptotic gene bcl-2 does not enable RGCs to regenerate axons in vivo or in culture unless additional factors are added (Chierzi et al., 1999; Goldberg et al., 2002a). In addition, retinas explanted from normal animals, or animals with lens injury or axotomy alone, show high survival within the first several days but fail to regenerate their axons.
Conclusion
After injury to the optic nerve, RGCs exposed to appropriate growth factors show changes in gene expression comparable with those seen in neurons undergoing regeneration in the PNS. Thus, at least one factor that normally limits axon regeneration in the CNS, the inherent growth state of neurons, can be radically altered using appropriate factors. RGCs in an active growth state can regenerate injured axons for considerable distances through the optic nerve, but their growth is still limited by inhibitory signals associated with myelin and the glial scar. Inactivating RhoA represents a general strategy to counteract many inhibitory signals at once: this led to a modest amount of regeneration by itself but greatly potentiated the amount of growth that occurred when the growth state of neurons was activated. These findings support the emerging concept that clinically successful regeneration requires a multi-pronged approach.
Footnotes
This work was supported by the National Institutes of Health (Grant EY05690 to L.I.B.), the German Research Foundation (Deutsche Forschungsgemeinschaft to D.F.), and the Paralyzed Veterans of America (D.F.). We are grateful to Fan Jiang (Children's Hospital, Boston, MA) for technical assistance, Alan Flint (Flow Cytometry Facility, Mental Retardation Research Center, Children's Hospital, Boston, MA) for help with cell sorting, Lloyd Greene (Columbia University, College of Physicians and Surgeons, New York, NY) for antibodies to MafK, Paul Worley (Johns Hopkins University, Baltimore, MD) for reagents to study Narp, and Stephen Strittmatter (Yale University, New Haven, CT) for antibodies to SPRR1A. We thank Paul Rosenberg (Children's Hospital, Harvard Medical School, Boston, MA) and Matthew Hemming (Harvard Medical School, Boston, MA) for helpful comments on this manuscript.
Correspondence should be addressed to Dr. Larry Benowitz, Children's Hospital, 300 Longwood Avenue, Boston, MA 02115. E-mail: larry.benowitz@childrens.harvard.edu.
Copyright © 2004 Society for Neuroscience 0270-6474/04/248726-15$15.00/0
References
- Aguayo AJ, Rasminsky M, Bray GM, Carbonetto S, McKerracher L, Villegas-Perez MP, Vidal-Sanz M, Carter DA (1991) Degenerative and regenerative responses of injured neurons in the central nervous system of adult mammals. Philos Trans R Soc Lond B Biol Sci 331: 337-343. [DOI] [PubMed] [Google Scholar]
- Aldskogius H, Kozlova EN (1998) Central neuron-glial and glial-glial interactions following axon injury. Prog Neurobiol 55: 1-26. [DOI] [PubMed] [Google Scholar]
- Bahr M, Vanselow J, Thanos S (1988) In vitro regeneration of adult rat ganglion cell axons from retinal explants. Exp Brain Res 73: 393-401. [DOI] [PubMed] [Google Scholar]
- Benowitz LI, Routtenberg A (1997) GAP-43: an intrinsic determinant of neuronal development and plasticity. Trends Neurosci 20: 84-91. [DOI] [PubMed] [Google Scholar]
- Benowitz LI, Apostolides PJ, Perrone-Bizzozero N, Finklestein SP, Zwiers H (1988) Anatomical distribution of the growth-associated protein GAP-43/B-50 in the adult rat brain. J Neurosci 8: 339-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkelaar M, Clarke DB, Wang YC, Bray GM, Aguayo AJ (1994) Axotomy results in delayed death and apoptosis of retinal ganglion cells in adult rats. J Neurosci 14: 4368-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry M, Carlile J, Hunter A (1996) Peripheral nerve explants grafted into the vitreous body of the eye promote the regeneration of retinal ganglion cell axons severed in the optic nerve. J Neurocytol 25: 147-170. [DOI] [PubMed] [Google Scholar]
- Bomze HM, Bulsara KR, Iskandar BJ, Caroni P, Skene JH (2001) Spinal axon regeneration evoked by replacing two growth cone proteins in adult neurons. Nat Neurosci 4: 38-43. [DOI] [PubMed] [Google Scholar]
- Bonilla IE, Tanabe K, Strittmatter SM (2002) Small proline-rich repeat protein 1A is expressed by axotomized neurons and promotes axonal outgrowth. J Neurosci 22: 1303-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Sapieha P, Kittlerova P, Hauswirth WW, Di Polo A (2002) TrkB gene transfer protects retinal ganglion cells from axotomy-induced death in vivo. J Neurosci 22: 3977-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chierzi S, Strettoi E, Cenni MC, Maffei L (1999) Optic nerve crush: axonal responses in wild-type and bcl-2 transgenic mice. J Neurosci 19: 8367-8376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung RS, Vickers JC, Chuah MI, West AK (2003) Metallothionein-IIA promotes initial neurite elongation and postinjury reactive neurite growth and facilitates healing after focal cortical brain injury. J Neurosci 23: 3336-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costigan M, Befort K, Karchewski L, Griffin RS, D'Urso D, Allchorne A, Sitarski J, Mannion JW, Pratt RE, Woolf CJ (2002) Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci 3: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dergham P, Ellezam B, Essagian C, Avedissian H, Lubell WD, McKerracher L (2002) Rho signaling pathway targeted to promote spinal cord repair. J Neurosci 22: 6570-6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Polo A, Aigner LJ, Dunn RJ, Bray GM, Aguayo AJ (1998) Prolonged delivery of brain-derived neurotrophic factor by adenovirus-infected Muller cells temporarily rescues injured retinal ganglion cells. Proc Natl Acad Sci USA 95: 3978-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domeniconi M, Cao Z, Spencer T, Sivasankaran R, Wang K, Nikulina E, Kimura N, Cai H, Deng K, Gao Y, He Z, Filbin M (2002) Myelin-associated glycoprotein interacts with the nogo66 receptor to inhibit neurite outgrowth. Neuron 35: 283. [DOI] [PubMed] [Google Scholar]
- Dubreuil CI, Winton MJ, McKerracher L (2003) Rho activation patterns after spinal cord injury and the role of activated Rho in apoptosis in the central nervous system. J Cell Biol 162: 233-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edsall LC, Pirianov GG, Spiegel S (1997) Involvement of sphingosine 1-phosphate in nerve growth factor-mediated neuronal survival and differentiation. J Neurosci 17: 6952-6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes KJL, Tetzlaff W (2001) Gene expression in axotomized neurons: identifying the intrinsic determinants of axon growth. In: Axonal regeneration in the central nervous system (Ingoglia NA, Murray M, eds), p 47. New York: Marcel Dekker.
- Fischer D, Pavlidis M, Thanos S (2000) Cataractogenic lens injury prevents traumatic ganglion cell death and promotes axonal regeneration both in vivo and in culture. Invest Ophthalmol Vis Sci 41: 3943-3954. [PubMed] [Google Scholar]
- Fischer D, Heiduschka P, Thanos S (2001) Lens-injury-stimulated axonal regeneration throughout the optic pathway of adult rats. Exp Neurol 172: 257-272. [DOI] [PubMed] [Google Scholar]
- Fischer D, He Z, Benowitz LI (2004) Counteracting the Nogo receptor enhances optic nerve regeneration if retinal ganglion cells are in an active growth state. J Neurosci 24: 1646-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier AE, GrandPre T, Strittmatter SM (2001) Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature 409: 341-346. [DOI] [PubMed] [Google Scholar]
- Fournier AE, Takizawa BT, Strittmatter SM (2003) Rho kinase inhibition enhances axonal regeneration in the injured CNS. J Neurosci 23: 1416-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo G, Letourneau PC (2004) Regulation of growth cone actin filaments by guidance cues. J Neurobiol 58: 92-102. [DOI] [PubMed] [Google Scholar]
- Goldberg JL, Espinosa JS, Xu Y, Davidson N, Kovacs GT, Barres BA (2002a) Retinal ganglion cells do not extend axons by default: promotion by neurotrophic signaling and electrical activity. Neuron 33: 689-702. [DOI] [PubMed] [Google Scholar]
- Goldberg JL, Klassen MP, Hua Y, Barres BA (2002b) Amacrine-signaled loss of intrinsic axon growth ability by retinal ganglion cells. Science 296: 1860-1864. [DOI] [PubMed] [Google Scholar]
- Goldberg JL, Vargas ME, Wang JT, Mandemakers W, Oster SF, Sretavan DW, Barres BA (2004) An oligodendrocyte lineage-specific semaphorin, Sema5A, inhibits axon growth by retinal ganglion cells. J Neurosci 24: 4989-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Zulueta M, Ensz LM, Mukhina G, Lebovitz RM, Zwacka RM, Engelhardt JF, Oberley LW, Dawson VL, Dawson TM (1998) Manganese superoxide dismutase protects nNOS neurons from NMDA and nitric oxide-mediated neurotoxicity. J Neurosci 18: 2040-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C, Brown M, Jacobs T, Ferrari G, Cann N, Teo M, Monfries C, Lim L (2001) Collapsin response mediator protein switches RhoA and Rac1 morphology in N1E-115 neuroblastoma cells and is regulated by Rho kinase. J Biol Chem 276: 43482-43486. [DOI] [PubMed] [Google Scholar]
- Hu H, Marton TF, Goodman CS (2001) Plexin B mediates axon guidance in Drosophila by simultaneously inhibiting active Rac and enhancing RhoA signaling. Neuron 32: 39-51. [DOI] [PubMed] [Google Scholar]
- Jung M, Petrausch B, Stuermer CA (1997) Axon-regenerating retinal ganglion cells in adult rats synthesize the cell adhesion molecule L1 but not TAG-1 or SC-1. Mol Cell Neurosci 9: 116-131. [DOI] [PubMed] [Google Scholar]
- Keller JN, Kindy MS, Holtsberg FW, St Clair DK, Yen HC, Germeyer A, Steiner SM, Bruce-Keller AJ, Hutchins JB, Mattson MP (1998) Mitochondrial manganese superoxide dismutase prevents neural apoptosis and reduces ischemic brain injury: suppression of peroxynitrite production, lipid peroxidation, and mitochondrial dysfunction. J Neurosci 18: 687-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeberle PD, Ball AK (1998) Effects of GDNF on retinal ganglion cell survival following axotomy. Vision Res 38: 1505-1515. [DOI] [PubMed] [Google Scholar]
- Kumagai N, Morii N, Fujisawa K, Nemoto Y, Narumiya S (1993) ADP-ribosylation of rho p21 inhibits lysophosphatidic acid-induced protein tyrosine phosphorylation and phosphatidylinositol 3-kinase activation in cultured Swiss 3T3 cells. J Biol Chem 268: 24535-24538. [PubMed] [Google Scholar]
- La Fleur M, Underwood JL, Rappolee DA, Werb Z (1996) Basement membrane and repair of injury to peripheral nerve: defining a potential role for macrophages, matrix metalloproteinases, and tissue inhibitor of metalloproteinases-1. J Exp Med 184: 2311-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M, Fournier A, Selles-Navarro I, Dergham P, Sebok A, Leclerc N, Tigyi G, McKerracher L (1999) Inactivation of Rho signaling pathway promotes CNS axon regeneration. J Neurosci 19: 7537-7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon S, Yin Y, Nguyen J, Irwin N, Benowitz LI (2000) Lens injury stimulates axon regeneration in the mature rat optic nerve. J Neurosci 20: 4615-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wong L (2001) Emerging patterns and gene expression data. Genome Inform Ser Workshop Genome Inform 12: 3-13. [PubMed] [Google Scholar]
- Li Y, Irwin N, Yin Y, Lanser M, Benowitz LI (2003) Axon regeneration in goldfish and rat retinal ganglion cells: differential responsiveness to carbohydrates and cAMP. J Neurosci 23: 7830-7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Fisher DA, Storm DR (1994) Intracellular sorting of neuromodulin (GAP-43) mutants modified in the membrane targeting domain. J Neurosci 14: 5807-5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson S, Ekstrom TJ, Elmer E, Kanje M, Ny L, Alm P (2000) Heme oxygenase-1, heme oxygenase-2 and biliverdin reductase in peripheral ganglia from rat, expression and plasticity. Neuroscience 95: 821-829. [DOI] [PubMed] [Google Scholar]
- Martin KR, Klein RL, Quigley HA (2002) Gene delivery to the eye using adeno-associated viral vectors. Methods 28: 267-275. [DOI] [PubMed] [Google Scholar]
- Monnier PP, Sierra A, Schwab JM, Henke-Fahle S, Mueller BK (2003) The Rho/ROCK pathway mediates neurite growth-inhibitory activity associated with the chondroitin sulfate proteoglycans of the CNS glial scar. Mol Cell Neurosci 22: 319-330. [DOI] [PubMed] [Google Scholar]
- Nakagomi S, Kiryu-Seo S, Kiyama H (2000) Endothelin-converting enzymes and endothelin receptor B messenger RNAs are expressed in different neural cell species and these messenger RNAs are coordinately induced in neurons and astrocytes respectively following nerve injury. Neuroscience 101: 441-449. [DOI] [PubMed] [Google Scholar]
- Niederost B, Oertle T, Fritsche J, McKinney RA, Bandtlow CE (2002) Nogo-A and myelin-associated glycoprotein mediate neurite growth inhibition by antagonistic regulation of RhoA and Rac1. J Neurosci 22: 10368-10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster SF, Bodeker MO, He F, Sretavan DW (2003) Invariant Sema5A inhibition serves an ensheathing function during optic nerve development. Development 130: 775-784. [DOI] [PubMed] [Google Scholar]
- Perrot V, Vazquez-Prado J, Gutkind JS (2002) Plexin B regulates Rho through the guanine nucleotide exchange factors leukemia-associated Rho GEF (LARG) and PDZ-RhoGEF. J Biol Chem 277: 43115-43120. [DOI] [PubMed] [Google Scholar]
- Petrausch B, Tabibiazar R, Roser T, Jing Y, Goldman D, Stuermer CA, Irwin N, Benowitz LI (2000) A purine-sensitive pathway regulates multiple genes involved in axon regeneration in goldfish retinal ganglion cells. J Neurosci 20: 8031-8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra Rao VL, Bowen KK, Dhodda VK, Song G, Franklin JL, Gavva NR, Dempsey RJ (2002) Gene expression analysis of spontaneously hypertensive rat cerebral cortex following transient focal cerebral ischemia. J Neurochem 83: 1072-1086. [DOI] [PubMed] [Google Scholar]
- Ramon y Cajal S (1991) Degeneration and regeneration of the nervous system. New York: Oxford UP.
- Rosenfeld J, Cook S, James R (1997) Expression of superoxide dismutase following axotomy. Exp Neurol 147: 37-47. [DOI] [PubMed] [Google Scholar]
- Selles-Navarro I, Ellezam B, Fajardo R, Latour M, McKerracher L (2001) Retinal ganglion cell and nonneuronal cell responses to a microcrush lesion of adult rat optic nerve. Exp Neurol 167: 282-289. [DOI] [PubMed] [Google Scholar]
- Shamah SM, Lin MZ, Goldberg JL, Estrach S, Sahin M, Hu L, Bazalakova M, Neve RL, Corfas G, Debant A, Greenberg ME (2001) EphA receptors regulate growth cone dynamics through the novel guanine nucleotide exchange factor ephexin. Cell 105: 233-244. [DOI] [PubMed] [Google Scholar]
- Shewan D, Berry M, Cohen J (1995) Extensive regeneration in vitro by early embryonic neurons on immature and adult CNS tissue. J Neurosci 15: 2057-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow DM, Watanabe M, Letourneau PC, Silver J (1991) A chondroitin sulfate proteoglycan may influence the direction of retinal ganglion cell outgrowth. Development 113: 1473-1485. [DOI] [PubMed] [Google Scholar]
- Spillmann AA, Bandtlow CE, Lottspeich F, Keller F, Schwab ME (1998) Identification and characterization of a bovine neurite growth inhibitor (bNI-220). J Biol Chem 273: 19283-19293. [DOI] [PubMed] [Google Scholar]
- Tanabe K, Bonilla I, Winkles JA, Strittmatter SM (2003) Fibroblast growth factor-inducible-14 is induced in axotomized neurons and promotes neurite outgrowth. J Neurosci 23: 9675-9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenfeld SM, Milekic MH, Monti B, Alberini CM (2001) The consolidation of new but not reactivated memory requires hippocampal C/EBP-beta. Nat Neurosci 4: 813-818. [DOI] [PubMed] [Google Scholar]
- Thibault C, Lai C, Wilke N, Duong B, Olive MF, Rahman S, Dong H, Hodge CW, Lockhart DJ, Miles MF (2000) Expression profiling of neural cells reveals specific patterns of ethanol-responsive gene expression. Mol Pharmacol 58: 1593-1600. [DOI] [PubMed] [Google Scholar]
- Torocsik B, Angelastro JM, Greene LA (2002) The basic region and leucine zipper transcription factor MafK is a new nerve growth factor-responsive immediate early gene that regulates neurite outgrowth. J Neurosci 22: 8971-8980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujino H, Kondo E, Fukuoka T, Dai Y, Tokunaga A, Miki K, Yonenobu K, Ochi T, Noguchi K (2000) Activating transcription factor 3 (ATF3) induction by axotomy in sensory and motoneurons: a novel neuronal marker of nerve injury. Mol Cell Neurosci 15: 170-182. [DOI] [PubMed] [Google Scholar]
- Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH (1990) Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci USA 87: 1663-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielmetter J, Lottspeich F, Stuermer CA (1991) The monoclonal antibody E587 recognizes growing (new and regenerating) retinal axons in the goldfish retinotectal pathway. J Neurosci 11: 3581-3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Koprivica V, Kim JA, Sivasankaran R, Guo Y, Neve RL, He Z (2002a) Oligodendrocyte-myelin glycoprotein is a Nogo receptor ligand that inhibits neurite outgrowth. Nature 417: 941-944. [DOI] [PubMed] [Google Scholar]
- Wang KC, Kim JA, Sivasankaran R, Segal R, He Z (2002b) p75 interacts with the Nogo receptor as a co-receptor for Nogo, MAG and OMgp. Nature 420: 74-78. [DOI] [PubMed] [Google Scholar]
- Xiao HS, Huang QH, Zhang FX, Bao L, Lu YJ, Guo C, Yang L, Huang WJ, Fu G, Xu SH, Cheng XP, Yan Q, Zhu ZD, Zhang X, Chen Z, Han ZG (2002) Identification of gene expression profile of dorsal root ganglion in the rat peripheral axotomy model of neuropathic pain. Proc Natl Acad Sci USA 99: 8360-8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, Tohyama M (2003) The p75 receptor acts as a displacement factor that releases Rho from Rho-GDI. Nat Neurosci 6: 461-467. [DOI] [PubMed] [Google Scholar]
- Yin Y, Cui Q, Li Y, Irwin N, Fischer D, Harvey AR, Benowitz LI (2003) Macrophage-derived factors stimulate optic nerve regeneration. J Neurosci 23: 2284-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelyaznik N, Schrage K, McCaffery P, Mey J (2003) Activation of retinoic acid signalling after sciatic nerve injury: up-regulation of cellular retinoid binding proteins. Eur J Neurosci 18: 1033-1040. [DOI] [PubMed] [Google Scholar]
- Zigmond RE, Sun Y (1997) Regulation of neuropeptide expression in sympathetic neurons. Paracrine and retrograde influences. Ann NY Acad Sci 814: 181-197. [DOI] [PubMed] [Google Scholar]
- Zuber MX, Strittmatter SM, Fishman MC (1989) A membrane-targeting signal in the amino terminus of the neuronal protein GAP-43. Nature 341: 345-348. [DOI] [PubMed] [Google Scholar]