The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112 (original) (raw)
Abstract
N6-methyladenosine (m6A) has recently been found abundantly on messenger RNA and shown to regulate most steps of mRNA metabolism. Several important m6A methyltransferases have been described functionally and structurally, but the enzymes responsible for installing one m6A residue on each subunit of human ribosomes at functionally important sites have eluded identification for over 30 years. Here, we identify METTL5 as the enzyme responsible for 18S rRNA m6A modification and confirm ZCCHC4 as the 28S rRNA modification enzyme. We show that METTL5 must form a heterodimeric complex with TRMT112, a known methyltransferase activator, to gain metabolic stability in cells. We provide the first atomic resolution structure of METTL5–TRMT112, supporting that its RNA-binding mode differs distinctly from that of other m6A RNA methyltransferases. On the basis of similarities with a DNA methyltransferase, we propose that METTL5–TRMT112 acts by extruding the adenosine to be modified from a double-stranded nucleic acid.
INTRODUCTION
Faithful synthesis of proteins from mRNA templates is performed by ribosomes. These are highly complex nanomachines composed, in eukaryotes, of four ribosomal RNAs (rRNAs) and 80 ribosomal proteins (1,2), assisted by translation factors and transfer RNAs (tRNAs). For optimal cell growth, human cells are estimated to synthesize about 7500 ribosomal subunits per minute, thanks to a highly coordinated choreography of more than 250 trans-acting factors that transiently associate with maturing ribosomal particles from the early step of rDNA transcription by RNA polymerases I and III in the nucleus to the final maturation steps in the cytoplasm (3,4). This plethora of trans-acting factors is required for the synthesis, maturation, export and assembly of the ribosomal subunits.
Dysfunction of several ribosomal proteins and ribosome assembly factors is associated with pathologies called ribosomopathies, often associated with cancers (5,6). While some factors have scaffolding or remodeling roles, others are directly involved in pre-rRNA processing (endonucleolytic cleavage, 5′ or 3′ trimming) or post-transcriptional modification (methylation, acetylation, …), two highly coordinated mechanisms (7). With recent technological developments such as next-generation sequencing, highly sensitive mass spectrometry and atomic-resolution cryo-electron microscopy (cryo-EM), our current knowledge on rRNA and ribosomal protein modification is rapidly increasing (8–11).
The complete set of rRNA modifications has been described very recently for the 80S human ribosome, revealing the existence of 228 modifications of 14 different types (10). Most of these modifications are pseudouridylations and 2’-OH methylations catalyzed, respectively, by H/ACA and C/D box snoRNP complexes (12), but other, generally highly conserved modifications (methylation and acetylation) occur on bases (13). The most highly conserved modifications cluster at, or in the vicinity of important functional sites on the ribosome, including the decoding center, the peptidyl transferase center, the mRNA- and tRNA-binding sites, the entry of the peptide exit tunnel and the subunit interface (13).
Nucleotide modification is emerging as an important source of ribosome heterogeneity (7), consistent with the idea that cells produce compositionally different ribosomes with specialized functions in translation (14,15). Yet the roles played by such modifications remain largely unclear, and identification of the enzymes responsible for depositing them lags far behind the discovery of the modifications themselves. Recently, progress has been made thanks to concerted efforts and the use of various model organisms such as Saccharomyces cerevisiae and archaea. In particular and very recently, a complete view has been obtained of the enzymes responsible for forming known 18S and 25S rRNA base modifications in S. cerevisiae (13,16). This is rapidly leading to the study of their human orthologs.
Despite recent advances, the enzymes responsible for a few base modifications on human rRNAs have continued to elude identification. This applies, for example, to the N6-methyl-adenine (m6A) modifications at positions 1832 on human 18S rRNA and position 4220 (formerly 4190) on human 28S rRNA, both specifically detected over 30 years ago by RNase T1 and RNase A digestion of rRNAs isolated from human cells and other vertebrates (17,18). The 18S rRNA m6A1832 modification is located in the 3′ minor domain of 18S, at the very base of helix h44, only a few nucleotides away from the decoding center (Figure 1A). The 28S rRNA m6A4220 modification is located in helix H81 of 28S rRNA domain V (Figure 1B). The enzyme involved in depositing the 28S rRNA m6A4220 mark has been identified very recently as ZCCHC4 (19), and we have confirmed it in this work. The enzymatic activity responsible for 18S rRNA m6A modification has escaped identification so far.
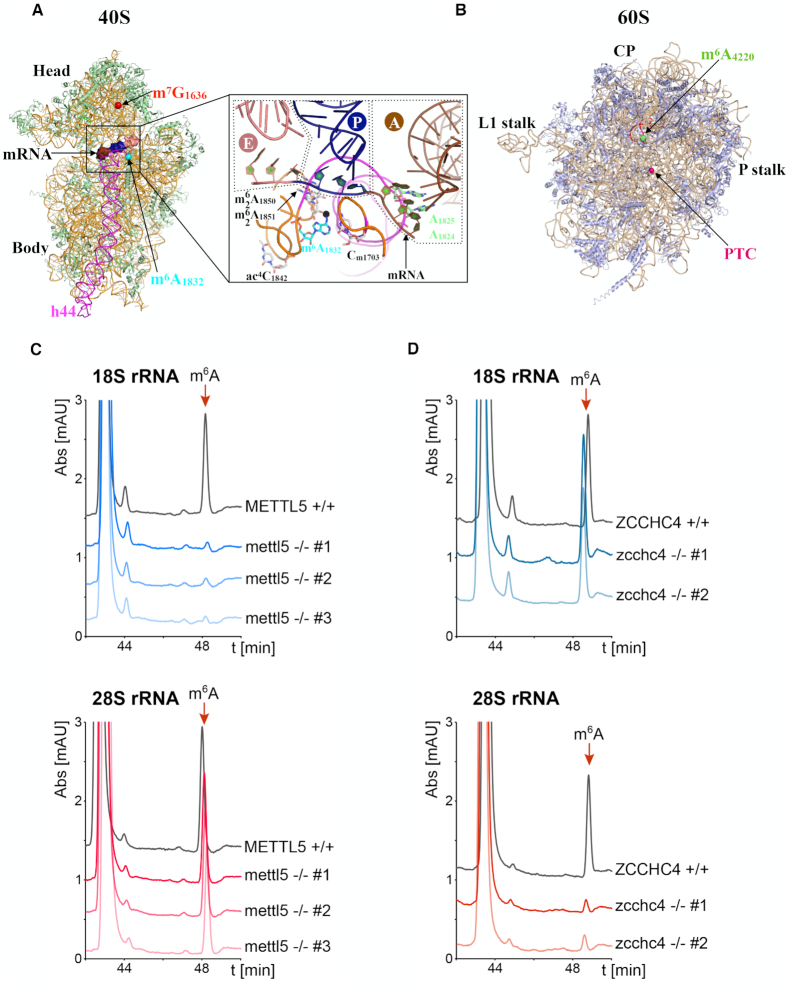
Figure 1.
METTL5 and ZCCHC4 are, respectively, the 18S and 28S rRNA m6A methyltransferases. (A) Ribbon representation of the human 40S subunit (11). Ribosomal proteins and 18S rRNA are colored light green and orange, respectively. Helix 44 (h44) of 18S rRNA is highlighted in magenta. The locations of m7G1636 and m6A1832 rRNA modifications are shown with red and cyan spheres, respectively. A 9-nt mRNA was modeled into the mRNA channel by superimposing the cryo-EM structure of mammalian ribosomal elongation complex bound to eEF1a and tRNAs (74). The mRNA codons corresponding to the A-, P- and E-sites are colored brown, blue and pink, respectively. Inset: Close-up view of the decoding site with some modified nucleotides highlighted. The anticodon loop of the tRNAs bound to the A-, P- and E-sites (delineated by dashed lines) are shown in brown, blue and pink, respectively. The methyl group of the m6A1832 nucleotide is shown as a black sphere. The tRNAs were modeled by superimposing the cryo-EM structure of the mammalian ribosomal elongation complex bound to eEF1a and tRNAs (74). (B) Ribbon representation of the human 60S subunit (11). Ribosomal proteins and 28S rRNA are colored light blue and beige, respectively. The respective locations of the m6A4220 rRNA modification and peptidyl transferase center (PTC) are shown as green and magenta spheres. CP: Central protuberance. Helix H81 of 28S rRNA is highlighted in red. (C) Human METTL5 is necessary for the formation of m6A on 18S but not on 28S rRNA. HPLC chromatograms (A254nm) of 18S rRNA (top panel) and 28S rRNA (lower panel) purified from mettl5 -/- clones and digested to nucleosides. The digested rRNAs purified from HCT116 parental cells (black curve) were used as controls. (D) Human ZCCHC4 is necessary for the formation of m6A on 28S but not on 18S rRNA. HPLC chromatograms (A_254 nm) of 18S rRNA (top panel) and 28S rRNA (lower panel) purified from z_cchc4 -/- clones and digested to nucleosides. The digested rRNAs purified from HCT116 parental cells (black curve) were used as controls.
In the present study, we have identified METTL5 as the enzyme responsible for the 18S rRNA m6A1832. We show that METTL5 works in close association with TRMT112 and is stabilized by it. We have established the atomic-resolution structure of the METTL5–TRMT112 complex, revealing that, in a manner similar to the action of DNA methyltransferases, it may act by extruding the substrate adenosine from a nucleic acid duplex.
MATERIALS AND METHODS
Human cell culture
Unless specified otherwise, the experiments were performed in HCT116 p53-positive colon carcinoma cells (ATCC, #CCL-247). The cell line was diagnosed by ATCC, by short tandem repeat (STR) analysis, prior to use. Cells were cultured in McCoy’s 5A Medium (Lonza™ BE12-168F) in a New Brunswick Galaxy 170R incubator at 37°C and under 5% CO2.
For recording growth curves, the cells were detached and counted in a hemocytometer 24, 48 and 72 h after staining with Trypan Blue (Sigma, #T8154). All measurements were performed in triplicate.
Generation of human cell lines deleted of METTL5 or ZCCHC4 by CRISPR–Cas9 genome editing
To delete from the genome the exon of METTL5 or ZCCHC4 carrying the methyltransferase signature, CRISPR guide RNAs (crRNA) were designed using http://crispr.mit.edu (20) near the exon–intron junctions. crRNAs with the best off-target scores (Supplementary Table S1), tracRNA (IDT, #1072532) and S.p. Cas9 (IDT, #1081058) were purchased from Integrated DNA Technologies (IDT). Cells were transfected with in vitro reconstituted crRNA:tracRNA:Cas9 complex (final concentration 3 μM) and electroporation enhancer (IDT, #1075915, final concentration 4 μM) in a nucleofector device (both versions IIb and 4D were used). Cells were incubated for 24 h to allow them to recover and then detached, and cloned (2D dilution series on 96-well plates). Cells from wells containing a single colony were detached, split into replicate 96-well plates and further incubated. Genomic DNA (gDNA) was extracted from one plate once cells reached confluence. By diagnostic PCR (Supplementary Table S2), each target sequence with a deletion was detected. The PCR products were sequenced to establish the exact junctions of each deletion (Supplementary Table S3).
Effect of TRMT112 on METTL5 metabolic stability in human cells
A human cell line expressing an inducible METTL5-Flag construct was generated. For this, the coding sequence of METTL5 (NM_014168.4) was cloned into a pcDNA5-based vector for expression of a C-terminally Flag-tagged protein. The plasmid was used to transfect HEK293 Flp-In T-Rex cells (Thermo Fisher Scientific) and stably transfected cells were selected according to the manufacturer’s instructions. In the selected cell line, METTL5-Flag was shown to be induced strictly upon addition of tetracycline. A tetracycline concentration between 0.01 and 0.1 μg/ml was sufficient to induce METTL5-Flag expression. To test the effect of TRMT112 on METTL5 metabolic stability in human cells, cells of the above-described cell line were depleted of TRMT112 for 3 days using an siRNA (siRNA LD068, Supplementary Table S1). METTL5-Flag expression from the tetracycline-inducible promoter was triggered by the addition of tetracycline (between 0.01 and 0.1 μg/ml was used) 24 h prior to cell harvesting.
Analysis of modified nucleosides by HPLC
The 18S and 28S rRNAs were purified from HCT116 cells and the mettl5 -/- and zcchc4 -/- clones by 10–50% sucrose velocity centrifugation. Briefly, 6 × 107 cells were grown, detached, washed in ice-cold PBS and resuspended in 500 μl Buffer A: 20 mM Tris-HCl, pH 7.4, 50 mM KCl, 1 mM DTT, 0.5% NP40, Complete™ Protease Inhibitor (Roche #COEDTAF-RO). The cell suspension was centrifuged at 20 000 × g for 20 min at 4°C, and the supernatant was stored at −80°C. Extracts were thawed on ice and loaded on a 10–50% sucrose density gradient (buffer A without protease inhibitor and DTT) and centrifuged at 23 000 × g for 20 h in a Beckman L-90K centrifuge with a SW41Ti rotor. The gradient was fractionated with an Isco density gradient fraction collector. RNA was extracted from peak fractions with TRIreagent (Sigma #T9424), and an aliquot of recovered RNA was loaded on a denaturing agarose gel to check for RNA integrity and purity. Pure RNA fractions were pooled and the concentration was determined with a NanoDrop instrument. About 30 μg of 18S rRNA or 60 μg 28S rRNA was denatured at 95°C and snap cooled on ice. The RNA was then digested to single nucleotides for 16 h at 37°C in 6 mM sodium acetate/20 mM ZnSO4, pH 5.4 with 2 U of Nuclease P1 (Sigma, #N8630) (total volume 50 μl). Subsequently, 10 μl Tris buffer (0.5M, pH 8.3) and 10 μl alkaline phosphatase (Sigma, #P4252) were added and further incubated at 37°C for 2 h. HPLC analysis was performed essentially as described previously (21). Briefly, 50 μl digested nucleotides were analyzed on an Agilent 1220 Infinity LC system with a Supelcosil LC-18S column (Sigma, #58298). The elution buffers were buffer A (0.01 M ammonium phosphate, pH 5.3, 2.5% methanol) and buffer B (0.01 M ammonium phosphate, pH 5.1, 20% methanol) and the elution program was (_A_[%]/_B_[%]/time[min]): 100/0/0, 100/0/12, 90/10/20, 75/25/25, 40/60/33, 36/64/37, 0/100/45, 100/0/60. Between each step, a continuous gradient was used. The m6A calibration control was purchased from Berry & Associates (#PR 3732).
Crystallization and structure determination
Two crystal forms of the human METTL5–TRMT112 complex were obtained in the same drops at 4°C by mixing 1 μl of protein complex (2.7 mg/ml in Buffer L200) pre-incubated with S-adenosyl-L-methionine (0.75 mM) and 1 μl of reservoir solution (25% PEG 4000 in 0.2 M ammonium sulfate, 100 mM Na citrate pH 5.6). Crystals were cryo-protected by transfer to their crystallization medium supplemented with 15% and then 30% ethylene glycol before being flash-frozen in liquid nitrogen.
Several datasets were collected at 100K on Proxima 1 and Proxima 2A at synchrotron SOLEIL (Saint-Aubin, France) and processed with the XDS and AIMLESS programs (22,23). Crystal forms I and II diffracted, respectively, down to 1.6 Å (one copy of the complex in the asymmetric unit) and 2.5 Å (two copies of the complex in the asymmetric unit).
The structure of the METTL5–TRMT112 complex was solved by molecular replacement with the program PHASER (24), using the dataset collected from crystal form I. Models of the METTL5 and TRMT112 protein structures were generated by the PHYRE2 server (25) using PDB codes, 1WY7 (39% sequence identity) and 5CM2 (35% sequence identity), as templates, respectively. The METTL5 and TRMT112 models were superimposed, respectively, onto the Bud23 and the Trm112 protein in the structure of the S. cerevisiae Bud23–Trm112 complex (26) and residues present in long loops were deleted to generate the search model used for molecular replacement. The final model for crystal form I was obtained by iterative cycles of building with COOT (27) and refinement with BUSTER (28) (for final statistics, see Supplementary Table S6). This model encompasses residues 3 to 190 and 198 to 209 (plus 2 histidine residues of the C-terminal His6-tag) for METTL5, and residues 1 to 118 for TRMT112, one SAM molecule, 5 ethylene glycol molecules, 2 sulfate ions and 320 water molecules. On the basis of electron density map analyses, METTL5 residues Cys38 and Cys110 and TRMT112 residue Cys33 were modeled, respectively, as s-hydroxycysteine, s-cysteinesulfinic acid and s-hydroxycysteine.
The structure of crystal form II was determined by molecular replacement, using the high-resolution structure of the METTL5–TRMT112 complex. The final model was obtained with the COOT and BUSTER programs, as described above. The model obtained contains residues 2 to 209 (plus 2 histidine residues of the C-terminal His6-tag) for METTL5 protomer A, residues 3–129, 136–188 and 198–209 for METTL5 protomer C, residues 1–12 and 20–118 (chain B) and 1–12 and 21–123 (chain D) for TRMT112, two SAM molecules, fragments of 3 PEG molecules, 2 ethylene glycol molecules, 7 sulfate ions and 28 water molecules. On the basis of electron density map analyses, TRMT112 Cys33 was modeled as s,s-(2-hydroxyethyl)thiocysteine, resulting from covalent adduction of 2-mercaptoethanol to the cysteine thiol in both TRMT112 molecules present in the asymmetric unit.
The atomic coordinates and structure factors have been deposited in the Brookhaven Protein Data Bank under accession numbers 6H2U (Form I) and 6H2V (Form II).
Transcriptome-wide single-nucleotide mapping of m6A by miCLIP
Total RNA from wild-type, _mettl5-_knockout and _zcchc4-_knockout HCT116 cells was extracted in Tri Reagent (AM9738, Thermo-Fisher Scientific). Any contaminating genomic DNA was degraded with DNase I, and poly(A) RNA was purified with an mRNA isolation kit (NEB #S1550) according to the manufacturer’s recommendations. Five micrograms of poly(A)-selected RNA was then used as input for single-nucleotide resolution m6A mapping according to the miCLIP protocol, as previously reported (29). Final libraries were amplified and subjected to 50-cycle paired-end sequencing on an Illumina HiSeq2500 at the Weill Cornell Medicine Epigenetic Core facility.
Additional materials and methods details are available in the Supplementary Data.
RESULTS
Identification of the methyltransferases responsible for deposition of m6A1832 and m6A4220 on human 18S and 28S rRNAs
Although the presence of one m6A modification on each ribosomal subunit in metazoans was described more than 30 years ago (17,18,30), the enzymes responsible for depositing these methylation marks have remained unknown until now. Indeed, although methyltransferases constitute one of the largest enzyme classes in nature, the cellular substrates and functions of over 200 candidates remain unknown.
To identify the methyltransferases responsible for depositing m6A at A1832 on 18S rRNA and A4220 on 28S rRNA, we searched the UniProt database (31) for human proteins harboring an N6-adenosine-specific DNA methyltransferase signature ([LIVMAC]-[LIVFYWA]-(DYP)-[DN]-P-P-[FYW]; Prosite entry number: PS00092; (32)). The rationale for doing so was that this signature is found in enzymes that methylate planar amino groups such as glutamine side chains in proteins (i.e.E. coli PrmC, eukaryotic Mtq2/HEMK2 (33–36)) and exocyclic amino groups on DNA and RNA (forming m2G, m4C and m6A modifications; (37)). This search resulted in seven hits including: (i) the HEMK1 and HEMK2 proteins, which methylate, respectively, the mitochondrial Mrf1 and cytoplasmic eRF1 translation termination factors on the side chain of a glutamine of a universally conserved GGQ motif (33,34,38); (ii) TRMT11, the human ortholog of the m2G10 tRNA methyltransferase; (iii) MGAM2, a membrane protein annotated as a probable maltase–glucoamylase and (iv) METTL4, METTL5 and ZCCHC4.
According to the Human Protein Atlas project (39), METTL4 is a mitochondrial protein, whereas in U2-OS human cells, METTL5 and ZCCHC4 localize to the nucleoli and nucleus, respectively (see www.proteinatlas.org), i.e. the cell compartments where the initial steps of ribosome biogenesis take place. Furthermore, human METTL5 has been shown to bind specifically to RNA but not DNA (40). On the basis of phylogenetic analyses, the HVO_1475 gene encoding the Haloferax volcanii METTL5 ortholog is proposed to be responsible for m6A formation at position 1432 on archaeal 16S rRNA (41,42), corresponding to A1832 in human 18S rRNA. Altogether, these facts led us to investigate whether METTL5 and ZCCHC4 might be the enzymes responsible for m6A deposition on human rRNAs.
To test this hypothesis, we obliterated part of the coding sequence of each of these proteins on both alleles of diploid human cells (HCT116) by CRISPR–Cas9 genome editing. Specifically, we removed exon 3 from METTL5 and exon 7 from ZCCHC4, as both exons encode the [N/D]PPF signature of the putative methyltransferase domain (see Supplementary Figure S1A and B). To avoid known problems associated with off-target effects, we choose to expose cells only transiently to in vitro assembled CRISPR-Cas9 ribonucleoprotein particles (RNPs) (see ‘Materials and Methods’ and ‘Discussion’ section). Three independent mettl5 -/- cell lines (#1, #2 and #3) and two independent zcchc4 -/- cell lines (#1 and #2) were isolated and the deletion of both alleles was validated by PCR on genomic DNA followed by genome sequencing in the edited area (Supplementary Figure S1A and B; Supplementary Tables S1–S3 for details).
To test for a role of METTL5 and ZCCH4 in ribosomal RNA modification, mature 18S and 28S rRNAs were purified by velocity centrifugation from the different mettl5- and _zcchc4-_deleted cell lines and from isogenic HCT116 control cells (see ‘Materials and Methods’ section). They were then digested to nucleosides and analyzed by quantitative HPLC (Figure 1C and D). In control cells, the m6A nucleoside was readily detected in both purified 18S and 28S, eluting just after 48 min (Figure 1C and D) as expected (Supplementary Figure S1C). In all three mettl5 -/- clones, interestingly, the 18S rRNA m6A peak was lost, while the 28S rRNA m6A peak remained unaffected (Figure 1C). Conversely, in both zcchc4 -/- clones, the 18S rRNA m6A peak was unchanged while the 28S rRNA m6A peak disappeared (Figure 1D).
We conclude that METTL5 and ZCCHC4 are the elusive human methyltransferases responsible, respectively, for the formation of m6A1832 on 18S rRNA and m6A4220 on 28S rRNA. This conclusion is further supported by the structural analysis of METTL5 (see below, Figures 2C,D and 3A,B) and by transcriptome-wide single-nucleotide-resolution mapping of the m6A modification in the mettl5- and _zcchc4_-knockout cell lines (see below, Figure 4C). Our identification of ZCCHC4 as the 28S rRNA m6A-modifying activity is fully consistent with a recent report (19).
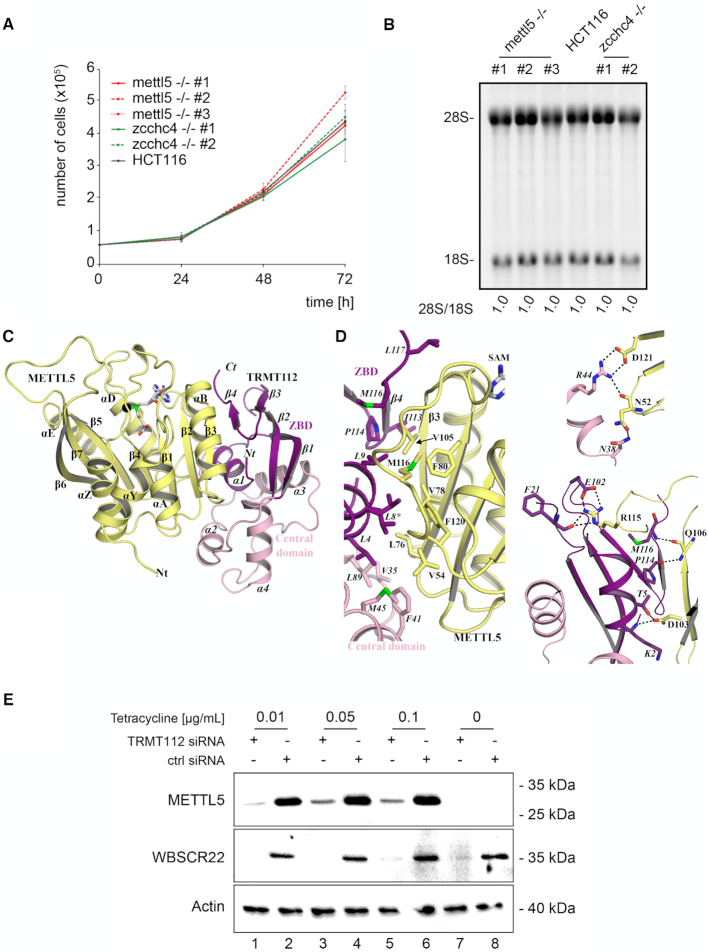
Figure 2.
METTL5 and ZCCHC4 are not essential to growth or ribosome biogenesis, and METTL5 gains metabolic stability by associating with human TRMT112. (A) Cell growth is not markedly affected in the absence of METTL5 or ZCCHC4. An identical number of cells of the indicated cell lines was seeded, and cell growth monitored every 24 h for 3 days by cell counting in an hemocytometer after staining with a vital stain. The experiment was performed in triplicate (s.d. shown in gray). (B) Mature rRNA production is not affected in the absence of METTL5 or ZCCHC4. Total RNA extracted from the indicated cell lines was resolved on a denaturing agarose gel and stained with ethidium bromide to reveal the mature 18S and 28S rRNAs. The 28S/18S ratio was extracted from Bioanalyzer electropherograms. (C) Atomic-resolution structure of the human METTL5–TRMT112 complex. The TRMT112 zinc-binding domain (ZBD) and its helical central domain are colored in purple and pink, respectively. The SAM molecule bound to METTL5 is shown as gray sticks. The methyl group transferred from SAM to the RNA substrate upon catalysis is depicted as a black sphere. Secondary structure elements of TRMT112 are labeled in italics. (D) Detailed representations of the METTL5–TRMT112 interface. Residues involved in the interface are shown as sticks. Residues of TRMT112 are labeled in italics. Asterisks (*) indicate residues adopting two alternative conformations according to the 2Fo-Fc electron density map. The top panel shows the hydrophobic core of the interface, while the two other panels show the electrostatic interactions. Hydrogen bonds and salt bridges are depicted by black dashed lines. Same color code as in panel (C). (E) The metabolic stability of METTL5 depends on its association with TRMT112. Western blot analysis of the steady-state accumulation of METTL5 in the presence and absence of TRMT112. Synthesis of Flag-tagged METTL5 was induced by adding tetracycline (see ‘Materials and Methods’ section). TRMT112 synthesis was knocked down with a specific siRNA. Duplicated blots were probed with anti-Flag (METTL5 detection) or anti-WBSCR22 antibodies. As a loading control, the blot was probed with an anti-actin antibody.
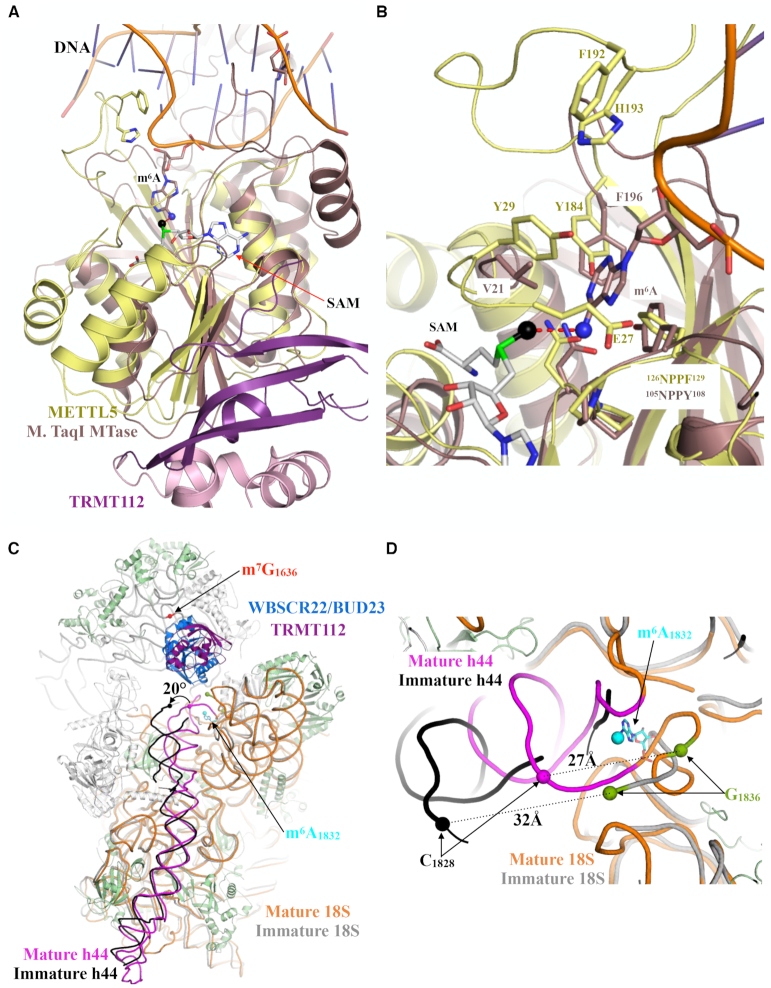
Figure 3.
Comparison of the active sites of METTL5–TRMT112 and of an N6-adenosine DNA methyltransferase from Thermus aquaticus, and structure of the m6A-methylated area in pre-40S and mature 40S subunits. (A) Superimposition of the METTL5–TRMT112 complex onto the M. TaqI–DNA complex (PDB code: 1G38). The m6A protruding into the active site of the M. TaqI methyltransferase is shown as sticks, and the N6 atom, on which the methyl group is grafted, is shown as a blue sphere. The DNA backbone is shown in orange. (B) Zoom-in on the active sites of METTL5 (yellow) and M. TaqI (light brown) (C) Superimposition of mature 40S (rRNA in orange; ribosomal proteins in green and assembly factors in gray; 11) and late pre-40S subunits (rRNA in gray, for the sake of clarity, the proteins and RNA corresponding to the head are not shown; 67). The WBSCR22–TRMT112 complex (blue for WBSCR22 and purple for TRMT112) as observed in state A of late 40S precursors is shown in ribbon representation (PDB code: 6G4W). Helix h44 is highlighted in magenta (mature 40S) and black (immature 40S). The m6A1832 as seen in the mature 40S is shown as cyan sticks and the N6 methyl group is shown as a sphere. The location of m7G1636 as seen in the cryo-EM structure of the mature ribosome is depicted by a red sphere. (D) Zoom-in on the region centered on position A1832, used to compare RNA conformations within the mature small subunit and late subunit precursors. Same color code as in panel (C).
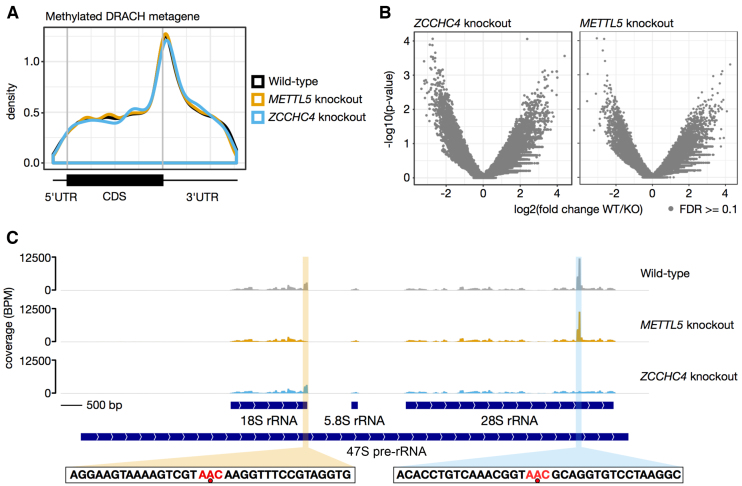
Figure 4.
Transcriptome-wide mapping of m6A by miCLIP reveals that METTL5 and ZCCHC4 are exclusively ribosomal RNA writers. (A) Metagene analysis of all m6A sites identified on mRNAs. The called m6A sites in each replicate were collapsed and plotted with the MetaPlotR pipeline (75). This shows the typical distribution of m6A, with enrichment at the stop codon and a high degree of overlap between the different conditions. DRACH, m6A modification consensus motif (D = A, G or U; R = A or G; H = A, C or U). (B) No other sites in the transcriptome are methylated by METTL5 or ZCCHC4. Coverage in sliding windows across each transcript was calculated and normalized per transcript. Differential bins between wild-type and each knockout were tested for significance with edgeR and plotted as a volcano plot. No sites in the transcriptome passed the false discovery rate (FDR) threshold of 0.1, indicating that no mRNA adenosines are methylated by METTL5 or ZCCHC4. (C) The genome track for the 47S pre-rRNA locus confirms that METTL5 and ZCCHC4 are the writers for m6A on the 18S and 28S rRNAs, respectively. C-to-T transitions are the most frequent substitution at m6A sites caused by antibody cross-linking. As both the 18S and 28S m6A marks are in a AAC context (red in the sequence shown below; the m6A is indicated by a red dot), reads were filtered for those containing CT to reduce background at this locus. The peaks for the 18S m6A and the 28S m6A (highlighted) are lost, respectively, in the mettl5- and _zcchc4-_knockout cells. The miCLIP sequencing data is available on the Gene Expression Omnibus database (accession number GSE128699).
METTL5 and ZCCHC4 are dispensable for cell growth and ribosome biogenesis
Having identified the enzymes responsible for human 18S and 28S rRNA m6A-methylation events, we examined whether these modifications might be important for cell growth and ribosome biogenesis.
First, cell proliferation was monitored for 3 days by manual counting after cell staining with a vital stain. The experiment, performed in triplicate, revealed that the absence of METTL5 or ZCCHC4 had no significant impact on cell growth (Figure 2A). Next, total RNA extracted from parental HCT116 cells and from all the independently isolated mettl5 -/- and zcchc4 -/- cell lines was subjected to denaturing agarose gel electrophoresis and ethidium bromide staining to reveal the mature 18S and 28S rRNAs (Figure 2B). Mature rRNAs were found to be produced normally in the absence of either METTL5 or ZCCHC4. This was confirmed by establishing the 28S-to-18S ratio from Bioanalyzer electropherograms: in all cases, this ratio was 1.0 (Figure 2B), as expected because the 18S and 28S rRNAs are produced by processing from a single long polycistronic transcript (see Supplementary Figure S2A). A detailed pre-rRNA processing analysis performed by quantitative high-resolution northern blotting revealed all the major pre-rRNA species with specific probes and showed no major differences (Supplementary Figure S2B and Supplementary Table S4). In the knockout clones, we noted a marginal increase in the steady-state level of the primary transcript (47S), but this had no impact on overall processing or mature rRNA production (Supplementary Figure S2B and Figure 2B).
In conclusion, METTL5 decorates the 18S rRNA with one m6A and ZCCHC4 installs another m6A on 28S rRNA. Neither enzyme is essential for cell growth or mature rRNA production nor is either of the deposited modifications.
Human METTL5 forms a heterodimer with TRMT112 to gain metabolic stability through the formation of a parallel β-zipper between main chain atoms
To obtain structural information on the two newly identified m6A methyltransferases, we attempted their heterologous expression in Escherichia coli. Despite much effort, we found no experimental conditions allowing their overexpression in soluble form (Supplementary Figure S3C and S3E). As a growing number of eukaryotic RNA methyltransferases have been found to act as multimeric holoenzymes (e.g. 26,43–47), we investigated whether METTL5 and ZCCHC4 might also form multi-protein complexes.
Interestingly, in our recent study aimed at characterizing the interaction network of the methyltransferase activator Trm112 in the archaeon H. volcanii, we found the METTL5 ortholog Hvo_1475 (hereafter called _Hvo_Mettl5, 35% sequence identity to METTL5), among the numerous methyltransferases co-purifying with _Hvo_Trm112 (48). This led us to test whether _Hvo_Mettl5 might interact directly with _Hvo_Trm112, and, by extension, whether human METTL5 might interact with TRMT112.
To test for the above-mentioned interactions, we co-expressed in E. coli, on the one hand, C-terminally His6-tagged _Hvo_Mettl5 with untagged _Hvo_Trm112 (Supplementary Figure S3A and Supplementary Table S5) and, on the other hand, His6-tagged human METTL5 with untagged human TRMT112 (Supplementary Figure S3C). As observed with Mtq2 (yeast)/HEMK2 (human), Trm9/ALKBH8, Bud23/WBSCR22 and several archaeal methyltransferases (34,48–53), co-expression of _Hvo_Mettl5 with _Hvo_Trm112 and of METTL5 with TRMT112 led to successful overexpression of the relevant methyltransferase as a soluble protein (Supplementary Figure S3A and C). This was a first indication of direct _Hvo_Mettl5–_Hvo_Trm112 and METTL5–TRMT112 interaction.
Next, we applied a stringent 3-step purification protocol, beginning with His-tag-targeting affinity purification, to extracts of E. coli cells co-expressing either His6-tagged _Hvo_Mettl5 with _Hvo_Trm112 or His6-tagged METTL5 with TRMT112 (for details, see ‘Materials and Methods’ section). At the end of this protocol, SDS-PAGE analysis of the main peak eluting from the size-exclusion chromatography (SEC) revealed two bands (illustrated for the human METTL5–TRMT112 complex in Supplementary Figure S4): one corresponding to the catalytic subunit (either _Hvo_Mettl5 or METTL5) and one corresponding to either _Hvo_Trm112 or TRMT112. The identities of these proteins were confirmed by mass spectrometry. We conclude that _Hvo_Mettl5 interacts physically with _Hvo_Trm112 and that human METTL5 associates directly with TRMT112. Size-exclusion chromatography coupled to multi-angle laser light scattering (SEC-MALLS) analysis of the purified proteins revealed the formation of heterodimers in solution in the cases of both the human (measured molecular weight of 41.6 kDa versus a theoretical value of 38.7 kDa) and archaeal (measured molecular weight of 29.9 kDa versus a theoretical value of 30 kDa) complexes (Supplementary Figure S3B and D). When ZCCHC4 was co-expressed with TRMT112, we observe no stabilizing effect (Supplementary Figure S3E). This is consistent with a recent study showing that ZCCHC4 can be successfully overexpressed in E. coli in the absence of any other human protein (see (19) and ‘Discussion’ section).
To learn more about the human METTL5–TRMT112 m6A rRNA methyltransferase complex, we determined its crystal structure at 1.6 Å resolution (final model with R and _R_free values of 18.6% and 21.5%, respectively; Supplementary Table S6). We also obtained another crystal form diffracting at lower resolution (2.5 Å). The lower resolution diffracting crystal was interesting, as it allowed us to model a METTL5 loop encompassing residues 191 to 197, which was not defined in the higher resolution structure. This METTL5 loop includes part of the putative active site and contains a strictly conserved signature (see below). Since the structures of the complexes observed in the two crystal forms are virtually identical (rmsd of 0.7–0.8 Å), merging them provided a complete model of the human METTL5–TRMT112 (Figure 2C).
Human TRMT112 comprises two domains: a zinc-binding domain (or ZBD) and a central domain (Figure 2C and Supplementary Figure S5A). The ZBD is thus named by reference to previously determined crystal structures of several eukaryotic Trm112 proteins, where it was shown to contain four cysteine residues coordinating a zinc atom (26,35,49,54). The ZBD is formed by residues of both the N- and C-terminal extremities of the protein and consists of an α-helix (α1) packed against a four-stranded anti-parallel β-sheet. In human TRMT112, remarkably, the four cysteines are not conserved and accordingly, the domain does not bind zinc (Supplementary Figures S5A and S7). The central domain consists of three α-helices (helices α2 to α4). The crystal structure of human TRMT112 superimposes on the structures of fungal Trm112 with rmsd values of 1.9 to 2.1 Å (over 110 Cα atoms; 35% sequence identity; 26,49,54) and on archaeal H. volcanii Trm112 with an rmsd value of 1.6 Å (over 50 Cα atoms; 37% sequence identity; 48).
The METTL5 protein consists of a single domain adopting a class I SAM-dependent methyltransferase fold composed of a central seven-stranded β-sheet flanked by four α-helices on one side (αY, αZ, αA and αB) and two on the other (αD and αE; Figure 2C and Supplementary Figure S5B). During refinement, an unambiguous electron density corresponding to SAM (the methyl donor) was observed in its expected binding site at the C-terminal face of the METTL5 central β-sheet, interacting exclusively with residues of METTL5.
The human METTL5 and TRMT112 proteins interact with each other through a large surface (area: 1160 Å2) formed by 29 METTL5 and 28 TRMT112 residues (Figure 2D). The interface is characterized by the presence of a large central hydrophobic core composed of residues V54, L76, V78, F80, M104, V105, M116, and F120 of METTL5, and L4, L8, L9, V35, F41, M45, L89, I113, P114, M116 and L117 of TRMT112 (Figure 2D; Supplementary Figure S5A and B). The hydrophobic interface is surrounded by polar residues involved in formation of eight hydrogen bonds and two salt bridges (Supplementary Table S7). Among these polar interactions, two hydrogen bonds are formed between the main-chain atoms of residues located in METTL5 strand β3 (Q106) and TRMT112 strand β4 (P114 and M116). They hence contribute to the formation of a parallel β-zipper between these two adjacent strands which, together, form a continuous eleven-stranded β-sheet (Figure 2C and D; Supplementary Figure S7). The two salt bridges involve the interaction of R115 and D121 of METTL5 with E102 and R44 of TRMT112, respectively (Figure 2D and Supplementary Table S7). All these characteristics of the METTL5–TRMT112 interface (hydrophobic core and β-zipper formation) are reminiscent of the previously described interaction mode through which eukaryotic Trm112 proteins contact their methyltransferase partners (namely Mtq2, Bud23, and Trm9; 26,35,47,54), and indeed, the structures of these complexes superimpose nicely on the structure of the METTL5–TRMT112 complex (rmsd values ranging from 2 to 2.5 Å over 230–250 Cα atoms; Supplementary Figure S7).
On METTL5, a very large hydrophobic surface, unfavorable in the hydrophilic cell environment, is masked by TRMT112 upon complex formation (Figure 2C and D; Supplementary Figure S5). As shown previously for its other partners (47), this observation suggests that TRMT112 may be required for METTL5 metabolic stability. This was indeed demonstrated by western blot analysis in a cell line expressing a C-terminal Flag-tagged construct of METTL5 under the control of a tetracycline-inducible promoter (Figure 2E). Flag-tagged METTL5 was detected only after the addition of tetracycline to the medium, and its steady-state accumulation was largely dependent on the presence of TRMT112 (compare odd and even lanes, with and without anti-TRMT112 siRNA). This indicates that TRMT112 is needed to stabilize METTL5. As a control, the blot was probed for another partner of TRMT112, WBSCR22, whose stability has been shown previously to depend on TRMT112 (Figure 2E; 53).
Comparison of METTL5–TRMT112 with DNA and RNA m6A methyltransferases: implications for ribosome biogenesis
Analysis of amino acid sequence conservation among METTL5 orthologs at the surface of the structure of the human METTL5–TRMT112 complex highlighted a large conserved surface around the SAM methyl group, very likely corresponding to the enzyme active site (Supplementary Figure S5C). This region is composed of the strictly conserved 126NPPF129 signature, a hallmark of methyltransferases that modify planar amino groups, plus the Glu27 and Tyr29 side chains of the [L/F]EQY motif present just upstream from helix αZ (Supplementary Figure S5B). Finally, the loop encompassing Phe192 and His193, which is disordered in our high-resolution crystal structure but visible in one of the two copies of the asymmetric unit in the low-resolution structure, is in close proximity to the putative active site and hence further extends it. This conserved region is also enriched in positively charged residues, in line with the view that the METTL5–TRMT112 complex functions as an RNA methyltransferase (Supplementary Figure S5D).
The mapping of sequence conservation at the surface of TRMT112 orthologs from multi-cellular eukaryotes revealed three conserved areas (Supplementary Figure S6): (i) the area involved in interaction with the methyltransferase subunit, (ii) a region overlooking the proposed METTL5 catalytic pocket (interestingly, in the context of yeast Mtq2–Trm112 and Trm11–Trm112 complexes, residues in this area have been shown to contribute to substrate binding, 35,55) and, finally, (iii) a third area on the solvent-exposed face of TRMT112, opposite the catalytic site of METTL5, which may be involved in interaction with yet to be identified factors.
To our knowledge, the structure of METTL5–TRMT112 is the fourth structure of a human m6A RNA methyltransferase to be determined, after those of METTL3–METTL14, METTL16 and CAPAM (cap-specific adenosine methyltransferase, also known as PCIF1; 45,46,56-60). The METTL3–METTL14 holoenzyme, where METTL3 is the catalytic subunit and METTL14 is important for RNA recognition, modifies eukaryotic mRNAs, thereby specifically affecting the metabolism of m6A-containing mRNAs, i.e. pre-mRNA splicing, nucleocytoplasmic export, translation and degradation (61–63). The human METTL16 enzyme methylates U6 spliceosomal RNA and interacts at least with several non-coding RNAs and pre-mRNAs (64,65). Interestingly, investigators have recently solved the crystal structure of human METTL16 bound to an RNA hairpin and with an adenine ring protruding into the active site (57). Lastly, CAPAM specifically methylates the base of the first transcribed nucleotide of eukaryotic mRNAs, when it is an adenine, to form m7Gpppm6Am (60). We therefore compared the structures of these three proteins (sharing between 7 and 16% sequence identity with METTL5) with that of METTL5–TRMT112. All three enzymes have the same overall architecture, although the topology of METTL3 differs from that of METTL5, CAPAM and METTL16 (rmsd values around 2.7–3.2 Å; Supplementary Figure S8A, C and E). Remarkably, the [N/D]PP[F/W] signatures of these different enzymes superimpose perfectly. Yet the other active-site residues conserved within the METTL5 family differ significantly from the corresponding residues in METTL3, METTL16 and CAPAM (Supplementary Figure S8B, D and F). Furthermore, a comparison with the RNA-bound structure of METTL16 shows that, with the exception of the NPPF signature, the residues coordinating the adenine ring in the METTL16 active site are not conserved in METTL5, and the METTL5 loop encompassing residues 184 to 200 sterically clashes with the RNA backbone. Altogether, this indicates that these enzymes might use distinct mechanisms to recognize the adenine base to be methylated.
Interestingly, when we compared the structure of METTL5–TRMT112 with those of m6A DNA methyltransferases bound to short double-stranded DNA fragments, we found unexpected commonalities (Figure 3A and B). Of particular interest is the structure of DNA-bound Thermus aquaticus M. TaqI (rmsd value of 2–2.1Å over 140 Ca atoms; 66), where the target adenine ring of one DNA strand extrudes from the double helix and points toward the active site, where it is coordinated by the 105NPPY108 signature and the Val21 and Phe196 side chains (Figure 3A and B). These correspond respectively to 126NPPF129, Tyr29 and Tyr184 of human METTL5, which we suggest might coordinate the target adenine in its active site as does M. TaqI, optimally positioning the adenine for transfer of the SAM methyl group onto its N6 atom (Figure 3B).
In the structure of the mature human 40S subunit (11), the m6A1832 nucleotide is located in the decoding center area, in a short region connecting 18S rRNA helices h44 and h45. It is base-paired with C1703, making it poorly accessible (Figure 1A). In the recent structures of late human 40S subunit maturation intermediates bound either to the WBSCR22–TRMT112 complex or to the NOB1, PNO1 and RIOK2 assembly factors (67 and see below), the decoding center is not completely matured, and A1832 is not defined in the cryo-EM electron density map due to increased flexibility (Figure 3C and D). In 40S precursors, the base-pairing involving nucleotides surrounding A1832 is similar to that observed in mature 40S, but the top of helix h44 is bent outward by 20° relatively to its position in mature subunits (Figure 3C). As a consequence, the distance between nucleotides C1828 and G1836 increases from 27 Å in the mature form to 32 Å in the immature form (Figure 3D). This conformation stretches this rRNA region and very likely results in melting of the hairpin formed in mature rRNA by nucleotides m6A1832 to G1836 leading, in the immature form, to the exposure of the A1832 base to the solvent. During the late processing steps, this nucleotide would then be located at the center of a region connecting helices h44 and h45, poised to be recognized by the METTL5–TRMT112 complex in a manner highly reminiscent of the recognition of double-stranded DNA by the M. TaqI enzyme (66) (Figure 3A and B).
Despite extensive efforts, we were unable to recapitulate METTL5–TRMT112 enzymatic activity in vitro with short single- or double-stranded RNAs corresponding to the sequences surrounding m6A1832 in the mature ribosome. This precludes further structural studies with RNA fragments.
METTL5 and ZCCHC4 are exclusively m6A ribosomal RNA writers
Finally, as cases of cross-talk are emerging where a particular RNA modification machinery appears to act on multiple classes of cellular and even viral RNAs (21,64,65,68–70), we wondered if METTL5–TRMT112 or ZCCHC4 might act on RNA substrates other than ribosomal RNAs.
To answer this question, we used the miCLIP technique to perform systematic single-nucleotide resolution mapping of m6A on the transcriptome of human cells lacking either METTL5 or ZCCHC4 (see ‘Materials and Methods’ section). Two independent knockout cell lines for each methyltransferases were tested in parallel. While 12 346 m6A sites were re-mapped on mRNAs of isogenic control cells, no position within either mRNAs or long non-coding RNAs was found to vary significantly in the absence of METTL5 or ZCCHC4 (Figure 4A and B). The miCLIP analysis further confirmed that METTL5–TRMT112 is responsible for installing m6A1832 on 18S rRNA and ZCCHC4 for depositing m6A4220 on 28S rRNA (Figure 4C). In conclusion, METTL5 and ZCCHC4 are highly specific, respectively, to the 18S and the 28S m6A modification.
DISCUSSION
Eukaryotic ribosomes are heavily modified. During ribosomal subunit biogenesis, many rRNA nucleotides, often in functionally important regions of the ribosome, are specifically selected to be modified either by snoRNP-based nanomachines or by more conventional protein-only enzymes (13). Yet the diversity of ribosomal RNA modifications appears rather limited, as yeast ribosomes have been found to contain only 12 types of RNA modification out of a so-far-uncovered natural repertoire of 163 (13). Apart from 2’-_O_-methylated sugars and pseudouridines, which constitute the bulk of rRNA modifications, yeast ribosomes carry only 12 base modifications, six on the small and six on the large subunit. Not until recently has the entire set of modification enzymes responsible for installing these rRNA modifications been identified in yeast (16). Because these rRNA base modifications are highly conserved throughout the eukaryotic kingdom, most of them being present also on human ribosomes, the human orthologs of the identified yeast enzymes are being characterized. Yet human ribosomes carry at least one additional RNA modification type: m6A, one on each subunit (Figure 1A and B). These modifications have no equivalent in yeasts, and the activities responsible for depositing them have remained unknown for 33 years (17,18).
In the present work we have identified METTL5 and ZCCHC4 as the m6A methyltransferases responsible, respectively, for 18S and 28S rRNA modification. We have identified these novel m6A methyltransferases by screening bioinformatically the human proteome for candidates harboring an N6-adenosine-specific DNA methyltransferase motif. Our quantitative HPLC data (Figure 1C and D) and single-nucleotide-resolution miCLIP analysis (Figure 4C) demonstrate that the activities identified are indeed responsible for 18S and 28S rRNA m6A methylation. To our knowledge, this is the first report of a function for METTL5. We also confirm the role of ZCCHC4 (19). Our miCLIP analysis further reveals, for the first time, that both enzymes act only on ribosomal RNAs (Figure 4A and B). For METTL5, we provide evidence that it must form a heterodimer with TRMT112 to gain metabolic stability in cells (Figure 2E). We have also determined the atomic-resolution structure of the METTL5–TRMT112 complex, highlighting the molecular nature of the interface between the two subunits: a parallel β-zipper between main-chains atoms (Figure 2C and D). The use of main-chain rather than side-chain atoms to form the interface enables TRMT112 to form such complexes with other methyltransferases sharing the same fold but displaying only limited sequence homology (see below and discussions in 26,47,54).
Upon comparing the structure of METTL5–TRMT112 with those of three recently described human m6A methyltransferases, namely METTL3–METTL14 (45,46,56), METTL16 (57–59) and CAPAM (60), we observed no major similarities (Supplementary Figure S7). This suggests that METTL5–TRMT112 likely operates in its own specific way. Interestingly, however, a comparison of METTL5–TRMT112 with an N6-adenosine DNA methyltransferase of Thermus aquaticus (M. TaqI) revealed that METTL5–TRMT112 might extrude the substrate adenosine from a nucleic acid duplex (Figure 3A and B). Consistently with this initial mechanistic insight, a comparison of the structure of the modified area in precursor and mature forms of 18S rRNA indicates that in maturing subunits, the RNA segment containing A1832 is ‘stretched’ by ∼5 Å. This may increase the accessibility of the target adenosine by ‘flipping it out’, thus exposing it to the catalytic site of the enzyme (Figure 3C and D).
Among the various m6A RNA methyltransferases identified so far, two appear to work as holoenzymes, METTL3–METTL14 and METTL5–TRMT112. The METTL3 mRNA m6A methyltransferase indeed strictly requires METTL14 as a coactivator. METTL14 has divergently evolved from a class I SAM-dependent methyltransferase fold, but it has lost both its ability to interact with SAM and its active site (45,46,56). It is assumed, however, to be essential for the binding and proper positioning of the mRNA substrate in the METTL3 active site. Thus, METTL5 is the second reported example of an m6A methyltransferase strictly requiring a coactivator, in this case TRMT112.
The folds adopted by METTL14 and TRMT112 are radically different, yet TRMT112 shares functional communalities with METTL14. On the basis of the striking similarities between the structures of the METTL5–TRMT112 complex and of previously characterized heterodimeric complexes formed between Trm112 and other methyltransferases, TRMT112 is expected not only to be important in stabilizing METTL5 but also to activate METTL5 by stimulating its SAM-binding capacity (35,55). However, a stimulatory role of TRMT112 on METTL5 catalysis remains to be formally demonstrated. In addition, if our proposed docking of the METTL5–TRMT112 complex onto pre-40S state ‘E’ is correct (see below, Figure 5), then TRMT112 appears to be in contact with the 40S ribosomal subunit, thereby directly contributing to the substrate-binding capacity of the catalytic subunit METTL5.
Figure 5.
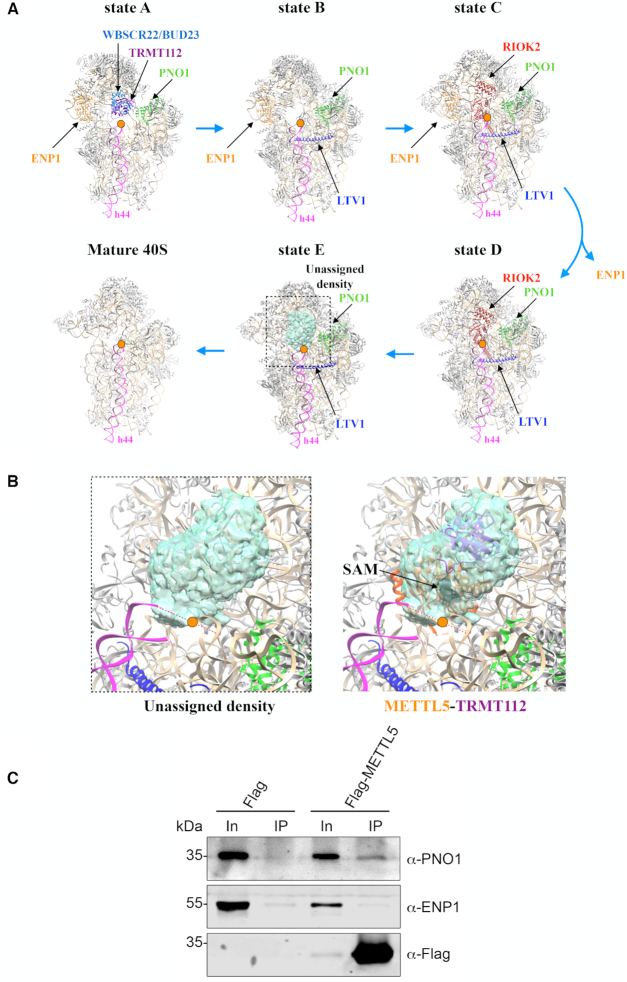
Tentative positioning of the METTL5–TRMT112 complex within the maturing ribosomal small subunit. (A) Five sequential stages (states ‘A’ to ‘E’) in the late maturation of human 40S subunit established by cryo-EM (67). Orange dot, site of m6A modification at the ribosomal decoding site. Only state ‘E’ exhibits in the A1832 area an unassigned density that could be ascribed to METTL5–TRMT112 (see panel B). In states ‘A’, ‘C’ and ‘D’, associated assembly factors (WBSCR22–TRMT112 in state ‘A’; RIOK2 in states ‘C’ and ‘D’) would sterically hinder the binding of METTL5–TRMT112. State ‘B’ shows no extra density in the area of interest. The assembly factors LTV1 (in blue), PNO1 (in green) and ENP1 (in orange), as well as helix 44 (h44, in pink) are shown for reference. PNO1 is detected in all five states while ENP1 is dissociated from pre-40S at the transition between states ‘C’ and ‘D’. The region delimited by the dotted box corresponds to the region shown in panel B. (B) Zoom-in on an unassigned density present in pre-40S state ‘E’ (PDB 6G53; EMDB 4351) in the m6A-modified area, and tentative docking of the METTL5–TRMT112 structure. This tentative docking results in SAM being properly positioned with respect to the substrate adenosine. Color scheme as in panel (A). (C) Flag-tagged METTL5 co-purifies with PNO1 but not with ENP1. Expression of Flag-METTL5 was induced in HEK293 cells by addition of tetracycline and complexes containing Flag-METTL5 were captured on anti-Flag beads (see ‘Materials and Methods’ section). Co-precipitated proteins were detected by western blotting using the antibodies indicated. As control, a cell line expressing only the Flag-tag was used.
Our identification of ZCCHC4 as the 28S rRNA m6A4220 methyltransferase is fully consistent with the results of a recent study (19), which additionally described a role of ZCCHC4 in HepG2 cell proliferation. We observed no comparable effect on the proliferation of HCT116 cells where the ZCCHC4 enzyme was inactivated by CRISPR–Cas9-mediated deletion of the active site signature (Figure 2A). It is unclear whether this discrepancy results from the different cell lines used in these studies or from the CRISPR–Cas9 protocol applied (in our case: transient exposure of cells to CRISPR–Cas9 complexes to minimize off-target effects).
When, during ribosome biogenesis, does METTL5–TRMT112 deposit the m6A mark on 18S rRNA?
A pathway of late maturation of human pre-40S ribosomal subunits has recently been described, in which five discrete species (states ‘A to E’) appear in sequential order (67, and summarized in Figure 5A). Prior to the present work, METTL5 was not known to be involved in human ribosome biogenesis, and to our knowledge, it had never been found associated with pre-40S human ribosomes. We therefore scrutinized the density maps of the cryo-EM structures of the five late pre-40S states to see if we might find, in the vicinity of position A1832, unattributed electron densities ascribable to METTL5–TRMT112.
Of the five pre-40S states only one, state ‘E’, retained our attention because it harbors, at the top of helix h44 in direct proximity to the m6A-modified residue A1832, an unassigned density that potentially corresponds to METTL5–TRMT112 (Figure 5A and B). The other states are either incompatible with METTL5–TRMT112 binding because of the presence of assembly factors that would sterically interfere with it (WBSCR22–TRMT112 in the case of state ‘A’; RIOK2, in the case of states ‘C’ and ‘D’, see Figure 5A), or simply because there is no density in the region of interest (state ‘B’).
In pre-40S state ‘E’, the local resolution of the unassigned density of interest on the cryoEM map is ∼8 Å. This precludes precise modeling of secondary structure elements. Nonetheless, we could tentatively dock the structure of the METTL5–TRMT112 complex in this density (Figure 5B), and this resulted in positioning its active site in close proximity to A1832 (orange dot in Figure 5B). Compatible with our hypothesis that METTL5–TRMT112 binds to pre-40S state ‘E’, Flag-tagged METTL5 co-purified PNO1, which is present in all five described pre-40S states, but not ENP1, which is dissociated from pre-40S ribosomes between states ‘C’ and ‘D’ (Figure 5C). Our results imply that m6A1832 formation is one of the last steps in 40S maturation and that it occurs after installation of m7G1636 by WBSCR22–TRMT112 (Figure 5A, state ‘A’). In line with this hypothesis, we wondered whether one of these modifications (18S rRNA m6A1832 or m7G1636) might depend on prior deposition of the other (Supplementary Figure S9). As we found no major reduction of 18S rRNA m6A methylation in wbscr22 -/- cells (Supplementary Figure S9A), lacking the m7G modification (71), and no reduction of 18S rRNA m7G methylation in mettl5 -/- cells (Supplementary Figure S9B), lacking the m6A modification, we conclude these two modifications are formed independently.
How many TRMT112 molecules on each pre-40S subunit?
TRMT112 is a very special protein because, in archaeal and eukaryal cells, it interacts with and activates multiple methyltransferases (at least 10 in archaea; 4 in budding yeast, and 5 in human cells) that modify factors (RNAs or proteins) involved in mRNA translation (see ‘Introduction’ and discussions in 47,48). In pre-40S ribosomes, remarkably, two complexes containing TRMT112, in association with either WBSCR22 or METTL5, act on neighboring areas (Figures 1A and 5A,B). Although quite tempting, we do not favor the idea that TRMT112, once bound to pre-40S, acts as a recruitment platform for the successive association of WBSCR22 and METTL5, because in the structure of the pre-40S state ‘A’, TRMT112 is exposed toward the solvent, without any residues in direct contact with the pre-ribosome. Since both WBSCR22 and METTL5 depend upon their association with TRMT112 to gain metabolic stability (Figure 2E and 53), we believe, rather, that both methyltransferases form heterodimers with TRMT112 independently and that these heterodimers coexist in cells. We therefore propose that the two methyltransferase heterodimers are recruited and act one after the other. Specifically, we suggest that WBSCR22–TRMT112 is recruited first, and that once it has performed its function and dissociated from pre-40S, METTL5–TRMT112 is recruited onto pre-40S.
On the importance of using multiple model organisms
Until recently, the identification of the human partners of TRMT112 was based solely on characterization of human orthologs of known yeast Trm112 partners (34, 51,53,72,73). Following this rationale, METTL5 would have escaped identification (yeast ribosomes are not m6A-modified at the position equivalent to human A1832, and yeast genomes do not encode METTL5 homologs). Here the approach was different: the present work on human cells stems from our recent characterization of the archaeal Trm112 interactome (48), which led us to identify _Hvo_Mettl5 (archaeal METTL5). This emphasizes the importance of conducting research on a range as diverse as possible of experimental models, including archaeal ones, which are sometimes neglected.
Supplementary Material
gkz619_Supplemental_File
ACKNOWLEDGEMENTS
We thank Dr P.-D. Coureux for his help with the visualization of cryo-EM density maps and preparation of Figure 5. We thank SOLEIL for provision of synchrotron radiation facilities. D.L.J.L. is affiliated with the Center for Microscopy and Molecular Imaging (CMMI), and the ULB Cancer Research Center (U-CRC).
Author Contributions: N.T.V., F.G.M.E., B.R.H., C.Z., N.U., P.H. and K.B. performed the experiments. N.T.V., F.G.M.E., B.R.H., M.T.B., S.R.J., D.L.J.L. and M.G. analyzed the data. D.L.J.L. and M.G. designed the research and wrote the manuscript. D.L.J.L. and M.G. contributed equally to this work.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
CNRS ATIP-AVENIR program, the Agence Nationale pour la Recherche [ANR-14-CE09-0016, ANR-16-CE11-0003]; CNRS and Ecole Polytechnique (to M.G.); CNRS PICS program [PICS07484 to M.G., D.L.J.L.]; Belgian Fonds de la Recherche Scientifique (F.R.S./FNRS) [CDR J.0095.19-33696322]; ULB; Région Wallonne (DGO6) [First Spin-Off n°1810070-RIBOcancer]; Fonds Jean Brachet (to D.L.J.L.); National Institutes of Health [NS111631, CA186702 to S.R.J.]; Deutsche Forschungsgemeinschaft [SPP1784: BO3442/2-2 to M.T.B.]; French Ministère de l’Enseignement Supérieur et de la Recherche (MESR) (to T.V.N.).
Conflict of interest statement. On behalf of all co-authors, D.L.J.L. declares that the scientists involved in this study have no financial or non-financial competing interests.
REFERENCES
- 1.Ben-Shem A., Jenner L., Yusupova G., Yusupov M.. Crystal structure of the eukaryotic ribosome. Science. 2010; 330:1203–1209. [DOI] [PubMed] [Google Scholar]
- 2.Bohnsack K.E., Bohnsack M.T.. Uncovering the assembly pathway of human ribosomes and its emerging links to disease. EMBO J. 2019; 38:e100278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassler J., Hurt E.. Eukaryotic ribosome assembly. Annu. Rev. Biochem. 2018; 88:8.1–8.26. [DOI] [PubMed] [Google Scholar]
- 4.Klinge S., Woolford J.L. Jr.. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 2019; 20:116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aspesi A., Ellis S.R.. Rare ribosomopathies: insights into mechanisms of cancer. Nat. Rev. Cancer. 2019; 19:228–238. [DOI] [PubMed] [Google Scholar]
- 6.Pelletier J., Thomas G., Volarevic S.. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat. Rev. Cancer. 2018; 18:51–63. [DOI] [PubMed] [Google Scholar]
- 7.Lafontaine D.L.J. Noncoding RNAs in eukaryotic ribosome synthesis and function. Nat. Struct. Mol. Biol. 2015; 22:11–19. [DOI] [PubMed] [Google Scholar]
- 8.Erales J., Marchand V., Panthu B., Gillot S., Belin S., Ghayad S.E., Garcia M., Laforets F., Marcel V., Baudin-Baillieu A. et al.. Evidence for rRNA 2'-O-methylation plasticity: Control of intrinsic translational capabilities of human ribosomes. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:12934–12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma S., Marchand V., Motorin Y., Lafontaine D.L.J.. Identification of sites of 2'-O-methylation vulnerability in human ribosomal RNAs by systematic mapping. Sci. Rep. 2017; 7:11490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taoka M., Nobe Y., Yamaki Y., Sato K., Ishikawa H., Izumikawa K., Yamauchi Y., Hirota K., Nakayama H., Takahashi N. et al.. Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Res. 2018; 46:9289–9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Natchiar S.K., Myasnikov A.G., Kratzat H., Hazemann I., Klaholz B.P.. Visualization of chemical modifications in the human 80S ribosome structure. Nature. 2017; 551:472–477. [DOI] [PubMed] [Google Scholar]
- 12.Watkins N.J., Bohnsack M.T.. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA. 2012; 3:397–414. [DOI] [PubMed] [Google Scholar]
- 13.Sharma S., Lafontaine D.L.J.. “View From A Bridge”: A new perspective on eukaryotic rRNA base modification. Trends Biochem. Sci. 2015; 40:560–575. [DOI] [PubMed] [Google Scholar]
- 14.Shi Z., Barna M.. Translating the genome in time and space: specialized ribosomes, RNA regulons, and RNA-binding proteins. Annu. Rev. Cell Dev. Biol. 2015; 31:31–54. [DOI] [PubMed] [Google Scholar]
- 15.Genuth N.R., Barna M.. Heterogeneity and specialized functions of translation machinery: from genes to organisms. Nat. Rev. Genet. 2018; 19:431–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sloan K.E., Warda A.S., Sharma S., Entian K.D., Lafontaine D.L.J., Bohnsack M.T.. Tuning the ribosome: the influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2016; 14:1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maden B.E. Identification of the locations of the methyl groups in 18 S ribosomal RNA from Xenopus laevis and man. J. Mol. Biol. 1986; 189:681–699. [DOI] [PubMed] [Google Scholar]
- 18.Maden B.E. Locations of methyl groups in 28 S rRNA of Xenopus laevis and man. Clustering in the conserved core of molecule. J. Mol. Biol. 1988; 201:289–314. [DOI] [PubMed] [Google Scholar]
- 19.Ma H., Wang X., Cai J., Dai Q., Natchiar S.K., Lv R., Chen K., Lu Z., Chen H., Shi Y.G. et al.. N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 2019; 15:88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. et al.. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biol. 2013; 31:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma S., Langhendries J.L., Watzinger P., Kotter P., Entian K.D., Lafontaine D.L.. Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 2015; 43:2242–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 1993; 26:795–800. [Google Scholar]
- 23.Evans P.R., Murshudov G.N.. How good are my data and what is the resolution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013; 69:1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015; 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Létoquart J., Huvelle E., Wacheul L., Bourgeois G., Zorbas C., Graille M., Heurgué-Hamard V., Lafontaine D.L.J.. Structural and functional studies of Bud23-Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E5518–E5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Sharff A., Smart O.S., Vonrhein C. et al.. BUSTER version 2.10.2. 2016; Cambridge: Global Phasing, Ltd. [Google Scholar]
- 29.Linder B., Grozhik A.V., Olarerin-George A.O., Meydan C., Mason C.E., Jaffrey S.R.. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 2015; 12:767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cecchini J.P., Miassod R.. Studies on the methylation of cytoplasmic ribosomal RNA from cultured higher plant cells. Eur. J. Biochem. 1979; 98:203–214. [DOI] [PubMed] [Google Scholar]
- 31.The UniProt, C. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sigrist C.J., de Castro E., Cerutti L., Cuche B.A., Hulo N., Bridge A., Bougueleret L., Xenarios I.. New and continuing developments at PROSITE. Nucleic Acids Res. 2013; 41:D344–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heurgue-Hamard V., Champ S., Mora L., Merkulova-Rainon T., Kisselev L.L., Buckingham R.H.. The glutamine residue of the conserved GGQ motif in Saccharomyces cerevisiae release factor eRF1 is methylated by the product of the YDR140w gene. J. Biol. Chem. 2005; 280:2439–2445. [DOI] [PubMed] [Google Scholar]
- 34.Figaro S., Scrima N., Buckingham R.H., Heurgue-Hamard V.. HemK2 protein, encoded on human chromosome 21, methylates translation termination factor eRF1. FEBS Lett. 2008; 582:2352–2356. [DOI] [PubMed] [Google Scholar]
- 35.Liger D., Mora L., Lazar N., Figaro S., Henri J., Scrima N., Buckingham R.H., van Tilbeurgh H., Heurgue-Hamard V., Graille M.. Mechanism of activation of methyltransferases involved in translation by the Trm112 ‘hub’ protein. Nucleic Acids Res. 2011; 39:6249–6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graille M., Heurgue-Hamard V., Champ S., Mora L., Scrima N., Ulryck N., van Tilbeurgh H., Buckingham R.H.. Molecular basis for bacterial class I release factor methylation by PrmC. Mol. Cell. 2005; 20:917–927. [DOI] [PubMed] [Google Scholar]
- 37.Bujnicki J.M. Phylogenomic analysis of 16S rRNA:(guanine-N2) methyltransferases suggests new family members and reveals highly conserved motifs and a domain structure similar to other nucleic acid amino-methyltransferases. FASEB J. 2000; 14:2365–2368. [DOI] [PubMed] [Google Scholar]
- 38.Polevoda B., Span L., Sherman F.. The yeast translation release factors Mrf1p and Sup45p (eRF1) are methylated, respectively, by the methyltransferases Mtq1p and Mtq2p. J. Biol. Chem. 2006; 281:2562–2571. [DOI] [PubMed] [Google Scholar]
- 39.Thul P.J., Akesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., Alm T., Asplund A., Bjork L., Breckels L.M. et al.. A subcellular map of the human proteome. Science. 2017; 356:eaal3321. [DOI] [PubMed] [Google Scholar]
- 40.Franke C., Grafe D., Bartsch H., Bachmann M.P.. Use of nonradioactive detection method for North- and South-Western Blot. Methods Mol. Biol. 2015; 1314:63–71. [DOI] [PubMed] [Google Scholar]
- 41.Kowalak J.A., Bruenger E., Crain P.F., McCloskey J.A.. Identities and phylogenetic comparisons of posttranscriptional modifications in 16 S ribosomal RNA from Haloferax volcanii. J. Biol. Chem. 2000; 275:24484–24489. [DOI] [PubMed] [Google Scholar]
- 42.Grosjean H., Gaspin C., Marck C., Decatur W.A., de Crecy-Lagard V.. RNomics and Modomics in the halophilic archaea Haloferax volcanii: identification of RNA modification genes. BMC Genomics. 2008; 9:470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leulliot N., Chaillet M., Durand D., Ulryck N., Blondeau K., van Tilbeurgh H.. Structure of the yeast tRNA m7G methylation complex. Structure. 2008; 16:52–61. [DOI] [PubMed] [Google Scholar]
- 44.Guy M.P., Phizicky E.M.. Two-subunit enzymes involved in eukaryotic post-transcriptional tRNA modification. RNA Biol. 2014; 11:1608–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sledz P., Jinek M.. Structural insights into the molecular mechanism of the m(6)A writer complex. eLife. 2016; 5:e18434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X., Feng J., Xue Y., Guan Z., Zhang D., Liu Z., Gong Z., Wang Q., Huang J., Tang C. et al.. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016; 534:575–578. [DOI] [PubMed] [Google Scholar]
- 47.Bourgeois G., Letoquart J., van Tran N., Graille M.. Trm112, a protein activator of methyltransferases modifying actors of the eukaryotic translational apparatus. Biomolecules. 2017; 7:E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Tran N., Muller L., Ross R.L., Lestini R., Letoquart J., Ulryck N., Limbach P.A., de Crecy-Lagard V., Cianferani S., Graille M.. Evolutionary insights into Trm112-methyltransferase holoenzymes involved in translation between archaea and eukaryotes. Nucleic Acids Res. 2018; 46:8483–8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heurgue-Hamard V., Graille M., Scrima N., Ulryck N., Champ S., van Tilbeurgh H., Buckingham R.H.. The zinc finger protein Ynr046w is plurifunctional and a component of the eRF1 methyltransferase in yeast. J. Biol. Chem. 2006; 281:36140–36148. [DOI] [PubMed] [Google Scholar]
- 50.Mazauric M.H., Dirick L., Purushothaman S.K., Bjork G.R., Lapeyre B.. Trm112p is a 15-kDa zinc finger protein essential for the activity of two tRNA and one protein methyltransferases in yeast. J. Biol. Chem. 2010; 285:18505–18515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Songe-Moller L., van den Born E., Leihne V., Vagbo C.B., Kristoffersen T., Krokan H.E., Kirpekar F., Falnes P.O., Klungland A.. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol. Cell Biol. 2010; 30:1814–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Figaro S., Wacheul L., Schillewaert S., Graille M., Huvelle E., Mongeard R., Zorbas C., Lafontaine D.L.J., Heurgue-Hamard V.. Trm112 is required for Bud23-mediated methylation of the 18S rRNA at position G1575. Mol. Cell Biol. 2012; 32:2254–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zorbas C., Nicolas E., Wacheul L., Huvelle E., Heurgue-Hamard V., Lafontaine D.L.J.. The human 18S rRNA base methyltransferases DIMT1L and WBSCR22-TRMT112 but not rRNA modification are required for ribosome biogenesis. Mol. Biol. Cell. 2015; 26:2080–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Letoquart J., van Tran N., Caroline V., Aleksandrov A., Lazar N., van Tilbeurgh H., Liger D., Graille M.. Insights into molecular plasticity in protein complexes from Trm9-Trm112 tRNA modifying enzyme crystal structure. Nucleic Acids Res. 2015; 43:10989–11002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bourgeois G., Marcoux J., Saliou J.M., Cianferani S., Graille M.. Activation mode of the eukaryotic m2G10 tRNA methyltransferase Trm11 by its partner protein Trm112. Nucleic Acids Res. 2017; 45:1971–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang P., Doxtader K.A., Nam Y.. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell. 2016; 63:306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Doxtader K.A., Wang P., Scarborough A.M., Seo D., Conrad N.K., Nam Y.. Structural basis for regulation of METTL16, an S-Adenosylmethionine homeostasis factor. Mol. Cell. 2018; 71:1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mendel M., Chen K.M., Homolka D., Gos P., Pandey R.R., McCarthy A.A., Pillai R.S.. Methylation of structured RNA by the m(6)A Writer METTL16 is essential for mouse embryonic development. Mol. Cell. 2018; 71:986–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ruszkowska A., Ruszkowski M., Dauter Z., Brown J.A.. Structural insights into the RNA methyltransferase domain of METTL16. Sci. Rep. 2018; 8:5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Akichika S., Hirano S., Shichino Y., Suzuki T., Nishimasu H., Ishitani R., Sugita A., Hirose Y., Iwasaki S., Nureki O. et al.. Cap-specific terminal N (6)-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science. 2019; 363:eaav0080. [DOI] [PubMed] [Google Scholar]
- 61.Fu Y., Dominissini D., Rechavi G., He C.. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat. Rev. Genet. 2014; 15:293–306. [DOI] [PubMed] [Google Scholar]
- 62.Meyer K.D., Jaffrey S.R.. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014; 15:313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu N., Pan T.. N6-methyladenosine-encoded epitranscriptomics. Nat. Struct. Mol. Biol. 2016; 23:98–102. [DOI] [PubMed] [Google Scholar]
- 64.Pendleton K.E., Chen B., Liu K., Hunter O.V., Xie Y., Tu B.P., Conrad N.K.. The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell. 2017; 169:824–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Warda A.S., Kretschmer J., Hackert P., Lenz C., Urlaub H., Hobartner C., Sloan K.E., Bohnsack M.T.. Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017; 18:2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goedecke K., Pignot M., Goody R.S., Scheidig A.J., Weinhold E.. Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. Nat. Struct. Biol. 2001; 8:121–125. [DOI] [PubMed] [Google Scholar]
- 67.Ameismeier M., Cheng J., Berninghausen O., Beckmann R.. Visualizing late states of human 40S ribosomal subunit maturation. Nature. 2018; 558:249–253. [DOI] [PubMed] [Google Scholar]
- 68.Safra M., Sas-Chen A., Nir R., Winkler R., Nachshon A., Bar-Yaacov D., Erlacher M., Rossmanith W., Stern-Ginossar N., Schwartz S.. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature. 2017; 551:251–255. [DOI] [PubMed] [Google Scholar]
- 69.Arango D., Sturgill D., Alhusaini N., Dillman A.A., Sweet T.J., Hanson G., Hosogane M., Sinclair W.R., Nanan K.K., Mandler M.D. et al.. Acetylation of cytidine in mRNA promotes translation efficiency. Cell. 2018; 175:1872–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ringeard M., Marchand V., Decroly E., Motorin Y., Bennasser Y.. FTSJ3 is an RNA 2'-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature. 2019; 565:500–504. [DOI] [PubMed] [Google Scholar]
- 71.Marchand V., Ayadi L., Ernst F.G.M., Hertler J., Bourguignon-Igel V., Galvanin A., Kotter A., Helm M., Lafontaine D.L.J., Motorin Y.. AlkAniline-Seq: Profiling of m(7) G and m(3) C RNA modifications at single nucleotide resolution. Angew. Chem. Int., Ed. Engl. 2018; 57:16785–16790. [DOI] [PubMed] [Google Scholar]
- 72.Fu D., Brophy J.A., Chan C.T., Atmore K.A., Begley U., Paules R.S., Dedon P.C., Begley T.J., Samson L.D.. Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol. Cell Biol. 2010; 30:2449–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ounap K., Leetsi L., Matsoo M., Kurg R.. The stability of ribosome biogenesis factor WBSCR22 is regulated by interaction with TRMT112 via Ubiquitin-Proteasome pathway. PLoS One. 2015; 10:e0133841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shao S., Murray J., Brown A., Taunton J., Ramakrishnan V., Hegde R.S.. Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes. Cell. 2016; 167:1229–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Olarerin-George A.O., Jaffrey S.R.. MetaPlotR: a Perl/R pipeline for plotting metagenes of nucleotide modifications and other transcriptomic sites. Bioinformatics. 2017; 33:1563–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gkz619_Supplemental_File