The Neuronal Apoptosis Inhibitory Protein Is a Direct Inhibitor of Caspases 3 and 7 (original) (raw)
Abstract
The neuronal apoptosis inhibitory protein (NAIP) was identified as a candidate gene for the inherited neurodegenerative disorder spinal muscular atrophy. NAIP is the founding member of a human protein family that is characterized by highly conserved N-terminal motifs called baculovirus inhibitor of apoptosis repeats (BIR). Five members of the human family of inhibitor of apoptosis proteins including NAIP have been shown to be antiapoptotic in various systems. To date, a mechanism for the antiapoptotic effect of NAIP has not been elucidated. To investigate NAIP function, we found cytoprotection of NAIP-expressing primary cortical neurons treated to undergo caspase-3-dependent apoptosis. The additional treatment of these neurons with the pancaspase inhibitor boc-aspartyl(OMe)-fluoromethylketone did not result in increased survival. Similar cytoprotective effects were obtained using HeLa cells transiently transfected with a NAIP N-terminal construct and treated to undergo a caspase-3-dependent cell death. To examine whether NAIP inhibits caspases directly, recombinant N-terminal NAIP protein containing BIR domains was overexpressed, purified, and tested for caspase inhibition potential. Our results demonstrate that inhibition of caspases is selective and restricted to the effector group of caspases, with _K_i values as low as ∼14 nm for caspase-3 and ∼45 nm for caspase-7. Additional investigations with NAIP fragments containing either one or two NAIP BIRs revealed that the second BIR and to a lesser extent the third BIR alone are sufficient to mediate full caspase inhibition.
Keywords: BIR domains, NAIP, caspase inhibition, neuronal apoptosis, inhibitor of apoptosis protein family, cytoprotection
With the discovery of biochemical and histological hallmarks of apoptosis, it has become increasingly evident that the intrinsic cellular suicide program is not only required for normal CNS development (Oppenheim, 1991) but also, if executed inappropriately, contributes to the pathology of neurodegenerative disorders such as Alzheimer's disease (Cotman, 1998), Parkinson's disease (Hartmann et al., 2000), or the childhood spinal muscular atrophy (SMA; Morrison, 1996). The defining characteristic of SMA is a progressive loss of motor neurons leading to wasting of the voluntary muscles. SMA patients are clinically heterogeneous and are classified into three types (types I–III) on the basis of age of onset, severity, and clinical progression (Morrison, 1996). The combined frequency for all three types of SMA is ∼1:10,000, making it one of the most common inherited neurodegenerative diseases (Emery, 1991). Analysis of the complex SMA locus that maps to chromosome 5q13 has led to the discovery of two candidate genes for SMA, the SMA-causing gene survival motor neuron (SMN) and the potential modifier neuronal apoptosis inhibitory protein (NAIP; Lefebvre et al., 1995; Roy et al., 1995). The NAIP gene coding region spans 4212 nucleotides encoding a 1403-amino acid 156 kDa protein with strong homology to the baculoviral inhibitor of apoptosis proteins (IAPs) Cp-IAP and Op-IAP. NAIP is the founding member of the human IAP protein family (Roy et al., 1995), which typically express one to three motifs in the N-terminal region termed baculovirus inhibitor of apoptosis repeat (BIR) domains that are defined by a CX2CX16HX6–8C consensus sequence (Uren et al., 1998).
Recent studies examining neuronal cell death indicate a key role for caspase-3 in the apoptotic process. For example, it has been shown that caspase-3 knock-out mice show severe alterations of brain structures in regions where neuronal apoptosis is predominantly believed to occur. The changes include an overall brain mass increase, disorganized cell deployment, and duplicated brain structures (Kuida et al., 1996). In addition, upregulation and activation of caspase-3 or a caspase-3-like protease has been shown to be a key mediator of neuronal death in neurons after excitotoxic and hypoxic injuries, as well as in the hippocampal CA1 sector after transient global ischemia (Yakovlev et al., 1997; Chen et al., 1998; Namura et al., 1998; Ni et al., 1998).
An antiapoptotic effect of NAIP and other members of the human IAP family has been shown in cell culture systems (Liston et al., 1996;Deveraux and Reed, 1999). Apoptotic hippocampal neurons can be rescued by stereotactic microinjection of NAIP-expressing adenovirus in the CA1 region, suggesting that the antiapoptotic activity of NAIP shown_in vitro_ extends to the in vivo situation (Xu et al., 1997). Recent evidence suggests that in addition to supraphysiologic levels of NAIP conferring neuronal protection, the loss of endogenous NAIP results in enhanced neuronal vulnerability (Holcik et al., 2000), indicating that NAIP plays an important role in regulation of neuronal apoptosis. However, a mechanism for antiapoptotic function of NAIP has not yet been identified. Although other IAP family members have been shown to inhibit caspases directly (Deveraux et al., 1997; Roy et al., 1997; Tamm et al., 1998), a similar interaction could not be demonstrated for NAIP (Roy et al., 1997), suggesting an alternate mechanism in mediating cytoprotection. The present study demonstrates the direct inhibition of effector caspases by NAIP BIR domains and thus provides a mechanistic explanation for the cytoprotective effect of NAIP and its function as an important regulator protein for neuronal apoptosis.
MATERIALS AND METHODS
Generation and cloning of NAIP BIR constructs. NAIP constructs encompassing one to three BIR domains (see Fig.2A) were generated by PCR using the following primer pairs: B123xt, 5′ forward primer GGATCC ATG GCC ACC CAG CAG AAA GCC and 3′ reverse primer CTCGAG CCA GAT GCC CAC AGA AAA GCT AT; B1, 5′ forward primer GGATCC GCA GTT CAG TTG GCA AGG and 3′ reverse primer CTCGAG CTC AGC CTG CTC TTC AGA TT; B2, 5′ forward primer GGATCC AGC AGG CTG AGA GAG GT and 3′ reverse primer CTCGAG GTA ATT TCC TCT GAG GAT TTC; B3, 5′ forward primer GGATCC TCC TCA GAG GAA ATT ACC and 3′ reverse primer CTCGAG ATA GGA CCA ACT GCA TTG AA; B12, 5′ forward primer GGATCC GCA GTT CAG TTG GCA AGG and 3′ reverse primer CTCGAG GTA ATT TCC TCT GAG GAT TTC; B23, 5′ forward primer GGATCC AGC AGG CTG AGA GAG GT and 3′ reverse primer CTCGAG ATA GGA CCA ACT GCA TTG AA; B2A, 5′ forward primer GGATCC GCA TCC TTC AGG AAC TGG and 3′ reverse primer CTCGAG TAT GTC AAC AAA TCC C; B2B, 5′ forward primer GGATCC ATG AGG TAC CAA GAA GAGG and 3′ reverse primer CTCGAG GGG AAC CAT TTG GCA TG; and B2C, 5′ forward primer GGATCC GGG ATA TCC CCT TGT GTG CTC and 3′ reverse primer CTCGAG ATC ATC TCC TTC TTC CCA. The PCR product was subcloned in pCR2.1 vector using the Invitrogen TOPO cloning kit (Invitrogen Canada Inc., Burlington, Ontario, Canada) according to the manufacturer's instructions. After sequencing, the constructs were digested with _Bam_HI and _Xho_I restriction enzymes and subcloned in pGEX-4T3 and pcDNA3.0-myc vectors.
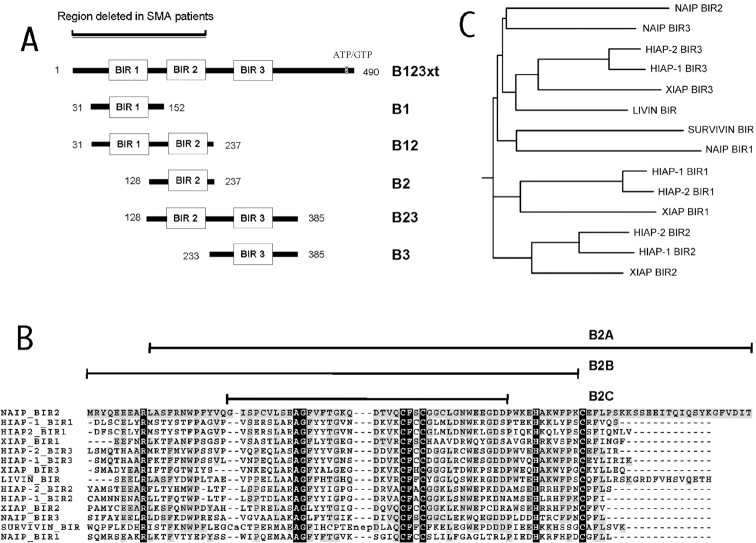
Fig. 2.
A, Illustration of the NAIP deletion constructs generated for caspase inhibition and cell death assays. B, Amino acid sequence alignment of BIR domains of human IAP family members known to have an antiapoptotic effect. The alignment was generated with the program CLUSTALW 1.8 (Thompson et al., 1994) with default settings (http://workbench.sdsc.edu/). The_black_ regions in highlight amino acids conserved in BIR motifs across species as determined by Uren et al. (1998); the gray regions indicate identity with NAIP BIR2 domain. The sequences of NAIP BIR2 deletion constructs (B2A, B2B, B2C) are illustrated above the NAIP BIR2 sequence. C, Phylogenetic tree diagram (rooted) of the BIR sequences aligned in_B_ generated with the program DRAWGRAM (Inference Package, version 3.5; Department of Genetics, University of Washington, Seattle, WA) using NAIP B2 as the leader sequence.
Overexpression and purification of NAIP BIR proteins. The recombinant glutathione _S_-transferase (GST)-NAIP proteins were overexpressed in Escherichia coli strain BL21 (Novagen Inc., Madison, WI). Briefly, overnight bacterial cultures were diluted 1:10 in Luria–Bertani medium. The culture was incubated at 30°C to an OD600 of 1.8, followed by isopropyl-β-d-thiogalactopyranoside induction (final concentration, 0.1 mm; B2, 0.5 mm). One hour after induction, the cells were harvested, and the pellets were resuspended in sodium Tris EDTA buffer [10 mm Tris-Cl, pH 8.0, 150 mm NaCl, 1 mm EDTA, 0.1% 3-[(3-cholamidopropyl)-dimethylammonio]-2-hydroxy-1-propanesulfonic acid (CHAPS), and 5 mm dithiothreitol (DTT)]. Purification was performed by adding glutathione-Sephadex 4B beads (Amersham Biosciences, Piscataway, NJ) to the soluble fraction, followed by washing in protein wash buffer (1× PBS, pH 8.0, containing 5 mm DTT and 0.1% CHAPS). The proteins were then eluted (50 mm Tris-Cl, pH 9.5, 15 mm reduced glutathione, and 0.1% CHAPS). Protein concentrations were measured using a protein assay (Bio-Rad, Hercules, CA) according to the manufacturer's protocol. The purity of the obtained protein was determined by SDS PAGE and was generally >90%.
Reversible caspase inhibition assay. The reversible caspase inhibition assay was performed as described previously (Thornberry et al., 1992; Nicholson et al., 1995) The inhibitory constants were derived by steady-state velocities of enzyme inhibitor complexes generated with a range of inhibitor (NAIP) concentrations and maintaining constant levels of both enzyme (caspase) and substrate (fluorogenic tetrapeptide) with the concentrations of the caspase and tetrapeptide in the range of the known_K_m for the caspase–tetrapeptide interaction with the following modifications. Ac-YVAD-AMC (Biomol, Plymouth Meeting, PA) was used as the substrate for caspase-1 and Ac-DEVD-AMC for caspase-3, caspase-7, and caspase-8. Before reading, the IAP protein was diluted 1:20 in assay buffer followed by serial dilution before addition of substrate (10 μmfinal) and enzyme (50 pm for caspase-3, 2.25 nm for caspase-8, 1.4 nmfor caspase-7, and 1 nm for caspase-1; all final concentrations; final assay volume, 200 μl). The assays were performed in 96-well microtiter plates at room temperature in continuous readings for 30 min [five readings per minute; Cytofluor II; PerkinElmer Life Sciences, Emeryville, CA; λ excitation (ex), 380 nm; λ emission (em), 460 nm]. IC50 values were calculated using the curve-fitting program PROFIT 5.5 for MacIntosh (QuantumSoft Inc., Zurich, Switzerland);_K_i values were calculated according to the formula developed by Cheng and Prusoff (1973). This formula is valid when the binding is competitive and reversible; we know that this is the case for capase-3 and XIAP given the recently published structural analyses of IAP and caspases (Chai et al., 2001; Huang et al., 2001; Riedl et al., 2001). Because substrate concentration and_K_m were kept constant in the assay (both at 10 μm), the_K_i should approximate half of the obtained IC50 value._K_i values were also calculated from the steady-state velocities of the enzyme inhibitor complexes using the equations developed by Morrison and Walsh (1988) for the analysis of slow and tight binding inhibitors. This was done in the event that the binding had a marked time dependence and was not wholly competitive and reversible, in which case the Cheng and Prusoff (1973) equation would not obtain. The fact that equivalent results were obtained by both statistical methods supports the accuracy of the inhibitory constants.
Culture and survival of primary cortical neurons. Mouse cortical neurons (CD1) were cultured from embryonic day 16 (E16) and grown in defined serum-free medium as described previously (Keramaris et al., 2000). For optimal infection of neurons with minimal cytotoxicity, recombinant adenoviral vectors (carrying lacZ,NAIP, or XIAP), at a multiplicity of infection (MOI) of 10 or 20 pfu/cell, were added to cell suspensions immediately before plating. Neurons were cultured up to day 3 and subsequently treated either with camptothecin (10 μm) alone or with camptothecin and boc-aspartyl(OMe)-fluoromethylketone (BAF; 100 μm). Cell survival was assessed by using the colorimetric (3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyltetrazolium bromide (MTT) survival assay (Cell Titer Kit; Promega, Madison, WI), which measures the conversion by mitochondrial enzymes of the tetrazolium salt to a blue formizan salt 24 hr after treatment in an adaptation of the method of Keramaris et al. (2000).
Cell culture and transfection. HeLa cells (CCL2; American Type Culture Collection, Manassas, VA) were grown in DMEM with 4.5 mg/ml d-glucose and pyridoxine hydrochloride (Invitrogen) containing 10% heat-inactivated fetal bovine serum, 2 mml-glutamine, 100 U/ml penicillin G, and 100 μg/ml streptomycin. Twenty-four hours before transfection, cells were plated at a density of 2 × 105 cells/well in 60 mm six-well culture dishes. Cells were transfected by applying the LipofectAMINE Plus system (Invitrogen) using 0.8 μg of each plasmid construct and 0.4 μg of pEGFP-N1 plasmid [expressing green fluorescent protein (GFP)] according to the manufacturer's protocol.
HeLa cell death assay. Twenty-four hours after transfection, the cells were treated with 100 μm etoposide in dimethylsulfoxide (DMSO) or with DMSO (control) for 8 hr before being returned to complete media. Seventy-two hours after treatment, the supernatant was removed and centrifuged to collect floating cells. The adherent cells were trypsinized and then combined with the floating cells by centrifugation and washed with cold PBS. After a second centrifugation, the cells were resuspended in 500 μl of cold PBS. Five minutes before flow cytometry analysis, 5 μl of 1 mg/ml propidium iodide (PI) was added to the cells. Flow cytometry was performed using an EPICS XL flow cytometry system (Beckman-Coulter Inc., Fullerton, CA), and analysis of cell viability was performed using the software WinMDI 2.8 (http://facs.scripps.edu). GFP- and PI-positive cells were counted in control and etoposide-treated cells transfected with the same construct. Specific apoptosis was calculated in an adaptation of the method of Scaffidi et al. (1998).
RESULTS
NAIP protects primary cortical neurons undergoing caspase-3-dependent apoptotic death
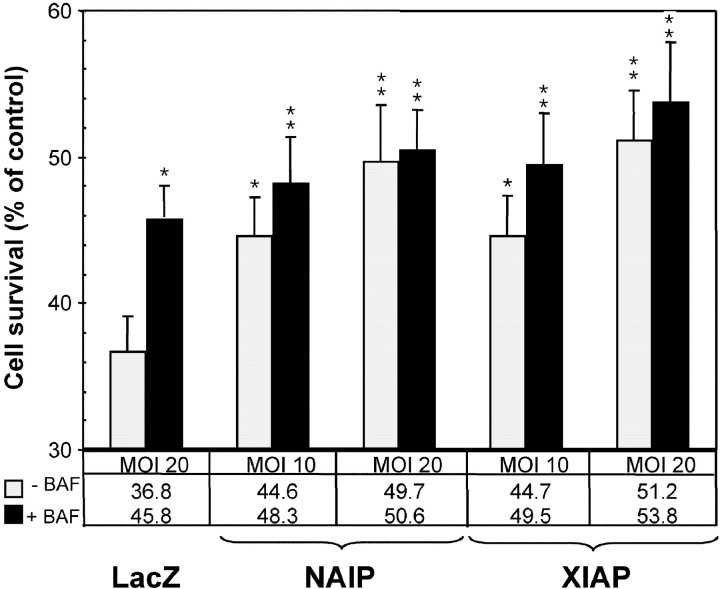
To elucidate the antiapoptotic mechanism mediated by NAIP, we sought to study its action in a physiologically relevant context. Previous work has shown that the genotoxic agent camptothecin evokes apoptotic death of cultured embryonic (E16) mouse cortical neurons through activation of caspase-3 (Stefanis et al., 1999; Keramaris et al., 2000). To clarify whether the chief means of neuroprotection by NAIP is mediated through a caspase-dependent or -independent mechanism, as suggested by the results of a previous in vitro study (Roy et al., 1997), an adenoviral vector was used to deliver full-length NAIP to cortical neurons undergoing apoptosis induced by the DNA-damaging agent camptothecin. The effect of NAIP in this system was compared with the IAP family member XIAP, shown previously to inhibit effector caspases directly (Deveraux et al., 1997), as well as with an adenovector carrying the lacZ gene (Ad-lacZ) used as a vector control. Cultured neuronal cells, infected with lacZ at an MOI of 20, exhibit 36.8 ± 2.3% survival 24 hr after treatment with camptothecin, as determined by MTT assay (Fig. 1). In contrast, neurons expressing NAIP exhibited a significantly prolonged survival in an MOI dose-dependent manner (Fig. 1); e.g., a significant delay in cell death was obtained with 44.6 ± 2.6% survival of these neurons when an MOI of 10 was used (Fig. 1) and 49.7 ± 3.8% with an MOI of 20 (Fig. 1). Comparable results were obtained with the caspase inhibitor XIAP (44.7 ± 2.8% at MOI 10 and 51.2 ± 3.5% at MOI 20) (Fig. 1). To establish whether the neuroprotective effect observed with NAIP is attributable to inhibition of caspases alone or via a caspase-independent mechanism, the identical experiments were conducted in the presence of the general caspase inhibitor BAF. The application of BAF (100 μm) alone to camptothecin-treated neurons expressing LacZ resulted in prolonged survival (45.8 ± 2.2% with an MOI of 20) (Fig. 1). However, when neurons were infected with Ad-NAIP at an MOI of 10 in addition to BAF, a small but significant (p < 0.05, Student's t test) increase in cell survival was observed (49.5 ± 3.2%) (Fig. 1). When Ad-NAIP was used at an MOI of 20, the addition of BAF resulted in no significant increase in cell survival (50.6 ± 2.7% with BAF treatment compared with 49.7 ± 3.8% without) (Fig. 1). Similar results were obtained when Ad-XIAP was used under identical conditions (Fig. 1). Together, the results of these studies indicate that NAIP expression can protect cortical neurons primarily through either direct inhibition of caspases or a caspase-dependent pathway.
Fig. 1.
Cytoprotection of Ad-NAIP in primary cortical neurons treated to undergo caspase-3-dependent cell death. The neurons were infected with Ad-lacZ, Ad-NAIP, or Ad-XIAP at the MOI indicated at the bottom. After 3 d of culture, the cells were treated with camptothecin alone (10 μm) or with addition of the pancaspase inhibitor BAF (100 μm). Control samples were kept untreated. A neuronal survival assay (MTT) was assessed 24 hr later. Data are expressed as number of surviving cells treated with camptothecin alone or with BAF versus nontreated cells (taken as 100%), and are the mean ± SEM of six independent experiments. *p < 0.05; **p < 0.01; Student's t test.
The N terminus of NAIP can protect HeLa cells undergoing caspase-3-dependent apoptosis
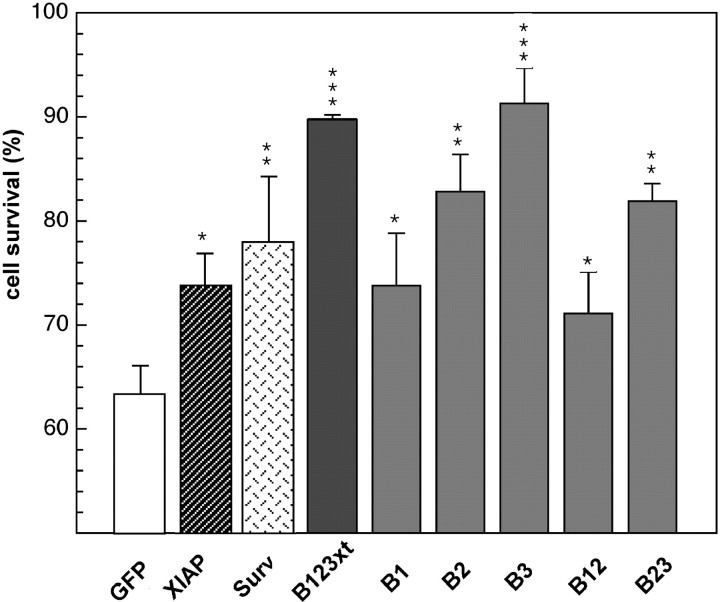
In view of the above findings, additional experiments to establish the results in an alternate cellular system were undertaken. When treated with the topoisomerase II inhibitor etoposide, HeLa cells are reported to undergo apoptosis characterized by elevated levels of caspase-3 (Liu, 1989; Mizukami et al., 1999). In these studies, we used a N-terminal construct of NAIP encompassing the three BIR domains (Fig.2A, B123xt), shown previously to be sufficient to mediate an antiapoptotic effect (Liston et al., 1996). HeLa cells were transiently transfected with pcDNA3.0-myc B123xt and treated with etoposide. The effect was compared with cells transfected with pcDNA3.0-myc-XIAP or pcDNA3.0-myc-Survivin, IAP family members shown previously to confer cytoprotection in a similar system by inhibiting caspase-3 and caspase-7 (Tamm et al., 1998). Cotransfection with pEGFP-N1 (expressing GFP) allows the identification of transfected cells. After an incubation period of 72 hr, cell viability for GFP-positive cells was determined by flow cytometry. The results of four experiments revealed significant cytoprotection by all IAP constructs applied in comparison with the samples transfected with the empty vector (63.3 ± 6.3%) (Fig.3), with the NAIP B123xt construct exhibiting the most profound cytoprotective effect (89.8 ± 0.4%), followed by Survivin (78.2 ± 6.3%) and XIAP (73.8 ± 3.2%) (all Fig. 3). Together, these results support the finding that expression of NAIP is cytoprotective when cells undergo caspase-dependent apoptosis and that this effect is mediated via its BIR domains.
Fig. 3.
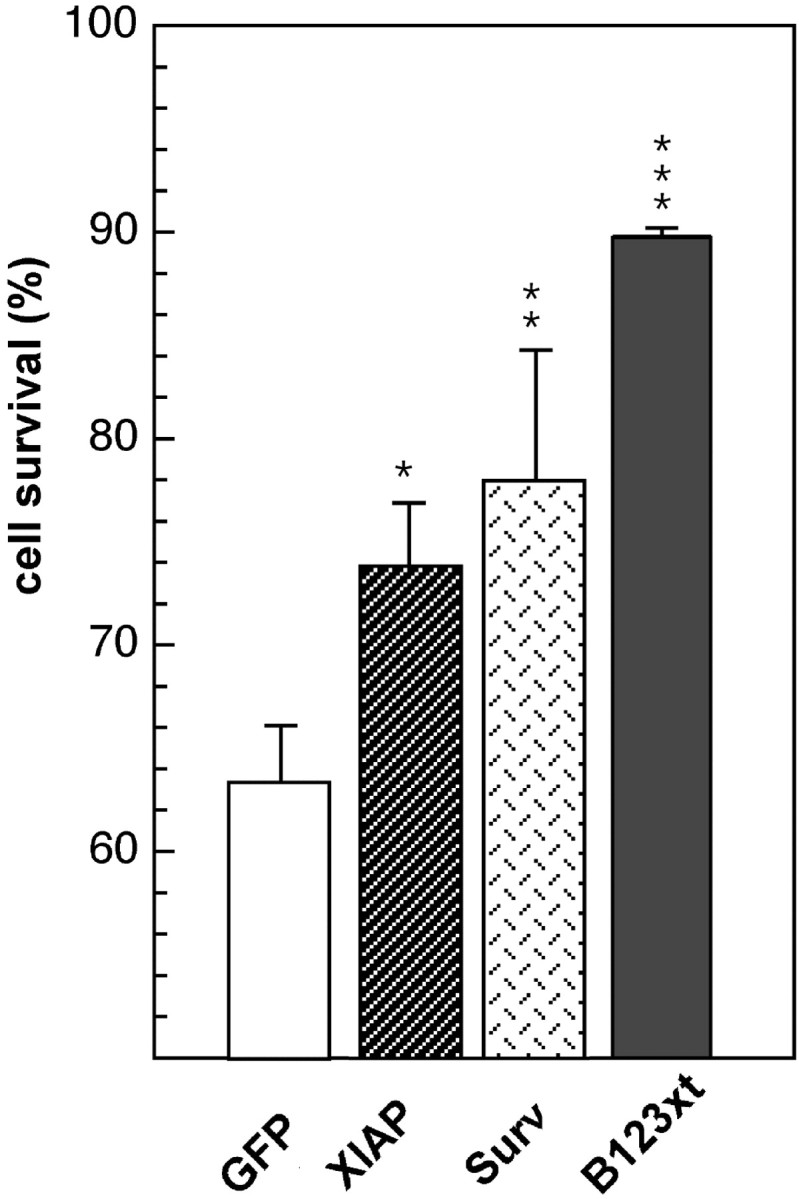
Cytoprotection assay of apoptotic HeLa cells transfected with IAP constructs. HeLa cells were transiently cotransfected with the pEGFP-N1 vector and either pcDNA3.0-myc vector alone (control) or pcDNA3.0-myc containing B123xt, XIAP, or Survivin (Surv) and treated with the topoisomerase II inhibitor etoposide for 8 hr (control samples were kept untreated). Seventy-two hours after treatment, cell death was assessed in GFP-positive cells by flow cytometry. Results (mean ± SEM) of four independently performed experiments are shown. Cell survival was calculated in an adaptation of the method of Scaffidi et al. (1998). *p < 0.05; **p < 0.01; ***p < 0.001; Student's _t_test.
Recombinant N-terminal NAIP protein selectively inhibits caspase-3 and caspase-7
We then overexpressed and purified a recombinant GST-NAIP fusion protein encompassing the three BIR domains (Fig. 2A,B123xt) to evaluate the possibility that NAIP directly inhibits caspases. Peptide aldehydes that reversibly bind and suppress caspase activity with high potency (Garcia-Calvo et al., 1998) as well as a recombinant GST-XIAP protein were used as enzyme inhibition controls. The GST-XIAP protein was purified and treated under conditions identical to those used for the NAIP B123xt protein.
On the basis of the classification of caspases (Thornberry et al., 1997), caspase-1 (interleukin-1 converting enzyme) was selected as a representative of group I caspases (caspase-1, caspase-4, and caspase-5) and tested for direct inhibition by purified GST-B123xt. This enzyme could not be inhibited by NAIP or XIAP, even when high concentrations of protein (NAIP, 3 μm; and XIAP, 5 μm) were used (Fig.4A). The same result was obtained when NAIP and XIAP were tested against caspase-8, a representative member of the group III caspases (Fig.4B). However, very potent and specific inhibition was obtained when NAIP was tested against members of the group II caspases, also termed effector caspases, because of the multitude of cellular and structural proteins that serve as their substrates (Nicholson, 1999). Figure 5A depicts the specific inhibition of caspase-7 by NAIP (_K_i, 50 nm), identical to that observed for XIAP (_K_i, 49 nm). Additional assays were then conducted to evaluate the potential for inhibition of the most prominent member of group II caspases, caspase-3. Figure 5B represents a typical experiment (n = 6) of caspase-3 inhibition by NAIP. As is the case for caspase-7, B123xt is able to inhibit caspase-3 but with a much higher affinity. The _K_i values of several independent experiments conducted to substantiate the potency of B123xt for caspase-3 inhibition ranged from 7 to 21 nm (14 ± 5 nm; Table1). Control proteins such as GST and SMN as well as the elution buffer itself showed no inhibitory effect on caspases (data not shown).
Fig. 4.
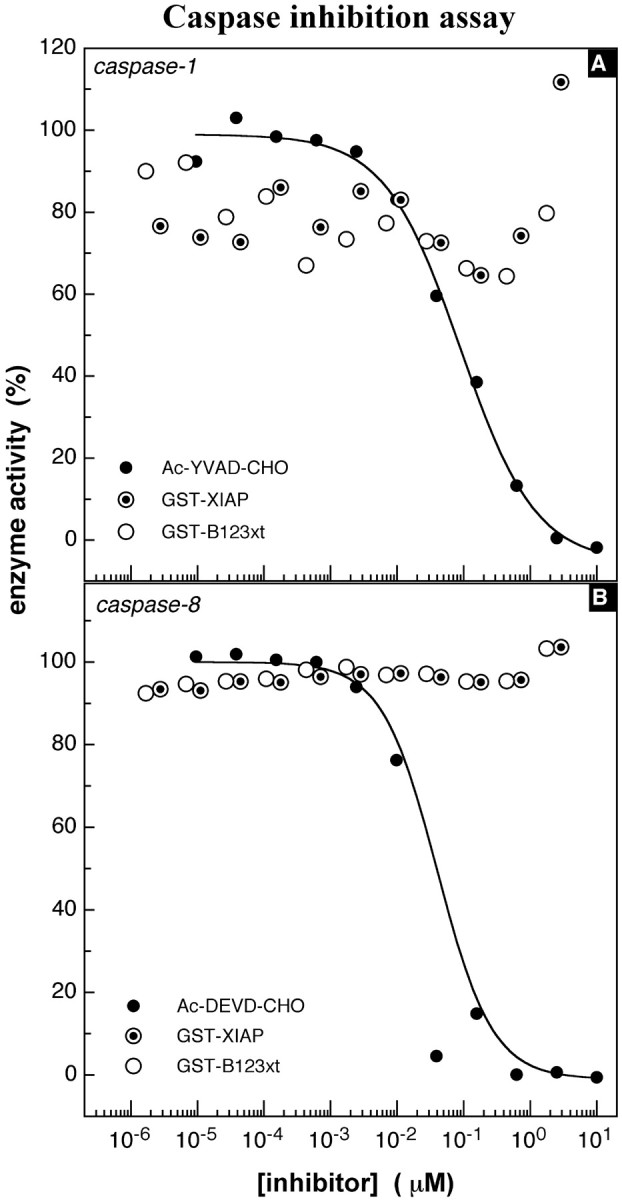
Caspase inhibition assay, group I and III caspases. A, Inhibition of caspase-1, a representative member of the group I caspases. The tetrapeptide Ac-YVAD-AMC (10 μm) served as the substrate. Enzyme concentration was held constant at 1 nm. The peptide aldehyde_Ac-YVAD-CHO_ was used as control for enzyme inhibition at an initial concentration of 10 μm. Continuous readings (λex, 380 nm; λem, 460 nm) were performed for 30 min at room temperature. The IC50 of the control inhibitor_Ac-YVAD-CHO_ was 74 nm. B, Inhibition of caspase-8, a representative member of group III caspases. The assay was performed as described above with a substrate concentration (Ac-DEVD-AMC) of 10 μm and an enzyme concentration of 2.25 nm. The peptide aldehyde_Ac-DEVD-CHO_ was applied as inhibition control, inhibiting the enzyme with an IC50 of 18 nm.
Fig. 5.
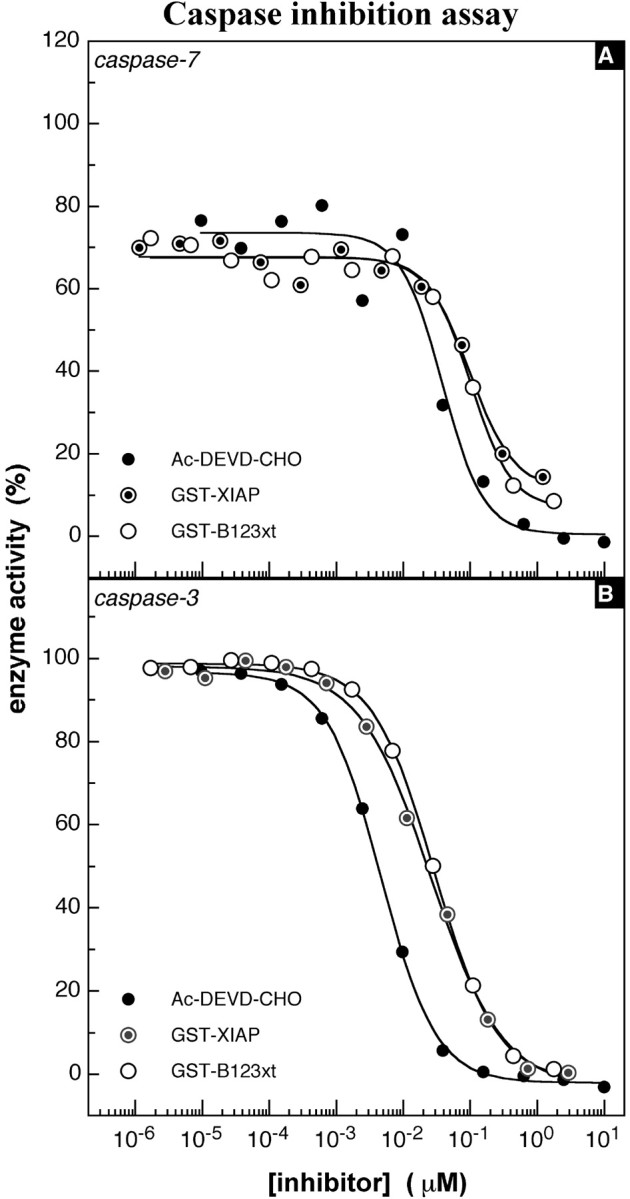
Caspase inhibition assay, group II caspases.A, Representative illustration of caspase-7 inhibition. The IC50 of NAIP (B123xt) was 101 nm in this experiment; XIAP was 99 nm; and_Ac-DEVD-CHO_ was 39 nm. The enzyme concentration in this experiment was 1.4 nm.B, Representative experiment of caspase-3 inhibition by NAIP (B123xt). The assay was performed as described above using caspase-3 at 50 pm. The IC50 values obtained from this experiment were 23 nm for_XIAP_, 28 nm for NAIP (B123xt), and 4 nm for the control inhibitor_Ac-DEVD-CHO_.
Table 1.
_K_i values of NAIP B123xt, B2, and XIAP obtained in caspase-3 inhibition assays
| XIAP (nm) | B123xt (nm) | B2 (nm) |
|---|---|---|
| 11 | 14 | 9 |
| 26 | 21 | 8 |
| 16 | 16 | 21 |
| 14 | 17 | 9 |
| 15 | 7 | 9 |
| 9 | 5 | |
| 16 ± 6 | 14 ± 5 | 10 ± 6 |
A single NAIP BIR domain is sufficient for caspase-3 inhibition
To investigate which BIR domains in combination or isolation inhibit caspase-3, NAIP deletion constructs comprising either single or dual BIR (Fig. 2A) were cloned into bacterial expression vectors, overexpressed as GST fusion proteins, purified, and tested for potential caspase-3 inhibition. In Figure6A, a representative result for caspase-3 inhibition using proteins containing two NAIP BIR domains in relation to control inhibitors, peptide aldehyde, or GST-XIAP is shown. The B12 protein shows no significant enzyme inhibition, whereas B23 inhibits caspase-3 with a low potency in comparison with full-length XIAP and B123xt. The IC50 value obtained from this experiment was 614 nm for B23, corresponding to a_K_i of 307 nm. When proteins comprising single NAIP BIR motifs were tested in inhibition assays, a wide range of inhibition potencies was observed. The GST-BIR1 (B1) protein did not inhibit caspase-3 activity (Fig. 6B) in contrast to GST fusion proteins containing either the BIR3 or BIR2 domain. GST-B3 shows a weaker inhibition profile, similar to that observed with the B23 protein (_K_i, 195 and 276 nm, respectively). Potent inhibition of caspase-3 was observed with the BIR2 construct (_K_i, 9) (Fig. 6B), similar to that seen for the protein containing all three BIR domains (Fig. 5B, B123xt). The profound inhibitory effect of the BIR2 protein was substantiated in additional experiments yielding K_i values of 5 and 21 nm (10 ± 6 nm; Table1), demonstrating that a single BIR domain of NAIP is sufficient to fully mediate the inhibitory effect. On the basis of these findings, we tested additional truncations of the BIR2 domain to delineate the minimum requirement for caspase-3 inhibition. Three constructs were generated (Fig, 2_B, B2A, B2B,B2C) with deletions of amino acids highly conserved within all known BIR motifs (Uren et al., 1998) (Fig. 2B). None of these BIR2 deletion proteins was able to inhibit caspase-3 (data not shown).
Fig. 6.
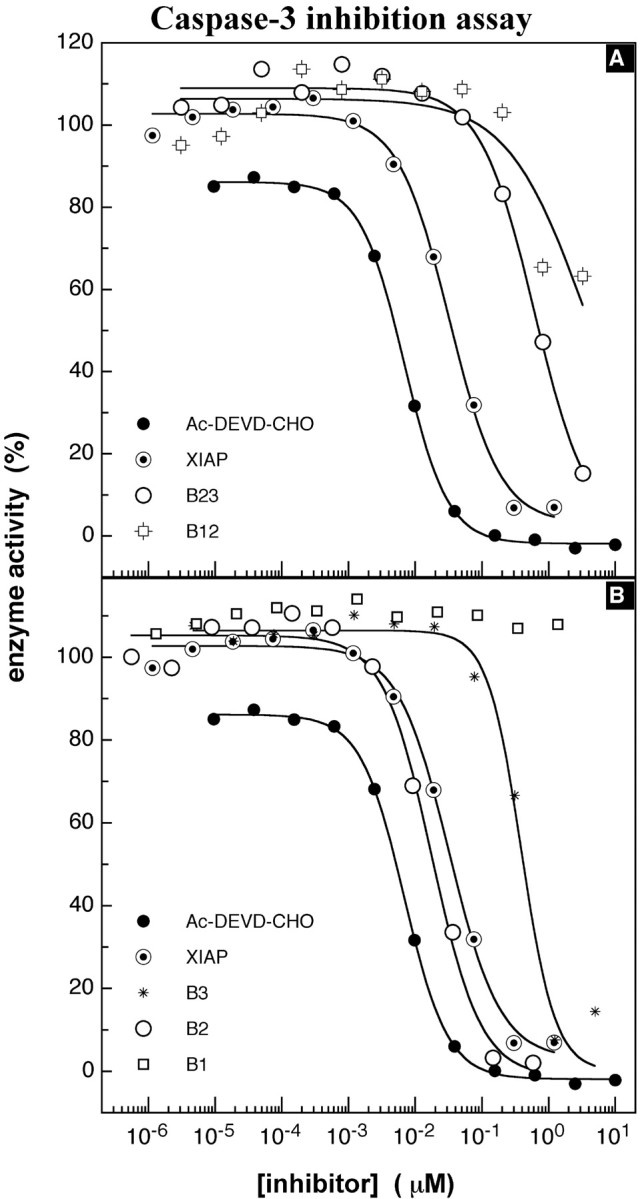
Caspase-3 inhibition assay, NAIP BIR deletion proteins. A, Caspase-3 inhibition assays of NAIP BIR deletion constructs using NAIP proteins with two BIR domains. The NAIP_B12_ protein does not inhibit caspase-3,B23, with an IC50 of 614 nm, ∼20-fold lower than XIAP, with an IC50 of 32 nm. B, Typical experiment of caspase-3 inhibition by NAIP proteins containing a single BIR domain. The experimental procedure is described above and revealed IC50values for B3 of 348 nm and for_B2_ of 17 nm; values of control inhibitors are as in A.
NAIP BIR deletion constructs show inhibition in cell death assays
After the demonstration that a single BIR domain is sufficient to inhibit caspases, a correlation between the in vitro_BIR-mediated caspase inhibition and cytoprotection was sought. The BIR deletion constructs were subcloned in a pcDNA3.0-myc vector and transiently transfected along with pEGFP-N1 into HeLa cells, applying the conditions described above. Expression of NAIP deletion proteins was confirmed by Western blot analysis using an anti-myc antibody (data not shown). The results of four experiments are illustrated in Figure7 and reveal cytoprotection by all NAIP BIR constructs used when compared with the control sample (63.3 ± 3.2%) (Fig. 7), with B3 showing the most profound cytoprotective effect (91.3 ± 3.4%), followed by B2 (82.8 ± 3.6%) and B1 (73.8 ± 5%). The proteins containing two BIR domains also conferred significant protection, with 71.2 ± 4% for B12 and 81.9 ± 1.7% for B23 (Fig. 7). These results show that NAIP proteins with the greatest capability for caspase inhibition_in vitro (B123xt, B3, B2, and B23) conferred the greatest cytoprotection in the HeLa cell death assays, consistent with a causal link between caspase inhibition and cellular survival.
Fig. 7.
Cytoprotection assay of apoptotic HeLa cells transfected with NAIP BIR deletion constructs. Description and procedure are as described in Figure 3. Surv, Survivin.
DISCUSSION
The first human IAP family member, NAIP, was cloned in 1995 as the result of a search for the genetic cause of the neurodegenerative disorder SMA. A number of laboratories have shown that NAIP is antiapoptotic and cytoprotective in a variety of cellular and in vivo models (Liston et al., 1996; Xu et al., 1997; Perrelet et al., 2000), suggesting that NAIP is an important regulator of neuronal apoptosis. However, a mechanism of NAIP action has not yet been described. In this report, we demonstrate that the BIR domains of NAIP are potent inhibitors of effector caspases, suggesting a mechanism for the antiapoptotic effect of NAIP.
Analysis of representative enzymes of all three groups of caspases demonstrate an inhibition of caspases that is apparently selective and restricted to group II caspases, with_K_i values as low as ∼15 nm for caspase-3 and ∼45 nm for caspase-7 (Fig. 5, Tables 1,2). The comparison of_K_i values obtained from our experiments reveals no difference between the caspase inhibitory properties of NAIP and XIAP (Figs. 4-6), an IAP family member shown previously to inhibit caspases (Deveraux et al., 1997). A previously published study assaying for an in vitro recombinant NAIP and caspase interaction failed to detect any binding to caspase (Roy et al., 1997) for reasons that are unclear. The N-terminal NAIP construct used in our study included the putative ATP and GTP binding site downstream from the third BIR (Fig. 2A,B123xt), adding 160 amino acids to the protein used in the previous study (Roy et al., 1997). The inclusion of these amino acids may permit proper protein folding. Generally, a strong tendency of NAIP BIR proteins toward oligomerization and precipitation was observed during the course of purification, especially with highly soluble proteins such as B23 and B3. The BIR–BIR interaction revealed in x-ray crystallographic analysis of the IAP family member Survivin (Verdecia et al., 2000) is consistent with a model of NAIP BIR domain self association.
Table 2.
_K_i values of NAIP peptides and XIAP in caspase-3 and caspase-7 inhibition assays
| Peptides | Caspase-3 (nm) | Caspase-7 (nm) |
|---|---|---|
| XIAP | 16 ± 6 | 49 |
| B123xt | 14 ± 5 | 50 |
| B1 | >1000 | |
| B2 | 10 ± 6 | |
| B3 | 185 ± 15 | |
| B12 | >1000 | |
| B23 | 287 ± 20 |
The effect of NAIP as a specific and potent inhibitor of group II caspases was also assessed in a context of greater physiological relevance. In view of the pivotal role of caspase-3 in mediating neuronal apoptosis, we investigated the effect of NAIP and caspase inhibition in apoptotic neuronal cells. When exposed to the DNA-damaging agent camptothecin, primary cortical neurons show characteristic features of apoptosis and elevated levels of caspase-3 (Stefanis et al., 1999; Keramaris et al., 2000). As shown in Figure 1, infection of camptothecin-treated neurons with an adenovirus expressing either NAIP or XIAP led to similar levels of survival of these neurons. This effect was dose-dependent, because the application of an MOI of 20 resulted in a higher protection than seen with an MOI of 10 (Fig. 1). The additional treatment of NAIP-expressing neurons with the pancaspase inhibitor BAF led to increased survival only when a low MOI was used (Fig. 1). The lack of an additional cytoprotective effect with BAF at a high MOI (Fig. 1) strongly suggests that a caspase-3-dependent apoptotic pathway is inhibited by NAIP. The results obtained are in accordance with cytoprotection studies using cerebellar granule neurons deprived of K+ and infected with identical adenoviral constructs (Simons et al., 1999). This previous study also showed caspase-3-dependent cell death inhibited by expression of NAIP or XIAP (Simons et al., 1999). These results, together with the inhibition data obtained from our in vitro assays, strongly suggest that the neuroprotective effect is mediated either in part or wholly as a consequence of direct inhibition of effector caspases by NAIP.
To define the motif(s) that are necessary and sufficient for caspase inhibition, constructs encompassing either single or dual BIR were generated. The purified recombinant GST fusion proteins were tested for caspase-3 inhibition. The experiments clearly demonstrate that the caspase-inhibiting domain can be reduced to the second or third BIR domain of NAIP or both. With B2, _K_ivalues were almost equal to those documented with the N-terminal NAIP protein B123xt and full-length XIAP, ranging from 5 to 21 nm (Fig. 6, Tables 1, 2). Therefore, the second BIR domain is sufficient to mediate the full inhibitory effect of NAIP on group II caspases. Deletions of highly conserved amino acids within this critical BIR (as shown in Fig. 2B) resulted in proteins unable to inhibit caspase-3 (data not shown), suggesting that the intact BIR motif is necessary to mediate the caspase inhibitory effect. A full potential for caspase-3 inhibition is also reported for the second BIR domain of XIAP (Takahashi et al., 1998), underlining the importance of this BIR in mediating an apoptotic resistance. However, recent publications revealing the structure of the XIAP complexed with caspase-3 and caspase-7 showed binding to effector caspases with the XIAP BIR1/2 linker region (Chai et al., 2001; Huang et al., 2001; Riedl et al., 2001). It is important to note that this caspase binding motif is not conserved among IAPs as is the BIR domain (Fig.2B,C); it will be of interest to delineate the caspase binding motif of the NAIP B2 protein. A study addressing the inhibition kinetics of linker region and BIR2 domain on caspase-3 is currently under way and shall hopefully shed more light on this issue. The recent crystallographic structural analyses have revealed unexpected details of the XIAP–caspase interaction. A DISD tetrapeptide lying outside and ∼11 amino acids upstream of the NH2 border of the XIAP BIR2 domain has been found to be critical for the interaction of both caspase-3 and caspase-7, binding to the catalytic groove of both proteases (Sun et al., 1999; Chai et al. 2001;Huang et al., 2001; Suzuki et al., 2001). This is the same groove to which the tetrapeptide DEVD caspase inhibitor binds. A scan of the NAIP sequence for a similar tetrapeptide mapping upstream of any of the three BIRs, assuming a similar mechanism of binding and inhibitory action, reveals a DIRV motif that maps 15 amino acids upstream of the BIR2. This domain, which binds most avidly to caspase 3, is included in all the BIR2-containing polypeptides used in this study. Thus it may be that the motif contributes to the caspase binding and inhibition of BIR2. The difference in the size and polarity of arginine (bulky and positive) from that of serine (small and uncharged) and charge dissimilarity between valine (uncharged) and aspartate (negative) makes such a possibility somewhat less likely, but such binding is currently being assessed.
The potency of NAIP B2 protein in inhibiting effector caspases correlates in its ability to protect cells from undergoing apoptosis. When HeLa cells are treated with the anticancer drug etoposide, apoptosis and elevated levels of activated caspase-3 are observed (Yang et al., 1997; Mizukami et al., 1999). Transfection of a single NAIP BIR2 construct in HeLa cells resulted in significant cytoprotection, with 83% cell survival compared with ∼60% of cells transfected with the control plasmid (Fig. 7). In Figure 2, B and_C_, sequence alignment of the BIR domains of human IAPs is illustrated, showing a close sequence identity between the second and third BIRs of NAIP. In this context, it might be expected that recombinant B3 protein would also inhibit caspase-3 strongly; however, the K_i values obtained from our_in vitro experiments are ∼20- to 30-fold lower than for the B2 protein (Fig. 6). In the cell death assays, B3 shows significantly higher protection than B2 (91 vs 83%; _p_= 0.0434) (Fig. 7). When evaluating the subcellular localization of B2 and B3 in transfected HeLa cells, we find a predominant staining of the hydrophobic B2 protein localizing to the membrane in contrast to the cytoplasmic and nuclear staining seen with B3 (data not shown). It may be that this membrane association impairs the ability of B2 to inhibit caspase activity when compared with the more soluble and accessible B3. Thus, cellular sublocalization in addition to caspase affinity appears to be an important factor in the cytoprotective potency of a given protein. Less cytoprotection was observed for the B12 and B1 proteins, correlating with their weaker caspase inhibition. Nonetheless, the cytoprotective effect of these constructs is significant (Fig. 7). Given the strong cytoprotective effects of BIR constructs that inhibit caspase-3 directly, such as B123xt, B23, B2, and B3 (Figs. 5, 6), we assume that the recombinant proteins (B1 and B12) used were misfolded but retain reduced capability for caspase inhibition when expressed in cells.
Together, the results from our study provide strong support for a model in which NAIP, more specifically the BIR2 domain, is playing a critical role in regulation of neuronal apoptosis by directly inhibiting effector caspases. It is perhaps of significance that two-thirds of SMA type I and a smaller fraction of type II and III patients have homozygous deletions in exons 5 and 6 of the NAIP gene (Roy et al., 1995) (Fig. 2A). Deletions of these exons would result in either the complete absence of NAIP or the generation of a protein lacking BIR2 domain (Fig. 2A). This could potentially result in an impaired or abolished ability to inhibit the effector caspases, resulting in enhanced levels of death in the SMN-deficient motor neurons, thus exacerbating the clinical severity of SMA.
Footnotes
This work was supported by grants from the Canadian Institutes of Health Research, the Muscular Dystrophy Association of Canada, and Aegera Therapeutics Inc. A.E.M. is a Burroughs-Wellcome clinical translation awardee.
Correspondence should be addressed to Dr. Alex E. MacKenzie, Children's Hospital of Eastern Ontario Research Institute, 401 Smyth Road, Ottawa, Ontario, Canada K1H 8L1. E-mail:alex@mgcheo.med.uottawa.ca.
REFERENCES
- 1.Chai J, Shiozaki E, Srinivasula SM, Wu Q, Dataa P, Alnemri ES, Shi Y. Structural basis of caspase-7 inhibition by XIAP. Cell. 2001;104:769–780. doi: 10.1016/s0092-8674(01)00272-0. [DOI] [PubMed] [Google Scholar]
- 2.Chen J, Nagayama T, Jin K, Stetler RA, Zhu RL, Graham SH, Simon RP. Induction of caspase-3-like protease may mediate delayed neuronal death in the hippocampus after transient cerebral ischemia. J Neurosci. 1998;18:4914–4928. doi: 10.1523/JNEUROSCI.18-13-04914.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng Y, Prusoff WH. Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50 percent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 4.Cotman CW. Apoptosis decision cascades and neuronal degeneration in Alzheimer's disease. Neurobiol Aging. 1998;19:S29–S32. doi: 10.1016/s0197-4580(98)00042-6. [DOI] [PubMed] [Google Scholar]
- 5.Deveraux QL, Reed JC. IAP family proteins—suppressors of apoptosis. Genes Dev. 1999;3:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 6.Deveraux QL, Takahashi R, Salvesen GS, Reed JC. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 7.Emery AE. Clinical and genetic heterogeneity in spinal muscular atrophy—the multiple allele model. Neuromuscular Dis. 1991;1:307–308. doi: 10.1016/0960-8966(91)90106-3. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Calvo M, Peterson EP, Leiting B, Ruel R, Nicholson DW, Thornberry NA. Inhibition of human caspases by peptide-based and macromolecular inhibitors. J Biol Chem. 1998;273:32608–32613. doi: 10.1074/jbc.273.49.32608. [DOI] [PubMed] [Google Scholar]
- 9.Hartmann A, Hunot S, Michel PP, Muriel MP, Vyas S, Faucheux BA, Mouatt-Prigent A, Turmel H, Srinivasan A, Ruberg M, Evan GI, Agid Y, Hirsch EC. Caspase-3: a vulnerability factor and final effector in apoptotic death of dopaminergic neurons in Parkinson's disease. Proc Natl Acad Sci USA. 2000;97:2875–2880. doi: 10.1073/pnas.040556597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holcik M, Thompson CS, Yaraghi Z, Lefebvre CA, MacKenzie AE, Korneluk RG. The hippocampal neurons of neuronal apoptosis inhibitory protein 1 (NAIP1)-deleted mice display increased vulnerability to kainic acid-induced injury. Proc Natl Acad Sci USA. 2000;97:2286–2290. doi: 10.1073/pnas.040469797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Y, Park YC, Rich RL, Segal D, Myszka DG, Wu H. Structural basis of caspase inhibition by XIAP: differential roles of the linker versus the BIR domain. Cell. 2001;104:781–790. [PubMed] [Google Scholar]
- 12.Keramaris E, Stefanis L, MacLaurin J, Harada N, Takaku K, Ishikawa T, Taketo MM, Robertson GS, Nicholson DW, Slack RS, Park DS. Involvement of caspase 3 in apoptotic death of cortical neurons evoked by DNA damage. Mol Cell Neurosci. 2000;15:368–379. doi: 10.1006/mcne.2000.0838. [DOI] [PubMed] [Google Scholar]
- 13.Kuida K, Zheng TS, Na S, Kuan C, Yang D, Karasuyama H, Rakic P, Flavell RA. Decreased apoptosis in the brain and premature lethality in CPP32-deficient mice. Nature. 1996;384:368–372. doi: 10.1038/384368a0. [DOI] [PubMed] [Google Scholar]
- 14.Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, Le Paslier D, Freza F, Cohen D, Weissenbach J, Munnich A, Melki J. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 15.Liston P, Roy N, Tamai K, Lefebvre C, Baird S, Cherton-Horvat G, Farahani R, McLean M, Ikeda JE, MacKenzie A, Korneluk RG. Suppression of apoptosis in mammalian cells by NAIP and a related family of IAP genes. Nature. 1996;379:349–353. doi: 10.1038/379349a0. [DOI] [PubMed] [Google Scholar]
- 16.Liu LF. DNA topoisomerase poisons as antitumor drugs. Annu Rev Biochem. 1989;58:351–375. doi: 10.1146/annurev.bi.58.070189.002031. [DOI] [PubMed] [Google Scholar]
- 17.Mizukami S, Kikuchi K, Higuchi T, Urano Y, Mashima T, Tsuruo T, Nagano T. Imaging of caspase-3 activation in HeLa cells stimulated with etoposide using a novel fluorescent probe. FEBS Lett. 1999;453:356–360. doi: 10.1016/s0014-5793(99)00755-3. [DOI] [PubMed] [Google Scholar]
- 18.Morrison JF, Walsh CT. The behavior and significance of slow-binding enzyme inhibitors. Adv Enzymol Relat Areas Mol Biol. 1988;61:201–301. doi: 10.1002/9780470123072.ch5. [DOI] [PubMed] [Google Scholar]
- 19.Morrison KE. Advances in SMA research: review of gene deletions. Neuromuscular Dis. 1996;6:397–408. doi: 10.1016/s0960-8966(96)00368-9. [DOI] [PubMed] [Google Scholar]
- 20.Namura S, Zhu J, Fink K, Endres M, Srinivasan A, Tomaselli KJ, Yuan J, Moskowitz MA. Activation and cleavage of caspase-3 in apoptosis induced by experimental cerebral ischemia. J Neurosci. 1998;18:3659–3668. doi: 10.1523/JNEUROSCI.18-10-03659.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ni B, Wu X, Su Y, Stephenson D, Smalstig EB, Clemens J, Paul SM. Transient global forebrain ischemia induces a prolonged expression of the caspase-3 mRNA in rat hippocampal CA1 pyramidal neurons. J Cereb Blood Flow Metab. 1998;18:248–256. doi: 10.1097/00004647-199803000-00003. [DOI] [PubMed] [Google Scholar]
- 22.Nicholson DW. Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ. 1999;6:1028–1042. doi: 10.1038/sj.cdd.4400598. [DOI] [PubMed] [Google Scholar]
- 23.Nicholson DW, Ali A, Thornberry NA, Vaillancourt JP, Ding CK, Gallant M, Gareau Y, Griffin PR, Labelle M, Lazebnik YA, Munday NA, Raju SM, Smulson ME, Yamin TT, Yu VL, Miller DK. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 24.Oppenheim RW. Cell death during development of the nervous system. Annu Rev Neurosci. 1991;14:453–501. doi: 10.1146/annurev.ne.14.030191.002321. [DOI] [PubMed] [Google Scholar]
- 25.Perrelet D, Ferri A, MacKenzie AE, Smith GM, Korneluk RG, Liston P, Sagot Y, Terrado J, Monnier D, Kato AC. IAP family proteins delay motoneuron cell death in vivo. Eur J Neurosci. 2000;12:2059–2067. doi: 10.1046/j.1460-9568.2000.00098.x. [DOI] [PubMed] [Google Scholar]
- 26.Riedl SJ, Renatus M, Schwarzenbacher R, Zhou Q, Sun C, Fesik SW, Liddington RC, Salvesen GS. Structural basis for the inhibition of caspase-3 by XIAP. Cell. 2001;104:791–800. doi: 10.1016/s0092-8674(01)00274-4. [DOI] [PubMed] [Google Scholar]
- 27.Roy N, Mahadevan MS, McLean M, Shutler G, Yaraghi Z, Farahani R, Baird S, Besner-Johnston A, Lefebvre C, Kang X, Salih M, Aubry H, Tamai K, Guan X, Ioannou P, Crawford TO, de Jong PJ, Surh L, Ikeda J, Korneluk RG, MacKenzie A. The gene for neuronal apoptosis inhibitory protein is partially deleted in individuals with spinal muscular atrophy. Cell. 1995;80:167–178. doi: 10.1016/0092-8674(95)90461-1. [DOI] [PubMed] [Google Scholar]
- 28.Roy N, Deveraux QL, Takahashi R, Salvesen GS, Reed JC. The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO J. 1997;16:6914–6925. doi: 10.1093/emboj/16.23.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scaffidi C, Fulda S, Srinivasan A, Friesen C, Li F, Tomaselli KJ, Debatin KM, Krammer PH, Peter ME. Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 1998;17:1675–1687. doi: 10.1093/emboj/17.6.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simons M, Beinroth S, Gleichmann M, Liston P, Korneluk RG, MacKenzie AE, Bahr M, Klockgether T, Robertson GS, Weller M, Schulz JB. Adenovirus-mediated gene transfer of inhibitors of apoptosis protein delays apoptosis in cerebellar granule neurons. J Neurochem. 1999;72:292–301. doi: 10.1046/j.1471-4159.1999.0720292.x. [DOI] [PubMed] [Google Scholar]
- 31.Stefanis L, Park DS, Friedman WJ, Greene LA. Caspase-dependent and -independent death of camptothecin-treated embryonic cortical neurons. J Neurosci. 1999;19:6235–6247. doi: 10.1523/JNEUROSCI.19-15-06235.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun C, Cai M, Gunaselcera AH, Meadows RP, Wang H, Chen J, Zhang H, Wu W, Xu N, Ng SC, Fesik SW. NMR structure and mutagenesis of the inhibitor-of-apoptosis protein XIAP. Nature. 1999;401:818–822. doi: 10.1038/44617. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki Y, Nakabayashi Y, Nakata K, Reed JC, Takahashi R. X-linked inhibitor of apoptosis protein (XIAP) inhibits caspase-3 and -7 in distinct modes. J Biol Chem. 2001;20:27058–27063. doi: 10.1074/jbc.M102415200. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi R, Deveraux Q, Tamm I, Welsh K, Assa-Munt N, Salvesen GS, Reed JC. A single BIR domain of XIAP sufficient for inhibiting caspases. J Biol Chem. 1998;273:7787–7790. doi: 10.1074/jbc.273.14.7787. [DOI] [PubMed] [Google Scholar]
- 35.Tamm I, Wang Y, Sausville E, Scudiero DA, Vigna N, Oltersdorf T, Reed JC. IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95) Bax caspases and anticancer drugs. Cancer Res. 1998;58:5315–5320. [PubMed] [Google Scholar]
- 36.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, Miller DK, Molineaux SM, Weidner JR, Aunins J, Elliston KO, Ayala JM, Casano FJ, Chin J, Ding GJF, Egger LA, Gaffney EP, Kimjuco G, Palyha OC, Raju SM, Rolando AM, Salley JP, Yamin TT, Lee TD, Shively JE. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature. 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 38.Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, Houtzager VM, Nordstrom PA, Roy S, Vaillancourt JP, Chapman KT, Nicholson DW. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 39.Uren AG, Coulson EJ, Vaux DL. Conservation of baculovirus inhibitor of apoptosis repeat proteins (BIRPs) in viruses, nematodes, vertebrates and yeasts. Trends Biochem Sci. 1998;23:159–162. doi: 10.1016/s0968-0004(98)01198-0. [DOI] [PubMed] [Google Scholar]
- 40.Verdecia MA, Huang H, Dutil E, Kaiser DA, Hunter T, Noel JP. Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement. Nat Struct Biol. 2000;7:602–608. doi: 10.1038/76838. [DOI] [PubMed] [Google Scholar]
- 41.Xu DG, Crocker SJ, Doucet JP, St-Jean M, Tamai K, Hakim AM, Ikeda JE, Liston P, Thompson CS, Korneluk RG, MacKenzie A, Robertson GS. Elevation of neuronal expression of NAIP reduces ischemic damage in the rat hippocampus. Nat Med. 1997;3:997–1004. doi: 10.1038/nm0997-997. [DOI] [PubMed] [Google Scholar]
- 42.Yakovlev AG, Knoblach SM, Fan L, Fox GB, Goodnight R, Faden AI. Activation of CPP32-like caspases contributes to neuronal apoptosis and neurological dysfunction after traumatic brain injury. J Neurosci. 1997;17:7415–7424. doi: 10.1523/JNEUROSCI.17-19-07415.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J, Liu X, Bhalla K, Kim CN, Ibrado AM, Cai J, Peng TI, Jones DP, Wang X. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]