Visualization of Cranial Motor Neurons in Live Transgenic Zebrafish Expressing Green Fluorescent Protein Under the Control of the Islet-1 Promoter/Enhancer (original) (raw)
Abstract
We generated germ line-transmitting transgenic zebrafish that express green fluorescent protein (GFP) in the cranial motor neurons. This was accomplished by fusing GFP sequences to _Islet-1_promoter/enhancer sequences that were sufficient for neural-specific expression. The expression of GFP by the motor neurons in the transgenic fish enabled visualization of the cell bodies, main axons, and the peripheral branches within the muscles. GFP-labeled motor neurons could be followed at high resolution for at least up to day four, when most larval neural circuits become functional, and larvae begin to swim and capture prey. Using this line, we analyzed axonal outgrowth by the cranial motor neurons. Furthermore, by selective application of DiI to specific GFP-positive nerve branches, we showed that the two clusters of trigeminal motor neurons in rhombomeres 2 and 3 innervate different peripheral targets. This finding suggests that the trigeminal motor neurons in the two clusters adopt distinct fates. In future experiments, this transgenic line of zebrafish will allow for a genetic analysis of cranial motor neuron development.
Keywords: zebrafish, neuron-specific promoter, transgenic, Islet-1, motor neuron, GFP, live visualization
Developing motor neurons in the vertebrate hindbrain are an attractive system in which to study mechanisms of segmentation and the processes regulating specific axonal pathfinding (Guthrie, 1996; Lumsden and Krumlauf, 1996; Pfaff and Kintner, 1998). Chick and mouse are the most frequently used experimental model organisms to study motor neuron development in vertebrates. Recently, the zebrafish has emerged as another useful model system. Several features of zebrafish make them especially amenable for developmental studies, including development of the hindbrain. The embryos are transparent, making it possible to readily visualize internal structures and cells in living zebrafish. The embryonic hindbrain is relatively simple and manipulable (Chandrasekhar et al., 1997, 1998), and the zebrafish is amenable to large-scale genetic studies (Driever et al., 1996; Haffter et al., 1996) that have generated interesting mutations in hindbrain motor neurons (Chandrasekhar et al., 1999).
Despite these potential advantages of zebrafish for the study of motor neuron differentiation in the hindbrain, this system has not yet been fully exploited. No one has ever successfully visualized motor neurons and their axons in live embryos, which would allow a dynamic analysis of migration by differentiating motor neurons and pathfinding by their extending growth cones. The ability to observe the dynamics of neuronal differentiation may facilitate the detection of the effects of genetic mutation or other molecular and cellular manipulations that might otherwise be missed. In Drosophila melanogaster and_Caenorhabditis elegans_, reproducible labeling of specific types of neurons in vivo by expression of GFP under the control of a cell-type-specific promoter has been used as a powerful tool to study development of those neurons (Murray et al., 1998; Zallen et al., 1998). In vertebrates, however, this approach has only been successful under limited conditions (Dynes and Ngai, 1998; Okada et al., 1999), partly because of a lack of appropriate promoters for cell-type-specific expression of GFP or because of difficulty in observation caused by opacity and complexity of the brain.
Recently, two groups have shown that transgenic zebrafish expressing GFP in specific tissues can be reliably generated (Higashijima et al., 1997b; Long et al., 1997). In the present study, we applied this approach to generate zebrafish in which cranial motor neurons could be selectively visualized to provide a new tool for studying motor neuron development. Our strategy was to use the Islet-1(Isl1) gene to drive GFP expression in cranial motor neurons. Isl1 is a member of the LIM/homeobox gene family and is expressed in all postmitotic motor neurons early in their development (Ericson et al., 1992; Korzh et al., 1993; Inoue et al., 1994; Tsuchida et al., 1994; Appel et al., 1995; Tokumoto et al., 1995;Varela-Echavarría et al., 1996; Osumi et al., 1997). Thus, we expected that the promoter/enhancer of the Isl1 gene would be able to drive GFP expression in cranial motor neurons. We cloned genomic fragments of the zebrafish Isl1 gene and identified DNA sequences that could direct expression of GFP in the cranial motor neurons of DNA-injected embryos. We then established a stable transgenic line (Isl1-GFP) of zebrafish that expressed GFP in the cranial motor neurons.
The intensity of GFP fluorescence by the cranial motor neurons in the Isl1-GFP line was sufficiently high to enable clear visualization of the cell bodies, main axons, and the peripheral branches in the target muscles. In addition to cranial motor neurons, GFP was expressed in the cranial sensory neurons, except for those in the trigeminal ganglion. The generation of this transgenic line and the characterization of the GFP-expressing neurons set the stage for a genetic dissection of the development of these neurons in future experiments.
As an initial illustration of the usefulness of the Isl1-GFP line, we characterized in detail the development of the trigeminal motor neurons. We asked whether the rhombomeric location of the trigeminal motor neurons correlated with the targets innervated by their axons. This was analyzed by backlabeling trigeminal motor neurons with selective DiI application to specific trigeminal nerve branches that could be seen because of their GFP fluorescence. We found that trigeminal neurons from rhombomeres 2 and 3 innervate different target muscles. This result, together with our developmental analysis of the trigeminal motor neurons, suggests that trigeminal neurons generated in r2 and r3 adopt distinct fates, supporting the idea that segmental origin dictates the fate of the two populations of trigeminal motor neurons.
MATERIALS AND METHODS
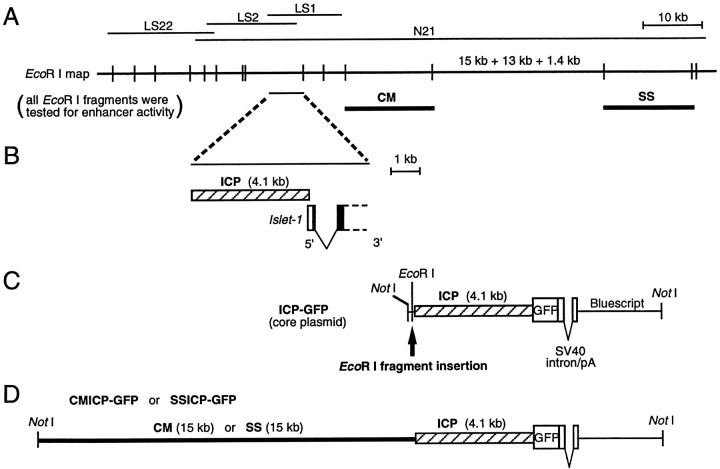
Cloning of the Isl1 genomic DNA, construction of plasmids, and microinjection of DNA into zebrafish embryos. The_Isl1_ cDNA was used to screen a series of zebrafish genomic phage libraries. An initial screening and two rounds of genome walking identified a genomic region of ∼40 kb in total (Fig.1A, LS1,LS2, LS22). Screening of a zebrafish BAC library was performed by Genome Systems, and one BAC clone (Fig.1A, N21), ∼90 kb in length, was isolated. To examine the putative transcriptional start site of the Isl1_gene, RACE (rapid amplification of cDNA ends)–PCR was performed as described previously (Higashijima et al., 1997a) using two_Isl1_-specific primers. The results suggested that the_Isl1 cDNA (Inoue et al., 1994) was almost full-length. Sequence analysis of the corresponding genomic DNA suggested that there are no further introns other than the one shown in Figure1B in the 5′ region of the Isl1 gene. A fragment of DNA corresponding to the 5′ half of LS1 was PCR-amplified. This fragment, termed the putative Isl1 core promoter (ICP), contained 4.1 kb of 5′ upstream region and ∼30 bp of 5′ untranslated region of the Isl1 gene. The fragment had, at the 5′ end, an_Eco_RI site derived from the λ DASHII (Stratagene, La Jolla, CA) phage vector sequence, and a _Kpn_I site at the 3′ end, which was introduced with the PCR primer. The ICP–GFP plasmid was generated in the following manner. The backbone plasmid was pBluescript II SK (Stratagene). The _Not_I site was converted to an_Nco_I site, and then, the _Kpn_I site was converted to an _Not_I site. Synthetic oligonucleotides were used in these experiments. A fragment containing an SV40 poly(A) signal with an intron was extracted from the pcDNA1 plasmid (Invitrogen, San Diego, CA) and introduced between the _Xba_I and _Nco_I sites of the modified pBluescript II SK. The resultant plasmid was digested with _Spe_I and _Xba_I and self-ligated to disrupt these sites. Then, the _Nco_I site at the 3′ end of the insert was converted to an _Xba_I site using a synthetic oligonucleotide. The _Hin_dIII–_Bgl_II fragment encoding the modified GFP (codon-humanized, S65A, Y145F; kindly provided by K. Umesono, Kyoto University) was PCR-amplified, and introduced between the _Hin_dIII site and _Bam_HI site of the resultant plasmid. This GFP-encoding fragment carried a_Kpn_I site upstream of the initiation codon. Then, the_Cla_I site within the remaining multiple cloning site of this plasmid was converted to an _Eco_RI site using a synthetic oligonucleotide. Finally, the ICP–GFP plasmid was created by inserting the ICP fragment between the _Eco_RI and _Kpn_I sites immediately upstream of the GFP-encoding region. The ICP–GFP plasmid therefore had unique _Not_I and _Eco_RI sites just 5′ to the ICP (Fig. 1C). All the recognizable _Eco_RI fragments derived from the cloned region of the Isl1 genomic DNA were individually subcloned into the _Eco_RI site of the ICP–GFP plasmid. Each plasmid DNA was linearized by _Not_I, which is located upstream of the _Eco_RI cloning site and used for microinjection. Preparation of DNA and microinjection was performed essentially as described previously (Higashijima et al., 1997b). Linearized plasmid DNA was extracted using phenol-chloroform and then chloroform, precipitated by ethanol, and dissolved in distilled water. For injection of DNA solution, a holder supporting a glass micropipette with a beveled opening was connected to a 10 ml disposable plastic syringe through a plastic tube. A DNA solution of ∼25 ng/ml was injected into the cytoplasm of a one-cell-stage zebrafish embryo with its chorion intact under dissecting microscope by manually adding air pressure to the syringe. We adjusted the injection volume so that the injected solution spread in a sphere with its diameter ranging from one quarter to one half that of the cytoplasm. For the transient expression assay, the injection volume was empirically adjusted such that, on average, approximately one-fourth of the injected embryos died or became malformed by the next day. When the injected embryos were raised to adulthood to generate germ line-transmitting fish, the injection volume was increased such that one-third to one-half of the injected embryos died or were malformed by the next day. Expression of GFP was examined on the first and second days of development. Three of 113 injected fish turned to be the founders for the transgenic progeny. The frequency of transgenic zebrafish successfully generated using the CMICP–GFP construct was significantly lower than in a previous study when we used constructs that drive GFP expression under the control of the α-actin promoter (20%; Higashijima et al., 1997b). Although the exact reason for this remains unknown, we may have missed transgenic lines with low-level GFP expression.
Fig. 1.
Genomic structure of the Islet-1(Isl1) gene and the plasmid constructs.A, _Eco_RI restriction map of the genomic DNA flanking the Isl1 gene. Lines over the map represent phage clones (LS1, LS2,and LS22) and a BAC clone (N21). The_thick line, CM, includes the enhancer elements for driving GFP expression in cranial motor neurons, whereas_SS includes the enhancer elements for driving GFP expression in the trigeminal ganglion cells and Rohon–Beard cells. Other Eco_RI fragments were negative for enhancer activity by our transient expression assay. B, Close-up view of the genomic region around the Isl1 promoter.Filled boxes are protein coding regions. The_hatched box represents the putative Isl1_promoter (ICP), which contains ∼4.1 kb of the 5′ upstream region and an ∼30 bp of the 5′ untranslated region of the_Isl1 gene. It was used in all the constructs as a core promoter. C, A map of the ICP–GFP, the core plasmid. Every genomic _Eco_RI fragment was inserted into the_Eco_RI site located in the immediate upstream of ICP._Not_I was used to linearize the plasmid DNA for microinjection. D, A map of the CMICP–GFP or SSICP–GFP (containing the SS-fragment) plasmid. The CMICP–GFP plasmid drove expression of GFP in the cranial motor neurons, and it was used for generating the Isl1-GFP line. The SSICP–GFP plasmid drove expression of GFP both in Rohon–Beard neurons and the trigeminal sensory neurons.
Embryos and staging. Zebrafish were maintained as described in Westerfield (1995). Most analyses were performed using homozygous transgenic embryos from the Isl1-GFP line. The embryos were collected after crossing homozygous parents. In the early phase of the experiments, homozygous embryos were obtained by crossing heterozygous parents. Homozygous fish exhibited no abnormality or retardation in the development of the nervous system. Embryos were collected within 30 min after fertilization and were allowed to develop at 28.5°C to the required age, in uncrowded conditions. The developmental age of the embryos corresponds to the number of hours elapsed since fertilization. Morphological features of the embryos in our experiments at a given time stage were, in general, consistent with the zebrafish standard staging table (Kimmel et al., 1995). Because repeated anesthetizations are suggested to retard development (Kimmel et al., 1995), each embryo was examined only once for most of the experiments. To reduce pigmentation, embryos were transferred to fish water containing 0.002% of phenylthiourea (PTU; Nakarai) between 12 and 22 hr (Burrill and Easter, 1994). The concentration of PTU was reduced to 0.001% at 48 hr, to 0.0003% at 72 hr, and to 0.0001% at 96 hr. At this concentration of PTU, no abnormality in development was observed.
Fluorescent microscopic observations. Initial examination was performed using an epifluorescence dissecting microscope with a GFP plus filter (Leica, Nussloch, Germany). A PlanApo 1.6× objective lens was used. For detailed examination, fluorescent embryos were anesthetized using 0.01% ethyl-_m_-aminobenzoate methanesulphonate (Sigma, St. Louis, MO) and mounted in 1.2% agar (Difco, Detroit, MI) on a depression slide glass, as described inWesterfield (1995). Embryos were either examined under a Zeiss(Oberkochen, Germany) upright epifluorescence microscope with an FITC or a GFP (Chroma 41015) filter cassette or under a Zeiss 510 confocal microscope (510-CLSM, upright) with 488 nm excitation and a 505–550 nm bandpass filter. Most observations, except for those of rhodamine–phalloidin or antibody-labeled samples, were performed using live material because visibility was best in living fish. A Plan-Neofluar 20× [numerical aperture (NA) 0.5] objective lens was used for most observations. For higher magnifications, water-immersion lenses, Achroplan 63× (NA 0.9) and 100× (NA 1.0), were used. For three-dimensional (3-D) reconstruction of the images, serial optical sections at ∼4 μm intervals (20× objective) or 0.5–1.5-μm intervals (63 and 100× objective) were taken, and 3-D images were reconstructed from the stacked confocal images by the software supplied with the 510-CLSM.
For DiI-, Cy3- and rhodamine-labeled samples, observations were performed using the 510-CLSM with 568 nm excitation and a 585 longpass filter. For 2-_p_-dimethylaminostyrylpyridylethyl iodide (DASPEI)-labeled samples, 488 nm excitation and a 585 longpass filter were used. A DASPEI signal was also apparent during the collection of images for GFP (505–550 nm bandpass). For 3-D reconstruction of images for signals from GFP and other dyes, serial optical sections were obtained either simultaneously or sequentially. Each image was pseudocolored (GFP, green; others, red) and compiled.
Mosaic analysis. Cells derived from the homozygous Isl1-GFP embryos were transplanted into wild-type hosts at the sphere stage as in Westerfield (1995). Donor cells were transplanted near the animal pole of host embryos to increase the chance that progeny of donor cells would be distributed in the cranial region of the host embryo. Mosaic embryos and larvae were examined between 48 and 90 hr.
Retrograde labeling of the trigeminal motor neurons. Larvae were anesthetized at ∼72–90 hr and embedded in 1.2% agar on a glass slide in an orientation that allowed access to the nerve to be labeled. The agar overlying the injection site was removed. The fluorescent lipophilic dye DiI (Molecular Probes, Eugene, OR), at 2 mg/ml in ethanol was pressure-injected (Eppendorf Transjector 5246) into a region around a labeled nerve under an epifluorescent dissection microscope with the longpass GFP filter (see above). Fluorescence signals derived from both GFP (green) and DiI (orange) were simultaneously observed under this condition. The injected larvae were allowed to develop at room temperature for 4 hr so that the DiI would retrogradely label the hindbrain neurons. DiI injection was also performed in fixed embryos. In this case, the injected embryos were incubated in 4% paraformaldehyde in PBS at room temperature for 12 hr.
In situ hybridization, antibody staining, rhodamine–phalloidin staining, and DASPEI live staining. In situ hybridization and antibody staining were performed according to standard methods (Westerfield, 1995). Isl1 cDNA (Inoue et al., 1994), zn5 antibody (Trevarrow et al., 1990), and Cy3-conjugated anti-mouse antibody (Jackson ImmunoResearch, West Grove, PA) were used. For rhodamine–phalloidin staining, larvae were fixed and treated in acetone as described previously (Westerfield, 1995). The larvae were washed in PBS containing 1% dimethylsulfoxide and 0.2% Triton-X 100, and incubated in 0.0002 U/ml rhodamine–phalloidin (Molecular Probes) in the same solution for 1 hr at room temperature with shaking. They were observed after washing. For DASPEI staining, larvae were immersed in 0.1 mm DASPEI (Molecular Probes) in E3 (in mm: 5 NaCl, 0.17 KCl, 0.33 CaCl2, and 0.33 MgSO4) for 20 min, as in Balak et al. (1990) and Whitfield et al. (1996), and observed after rinsing.
RESULTS
Identification of the Isl1 promoter/enhancer region and generation of transgenic zebrafish
The Isl1 gene and its protein product have been used as early markers for motor neurons in vertebrates. However, the promoter/enhancer region that directs gene expression in motor neurons has not yet been identified in any vertebrate species. To isolate the promoter/enhancer of the Isl1 gene that is sufficient for expression in zebrafish motor neurons, ∼100 kb genomic sequences flanking the Isl1 gene were isolated by screening both λ phage and BAC zebrafish genomic libraries (Fig. 1A). The structure of the 5′ region of the Isl1 gene is shown in Figure 1B. The core plasmid, ICP–GFP, was generated by fusing 4.1 kb of the Isl1 promoter, ICP, to the gene encoding a modified GFP (Fig. 1C). When the ICP–GFP DNA was injected into embryos, GFP was expressed by hatching gland cells (data not shown) whose precursors normally expresses the Isl1 gene (Inoue et al., 1994), but not by motor neurons. This suggests that ICP includes enhancer elements that regulate hatching gland expression of the Isl1 gene.
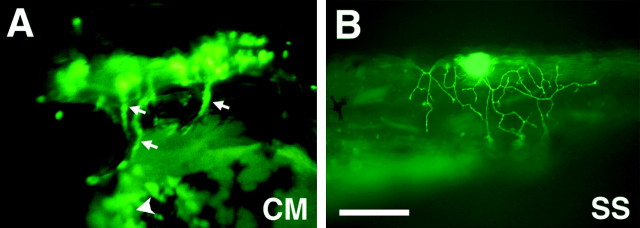
To identify enhancer elements that control expression in motor neurons, a series of constructs were generated by introducing genomic_Eco_RI fragments into the ICP–GFP plasmid (Fig.1C). All recognizable _Eco_RI fragments in the cloned region (Fig. 1A) were tested for their enhancer activity by injecting each construct into embryos. In the injected embryos, GFP expression was frequently observed in the hatching gland cells presumably because of the core promoter (ICP) activity that was common to all the constructs (Fig.2A, arrowhead). Thus, hatching gland expression of GFP served as a positive control for the injections. At least 50 surviving embryos were examined for each construct. One construct, CMICP–GFP (Fig. 1D), which carries a 15 kb _Eco_RI fragment (Fig. 1A, CM), was found to drive GFP expression in branchiomotor neurons innervating the pharyngeal arches in injected embryos (Fig.2A, arrows). Additionally, we identified another enhancer fragment (Fig. 1A, SS) that was capable of driving GFP expression in the trigeminal ganglion cells and Rohon–Beard cells in the spinal cord (Fig. 2B).Isl1 is normally expressed by both of these cell types as well as the motor neurons and the hatching gland (Inoue et al., 1994). All of the other _Eco_RI fragments were negative: we carefully looked for GFP expression in primary motor neurons in the spinal cord, but expression was not detected in these cells that normally express Isl1. Furthermore, Isl1 is expressed in many other cells such as those in the epiphysis and the nucleus of the posterior commissure, but no constructs gave GFP expression in these cells. It should be noted that because transgenes are mosaically expressed in the injected embryos, our negative results should not be construed as conclusive.
Fig. 2.
Zebrafish _Isl1_promoter/enhancer activity in transient expression assays.A, GFP expression in the head of a 40 hr embryo injected with the CMICP–GFP plasmid. The arrows indicate GFP expression in branchial motor nerves. The _arrowhead_indicates GFP expression in the hatching gland cells. Anterior is to the left, dorsal is up. _B,_A lateral view of GFP expression in a Rohon–Beard cell in the spinal cord of a 36 hr embryo injected with the SSICP–GFP plasmid. Scale bar:A, 200 μm; B, 230 μm.
To establish stable transgenic zebrafish, CMICP–GFP-injected embryos were raised to sexual maturity, and either incrossed with each other or outcrossed with noninjected adult fish to generate F1 progeny. Of 113 fish, three produced embryos expressing GFP in a subset of_Isl1-_expressing cells, including the cranial motor neurons (Fig. 3A,B). The expression patterns of GFP in embryos derived from the three founder fish were identical, although expression levels varied. The line with the highest GFP expression, referred to as Isl1-GFP hereafter, was chosen for further analysis. In this study, we focused on GFP expression patterns of the Isl1-GFP line in the cranial region. As predicted by the transient expression analysis, GFP was also expressed in the hatching gland cells (Fig. 3A, arrow). Additionally, some secondary motor neurons in the spinal cord and a small number of interneurons in the CNS also expressed GFP (data not shown).
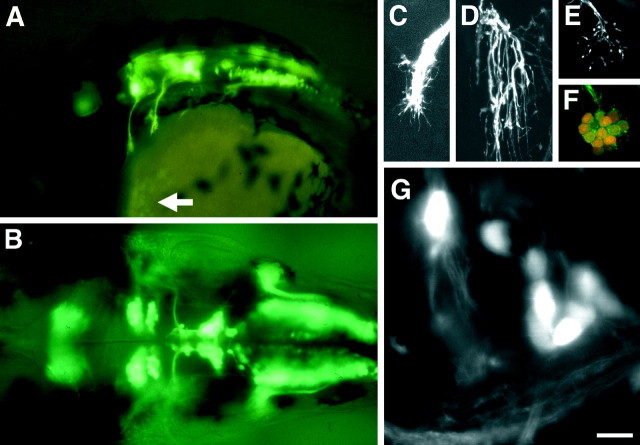
Fig. 3.
GFP expression in cranial motor neurons in the Isl1-GFP line. A, Lateral view of the head region of a 34 hr embryo. The arrow indicates GFP expression in the hatching gland cells. B, Dorsal view of the midbrain and the hindbrain of a 42 hr embryo. In A and_B_, pictures were taken under a conventional epifluorescence microscope. C–G, Close-up views of the GFP-positive nerve endings and neurons in the Isl1-GFP line using a confocal microscope. In panels C–F, composite pictures were generated from the stacked confocal images. C,Lateral view of the distal tips of the trigeminal motor axons at 30 hr. Note the fine filopodial structures extending from the distal ends of the axons. D, Lateral view of the neuromuscular junction in the levator arcus palatini (lap) muscle at 96 hr. Note the fine arborization of the nerve branches innervating individual muscle fibers. E, F, GFP expression in the efferent component of the lateral line nerves in a 96 hr embryo. Close-up view of their endings at the neuromasts. The position of the neuromast corresponds to_h in Figure 7C. In F, the larva was treated with the vital dye DASPEI, which stains hair cells. DASPEI signals appear as orange. Note that the nerve endings appear to terminate within the hair cells. G, A confocal optical section showing the trigeminal motor neurons at 105 hr. Dorsal view with medial to the top. A cluster of neurons situated on the_right correspond to those in the posterior (Vp) cluster of the trigeminal motor neurons (Figs. 5-7). Note that individual GFP-positive neurons are readily visualized. In all figures, anterior is to the left. In lateral views, dorsal is_up_. Scale bar: A, 110 μm;B, 60 μm; C–F, 20 μm;G, 10 μm.
The Isl1-GFP line enables clear visualization of cranial motor neurons
To examine how faithfully GFP expression reflects the expression of the intrinsic Isl1 mRNA in the Isl1-GFP line, we compared the distribution of GFP mRNA and Isl1 mRNA.GFP mRNA (Fig.4A,C,E) is expressed in a majority of cranial motor neurons in a similar manner to Isl1 mRNA (Fig. 4B,D,F). Outside the CNS, GFP mRNA is also expressed in all the branchiomeric sensory ganglion cells except for the trigeminal (V) ganglion cells (Fig. 4E, fs, gs, vs). Branchiomeric nerves (V, VII, IX, and X) are nerves innervating derivatives of the pharyngeal arches and are known to include both motor and sensory components. Besides, GFP is also expressed in the efferent neurons for the lateral line and vestibuloacoustic nerves, as described in more detail later.
Fig. 4.
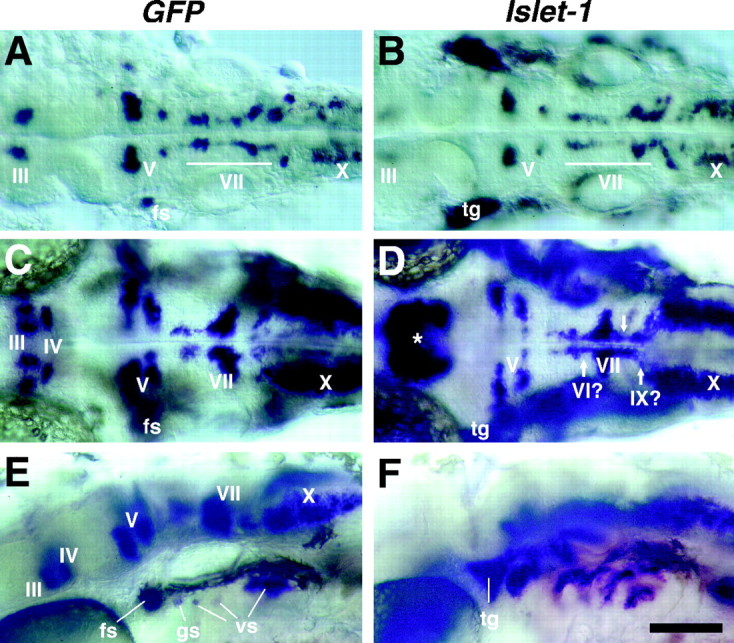
Comparison of the expression patterns of_Islet-1_ (Isl1) and _GFP_mRNAs in the cranial motor and sensory neurons. A,C, E, GFP mRNA expression in embryos from the Isl1-GFP line. B, D,F, Isl1 mRNA expression in wild-type embryos. A, B, Dorsal views of 28 hr embryos.GFP and Isl1 mRNAs are both expressed in presumptive cranial motor neurons. Whereas Isl1 mRNA is expressed in the trigeminal ganglion cells (tg),GFP mRNA is not expressed in these cells. C, D, Dorsal views of 40 hr embryos. Isl1 mRNA expression in the nIII and nIV neurons is difficult to see because many other neighboring cells also express Isl1 mRNA (D, asterisk). Arrows in _D_indicate cells that do not express GFP mRNA in the Isl1-GFP line. The nVI and nIX neurons do not express GFP in the Isl1-GFP zebrafish (see the following figures). _E, F,_Lateral views of 40 hr embryos. GFP is expressed in cells in the facial sensory ganglion (fs), glossopharyngeal sensory ganglion (gs), and vagus sensory ganglions (vs), but is not expressed in cells in the trigeminal sensory ganglion (tg). Scale bar, 100 μm.
We examined GFP expression in live zebrafish most often with a confocal microscope (see Materials and Methods). After obtaining a series of optical sections, composite pictures were generated from the stacked images. The expression levels of GFP were high enough to visualize both fine neuronal processes during development such as filopodia (Fig.3C) and fine terminal arbors of the motor axons (Fig.3D), suggesting that neuronal processes were visualizable in their entirety in the Isl1-GFP line.
The expression of Isl1 in motor neurons begins early in their development. Likewise, GFP expression was detected early in cranial motor/efferent neurons (Fig.5A, see GFP expression in a 21 hr embryo). The transparent nature of zebrafish embryos and larvae enabled visualization of GFP-labeled neurons and their processes at high resolution in living animals. This was expected at early stages such as 24 hr because the early hindbrain is relatively simple (Chandrasekhar et al., 1997), but was also the case at much later stages, such as day 4 when the hindbrain is much larger and much more complex, and the larvae begin complex behaviors such as swimming. An example is shown in Figure 3G (an optical section) in which individual neurons in the trigeminal motor nucleus, which lie deep in the hindbrain, were visualized even in a 4 d, live larva. Thus, it is possible to observe development of the cranial motor neurons in intact animals with a high level of resolution from early stages to relatively mature stages in the Isl1-GFP line.
Fig. 5.
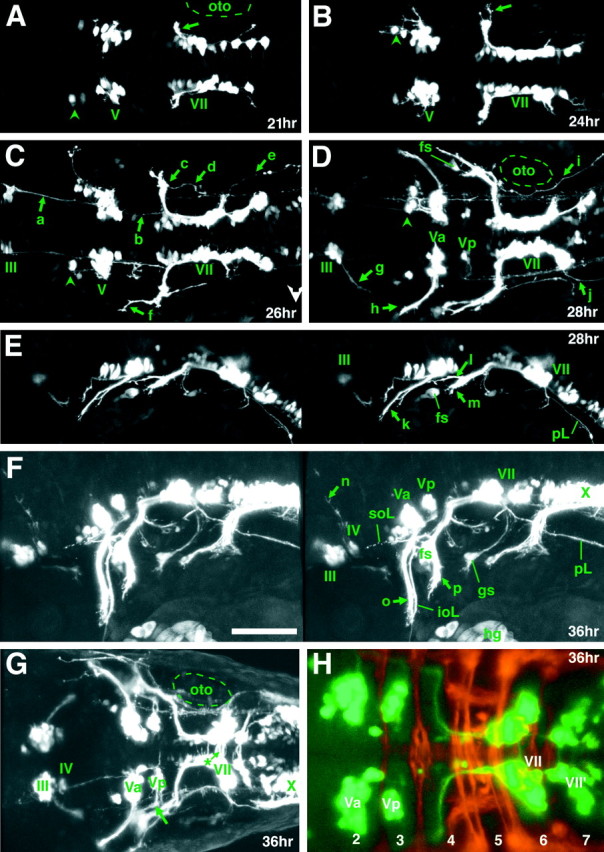
GFP expression early in the development of the cranial motor neurons (21–36 hr) in the Isl1-GFP line. All micrographs are confocal composite images generated from a series of optical sections. E and F are stereographic images. A, Dorsal view of a 21 hr embryo. The arrow indicates the nVII/OLe axons. At this stage most of the GFP-positive nVII/OLe neurons are located in r4 and r5. Rhombomere boundaries were estimated by designating the hindbrain region spanning the central half of the otocyst (oto) as r5, and those spanning the anterior and posterior quarters of the otocyst as parts of r4 and r6, respectively. B, Dorsal view of a 24 hr embryo. The pioneering axons from the nVII/OLe neurons are seen outside the hindbrain (arrow).C, Dorsal view of a 26 hr embryo. *Longitudinal processes that connect the nIII, nV, and nVII motor nuclei are visible (for example, a and b). *The significance of these early-forming axons is unknown. The majority of the nVII/OLe axons exit from the hindbrain (c).f indicates anteriorly projecting axons that subsequently extend into the lateral line system (corresponding to_l_ in E). D, Dorsal view of a 28 hr embryo. g indicates axons from the nIII neurons.h indicates the peripherally extending axons from the Va cluster of the nV neurons. d in C and_i_ in D indicate efferent axons extending into the posterior lateral line. *Efferent axons for the posterior lateral line also exit from the hindbrain via a posteriorly located exit point (e in C and j_in D; Metcalfe et al., 1985). *Arrowheads_in A–D indicate GFP-positive cells located anteriorly to the main nV cluster. *It is not clear whether these cells constitute a part of the nV neurons. E, Lateral view of a 28 hr embryo. k indicates the peripherally extending axons from the nV neurons. l indicates diverging axons extending into the lateral line system, whereas m_indicates the main nVII/OLe axons (mostly, the facial motor axons). It should be noted that, viewed laterally, the efferent axons for the posterior lateral line appear interrupted by the otocyst because of the opaque nature of the otolith (also in Figs. 5F, 6_B,E, 7_C).F, Lateral view of a 36 hr embryo. n_indicates dorsally extending axons from the nIV neurons.o and p indicate the peripherally extending axons from the nV and nVII/OLe neurons, respectively.G, Dorsal view of a 36 hr embryo. The thick arrow indicates axons from the Vp cluster of the nV neurons. *Contralaterally projecting neurites from the nVII/OLe neurons are visible (asterisk). H, A confocal composite image of a 36 hr embryo in which hindbrain commissural axons that are located at the rhombomere boundary are labeled with the zn5 antibody. Dorsal view. Numbers listed are rhombomere numbers. Green is the GFP signal, whereas_red is the zn5 signal. VII and_VII′, VII/OLe complex; oto, otocyst;Va, Vp, anterior and posterior clusters of nV neurons, respectively; fs, facial sensory ganglion;pL, posterior lateral line; soL, supraorbital lateral line; ioL, infraorbital lateral line; gs, glossopharyngeal sensory ganglion;hg, hatching gland. The sentences denoted by an_asterisk_ in this and all other legends describe observations not detailed in Results. In lateral views, dorsal is to the top. Scale bar, _A–G,_100 μm; H, 50 μm.
Overall development of the trigeminal motor (nV) neurons
Although axonal outgrowth by the nV neurons in zebrafish has been examined by a variety of methods, including retrograde labeling or immunohistochemistry (Hatta et al., 1990; Chandrasekhar et al., 1997;Schilling and Kimmel, 1997), their overall development is still largely obscure because of the limitation of these methods. Given below is a description of the overall development of the nV neurons as gleaned from observations of the Isl1-GFP line. Some of the figures (Figs.5-7) present micrographs in stereographic pairs.
Fig. 6.
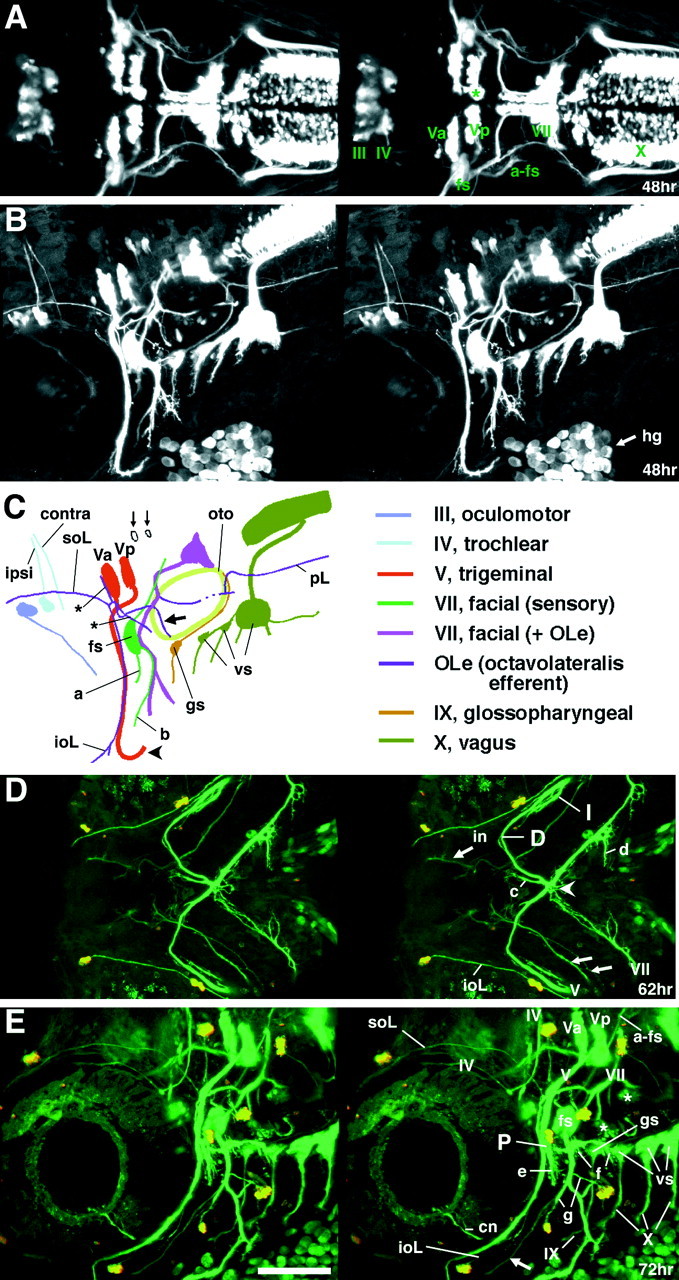
GFP expression in the Isl1-GFP line on days 2 and 3 of development. All panels except C are confocal stereographic images. A, Dorsal view of a 48 hr embryo. The asterisk indicates medially located nV neurons that are presumably born later than laterally located nV neurons. B, Lateral view of a 48 hr embryo.C, Schematic illustration of GFP-labeled nerves in_B_. ipsi and contra_indicate fourth nerves from the nIV neurons on the ipsilateral and the contralateral sides, respectively. a and_b indicate the peripherally projecting axons from the facial sensory ganglion cells. The axons of a take an internal pathway, whereas those of b take an external pathway. The axons indicated by a and b,respectively, correspond to those indicated by in and_ex in Figure 9A–C. Thin arrows show GFP-expressing cells that presumably do not correspond to motor neurons. The thick arrow indicates an OLe nerve branch extending into the hair cells in the otocyst. The_asterisk indicates branching processes of the OLe nerves, which are added later to the major branches. The_arrowhead_ indicates the distal tip of the fifth motor axons. D, Ventral view of a 62 hr embryo.E, Lateral view of a 72 hr larva. The embryo in_D_ and the larva in E were treated with the vital dye DASPEI, which stains hair cells in lateral line neuromasts. DASPEI signals appear as yellow.I, D, and P indicate the intermediate, distal, and proximal branches of the fifth motor nerve, respectively. Arrows in D and_E_ indicate peripherally projecting axons from the facial sensory ganglion cells (equivalent to a and_b_ in C). Among them, in_indicates the axons that are distal tip of the axons indicated as_a in C. They take an internal pathway. Most part of their tract is out-of-focus. The _arrowhead_indicates the junction of the four motor nerves (both sides of the fifth and seventh motor nerves). c and _d_indicate nerve branches that innervate the intermandibularis anterior and posterior (ima and imp), and the hyohyal (hh), respectively (Fig. 7E).e indicates the neuromuscular junction at the levator arcus palatini (lap), whereas f indicates the neuromuscular junctions at the abductor hyomandibulae (ah) and the abductor operculi (ao) (Fig.7D). g indicates distal branches of OLe nerves from the main nVII/OLe nerve. Asterisks indicate nerve endings of the OLe system in the ear (Fig. 7C). *cn is likely to be the ciliary nerve innervating the lens muscle for visual accommodation. *The lens muscle in teleost was shown to be controlled by the postganglionic fibers of the oculomotor (parasympathetic) nerve (Somiya, 1987), and thus, GFP should be expressed in the parasympathetic ganglion cells corresponding to the ciliary nerve. a-fs, centrally projecting afferent facial sensory axons; pL, posterior lateral line;vs, vagus sensory ganglion. Scale bar, 100 μm.
Fig. 7.
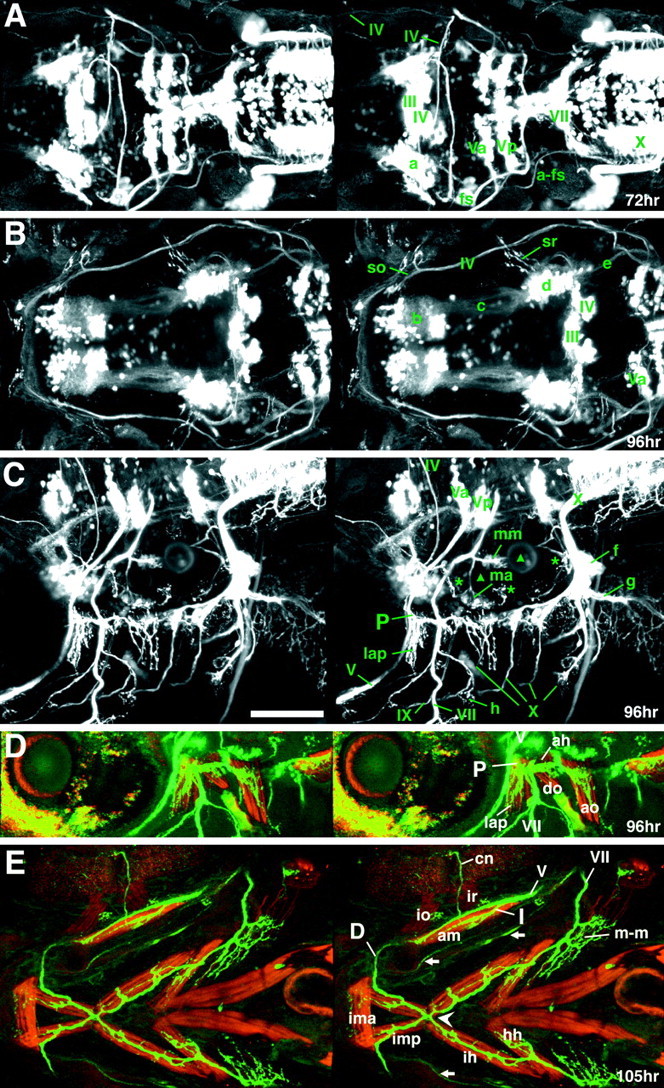
GFP expression in the Isl1-GFP line on days 3 and 4 of development. All figures are stereographic pictures reconstructed from the stacked confocal images. A,Dorsal view of a 72 hr larva. B, Dorsal view of a 96 hr larva. GFP is also expressed in cells other than motor neurons in the brain. The identities of these cells are not known. They are a cluster of cells in the telencephalon (b) and a cluster of cells in the mesencephalic region (d). Tracts are formed between these two clusters (c). The cluster of the mesencephalic region (d) also sends processes posteriorly (e). a_in A corresponds to d in_B. For these cells and tracts, see also the legend to Figure 9E. C, Lateral view of a 96 hr larva. Five terminal-like structures are observed in the ear. These appear to correspond to the anterior macula (ma) accompanied by the anterior otolith (the anteriorly located triangle), the medial macula (mm) accompanied by the posterior otolith (the posteriorly located triangle), and the three crista ampullarises (asterisks) associated with the anterior, lateral, and posterior canals. Nomenclatures are according to Whitfield et al. (1996). f indicates the largest vagus sensory ganglion, which is likely to include neurons innervating visceral organs. g indicates the vagus nerve extending into the visceral organs. The nerve is likely to contain sensory components and visceral motor (parasympathetic) components. h indicates the endings of the OLe nerve at a lateral line neuromast. Higher magnification views of the corresponding structures are shown in Figure3, E and F. In B and_C, elaborated fine branches are visible at neuromuscular junctions (for example, so and sr in_B, lap in C). D, E, Fixed larvae were treated with rhodamine–phalloidin to reveal actin filaments of muscles. Green is the GFP signal, whereas red is the rhodamine–phalloidin signal.D, Lateral view of a 96 hr larva. E,Ventral view of a 105 hr larva. P, I, and_D in C–E indicate the proximal, intermediate, and distal branches of the fifth motor nerve, respectively. The proximal branch of the fifth nerve innervates the_lap and do muscles, the intermediate branch innervates the am muscle, and the distal branch innervates the ima and imp muscles. The configuration is schematically summarized in Figure 8A. Arrows indicate peripherally projecting axons from the facial sensory ganglion cells. *The seventh motor nerves innervate surfacial membranous muscles (indicated as m-m), which presumably corresponds to the platysma in higher vertebrates. The_arrowhead_ indicates the junction of the four motor nerves (both sides of the fifth and seventh motor nerves). Abbreviations for muscles (so, sr,io, ir, lap,do, am, ima,imp, ah, ao,ih, and hh) are shown in Table 1. *cn is likely to be the ciliary nerve (Fig.6E, legend). Scale bar, 100 μm.
GFP is expressed in the nV neurons, but not in the trigeminal sensory ganglion cells (Isl1 is expressed in both; compare Fig.4A,C,E with_B_,D,F). Thus, all the GFP-labeled axons in cranial nerve V are motor axons. GFP-positive nV neurons are evident as early as 21 hr (Fig.5A) and progressively increase in number with further development (Fig. 5A–C). Thus, GFP is expressed early enough so that the entire time course of axonal outgrowth by the nV neurons could be observed. By 28 hr, the motor axons derived from the nV neurons (Fig. 5D, Va) have extended laterally to exit from the hindbrain into the periphery (Fig. 5D, h, E, k). At about the same time, an additional more posterior cluster of nV neurons begins to express GFP (Fig. 5D, Vp). GFP-positive neurons in the Vp cluster increase in number as development proceeds. At ∼36 hr, their axons (Fig. 5G, thick arrow) have extended laterally and then anteriorly to merge with the axons from the Va cluster. Our observations showed that, throughout development, the Va and Vp clusters lie discretely apart, and that their axons follow their own pathways within the hindbrain until joining together at a common hindbrain exit point (Figs. 5F,G,6A–C,E, 7A,8C,F,I).
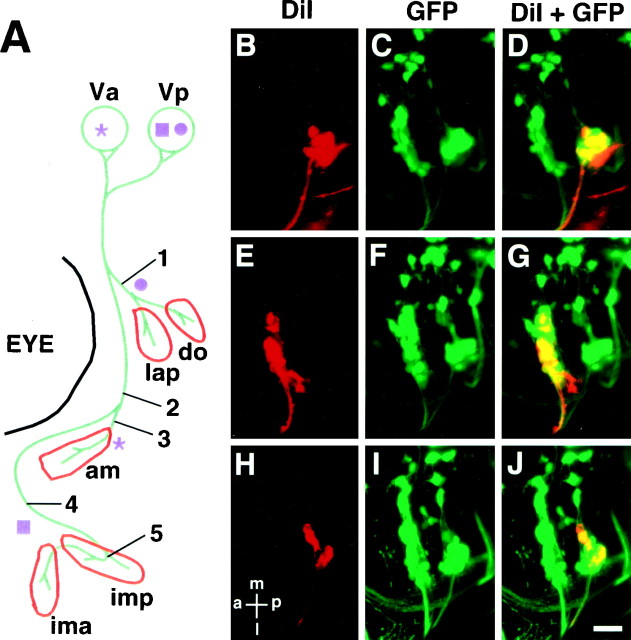
Fig. 8.
Position of the trigeminal motor (nV) neurons with respect to the peripheral nerve branches.A, A diagram showing dye application sites and a summary of the results. Trigeminal motor neurons were retrogradely labeled with DiI. DiI application was performed at positions 1–5. The results showed that Va solely consists of neurons projecting to the intermediate branch (asterisk), whereas Vp consists of neurons projecting to the either proximal (circle) or distal branch (square). Abbreviations for the muscles are the same as those shown in Table 1. In B–J, all pictures are dorsal views (composite pictures made from stacked confocal images) of larvae at ∼90 hr with anterior to the_left_ and medial to the top.B–D, DiI was applied at position 1. Neurons belonging to the posterior cluster (Vp) of the nV neurons were labeled with DiI. E–G, DiI was applied at position 3. Neurons belonging to the anterior cluster (Va) were labeled with DiI. H–J, DiI was applied at position 4. Neurons belonging to the posterior cluster (Vp) were labeled. Scale bar, 20 μm.
We examined the locations of the Va and Vp clusters with respect to the hindbrain rhombomeres using zn5 antibody, which labels hindbrain commissural axons that are located at the rhombomere boundaries (Trevarrow et al., 1990). Figure 5H shows that the Va and Vp lie in r2 and r3, respectively. The hindbrain exit point of the fifth motor nerve lies in r2 (Fig. 5G). The locations and distributions of the nV neurons are, in general, consistent with those revealed by retrograde labeling with DiI (Chandrasekhar et al., 1997). However, the number of neurons that are labeled at a given stage is much larger in our analysis. Presumably, this is attributable to the fact that DiI backlabeling from the target pharyngeal arch only labels those neurons whose axons have reached the arch and that backlabeling may be incomplete, whereas in the Isl1-GFP line, all the motor neurons, even though they have not reached their targets, are labeled. Furthermore, we found that cells in a medial position were labeled in our Isl1-GFP line (Fig. 6A, asterisk), whereas at similar stages backlabeling with DiI only labeled cells in a lateral position (Chandrasekhar et al., 1997). Thus, it could be that the earliest nV neurons are born medially and migrate laterally. This is consistent with the possibility that nV neurons are arranged medial to lateral within the hindbrain according to the time they are born and differentiate with early ones laterally and later ones more medially. A similar pattern of development of trigeminal motor neurons has also been described in the chick embryo (Heaton and Moody, 1980; Simon et al., 1994).
Once outside of the hindbrain, axons of the nV neurons extend ventrally along the posterior edge of the eye (Fig. 5F, o), and by 48 hr the axons turn posteriorly (Fig. 6C, arrowhead). At 62 and 72 hr, nerve branches innervating the mandibular arch muscles are clearly visible (I, D in Fig. 6D; P in Fig. 6E). Among them, the distal branch of the fifth nerve (D in Fig. 6D) runs in the posteromedial direction to the midline, where it merges with both the contralateral fifth nerve and the distal tip of the seventh (facial) nerve (Figs. 6D, 7_E_,arrowhead). As the zebrafish developed, the labeled fifth axons progressively shifted anteriorly because of extension of the jaw (compare Fig. 6D with 7_E_).
Correspondence between peripheral branches of the fifth nerve and target muscles was determined by labeling of fixed sample with rhodamine–phalloidin to show the configuration of the motor nerves and their target mandibular muscles (Fig.7D,E; see Table1 for summary of the nomenclature of the mandibular arch muscles and the extraocular muscles, the abbreviated forms of their names, and their innervation patterns). The proximal branch of the fifth nerve (P in Figs. 6E,7C,D) innervates the levator arcus palatini (lap) and dilator operculi (do) muscles. The intermediate branch (I in Figs. 6D,7E) innervates the abductor mandibulae (am) muscle. Finally, the distal branch (D in Figs.6D, 7E) innervates the intermandibularis anterior and posterior (ima and imp) muscles. This configuration is schematically shown in Figure8A.
Table 1.
Extraocular and jaw muscles in the zebrafish
| Region and muscles | Abbreviation | Innervation |
|---|---|---|
| Extraocular | ||
| Superior oblique | so | IV |
| Inferior oblique1-a | io1-a | III |
| Superior rectus | sr | III |
| Inferior rectus | ir | III |
| Medial rectus | mr | III |
| Lateral (posterior) rectus1-b | lr1-b | VI |
| Mandibular arch | ||
| Intermandibularis anterior | ima | V |
| Intermandibularis posterior | imp | V |
| Abductor mandibulae | am | V |
| Levator arcus palatini | lap | V |
| Dilator operculi | do | V |
| Hyoid arch | ||
| Interhyal | ih | VII |
| Hyohyal | hh | VII |
| Abductor hyomandibulae | ah | VII |
| Abductor operculi | ao | VII |
Trigeminal motor axons from different rhombomeres innervate distinct target muscles
Because the trigeminal motor neurons innervate several different muscles and they are organized into two clusters, Va in r2 and Vp in r3, we wondered whether the two clusters of trigeminal motor neurons innervated different target muscles. To examine this question, we took advantage of the fact that nV nerve and branches are readily visible in the Isl1-GFP line. We selectively backlabeled those motor neurons that supply axons to a specific nerve branch by application of DiI to that branch. In the periphery, there are three major branches, and DiI was applied at five different positions along the branches in 72–90 hr larvae (Fig. 8A, 1–5). DiI application at position_1_, which contains axons innervating the lap and_do_ muscles, labeled Vp neurons (Fig.8B–D; n = 5). DiI application at position 2, which contains axons innervating the_am_, ima, and imp muscles, labeled Va and Vp neurons (data not shown; n = 2). Application of DiI at position 3, which contains axons innervating the_am_ muscle, labeled Va neurons (Fig. 8E–G;n = 6). Finally, DiI application at position_4_ or 5, which contains axons innervating the_ima_ and imp muscles, labeled Vp neurons (Fig.8H–J; n = 7 for 4 and_n_ = 4 for 5). These results indicate that the Va neurons exclusively innervate the am muscle, whereas the Vp neurons innervate the lap, do,ima, and imp muscles. Thus, the two clusters of trigeminal motor neurons innervate distinct target muscles, and this can be seen at early larval stages. These results confirm the pattern of innervation found in adult fish (Song and Boord, 1993). Because Va and Vp are located in different rhombomeres and appear not to intermingle during development, our results are consistent with the notion that nV neurons born in r2 and r3 adopt distinct fates, and this suggests that the segment of origin determines the identity of the nV neurons.
Characterization of GFP-positive cranial nerves other than the trigeminal nerve
Below we describe the GFP-positive cranial motor and sensory neurons because many of them have not yet been fully characterized in developing zebrafish. To keep the text concise, some minor observations are described only in the figure legends (Figs. 5-7, see the comments denoted by asterisks). Some of our findings overlap with previously reported results (Chandrasekhar et al., 1997).
Facial (VII) and octavolateralis efferent (OLe) nerves
In addition to motor neurons, GFP is expressed in the efferent neurons for the lateral line and the vestibuloacoustic nerves. The lateral line, which is unique to aquatic vertebrates, is a sensory system mainly responsible for detection of water displacement (Coombs et al., 1989). The lateral line nerve as well as the vestibulo-acoustic nerve terminates at hair cells that are innervated by both sensory and efferent (octavolateralis efferent, OLe) nerves (Roberts and Meredith, 1989; 1992; Highstein, 1991). The facial motor (nVII) and OLe neurons are located in close proximity, and the efferent axons from both cell types extend together in the hindbrain. Therefore, we have combined the description of axonal outgrowth by both sets of efferent neurons.
GFP-positive nVII/OLe neurons (Fig. 5A, VII) and their axons (Fig. 5A, arrow) are evident by 21 hr. At ∼24 hr, the pioneering axons among the nVII/OLe axons exit from the hindbrain (Fig. 5B, arrow), and by 26 hr, most of the nVII/OLe axons have left the hindbrain (Fig. 5C, c). The thick, main bundle of nVII/OLe axons (Fig. 5E, m) grows toward the facial ganglion (Fig. 5E, fs), which becomes discernible at ∼27–28 hr. By the facial ganglion, the main bundle grows ventrally (Fig. 5F, p). At 48 hr, the main bundle (Fig. 6A–C, purple) is mainly composed of the facial motor axons (see below for the sensory and OLe fibers). Projection of motor nerves to target muscles was confirmed by labeling muscles with rhodamine–phalloidin at later stages (Fig.7D,E). The labeled seventh motor nerve innervates the ah, ao, ih, and_hh_ muscles (see Table 1 for abbreviations for the names of the muscles).
The nVII/OLe motor neurons appear to migrate caudally during development. At early stages (21 hr), most of the nVII/OLe neurons are localized in r4 and r5, as judged by comparison with the location of the otocyst (Fig. 5A, see its legend for assignment of rhombomere). In the next 15 hr, the nVII motor neurons were seen to migrate caudally such that by 36 hr most of the motor neurons (Fig.5H, VII, VII′) were located in r6 with some in r7. Our findings confirm the hypothesis that the nVII/OLe motor neurons migrate caudally, as first suggested by Chandrasekhar et al. (1997)based on a caudal shift of cells expressing the Isl1 protein and_tag1_ mRNA (cranial motor neuron markers) from r4 and r5 to r6 and r7. In this earlier study, however, there remained the possibility that the apparent caudal shift of the Isl1/_tag1_-positive cells was not caused by migration of neurons, but was caused by changes in gene expression by neurons in these rhombomeres. Because the nVII/OLe neurons (identified by their axonal trajectories) were seen to shift caudally in Isl1-GFP embryos by observing them at several different time points, it is clear that these neurons indeed do migrate caudally.
To get a clearer description of axonal outgrowth by the motor and sensory components, we made mosaic embryos, by isochronically transplanting cells of the Isl1-GFP line into wild-type embryos at the sphere stage. In the resultant embryos, GFP was expressed by only a small number of cranial motor and sensory neurons, making it possible to unambiguously follow axonal outgrowth in these cells. Interestingly, observations of facial sensory neurons in mosaic embryos showed that their centrally projecting axons (a-fs in Figs.6A,E, 7A,9A, C,D; green in Fig. 6C) take a distinct pathway from the outgrowing nVII/OLe motor axons. In amniotes, the motor/efferent axons and the centrally projecting sensory axons follow the same pathway. The centrally projecting axons initially extend dorsoposteriorly, turn medially into the hindbrain, and then posteriorly within the hindbrain (Figs. 7_A, a-fs;9_C,D) to terminate in the dorsoposterior part of the hindbrain by day 3 (Fig. 9C,D, arrowheads). The peripherally extending axons of the sensory nVII neurons diverge into two nerves as they extend in a ventroanterior direction (Fig. 6C, a, b), but they all eventually terminate around the mouth (Figs.6D,E, 7_E, arrows;9_A,B). This outgrowth pattern is consistent with the notion that the facial sensory nerves in vertebrates mostly convey gustatory information (Fig. 9_A–C,_see the legend to for further details on the peripheral trajectories).
Fig. 9.
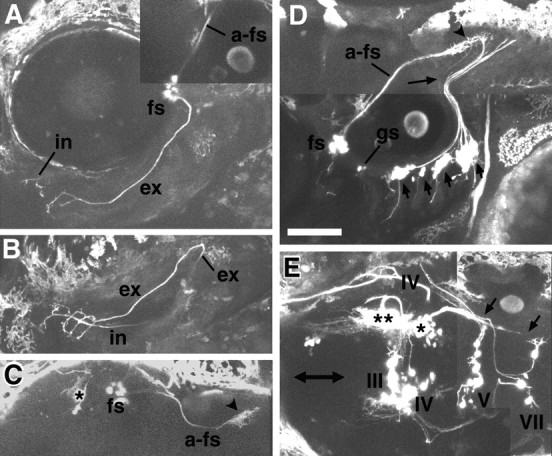
Mosaic analysis of the Isl1-GFP line. Mosaic animals were made by isochronically transplanting cells of the Isl1-GFP line into the normal embryo at the sphere stage to obtain stochastic labeling of cranial motor and sensory neurons. All images except for C are taken from larvae at ∼75 hr.C is from a larva at ∼90 hr. All figures are composite pictures made from the stacked confocal images. A,Lateral (slightly ventral) view of a mosaic larva in which GFP is expressed in the facial sensory ganglion cells (fs). B, C, Ventral (B) and dorsal (C) views of the same larva shown in A. a-fs shows the centrally projecting afferent facial sensory axons, which terminate in the dorsoposterior part of the hindbrain (arrowhead). The asterisk indicates signals derived from GFP-expressing cells in the mesencephalic region (Fig. 7A, a; B, d; see also single and double asterisks in E). Peripherally extending axons from the facial sensory ganglion cells take internal (in) and external (ex) pathways. Axons of both internal and external pathways terminate near the mouth. The internal fibers (correspond to a in Fig.6C) take a deep route, and most of the parts of the fibers are not visible, partly because they run out of the focal plane, and partly because of the opaque nature of the eye (lateral view) and cartilages (ventral view). The external fibers initially take the same superficial pathway as the seventh motor nerve and then diverge (Fig.6C, b; D,E, 7_E, arrows). D, Lateral view of a mosaic larva in which GFP is expressed in neurons in the facial sensory ganglion (fs), in the glossopharyngeal sensory ganglion (gs), and the vagus sensory ganglion (short arrows). a-fs shows the centrally projecting afferent facial sensory axons, which terminate in the dorsoposterior part of the hindbrain (arrowhead). The_long thin arrow indicates the faintly fluorescent centrally projecting axons from the glossopharyngeal sensory neurons. Terminals of the centrally projecting axons from the facial, glossopharyngeal, and vagus ganglion cells are located near one another in the dorsoposterior part of the hindbrain. The terminal region is likely to correspond to the nucleus tractus solitarii in the adult fish. We have examined a number of mosaic embryos in which GFP was expressed in the facial sensory neurons (n > 15). In all cases, centrally projecting fibers took the same pathway (Figs.6A,E, 7_A_,a-fs; 6_C, green_), and peripherally projecting fibers eventually terminated around the mouth.E, Dorsal view of a mosaic embryo in which GFP is expressed in a number of cells other than the motor neurons in the brain. The double-headed arrow marks the midline. The_double asterisks_ and the _single asterisk_indicate the GFP-positive cells in the mesencephalic region (Fig.7A, a; B, d). Anteriorly projecting processes from these cells (probably from the cells marked by the double asterisk) correspond to the tracts marked by _c_in Figure 7B, whereas posteriorly projecting processes from these cells (probably from the cells marked by the single asterisk) correspond to the tracts marked by _e_in Figure 7B. The posteriorly projecting axons may terminate around the trigeminal motor nucleus and facial motor nucleus (arrows). Scale bar, 100 μm.
Because GFP is not expressed in the sensory neurons related to the octavolateralis system, all of the GFP-labeled octavolateralis nerves in the periphery are efferent axons. Thus, it was possible to systematically follow the axonal outgrowth of the lateral line efferents. As described earlier, the pioneering axons among the nVII/OLe axons exit from the hindbrain at ∼24 hr (Fig. 5B, arrow). In vivo, time-lapse microscopic observations of these embryos suggested that these pioneering axons correspond to the OLe axons (data not shown). These axons extend both anteriorly (Fig.5C, f ) and posteriorly (Fig. 5C, d; D, i) from the exit point. Posteriorly extending axons are likely to project to the posterior lateral line (Metcalfe et al., 1985). The anteriorly extending nerve takes a superficial pathway (Fig. 5E, l), and subsequently, branches with one branch extending anteriorly [supraorbital lateral line (soL)] and the other extending ventrally [infraorbital lateral line (ioL)] (Figs. 5F,6B,C). Later in development, other branching processes can also be recognized (Fig. 6C, asterisks; E, g). Vital staining of hair cells in the lateral line neuromasts by DASPEI revealed that the OLe axons were found near the neuromasts as expected (Fig.6D,E). Their endings spread into fine branches terminating in the circumference of the individual hair cells (Fig. 3E,F). Branches of the OLe nerves were also seen to project to the hair cells in the otocyst (Fig. 7C, mm, ma, asterisks). Thus, OLe nerves project to hair cells in the lateral line and otocyst quite early in their development, suggesting that hair cells receive efferent input from the onset of their function.
Other cranial nerves
Among the motor neurons innervating the extraocular muscles, GFP is expressed in the oculomotor (nIII) and the trochlear motor (nIV) neurons, but not in the abducens motor (nVI) neurons. GFP-positive nIII neurons are visible at 26 hr (Fig. 5C), and subsequently, they extend axons toward the extraocular muscles (Fig. 5D, g; 6_B_,C). The projection pattern of the GFP-labeled axons to target extraocular muscles are described in Table 1. GFP-positive nIV neurons appear at 30–32 hr (data not shown). Their axons extend dorsally within the midbrain (Fig.5F, n), cross the dorsal midline (Fig. 7A) before they exit the CNS, extend ventrally in the periphery (Fig.6B,C), and then turn anteriorly toward their target, the superior oblique (so) muscle (Fig.7A,B).
In the glossopharyngeal (IX) and vagus (X) cranial systems, GFP is expressed in the glossophayngeal sensory ganglion cells (Fig.4E, gs), the vagus motor (nX) neurons, and the vagus sensory ganglion cells, which form clusters associated with each gill (vs; Figs. 4E, 9D). No GFP expression is detected in the glossopharyngeal motor (nIX) neurons. By 36 hr, GFP-expressing cells in the glossopharyngeal ganglion along with their centrally projecting axons can be observed (Fig. 5F, gs). Later in development, peripherally extending axons into the first gills are also observed (Figs. 6B,C,E,7C, IX ). The nX motor neurons form a large cluster in the posterior part of the hindbrain (Fig. 4C,E;Chandrasekhar et al., 1997) and extend thick axon bundles into the periphery (Fig. 5F). The pathways of the vagus sensory axons extending in both central and peripheral directions appear to coincide with the branchial (to gills) and visceral motor pathways (Fig. 7C; g shows fibers extending into the visceral organs). Thus, the labeled tenth nerves are likely to include both motor and sensory components. Later in development, GFP is also expressed in cells other than motor neurons in the brain (Figs.6C, thin arrows; 7_A_, a, B, b, d; 9_E, single_ and double asterisks).
DISCUSSION
This study reports the generation of a line of transgenic zebrafish, the Isl1-GFP fish, that expresses GFP in the cranial motor neurons, some of the cranial sensory neurons, and several other groups of cells. The transparency of zebrafish embryos and larvae enabled us to obtain high-resolution images of cranial motor neurons and their axonal trajectories in intact animals throughout development. Observations of the development of the cranial motor and sensory neurons in the transgenic zebrafish were easy and reproducible. Taking advantage of these features, we first followed overall development of the nV neurons. Next, we have examined whether the segmental origin of the nV neurons correlates with target selection. Additionally, we characterized several other groups of GFP-positive neurons. The generation of this transgenic line and the description of axonal outgrowth by the GFP-positive neurons derived from analysis of this line set the stage for future experimental and genetic examination of the development of these neurons.
Identification of the promoter/enhancer region of the Isl1 gene and generation of the transgenic fish
In transient expression assays, we identified two enhancer elements of the Isl1 gene: one for expression in cranial motor neurons and the other for expression in Rohon–Beard cells and the trigeminal ganglion cells. Using the former element, we generated a stable transgenic line. Our strategy in dissecting the enhancer regions was as follows. First, we prepared a core plasmid that has a minimum promoter of the Isl1 gene sufficient only for expression in hatching gland cells. Next, we generated a series of DNA constructs in which different genomic fragments near the Isl1 gene were inserted into the core plasmid and examined enhancer activity by injecting each construct into zebrafish embryos. The ease of transient expression assays in zebrafish embryos made it feasible to test a number of DNA constructs. This approach should be generally applicable to the identification of enhancer elements of any gene that controls tissue-specific gene expression and to the subsequent generation of stable transgenic fish lines.
Segmental origins within the nV neurons define their target fields
Our analysis of the fifth nerve by branch-specific DiI labeling revealed that neurons supplying axons into an intermediate branch (innervating the am) are located in the anterior cluster of the nV, and neurons supplying axons into proximal (innervating the lap and do) and distal (innervating the ima and imp) branches are located in the posterior cluster of the nV. The am, the only muscle innervated by the anteriorly located neurons, functions as a jaw “closer” acting antagonistically with the rest of jaw “openers”. Thus, the closer and opener motor neurons are located in different rhombomeres.
The organization of the trigeminal motor neurons in the CNS with respect to the peripheral nerve branches has been examined in many adult vertebrates (for references, see Song and Boord, 1993). These studies suggested that, despite the wide range of morphological differences, the organization of the trigeminal system is conserved among vertebrates. All vertebrates examined so far contain equivalent proximal, intermediate, and distal branches of nV, with motor neurons that project into the intermediate branch located as a discrete cluster at the anterior part of the trigeminal nucleus. Our results support such previous observations and reveal that the stereotyped connections between subpopulations of the trigeminal motor neurons and the opener and closer muscles are established early in development. Because GFP-positive clusters of trigeminal neurons, from the beginning, start out as discrete clusters in r2 and r3, it is likely that the neurons of the anterior and posterior clusters are born in r2 and r3, respectively. Thus, the differences between the two clusters of trigeminal motor neurons can be attributed to the segmental origin of the motor neurons; i.e., neurons born in r2 and r3 will eventually project to distinct target muscles of different physiological functions. It is possible that the anterior cluster (r2) and the posterior cluster (r3) are distinct in nature at birth, presumably under the control of the regulatory genes expressed in a rhombomere-specific manner (Lumsden and Krumlauf, 1996). Conservation of both the adult trigeminal pattern (Song and Boord, 1993) and hindbrain segmentation processes during development among vertebrates (Gilland and Baker, 1993; Lumsden and Krumlauf, 1996) suggests that this idea applies to other vertebrates, including chick and mouse.
Potential use of the Isl1-GFP line for neurobiology
The Isl1-GFP zebrafish will be useful for several types of studies. For instance, it could be used to analyze the function of genes regulating motor neuron development. By injecting DNA constructs, such as dominant negative alleles of genes normally expressed by the cranial motor neurons into Isl1-GFP embryos, one could potentially examine loss of function effects on outgrowth and pathfinding by the motor axons. Live visualization of motor neurons and their axons would allow for a dynamic analysis of the effects of such molecular manipulations on pathfinding by growth cones. The ability to assay effects dynamically would increase the detectability of phenotypes because the effect of some molecular manipulations may be most apparent and informative in a dynamic analysis. Similarly, the Isl1-GFP line would be useful for cellular manipulations such as laser ablation experiments, which could ablate specific groups of trigeminal neurons or other cell types. Furthermore, the transgenic fish might be also useful for electrophysiological studies because it would allow researchers to record the activity of a GFP-labeled neuronal type reproducibly.
Because the zebrafish is genetically manipulable, the Isl1-GFP zebrafish provides an opportunity for genetic analyses of the development of neurons such as the cranial motor neurons. Recent large-scale mutant screens have identified many mutations that cause defects in CNS or craniofacial development (Driever et al., 1996;Haffter et al., 1996). The Isl1-GFP fish will enable further examination of these mutations after introduction of the Isl1-GFP transgene into the mutant background by crossing mutant with Isl1-GFP lines. Our detailed characterization of GFP-labeled motor and sensory neurons along with their fibers provides an anatomical basis for these studies. Finally, the Isl1-GFP line can be used as a starting strain for mutational analysis of GFP-positive neurons. The line affords the possibility of screening for new mutations that affect hindbrain segmentation and determination of specific neuronal types. Furthermore, the fact that axon outgrowth can be so readily assayed in these embryos should make it possible to screen directly for mutations that affect outgrowth, pathfinding, and synapse formation by the GFP-positive neurons.
Footnotes
This work was supported by grants from the Japan Science and Technology Corporation, the Ministry of Education, Science, and Culture of Japan, and Special Coordination Fund from Science and Technology Agency of Japan. We especially thank N. Ueno, in whose laboratory the majority of this study was performed. We thank G. Eguchi and G. Mandel for generous support. We thank Y. Ishikawa, K. Kawamura, and S. Kuratani for their suggestions on the anatomy of cranial motor and sensory neurons. We thank G. Mandel, J. Fetcho, K. Cho, A. Thomson, Y. Ishikawa, and J. Y. Kuwada for comments on this manuscript. We thank M. Sugiura for assistance with the experiments; K. Takamatsu and Y. Katoh for maintaining our fish colony; M. Nikaido for instruction in the technique of mosaic analysis; K. Umesono for the donation of a modified GFP; M. Petkovich, A. Picker, and H. Takeda for the donation of zebrafish genomic libraries; and members of the Ueno laboratory for encouragement and discussion. The zn8 (= zn5) antibody was obtained from the Developmental Studies Hybridoma Bank maintained by the Department of Biology at the University of Iowa.
Correspondence should be addressed to Hitoshi Okamoto, Laboratory for Developmental Gene Regulation, Brain Science Institute, RIKEN (The Institute of Physical and Chemical Research), 2–1 Hirosawa, Wako, Saitama 351–0198, Japan. E-mail: hitoshi@brain.riken.go.jp.
Dr. Higashijima's present address: Department of Neurobiology and Behavior, State University of New York at Stony Brook, Stony Brook, NY 11794. E-mail: shigashijima@notes.cc.sunysb.edu.
REFERENCES
- 1.Appel B, Korzh V, Glasgow E, Thor S, Edlund T, Dawid IB, Eisen JS. Motoneuron fate specification revealed by patterned LIM homeobox gene expression in embryonic zebrafish. Development. 1995;121:4117–4125. doi: 10.1242/dev.121.12.4117. [DOI] [PubMed] [Google Scholar]
- 2.Balak KJ, Corwin JT, Jones JE. Regenerated hair cells can originate from supporting cell progeny: evidence from phototoxicity and laser ablation experiments in the lateral line system. J Neurosci. 1990;10:2502–2512. doi: 10.1523/JNEUROSCI.10-08-02502.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burrill JD, Easter S., Jr Development of the retinofugal projections in the embryonic and larval zebrafish (Brachydanio rerio). J Comp Neurol. 1994;346:583–600. doi: 10.1002/cne.903460410. [DOI] [PubMed] [Google Scholar]
- 4.Chandrasekhar A, Moens CB, Warren J, Jr, Kimmel CB, Kuwada JY. Development of branchiomotor neurons in zebrafish. Development. 1997;124:2633–2644. doi: 10.1242/dev.124.13.2633. [DOI] [PubMed] [Google Scholar]
- 5.Chandrasekhar A, Warren JT, Jr, Takahashi K, Schauerte HE, van Eeden FJ, Haffter P, Kuwada JY. Role of sonic hedgehog in branchiomotor neuron induction in zebrafish. Mech Dev. 1998;76:101–115. doi: 10.1016/s0925-4773(98)00101-4. [DOI] [PubMed] [Google Scholar]
- 6.Chandrasekhar A, Schauerte HE, Haffter P, Kuwada JY. The zebrafish detour gene is essential for cranial but not spinal motor neuron induction. Development. 1999;126:2727–2737. doi: 10.1242/dev.126.12.2727. [DOI] [PubMed] [Google Scholar]
- 7.Coombs S, Görner P, Münz H. The mechanosensory lateral line: neurobiology and evolution. Springer; New York: 1989. [Google Scholar]
- 8.Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, Belak J, Boggs C. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- 9.Dynes JL, Ngai J. Pathfinding of olfactory neuron axons to stereotyped glomerular targets revealed by dynamic imaging in living zebrafish embryos. Neuron. 1998;20:1081–1091. doi: 10.1016/s0896-6273(00)80490-0. [DOI] [PubMed] [Google Scholar]
- 10.Ericson J, Thor S, Edlund T, Jessell TM, Yamada T. Early stages of motor neuron differentiation revealed by expression of homeobox gene Islet-1. Science. 1992;256:1555–1560. doi: 10.1126/science.1350865. [DOI] [PubMed] [Google Scholar]
- 11.Gilland E, Baker R. Conservation of neuroepithelial and mesodermal segments in the embryonic vertebrate head. Acta Anat. 1993;148:110–123. doi: 10.1159/000147530. [DOI] [PubMed] [Google Scholar]
- 12.Guthrie S. Patterning the hindbrain. Curr Opin Neurobiol. 1996;6:41–48. doi: 10.1016/s0959-4388(96)80007-9. [DOI] [PubMed] [Google Scholar]
- 13.Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, Kelsh RN, Furutani-Seiki M, Vogelsang E, Beuchle D, Schach U, Fabian C, Nüsslein-Volhard C. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- 14.Hatta K, Schilling TF, BreMiller RA, Kimmel CB. Specification of jaw muscle identity in zebrafish: correlation with engrailed-homeoprotein expression. Science. 1990;250:802–805. doi: 10.1126/science.1978412. [DOI] [PubMed] [Google Scholar]
- 15.Heaton MB, Moody SA. Early development and migration of the trigeminal motor nucleus in the chick embryo. J Comp Neurol. 1980;189:61–99. doi: 10.1002/cne.901890105. [DOI] [PubMed] [Google Scholar]
- 16.Higashijima S, Nose A, Eguchi G, Hotta Y, Okamoto H. Mindin/F-spondin family: novel ECM proteins expressed in the zebrafish embryonic axis. Dev Biol. 1997a;192:211–227. doi: 10.1006/dbio.1997.8760. [DOI] [PubMed] [Google Scholar]
- 17.Higashijima S, Okamoto H, Ueno N, Hotta Y, Eguchi G. High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev Biol. 1997b;192:289–299. doi: 10.1006/dbio.1997.8779. [DOI] [PubMed] [Google Scholar]
- 18.Highstein SM. The central nervous system efferent control of the organs of balance and equilibrium. Neurosci Res. 1991;12:13–30. doi: 10.1016/0168-0102(91)90096-h. [DOI] [PubMed] [Google Scholar]
- 19.Inoue A, Takahashi M, Hatta K, Hotta Y, Okamoto H. Developmental regulation of islet-1 mRNA expression during neuronal differentiation in embryonic zebrafish. Dev Dyn. 1994;199:1–11. doi: 10.1002/aja.1001990102. [DOI] [PubMed] [Google Scholar]
- 20.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 21.Korzh V, Edlund T, Thor S. Zebrafish primary neurons initiate expression of the LIM homeodomain protein Isl-1 at the end of gastrulation. Development. 1993;118:417–425. doi: 10.1242/dev.118.2.417. [DOI] [PubMed] [Google Scholar]
- 22.Long Q, Meng A, Wang H, Jessen JR, Farrell MJ, Lin S. GATA-1 expression pattern can be recapitulated in living transgenic zebrafish using GFP reporter gene. Development. 1997;124:4105–4111. doi: 10.1242/dev.124.20.4105. [DOI] [PubMed] [Google Scholar]
- 23.Lumsden A, Krumlauf R. Patterning the vertebrate neuraxis. Science. 1996;274:1109–1115. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- 24.Metcalfe WK, Kimmel CB, Schabtach E. Anatomy of the posterior lateral line system in young larvae of the zebrafish. J Comp Neurol. 1985;233:377–389. doi: 10.1002/cne.902330307. [DOI] [PubMed] [Google Scholar]
- 25.Murray MJ, Merritt DJ, Brand AH, Whitington PM. In vivo dynamics of axon pathfinding in the Drosophila CNS: a time- lapse study of an identified motorneuron. J Neurobiol. 1998;37:607–621. [PubMed] [Google Scholar]
- 26.Okada A, Lansford R, Weimann JM, Fraser SE, McConnell SK. Imaging cells in the developing nervous system with retrovirus expressing modified green fluorescent protein. Exp Neurol. 1999;156:394–406. doi: 10.1006/exnr.1999.7033. [DOI] [PubMed] [Google Scholar]
- 27.Osumi N, Hirota A, Ohuchi H, Nakafuku M, Iimura T, Kuratani S, Fujiwara M, Noji S, Eto K. Pax-6 is involved in the specification of hindbrain motor neuron subtype. Development. 1997;124:2961–2972. doi: 10.1242/dev.124.15.2961. [DOI] [PubMed] [Google Scholar]
- 28.Pfaff S, Kintner C. Neuronal diversification: development of motor neuron subtypes. Curr Opin Neurobiol. 1998;8:27–36. doi: 10.1016/s0959-4388(98)80005-6. [DOI] [PubMed] [Google Scholar]
- 29.Roberts BL, Meredith GE. The efferent system. In: Coombs S, Görner P, Münz H, editors. The mechanosensory lateral line. Springer; New York: 1989. pp. 445–460. [Google Scholar]
- 30.Roberts BL, Meredith GE. The efferent innervation of the ear: variations on an enigma. In: Webster DB, Fay RR, Popper AN, editors. The evolutionary biology of hearing. Springer; New York: 1992. pp. 185–210. [Google Scholar]
- 31.Schilling TF, Kimmel CB. Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development. 1997;124:2945–2960. doi: 10.1242/dev.124.15.2945. [DOI] [PubMed] [Google Scholar]
- 32.Simon H, Guthrie S, Lumsden A. Regulation of SC1/DM-GRASP during the migration of motor neurons in the chick embryo brain stem. J Neurobiol. 1994;25:1129–1143. doi: 10.1002/neu.480250908. [DOI] [PubMed] [Google Scholar]
- 33.Somiya H. Dynamic mechanism of visual accommodation in teleosts: structure of the lens muscle and its nerve control. Proc R Soc Lond B Biol Sci. 1987;230:77–91. doi: 10.1098/rspb.1987.0010. [DOI] [PubMed] [Google Scholar]
- 34.Song J, Boord RL. Motor components of the trigeminal nerve and organization of the mandibular arch muscles in vertebrates. Phylogenetically conservative patterns and their ontogenetic basis. Acta Anat. 1993;148:139–149. doi: 10.1159/000147533. [DOI] [PubMed] [Google Scholar]
- 35.Tokumoto M, Gong Z, Tsubokawa T, Hew CL, Uyemura K, Hotta Y, Okamoto H. Molecular Heterogeneity among primary motoneurons and within myotomes revealed by the differential mRNA expression of novel Islet-1 homologs in embryonic zebrafish. Dev Biol. 1995;171:578–589. doi: 10.1006/dbio.1995.1306. [DOI] [PubMed] [Google Scholar]
- 36.Trevarrow B, Marks DL, Kimmel CB. Organization of hindbrain segments in the zebrafish embryo. Neuron. 1990;4:669–679. doi: 10.1016/0896-6273(90)90194-k. [DOI] [PubMed] [Google Scholar]
- 37.Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 38.Varela-Echavarría A, Pfaff SL, Guthrie S. Differential expression of LIM homeobox genes among motor neuron subpopulations in the developing chick brain stem. Mol Cell Neurosci. 1996;8:242–257. doi: 10.1006/mcne.1996.0061. [DOI] [PubMed] [Google Scholar]
- 39.Westerfield M. The zebrafish book. University of Oregon; Eugene, OR: 1995. [Google Scholar]
- 40.Whitfield TT, Granato M, van Eeden FJ, Schach U, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg CP, Jiang YJ, Kane DA, Kelsh RN, Mullins MC, Odenthal J, Nüsslein-Volhard C. Mutations affecting development of the zebrafish inner ear and lateral line. Development. 1996;123:241–254. doi: 10.1242/dev.123.1.241. [DOI] [PubMed] [Google Scholar]