Prion disease and the ‘protein-only hypothesis’ (original) (raw)
. Author manuscript; available in PMC: 2019 Sep 25.
Published in final edited form as: Essays Biochem. 2014;56:181–191. doi: 10.1042/bse0560181
Abstract
Prion disease is the only naturally occurring infectious protein misfolding disorder. The chemical nature of the infectious agent has been debated for more than half a century. Early studies on scrapie suggested that the unusual infectious agent might propagate in the absence of nucleic acid. The ‘protein-only hypothesis’ provides a theoretical model to explain how a protein self-replicates without nucleic acid, which predicts that prion, the proteinaceous infectious agent, propagates by converting its normal counterpart into the likeness of itself. Decades of studies have provided overwhelming evidence to support this hypothesis. The latest advances in generating infectious prion with bacterially expressed recombinant prion protein in the presence of cofactors not only provide convincing evidence supporting the ‘protein-only hypothesis’, but also indicate a role of cofactors in forming prion infectivity and encoding prion strains. In the present chapter, we review the literature regarding the chemical nature of the infectious agent, describe recent achievements in proving the ‘protein-only hypothesis’, and discuss the remaining questions in this research area.
Keywords: infectious protein, prion infectivity, prion protein conversion, prion strain, ‘protein-only hypothesis’, recombinant prion, transmissible spongiform encephalopathy
Introduction
TSEs (transmissible spongiform encephalopathies), also known as prion diseases, are a group of fatal neurodegenerative disorders that can be manifested as sporadic, inherited or acquired forms [1,2]. Prion disease affects a wide variety of mammals including kuru disease or vCJD (variant Creutzfeldt–Jacob disease) in humans, scrapie in sheep, BSE (bovine spongiform encephalopathy) in cattle and CWD (chronic wasting disease) in deer and elk [1,3]. It shares the characteristics of late-age onset, accumulation of misfolded protein aggregates in the central nervous system, and neurodegeneration with a large group of disorders including Alzheimer’s and Parkinson’s diseases. Despite the similarities, prion disease is the only naturally occurring infectious protein misfolding disorder that can be transmitted within and, in rare occasions, between species [4].
The infectious agent in TSEs has been intensely investigated and overwhelming evidence supports that ‘prion’, a proteinaceous infectious particle, is responsible for the transmissibility. The ‘protein-only hypothesis’ predicts that a prion conveys its infectious structural information to its normally folded non-infectious counterpart, leading to the transmission of disease. The following sections summarize studies regarding the chemical nature of the infectious agent, describe the latest advances in generating infectious prion with bacterially expressed recPrP (recombinant prion protein), and discuss the potential role of cofactors in forming highly infectious prions and in enciphering the baffling prion strain phenomenon.
Exploring the chemical nature of the scrapie agent
Scrapie is the prototype of prion disease affecting sheep and goats, and was the most studied prion disease before rodents were introduced as disease models. Although scrapie was suspected as a contagious disease as early as the mid-18th Century, experimental evidence that scrapie could be transmitted to healthy sheep or goats by inoculating with brain homogenate from sick animals was not attained until the 1930s [5,6]. After establishing that scrapie is a transmissible disease, scientists started exploring the chemical nature of the infectious agent. A bacterium was first excluded because the agent was able to pass an antibacterial filter. A ‘slow virus’ was speculated because of the extraordinarily long incubation times of scrapie (>14 months in sheep and goats). Although extensive attempts failed to identify such a viral agent, those studies revealed unexpected properties of the agent that survives many common procedures to inactivate viruses, including formalin treatment, boiling in water, extracting with organic solvents, digesting with nucleases, UV and ionizing radiations. These unusual properties led to alternative theories positing that the infectious agent could be a protein, a polysaccharide or a fragment of lipid membrane [7].
The ‘protein-only hypothesis’
Despite the unusual characteristics, the mainstream thought remained that scrapie was caused by a novel viral agent containing nucleic acids (DNA or RNA) as the genetic information carrier. British scientist Tikvah Alper et al. [8] used ionizing radiation to determine the size of the scrapie agent based on the idea that the target size can be calculated from the dosage of electron beam used to inactivate the biological activity. Extrapolated from the exceptionally high electron dose required to inactivate scrapie infectivity, they concluded that the size of the scrapie agent was extraordinarily small, much smaller than bacteriophage, the smallest known virus at that time. More importantly, they found that scrapie infectivity remained after a high dosage of UV irradiation that would destroy nucleic acids, suggesting that the agent may replicate without nucleic acid [8]. On the basis of on the unusual characteristics and the radiation results, Pattison and Jones [9] proposed that the scrapie agent could be a self-replicating protein, and Griffith [10], a mathematician without any biological science background, proposed three models to explain how a protein is capable of self-replicating in the absence of nucleic acid. The second model, currently known as the ‘protein-only hypothesis’, was derived from the following thermodynamic equations (eqns 1, 2 and 3) and assumptions.
Equations
Combining eqns (1) and (2), one can derive eqn (3):
Assumptions:
- a′ is the normal cellular protein with a stable structure
- a is in the reactive state bearing a different conformation from a′
- ΔF1 is so large that a′-to-a conversion hardly occurs
- Without pre-existing a2 (the hypothetic infectious protein structure), eqn 2 cannot take place even the reaction is thermodynamically favourable.
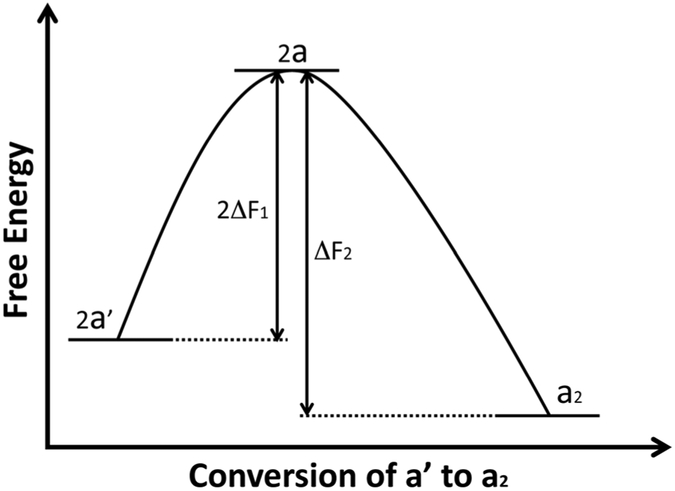
However, when ΔF2 is larger than 2ΔF1 and pre-formed a2 is present as a template, normal cellular protein a′ could proceed to form new a2 (eqn 3), completing the self-replication of a2. Therefore if the scrapie agent is composed of protein and acts like a2, this model readily explains how a proteinaceous agent, without nucleic acid, could self-replicate after being introduced to the healthy animals where normal cellular protein a′ is present. Figure 1 illustrates a simplified thermodynamic diagram of a′-to-a2 conversion.
Figure 1. Conversion from a′ to a2.
When ΔF2 is larger than 2ΔF1, the conversion from a′ into a2 is a thermodynamically favourable reaction. According to the ‘protein-only hypothesis’, in the presence of pre-existing a2 template, the reaction can take place indefinitely, completing the self-replication of a2 in the absence of nucleic acid. a′, the stable normal cellular protein; a, the reactive state of a′; a2, dimer formed from two units of a.
This model describes the thermodynamic feasibility for an infectious proteinaceous agent to self-replicate via propagating its conformation. However, this putative infectious protein remained elusive for a long period of time.
Prion: the proteinaceous infectious particle
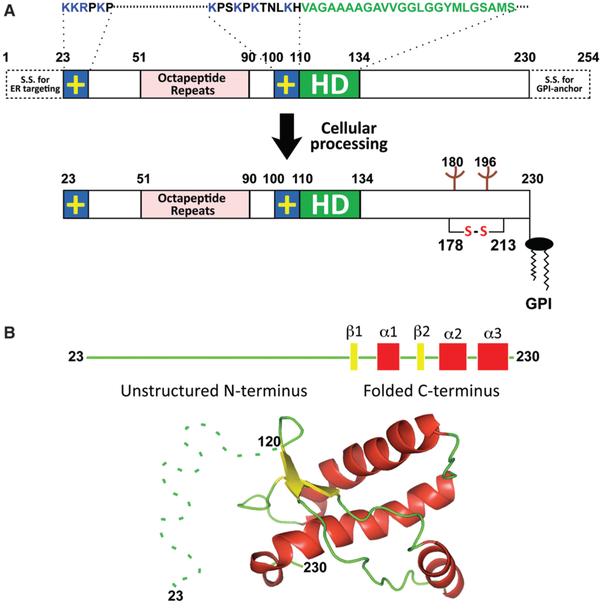
In an effort to isolate the scrapie infectivity using differential sedimentation, detergent extraction and enzymatic digestion, Stanley Prusiner et al. [11] discovered that the infectivity in diseased hamster brain homogenates could be enriched in a fraction that was mostly composed of a partially protease-resistant protein with apparent molecular mass of 27–30 kDa. In 1982, Prusiner postulated that the agent is a ‘prion’, the ‘small proteinaceous infectious particles that are resistant to inactivation by most procedures that modify nucleic acids’ [11]. Purification of the PrP (prion protein) of 27–30 kDa, also known as PrP27–30, led to the identification of the PrP gene, Prnp, a single copy chromosomal gene that is highly conserved in mammals. The Prnp gene encodes a PrPC (normal cellular prion protein), which is primarily expressed in the central nervous system and, at much lower levels, in several peripheral tissues. The primary translation product of Prnp contains an N-terminal signal sequence targeting PrP to the secretory pathway, five octapeptide repeats, a highly conserved central hydrophobic domain, a globular C-terminal domain consisting of three α-helices and a short stretch of β-strands, and a signal sequence for adding a GPI (glycosylphosphatidylinositol) anchor (Figure 2). After removing N- and C-terminal signal sequences, adding N-linked sugars to two asparagine residues, and forming the single disulfide bond between two cysteine residues, the mature PrPC localizes at the cell surface and attaches to the plasma membrane by its GPI anchor. Although PrPC is expressed in both healthy and scrapie-affected animals, the scrapie associated PrP27–30 can only be isolated from diseased brain homogenates after limited protease digestion. The disease-specific conformation of PrP is denoted as PrPSc, which shares the same primary amino acid sequence of PrPC, but differs drastically in protein conformation, resulting in distinct properties (Table 1). PrPC is highly α-helical, soluble in mild detergents and sensitive to protease digestion. In contrast, PrPSc is mainly β-sheet, highly aggregated and partially resistant to PK (proteinase-K) digestion.
Figure 2. Schematic illustrations of mouse PrP.
(A) Mouse PrP contains two positively charged (+) amino acid clusters (blue), five octapeptide repeats (magenta) and a hydrophobic domain (green). S.S., signal sequence. After cellular processing, mature mouse PrP23–230 attaches to the cell membrane by GPI anchor after N- and C-terminal signal sequences are removed, N-linked carbohydrates are added to Asn180 and Asn196, and a single disulfide bond forms between Cys178 and Cys213. (B) Mouse PrP is composed of an unstructured N-terminus and a globular C-terminus consisting of three α-helices and two short β-strands. The image of tertiary structure of mouse PrP120–230 (PDB code 1AG2) was generated in PyMOL. Dashed line is added to represent the unstructured N-terminus.
Table 1.
Differences between PrPC and PrPSc
| PrPC | PrPSc |
|---|---|
| Non-infectious | Infectious |
| Rich in α-helical content | Highly β-sheeted |
| Soluble in mild detergents | Aggregated in mild detergents |
| Sensitive to protease digestion | Partially resistant to protease digestion |
Conversion of non-infectious PrPC into infectious PrPSc
After identifying the scrapie-infectivity-associated prion, Prusiner further proposed that the prion, composed entirely or principally of PrPSc, the misfolded isoform of normal PrPC, is the proteinaceous infectious agent and self-replication of PrPSc involving PrPC-to-PrPSc conversion induces prion diseases in hosts. The observation that PrP-knockout mice were resistant to prion infection strongly supports that PrP is essential for disease pathogenesis [12,13].
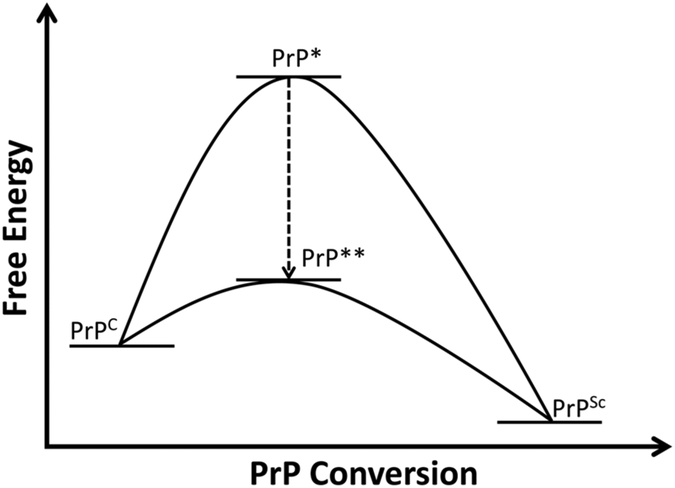
Given the aggregated nature of PrPSc, it is almost impossible to purify it to homogeneity to prove that PrPSc is the infectious agent. Based on the idea that PrPSc self-replication, the catalytic conversion of PrPC into PrPSc, would produce an unlimited amount of newly formed PrPSc, a series of in vitro studies was carried out to correlate PrPSc with prion infectivity. According to the thermodynamic model [8], PrPC with a stable structure needs to reach the reactive state, PrP*, before it converts into PrPSc. As PrPSc and PrPC represent two distinct conformations of the same protein (Table 1), the reactive PrP* must represent at least partially unfolded intermediate PrP species between the mainly α-helical PrPC and the β-sheet-rich PrPSc (Figure 3). For PrPC to reach this PrP* conformational state, exogenous energy is required to overcome the large energy barrier between PrPC and PrPSc. Alternatively, other facilitations, such as denaturants or PrP-binding molecules, may lower the energy barrier and allow the conversion to occur.
Figure 3. PrP conversion.
To fulfil self-replication of PrPSc, the stable normal PrPC needs to reach the reactive state, PrP* or PrP**, to further convert into PrPSc. Factors that lower the energy barrier would allow the reaction to go through the PrP** state, facilitating the PrPC-to-PrPSc conversion.
The first cell-free conversion assay used a denaturant, GuHCl (guanidine hydrochloride), at low concentration to facilitate the PrPSc-seeded conversion [14]. Mixing partially purified PrPSc with purified 35S-labelled PrPC led to the formation of PK-resistant radioactive PrP species, which co-aggregated with unlabelled PrPSc seed. This cell-free conversion assay undoubtedly demonstrated the seeding capability of PrPSc, yet the low conversion efficiency resulted in an excessive amount of unlabelled PrPSc seed in the final product, making it difficult to assess the infectivity of newly generated radioactive PrPSc. To enhance the conversion efficiency, a new technique named PMCA (protein misfolding cyclic amplification) was developed, in which a mixture of a limited amount of crude diseased brain homogenate containing PrPSc and excess normal brain homogenate containing PrPC was subjected to successive sonication and incubation cycles. After reaction, newly formed PrPSc can be detected by PK-digestion assay and used to seed a new round of PrPC conversion, allowing indefinite propagation of PrPSc [15]. More importantly, after sufficient PMCA rounds that dilute out the original PrPSc seed from diseased brain homogenates, the newly formed PrPSc is capable of inducing bona fide prion disease in wild-type animals [16].
Compared with the low efficiency of the cell-free conversion assay, the ability of PMCA to efficiently propagate PrPSc and prion infectivity might be attributed to the following aspects. Sonication in PMCA is believed to fragment large PrPSc aggregates into smaller particles, increasing the PrPSc–PrPC contacting surfaces and resulting in more efficient conversion. On the other hand, sonication can certainly transfer energy to PrPC helping it to reach the reactive PrP* state, or partially unfold PrPC to facilitate the conversion. Therefore it is likely that with proper setups (in the form of power and duration of sonication, the incubation time and temperature), a certain amount of the reactive PrP intermediate, PrP*, could be generated from PrPC, which is further converted into PrPSc under the catalytic influence of pre-existing PrPSc seed.
The concomitant propagation of PrPSc and the prion infectivity by PMCA strongly supports the ‘protein-only hypothesis’. However, due to the use of crude brain homogenates in this assay, it is still difficult to conclusively pinpoint that PrPSc is the infectious agent. One can argue that other components in the brain homogenates, such as a small fragment of nucleic acid, may have been propagated during the PMCA, which might be the culprit in transmitting the disease. Notably, the ‘Virino theory’ postulates that the scrapie agent could be a nucleic acid–PrPSc complex. Although the nucleic acid carries the genetic information for transmitting the disease, PrPSc serves as the protective coat allowing the disease-causing nucleic acid to survive all the harsh treatments [17].
It is widely accepted that the most stringent proof for the ‘protein-only hypothesis’ would be the generation of infectious PrPSc from pure non-infectious PrPC. Owing to its denaturation/refolding purification procedures and remarkably similar tertiary structure to PrPC [18], purified bacterially expressed recPrP has been regarded as the purest available PrP species and is widely used in PrP conversion studies. Because scrapie-associated PrP27–30 forms short amyloid fibres after detergent extraction and protease digestion, it was reasoned that the _in vitro_-formed PrP amyloid fibrils might possess prion infectivity. Soluble, monomeric and mainly α-helical recPrP has been successfully converted into amyloid fibrils in the presence of denaturing chaotropic agents, such as GuHCl or urea [19,20]. Despite the similarities to PrPSc (in being highly aggregated, rich in β-sheet and with strong in vitro seeding capability), recPrP amyloid fibrils induced prion disease only in transgenic mice overexpressing PrP after a prolonged incubation period (an indication of minimal infectivity), but failed to cause disease in wild-type animals [21,22].
The minimal infectivity associated with recPrP amyloid fibrils may suggest that the _in vitro_-generated recPrP amyloid fibrils still have large structural differences from the infectious PrPSc. Alternatively, other non-PrP cofactors, which are not present in the pure recPrP amyloid fibril system, might be essential for generating the infectious PrPSc conformer. The latter possibility is consistent with a significant difference in PrP conversion efficiency between the cell-free and PMCA assays. These non-PrP cofactors in brain homogenates used in the PMCA reaction may interact with PrPSc and/or PrPC to facilitate the conversion. Notably, polyanions, such as proteoglycans and nucleic acids have been shown to bind PrP, induce conformational changes of PrP and promote PrP conversion in vitro [23-25].
In addition to polyanions, lipids are also a plausible candidate for facilitating PrP conversion. The GPI-anchored PrPC is in the vicinity of lipid membranes and PrP–lipid interactions have long been implicated in PrPC-to-PrPSc conversion. PrPC can be released from lipid membranes after PI-PLC (phosphoinositide-specific phospholipase C) cleavage of the GPI anchor. However, PI-PLC digestion failed to release PrPSc from lipid membranes, indicating an additional mode of interaction between PrPSc and lipid membranes. Moreover, it has been shown that a direct PrP–lipid interaction is required for PrP conversion in the presence of lipid membranes in a modified cell-free PrP conversion assay [07].
The PrP–lipid interaction has been verified by experiments showing that bacterially expressed recPrP binds to synthetic liposomes and the binding destabilizes the well-structured C-terminal domain of recPrP [26]. The recPrP–lipid interaction is initiated by the electrostatic binding between positively charged amino acid residues of recPrP and negatively charged anionic phospholipid headgroups, which is followed by the hydrophobic interactions between recPrP hydrophobic domain and lipid acyl chains. The lipid interaction converts α-helical structured recPrP into a β-sheet-rich, C-terminal PK-resistant conformation, both of which are biochemical hallmarks of PrPSc [27]. These observations indicate that similar to denaturant treatment, PrP–lipid interactions are able to unfold recPrP to another stable conformational state.
The similar biochemical properties of lipid-bound recPrP to those of infectious PrPSc led to the hypothesis that recPrP-lipid interactions may lead recPrP to reach the reactive PrP* state and thereby lower the energy barrier for the conversion into PrPSc (Figure 3). This hypothesis was tested by PMCA using a substrate mixture of bacterially expressed recPrP plus two cofactors: a negatively charged phospholipid [POPG (1-palmitoyl-2-oleoyl-_sn_-glycero-3-phospho-(10-rac-glycerol)] and polyanions (total RNA isolated from normal mouse liver). Indeed, recPrPSc generated by this approach not only possesses all the hallmarks of diseased brain-derived PrPSc (aggregated, C-terminal PK-resistant, capable of seeding the conversion of PrPC in normal brain homogenate by PMCA and converting PrPC in cultured cells to create a chronic infected state), but also induces prion disease in wild-type mice after a short incubation period and with a relatively synchronized onset, indicating a high degree of specific prion infectivity [28]. When the total mouse liver RNA is replaced by synthetic poly(rA) (polyriboadenylic acid), the resulting recPrPSc is equally infectious and causes prion disease in wild-type animals with a 100% attack rate. Since poly(rA) does not contain meaningful genetic information, the latter experiment reveals that the role of poly(rA) is to facilitate PrP conformational change instead of providing genetic information for the infectivity, and disapproves the ‘Virino theory’ [29]. Generation of recPrPSc in vitro with defined cofactors strongly supports that the pathogen in prion disease is a protein-conformation-based infectious agent [28-30].
Cofactors: possible roles in prion infectivity and prion strains
Thus far, it remains unclear whether cofactors are essential for the infectivity or just contribute as a chaperone to facilitate PrP to reach the infectious conformation. The most recent success in generating infectious recPrPSc with only a single cofactor, synthetic PE (phosphatidylethanolamine), indicates that polyanions such as RNA are not essential for prion infectivity [31]. However, whether lipid is required for prion infectivity remains unanswered. Early ionizing radiation studies indicated a role of lipids in maintaining high prion infectivity [32]. Recent attempts to generate infectious prions with bacterially expressed recPrP showed that the infectivity of pure recPrP amyloid fibrils or recPrPSc formed by PMCA [33,34] in the absence of any cofactor is very low, but the infectivity of recPrPSc generated by PMCA in the presence of a lipid cofactor is much higher. These results suggest that lipid cofactors might be important for PrP to gain and/or maintain the highly infectious conformation.
A puzzling observation in prion disease is the presence of multiple strains. The ‘protein-only hypothesis’ explains the prion strain phenomenon by variations in PrPSc conformation. However, the fact that a single protein can stably exist in multiple conformations (>20 prion strains in mouse) is difficult to reconcile with the thermodynamic rules of protein folding. If the stable infectious PrPSc conformation is maintained by forming a PrPSc–cofactor complex, it is not difficult to envision that different cofactors (e.g. phospholipids with different headgroups) or different PrP/cofactor ratios would result in multiple stable infectious PrPSc conformations. A recent study showed that propagating three prion strains to recPrP with PE as the sole cofactor led to the convergence of three strains to a single new strain, supporting a role of cofactors in modulating prion strain phenotype [35]. If cofactors indeed contribute to the formation of prion strains, it would bring the peculiar prion strain phenomenon back to the protein-folding paradigm.
Conclusions
After decades of intense research and heated debate, the latest studies provide unequivocal evidence supporting that a protein-conformation-based infectious agent is responsible for the transmissibility of prion disease (or TSEs). If cofactors are essential for prion infectivity, does it disapprove the ‘protein-only hypothesis’? If one interprets ‘protein-only’ in the strictest manner that no other factors are required in the propagation of the infectious PrPSc conformers, then the requirement of a cofactor, even as a chaperone, would be inconsistent with the hypothesis. However, it might be more plausible to interpret the ‘protein-only hypothesis’ as the genetic information of prion infectivity is carried only by protein conformation. In this case, even if cofactors are required for the infectivity, the information of infectivity is still governed by protein conformation, which is consistent with the ‘protein-only hypothesis’. Further studies to elucidate the role(s) of cofactors in prion infectivity and the formation and evolution of diverse prion strains would lead to a better understanding of the enigmatic agent in prion diseases. Moreover, the generation of recombinant prions in vitro makes it possible to study the high-resolution three-dimensional structure of the infectious PrPSc, which would provide a molecular basis for explaining the puzzling biological observations and for developing diagnostic and therapeutic tools. The clean recombinant prion system also offers a valuable platform to investigate the molecular mechanism of prion propagation and to screen for compounds that inhibit prion propagation. These studies are not only important for us to combat the devastating prion diseases, they may also shed light on the mechanism of recently discovered ‘prion-like’ propagation of misfolded proteins in a variety of more common neurodegenerative disorders [36].
Summary.
- Prion diseases are a group of infectious illnesses affecting humans and animals.
- The infectious agent in prion disease has been proposed as prion, an infectious protein that is capable of self-propagating in the absence of nucleic acid.
- The ‘protein-only hypothesis’ posits that prion self-replicates by conveying the infectious protein conformation to its normally folded counterpart.
- Various PrP conversion studies have provided unequivocal evidence supporting the ‘protein-only hypothesis’.
- Generating highly infectious recombinant prions with bacterially expressed recPrP in the presence of defined cofactors supports that a protein-conformation-based infectious agent is responsible for the infectivity in prion disease.
- Experiments suggest that cofactors may play a role in maintaining the highly infectious prion conformation and encoding various prion strains.
References
- 1.Prusiner SB (1998) Prions. Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguzzi A, Baumann F and Bremer J (2008) The prion’s elusive reason for being. Annu. Rev. Neurosci. 31, 439–477 [DOI] [PubMed] [Google Scholar]
- 3.Sigurdson CJ and Aguzzi A (2007) Chronic wasting disease. Biochim. Biophys. Acta 1772(6), 610–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kraus A, Groveman BR and Caughey B (2013) Prions and the potential transmissibility of protein misfolding diseases. Annu. Rev. Microbiol. 67, 543–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curril J and Chelle PL (1936) Is the disease of scrapie inoculable? Comptes Rendus Hebdomadaires Des Seances De L Academie Des Sciences 203, 1552–1554 [Google Scholar]
- 6.Cuille J and Chelle PL (1939) Experimental transmission of trembling to the goat. C.R. Seances Acad. Sci. 208, 1058–1160 [Google Scholar]
- 7.Wang F and Ma J (2013) Role of lipid in forming an infectious prion? Acta Biochim. Biophys. Sin. (Shanghai). 45, 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alper T, Cramp WA, Haig DA and Clarke MC (1967) Does the agent of scrapie replicate without nucleic acid? Nature 214, 764–766 [DOI] [PubMed] [Google Scholar]
- 9.Pattison IH and Jones KM (1967) The possible nature of the transmissible agent of scrapie. Vet Rec. 80, 2–9 [DOI] [PubMed] [Google Scholar]
- 10.Griffith JS (1967) Self-replication and scrapie. Nature 215, 1043–1044 [DOI] [PubMed] [Google Scholar]
- 11.Prusiner SB (1982) Novel proteinaceous infectious particles cause scrapie. Science 216, 136–144 [DOI] [PubMed] [Google Scholar]
- 12.Bueler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M and Weissmann C (1993) Mice devoid of PrP are resistant to scrapie. Cell 73, 1339–1347 [DOI] [PubMed] [Google Scholar]
- 13.Sailer A, Bueler H, Fischer M, Aguzzi A and Weissmann C (1994) No propagation of prions in mice devoid of PrP Cell 77, 967–968 [DOI] [PubMed] [Google Scholar]
- 14.Kocisko DA, Come JH, Priola SA, Chesebro B, Raymond GJ, Lansbury PT and Caughey B (1994) Cell-free formation of protease-resistant prion protein. Nature 370, 471–474 [DOI] [PubMed] [Google Scholar]
- 15.Saborio GP, Permanne B and Soto C (2001) Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 411, 810–813 [DOI] [PubMed] [Google Scholar]
- 16.Castilla J, Saa P, Hetz C and Soto C (2005) In vitro generation of infectious scrapie prions. Cell 121, 195–206 [DOI] [PubMed] [Google Scholar]
- 17.Kimberlin RH (1982) Scrapie agent: prions or virinos? Nature 297, 107–108 [DOI] [PubMed] [Google Scholar]
- 18.Hornemann S, Schorn C and Wuthrich K (2004) NMR structure of the bovine prion protein isolated from healthy calf brains. EMBO Rep. 5, 1159–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swietnicki W, Petersen R, Gambetti P and Surewicz WK (1997) pH-dependent stability and conformation of the recombinant human prion protein PrP(90–231). J. Biol. Chem. 272, 27517–27520 [DOI] [PubMed] [Google Scholar]
- 20.Baskakov IV, Legname G, Baldwin MA, Prusiner SB and Cohen FE (2002) Pathway complexity of prion protein assembly into amyloid. J. Biol. Chem. 277, 21140–21148 [DOI] [PubMed] [Google Scholar]
- 21.Legname G, Baskakov IV, Nguyen HO, Riesner D, Cohen FE, DeArmond SJ and Prusiner SB (2004) Synthetic mammalian prions. Science 305, 673–676 [DOI] [PubMed] [Google Scholar]
- 22.Colby DW, Wain R, Baskakov IV, Legname G, Palmer CG, Nguyen HO, Lemus A, Cohen FE, DeArmond SJ and Prusiner SB (2010) Protease-sensitive synthetic prions. PLoS Pathog. 6, e1000736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong C, Xiong LW, Horiuchi M, Raymond L, Wehrly K, Chesebro B and Caughey B (2001) Sulfated glycans and elevated temperature stimulate PrP(Sc)-dependent cell-free formation of protease-resistant prion protein. EMBO J. 20, 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deleault NR, Lucassen RW and Supattapone S (2003) RNA molecules stimulate prion protein conversion. Nature 425, 717–720 [DOI] [PubMed] [Google Scholar]
- 25.Deleault NR, Harris BT, Rees JR and Supattapone S (2007) Formation of native prions from minimal components in vitro. Proc. Natl. Acad. Sci. U.S.A. 104, 9741–9746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morillas M, Swietnicki W, Gambetti P and Surewicz WK (1999) Membrane environment alters the conformational structure of the recombinant human prion protein. J. Biol. Chem. 274, 36859–36865 [DOI] [PubMed] [Google Scholar]
- 27.Wang F, Yang F, Hu Y, Wang X, Jin C and Ma J (2007) Lipid interaction converts prion protein to a PrPSc-like proteinase K-resistant conformation under physiological conditions. Biochemistry 46, 7045–7053 [DOI] [PubMed] [Google Scholar]
- 28.Wang F, Wang X, Yuan CG and Ma J (2010) Generating a prion with bacterially expressed recombinant prion protein. Science 327, 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang F, Zhang Z, Wang X, Li J, Zha L, Yuan CG, Weissmann C and Ma J (2012) Genetic informational RNA is not required for recombinant prion infectivity. J. Virol. 86, 1874–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z, Zhang Y, Wang F, Wang X, Xu Y, Yang H, Yu G, Yuan C and Ma J (2013) De novo generation of infectious prions with bacterially expressed recombinant prion protein. FASEB J. 27, 4768–4775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deleault NR, Piro JR, Walsh DJ, Wang F, Ma J, Geoghegan JC and Supattapone S (2012) Isolation of phosphatidylethanolamine as a solitary cofactor for prion formation in the absence of nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 109, 8546–8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alper T, Haig DA and Clarke MC (1978) The scrapie agent: evidence against its dependence for replication on intrinsic nucleic acid. J. Gen. Virol. 41, 503–516 [DOI] [PubMed] [Google Scholar]
- 33.Makarava N, Kovacs GG, Bocharova O, Savtchenko R, Alexeeva I, Budka H, Rohwer RG and Baskakov IV (2010) Recombinant prion protein induces a new transmissible prion disease in wild-type animals. Acta Neuropathol. 119, 177–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JI, Cali I, Surewicz K, Kong Q, Raymond GJ, Atarashi R, Race B, Qing L, Gambetti P, Caughey B and Surewicz WK (2010) Mammalian prions generated from bacterially expressed prion protein in the absence of any mammalian cofactors. J. Biol. Chem. 285, 14083–14087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deleault NR, Walsh DJ, Piro JR, Wang F, Wang X, Ma J, Rees JR and Supattapone S (2012) Cofactor molecules maintain infectious conformation and restrict strain properties in purified prions. Proc. Natl. Acad. Sci. U.S.A. 109, E1938–E1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prusiner SB (2012) Cell biology: a unifying role for prions in neurodegenerative diseases. Science 336, 1511–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]