Integrin Subunit Gene Expression Is Regionally Differentiated in Adult Brain (original) (raw)
Abstract
Integrins are a diverse family of heterodimeric (αβ) adhesion receptors recently shown to be concentrated within synapses and involved in the consolidation of long-term potentiation. Whether neuronal types or anatomical systems in the adult rat brain are coded by integrin type was studied in the present experiments by mapping the relative densities of mRNAs for nine α and four β subunits. Expression patterns were markedly different and in some regions complementary. General results and areas of notable labeling were as follows: α1—limited neuronal expression, neocortical layer V, hippocampal CA3; α3 and α5—diffuse neuronal and glial labeling, Purkinje cells, hippocampal stratum pyramidale, locus coeruleus (α3); α4— discrete limbic regions, olfactory cortical layer II, hippocampal CA2; α6—most prominently neuronal, neocortical subplate, endopiriform, subiculum; α7—discrete, all neocortical layers, hippocampal granule cells and CA3, cerebellar granule and Purkinje cells, all efferent cranial nerve nuclei; α8—discrete neuronal, deep cortex, hippocampal CA1, basolateral amygdala, striatum; αV—all cortical layers, striatum, Purkinje cells; β4—dentate gyrus granule cells; β5—broadly distributed, neocortex, medial amygdala, cerebellar granule and Purkinje cells, efferent cranial nerve nuclei; α2, β2, and β3—mRNAs not detected. These results establish that brain subfields express different balances of integrin subunits and thus different integrin receptors. Such variations will determine which matrix proteins are recognized by neurons and the types of intraneuronal signaling generated by matrix binding. They also could generate important differences in synaptic plasticity across brain systems.
Keywords: adhesion molecules, extracellular matrix, hippocampus, cortex, brainstem, in situ hybridization
Integrins are membrane-spanning, noncovalently bound αβ heterodimers that act as cell–matrix and cell–cell adhesion receptors throughout the body (Hynes, 1992). They also activate intracellular signaling cascades, including tyrosine, serine/threonine, and mitogen-activated protein kinase activities associated with neurotrophic factors and cytokines (Clark and Brugge, 1995). A large body of work indicates that the integrins exert potent influences over a diverse array of cellular functions, including proliferation, differentiation, process outgrowth, gene expression, and survival (Damsky and Werb, 1992; Yamada and Miyamoto, 1995; Giancotti, 1996; Katz and Yamada, 1997). Cell migration, axon guidance, and synapse formation in the developing nervous system are likely examples of integrin-dependent operations (Dodd and Jessell, 1988; Sanes, 1989;Reichardt and Tomaselli, 1991; Kil et al., 1996). Modifying synapses in the adult brain may be another. Specifically, hippocampal slice experiments have shown that peptides that block ligand binding by a major subclass of integrins prevent the stabilization of long-term potentiation (LTP) and the formation of in vitro kindling while having no detectable effects on baseline physiology (Staubli et al., 1990; Xiao et al., 1991; Grooms and Jones, 1997). LTP can be partially disrupted by integrin antagonists several minutes_after_ its induction (Bahr et al., 1997; Staubli et al., 1998), as expected from the hypothesis that stabilization involves the activation and engagement (matrix binding, attachment to cytoskeleton) of latent integrins. Memory consolidation across many paradigms has a similar time course, but there are as yet no tests for integrin involvement in this phenomenon. However, Drosophila with a mutation in a synapse-associated integrin α subunit (vol) have retention deficits that are alleviated by_vol_ transgene expression (Grotewiel et al., 1998).
Sixteen α and eight β subunits have been identified (Aplin, 1998), which form a large number of heterodimeric receptor combinations. As a consequence of differences in subunit composition, integrin receptors differ in the specific matrix proteins they recognize (e.g., laminins, collagens, vitronectin, etc.) and the intracellular cascades they initiate (Hynes, 1992; Clark and Brugge, 1995; Jewell et al., 1995;Ruoslahti, 1996; Katz and Yamada, 1997; Gong et al., 1998) and, in some instances, can have opposing effects on gene expression (Huhtala et al., 1995). Whether regions or neuron classes in adult brain are coded by integrin type has not been studied. Biochemical experiments indicate that synaptic fractions are greatly enriched in integrin receptors (Bahr and Lynch, 1992; Capaldi et al., 1997; Nishimura et al., 1998), whereas immunoelectron microscopy has identified two subunits (α8, β8) concentrated in postsynaptic densities (Einheber et al., 1996;Nishimura et al., 1998). Regional differences in levels of α8 and β8 immunoreactivities (Einheber et al., 1996; Nishimura et al., 1998) and β1 mRNA (Pinkstaff et al., 1998) have been described, but there are no data addressing the possibilities of regional patterns associated with particular integrin classes or regional differences in α-subunit gene expression in brain. Given the diversity of integrin effects, regional differences in receptor expression could be involved in a wide range of neuronal specializations, including variants of activity-dependent synaptic plasticity (Maren and Baudry, 1995). To examine these possibilities, the studies reported here used in situ hybridization to map the regional and cellular localization of gene expression for nine α and four β integrin subunits in adult rat brain.
MATERIALS AND METHODS
In situ hybridization. Adult male Sprague Dawley rats (250–350 gm; Simonsen labs, Gilmore, CA) were used for all procedures. For in situ hybridization analyses the rats (n = 8) were killed with an overdose of sodium pentobarbital (100 mg/kg) and perfused transcardially with 4% paraformaldehyde in 0.1 m phosphate buffer (PPB). The brains were post-fixed for 24–48 hr in PPB, cryoprotected in PPB/20% sucrose for 24–36 hr, and sectioned on a freezing microtome (coronal, 25 μm) into cold PPB. Spaced series of tissue sections were processed free-floating for in situ hybridization, as described in detail elsewhere (Guthrie et al., 1995). This entailed hybridization with 35S-labeled antisense riboprobes (1 × 107 cpm/ml) at 60°C for 36–48 hr in buffer containing 50% deionized formamide, 35× Denhardt’s reagent (0.7% polyvinylpyrrolidone, 0.7% bovine serum albumin, and 0.7% Ficoll), and 0.14× SSC (1× SSC = 0.15 m NaCl/0.015m sodium citrate). After hybridization the tissue was treated with RNase A and washed through increasingly dilute SSC solutions to a final wash in 0.1× SSC at 60°C. Then the tissue was mounted onto gelatin-coated slides, air-dried, processed for film (Amersham β-Max, Arlington Heights, IL) and then emulsion (Kodak NTB-2, Rochester, NY) autoradiography, stained with cresyl violet or hematoxylin, and coverslipped with Permount (Fisher Scientific, Fair Lawn, NJ). As a control for specificity, alternate sections were processed (as above) for hybridization with 35S-labeled sense transcripts transcribed from each of the cDNA templates used to generate the antisense probes. No regional or cellular labeling was obtained with sense probe hybridizations. The amino acid sequences for the β integrins are from ∼30% (β4 vs others) to 60% (β3, β5, β6, and β1, β2) identical (Moyle et al., 1991), whereas the α integrins are more divergent with <30% (α2, α4 vs others) to <50% (all comparisons) identity (Hynes, 1992). Nevertheless, the subunit specificity of hybridization was demonstrated by the fact that the integrin subunit cRNAs used here yielded labeling patterns that differed from each other in both positive and negative directions. Moreover, all labeling patterns differed from those obtained with a variety of other cRNAs, including those for integrin β1, brain-derived neurotrophic factor, basic fibroblast growth factor, and AMPA receptor subunit transcripts (data not shown).
To evaluate the potential colocalization of integrin α7 and β1 mRNAs, we processed sections through cerebellum and lower brainstem for simultaneous in situ hybridization with digoxigenin-labeled α7 cRNA and 35S-labeled β1 cRNA. Labeling was localized by using anti-digoxigenin immunohistochemistry with alkaline phosphatase as chromagen (DIG-Nucleic Acid Detection Kit; Boehringer Mannheim, Indianapolis, IN) for α7 cRNA labeling and Ilford K5.D emulsion (Polysciences, Warrington, PA) autoradiography for β1 cRNA labeling, as described in full detail elsewhere (Bizon et al., 1995).
The distribution of hybridization of each cRNA probe was evaluated by microscopic examination of the emulsion autoradiograms by two investigators working independently; all labeling patterns that were reported were verified by both investigators for at least three brains. Regions were identified with reference to the atlases of Paxinos and Watson (1986) and Swanson (1992).
Riboprobes. The radiolabeled cRNA probes were prepared by transcription from linearized cDNA templates in the presence of35S-UTP for in situ hybridization or32P-CTP for RNA blots. Table1 summarizes the information on plasmids and resultant cRNAs excepting those for α3 and αV (see below). Restriction enzymes and polymerases were obtained from Boehringer Mannheim and Stratagene (La Jolla, CA), respectively.
Table 1.
List of cDNAs
| cDNA | Species | Length (kb) | Reference |
|---|---|---|---|
| α 1 | Rat | 4.0 | (Ignatius et al., 1990) |
| α 2 | Mouse | 0.843 | (Wu and Santoro, 1996) |
| α 3 | Rat | 0.516 | GenBank accession number S66292 |
| α 4 | Mouse | 3.7 | (De Meirsman et al., 1994) |
| α 5 | Mouse | 0.860 | (Yang et al., 1993) |
| α 6 | Mouse | 0.800 | (Cooper et al., 1991) |
| α 7 | Rat | 2.3 | (Song et al., 1992) |
| α 8 | Mouse | 3.0 | (Denda et al., 1998) |
| α V | Rat | 0.516 | GenBank accession number S58528 |
| β 1 | Rat | 2.8 | (Malek-Hedayat and Rome, 1994) |
| β 2 | Mouse | 1.0 | (Wilson et al., 1989) |
| β 3 | Mouse | 2.6 | (Chiba et al., 1996) |
| β 4 | Mouse | 0.355 | (Falcioni et al., 1994) |
| β 5 | Mouse | 0.444 | GenBank accession number W14823 |
The cDNAs for α3 and αV were created by using the reverse transcriptase PCR (RT-PCR). First-strand cDNA synthesis consisted of reverse transcription of 1 μg of hippocampal total RNA, using Maloney murine leukemia virus (MMLV) reverse transcriptase (Promega, Madison, WI) and random primers. The complementary DNA was amplified with 100 pmol of the appropriate primers and TAQ polymerase (Promega). For α3, the primers 5′CACGGGTGTGGAACAGCAC3′ and 5′-CAGTGCTTCTTGGTGGGTAG-3′ corresponding to bases 62–578 of the mouse α3 mRNA sequence (Tamura et al., 1991) were used. The primers for αV were 5′-AAGGCGCAGAATCAAGGAG-3′ and 5′-TAAGGCCACTGGAGGTTCAG-3′, corresponding to bases 76–592 of the rat αV mRNA sequence (Shinar et al., 1993). For both cDNAs the PCR consisted of 35 cycles at 94, 60, and 72°C for denaturing, annealing, and elongation steps, respectively. After this PCR amplification the reaction products were purified by the Boehringer Mannheim PCR Purification Kit and ligated into PGEM-T vector (Promega). Clones were verified by dideoxynucleotide sequencing (Sequenase 2.0, United States Biochemical, Cleveland, OH).
RNA blot analysis. Northern blot hybridizations were conducted to verify that cRNAs for α1–α8 and αV and β2–β5 detected transcripts corresponding to the predicted sizes of the target subunit mRNAs. Untreated adult male rats (n = 2) were anesthetized with isoflurane and decapitated; their brains were removed for subfield dissections. Total RNA was extracted by homogenization in Trizol reagent (Pharmacia, Piscataway, NJ), quantified by UV spectrophotometry, and processed by RNA blotting techniques as previously described (Pinkstaff et al., 1998). Each of the cRNAs hybridized with mRNAs of the predicted size from total RNA extracts of hippocampus or cortex, with the exception of transcripts for α2, β3, and β4, which were not detected in hippocampus or cortex of control brains.
RESULTS
As summarized in Tables 2 and3, in situ hybridization analyses demonstrated highly region-specific patterns of integrin subunit mRNA expression in adult rat brain. Because the distributions and character of labeling differed dramatically across probes, the descriptions that follow will focus on the cRNAs separately, with the exception of α2, β2, and β3. Although each of these cRNAs detects transcripts in non-neuronal systems (Wilson et al., 1989; Chiba et al., 1996; Wu and Santoro, 1996) and, for α2 and β3 in experimental brain tissue (J. Pinkstaff and C. Gall, unpublished observations), they did not yield detectable in situ hybridization labeling in tissue sections from untreated adult rat brain. For the remaining cRNAs the distributions of hybridization are described for tissue sections from the most rostral levels of frontal cortex through lower brainstem or high cervical spinal cord. Autoradiographic exposure times varied across probes to best visualize the distributions of abundant (e.g., α7 mRNA) and rarer transcripts; as a consequence of this and of differences in cRNA lengths, comments on the relative densities of labeling between brain areas pertain to the cRNA at hand and are not intended to connote differences in the abundance of the different transcripts.
Table 2.
Distribution of integrin transcripts in telencephalon
| α1 | α3 | α4 | α5 | α6 | α7 | α8 | αV | β12-a | β4 | β5 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Olfactory | |||||||||||
| Tubercle | − | +/− | − | +/− | − | +dp | − | +++ | − | − | ++ |
| AON | − | + | + | + | +dp | + | ++ | ++ | − | − | + |
| NLOT | − | +/− | + | +/− | − | − | − | +BAOT | − | − | − |
| Cortex | |||||||||||
| NC 1 | − | − | − | − | − | − | − | − | − | − | + |
| NC 2 | − | + | +/−T | +/− | − | +/− | − | +++ | − | − | + |
| NC 3 | − | + | +/−T | +/− | − | +/− | − | ++ | − | − | + |
| NC 4 | − | + | − | +/− | − | +/− | − | + | − | − | ++ |
| NC 5 | ++ | + | − | +/− | − | + | − | ++ | − | − | ++ |
| NC 6 | − | + | − | +/− | + | +/− | ++ | ++ | − | − | − |
| Subplate | − | + | − | + | ++C | − | +++ | − | − | − | − |
| Piriform | − | ++2 | +++ | + | +C | +/−R | +D | +++2 | − | − | +/− |
| Entorhinal | +C | ++2 | +++ | +/− | ++ | − | +D | +++2 | − | − | +/− |
| Perirhinal | − | + | +/− | +/− | − | − | + | + | − | − | +/− |
| Orbital | + | + | +/− | +/− | − | − | +dp | − | − | − | + |
| Cingulate | ++ | + | − | +/− | − | − | +dp | + | − | − | ++ |
| Endopiriform | − | + | − | +/− | +++ | − | +++ | +C | − | − | +/− |
| Claustrum | + | + | − | +/− | ++ | +/− | +++ | + | − | − | +/− |
| Subfornical organ | + | + | − | − | ++ | ++ | − | − | ++ | ++ | + |
| Amygdala | |||||||||||
| Basolateral | − | + | − | +/− | +/− | − | +++ | − | − | − | +/− |
| Central | − | +/− | − | +/− | − | +/− | − | − | − | − | +/− |
| Cortical | − | + | + | +/− | +/− | − | +/− | ++ | − | − | +/− |
| Medial | − | + | − | +/− | +PA | +/−PD | +++PA | − | − | − | ++ |
| Hippocampus | |||||||||||
| Str. granulosum | − | − | +C | + | +/−C | +++ | +/−T | + | − | +++ | +/− |
| CA1, sp | +C | ++ | − | ++ | − | − | +++ | +/− | + | − | +T |
| CA2, sp | − | ++ | ++ | ++ | + | +++ | − | − | + | − | − |
| CA3, sp | +++ | +++ | +/−C | ++ | − | ++T | +T | − | + | − | +T |
| Subiculum, sp | − | + | +/− | + | ++T | +T | +++T | − | − | − | +T |
| Hilus | +R | ++ | − | − | ++C | +T | ++T | − | + | − | + |
| Tenia tecta | + | +/− | +++ | + | +++ | ++ | − | + | − | − | + |
| Septum | ++M | +L | +L | ++M | ++M, CL | ||||||
| HDB/MCPO | + | ++ | − | + | − | − | − | − | + | − | ++ |
| Stria terminalis | + | +/− | − | +/− | − | − | + | − | − | − | ++ |
| Caudate putamen | − | +/− | − | +/− | − | − | ++V | ++ | − | − | − |
Table 3.
Integrin transcript distribution in diencephalon and lower centers
| α1 | α3 | α4 | α5 | α6 | α7 | α8 | αV | β13-a | β4 | β5 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Thalamus | |||||||||||
| Reticular | ++R | +/− | − | +/− | ++ | − | − | − | − | − | + |
| MGB | − | + | − | +/− | − | +/− | − | +/− | − | − | +/− |
| Intralaminar | + | + | − | +/− | − | − | − | − | − | − | − |
| Other | ++AVL | +AD | − | − | +/−VL | +/−AV | +VM | − | − | − | ++VP |
| Hypothalamus | |||||||||||
| Paraventricular | ++ | ++ | − | + | + | − | − | + | ++ | − | +/− |
| Supraoptic | + | ++ | − | + | − | + | − | − | ++ | − | +/− |
| Ventromedial | − | +/− | − | +/− | + | − | +/− | − | − | − | + |
| Arcuate | + | + | − | +/− | + | − | ++ | − | − | − | +/− |
| Med. eminence | + | +/− | − | +/− | + | − | − | + | + | − | ++ |
| Suprachiasmatic | − | + | − | +/− | − | − | ++ | − | − | − | − |
| Other | − | +PO | − | − | − | − | +MB | − | − | − | − |
| Midbrain | |||||||||||
| Sup. colliculus | − | +/− | − | − | − | +dp | − | +S | − | − | + |
| SN/VTA | + | + | − | + | − | − | − | − | + | − | ++ |
| Red nucleus | − | − | − | − | − | Sub | − | + | ++ | − | ++ |
| Midbrain RF | − | + | − | +/− | − | + | − | − | + | − | +/− |
| Oculomotor | − | + | − | − | + | +++ | − | + | + | − | ++ |
| Trochlear | − | − | − | − | − | +++ | − | − | + | − | ++ |
| Cerebellum | |||||||||||
| Purkinje | − | +++ | − | +++ | ++ | + | − | +++ | +++ | − | ++ |
| Granule | − | − | − | − | − | +++ | − | − | − | − | +++ |
| Lower brainstem | |||||||||||
| Raphe | − | + | − | − | − | − | − | − | − | − | − |
| Inferior olive | − | +/− | − | +/− | − | + | − | − | − | − | +/− |
| Motor 53-b | − | +++ | − | − | +/− | +++ | − | − | +++ | − | +++ |
| Chief 5 | − | − | − | − | +/− | − | − | − | − | − | + |
| Spinal 5 | − | − | − | − | − | + | + | − | − | − | ++ |
| Mes. 5 | + | − | − | − | +/− | +++ | − | − | ++ | − | ++ |
| Abducens | − | + | − | − | − | ++ | − | − | + | − | ++ |
| Facial motor | − | + | − | +/− | + | +++ | − | + | +++ | − | ++ |
| Salivatory | − | − | − | − | − | + | − | − | ++ | − | ++ |
| Ambiguus | − | ++ | − | +/− | − | +++ | − | − | ++ | − | ++ |
| DM vagus | − | ++ | − | +/− | − | + | − | − | ++ | + | ++ |
| Hypoglossal | + | + | − | +/− | + | +++ | − | + | ++ | − | ++ |
| Cochlear | − | − | − | − | − | + | − | + | − | − | |
| Vestibular | − | ++ | − | +/− | − | ++ | Sub | − | + | − | + |
| Solitarius | − | − | − | − | − | − | − | − | − | − | |
| Area postrema | ++ | ++ | − | +/− | ++ | + | − | +++ | + | ++ | + |
| Ventral horn3-c | − | ++ | − | +/− | − | Sub | − | − | + | − | + |
| Other | +SO | +++LC | − | − | − | − | − | − | +LC | − | − |
| Pia mater | ++ | − | − | − | +/−VE | +VE | ++ | − | ++ | +++VE | − |
| Blood vessels | + | − | − | − | − | − | − | − | − | − | − |
α1 mRNA
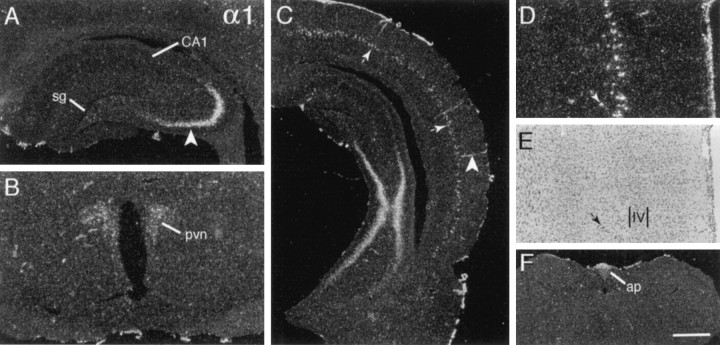
Like many of the transcripts that were evaluated, the α1 cRNA generated low-density and faintly patchy labeling throughout gray and white matter, giving the impression of low expression levels by glial cells. In addition, as summarized in Tables 2 and 3, integrin α1 mRNA was highly expressed in a few discrete regions distributed across the neuraxis; in these regions the hybridization appeared to be localized to neurons on the basis of the sizes and cytoarchitectonic distributions of labeled cells. In rostral telencephalon the labeling was limited to a few cells in deep orbital frontal cortex and intermediate layers of medial allocortex. Labeling in the latter field continued caudally through prelimbic, cingulate, and retrosplenial cortex. At the level of the hippocampal commissure a second population of well labeled cortical cells was evident within the suprarhinal region, and at progressively more caudal planes the population extended dorsal from the rhinal fissure to encompass the medial-to-ventral arc of neocortex fully (Fig.1C). As shown in Figure 1,D and E, these large, heavily labeled cells were distributed within a superficial sublamina of layer 5.
Fig. 1.
Integrin α1 mRNA is expressed by a few discrete neuronal groups. Photomicrographs show the autoradiographic localization of integrin α1 cRNA labeling in coronal tissue sections.A, C, Sections through rostral (A) and caudal (C) hippocampus show dense labeling in CA3a and CA3b stratum pyramidale (arrowhead in A) and the absence of hybridization in stratum granulosum (sg) and CA1 stratum pyramidale (CA1). C also shows the labeling of a discrete intermediate layer of neocortical cells (thin arrows), cortical blood vessels (arrowhead), and the pia mater. D,E, Higher magnification photomicrographs show the same field of primary visual neocortex with dark-field (D) and bright-field (E) illumination (hematoxylin counterstain); in comparing these panels one can see that the labeled cells (D) are distributed within the more superficial aspect of cortical layer 5 (E). (Small arrows indicate the same labeled cell in D and E; the position of cortical layer 4 is shown in E.)B, F, Sections through hypothalamus (B) and lower brainstem (F) show labeling within the paraventricular hypothalamic nucleus (pvn) and area postrema (ap). Scale bar (shown in F): A, 625 μm; B, D, E, 500 μm;C, 1 mm; F, 720 μm.
A few neuronal groups in the limbic telencephalon (Table 2) also expressed α1 mRNA. There was dense labeling of hippocampal CA3b and CA3a stratum pyramidale (Fig. 1A,C) and light labeling of cells in the bed nucleus of stria terminalis extending into the amygdala, the amygdalohippocampal transition area, and the horizontal limb of the diagonal bands of Broca/magnocellular preoptic area (data not shown). In diencephalon the cRNA labeled cells in the reticular and anteroventral ventrolateral thalamic nuclei, the paraventricular (Fig. 1B), supraoptic, and arcuate hypothalamic nuclei, and the median eminence at moderate-to-low levels. There was little regionally differentiated hybridization at lower levels of the neuraxis (Table 3), with lightly labeled cells detected in the substantia nigra/ventral tegmental area, the superior olivary complex, the trigeminal mesencephalic nucleus, and the hypoglossal nucleus. Finally, labeling was distributed within the circumventricular organs (i.e., area postrema and the subfornical organ; Fig. 1F) and associated with blood vessels and the pia mater (Fig. 1C,D).
α3 mRNA
As illustrated in Figure 2, α3 mRNA was distributed broadly with both low-density labeling throughout the neuropil in regions of gray and white matter (see hippocampal molecular layers, Fig. 2A) and broadly distributed, but nevertheless selective, labeling of neuronal groups. As shown in Figure 2B, labeling was associated nonpreferentially with neuronal fields in neocortex, olfactory cortex, claustrum, and the endopiriform nuclei. Labeling extended from superficial olfactory cortex into layer 2 of olfactory tubercle (Fig. 2B); at these same planes the islands of Calleja and the tenia tecta were not labeled. Labeling was light to moderately dense in corticomedial and basolateral amygdala and less dense in the central and lateral nuclear groups. Hybridization was relatively dense in cholinergic basal forebrain regions, including the medial septal nucleus, the diagonal bands of Broca (Fig. 2C), and nucleus basalis (data not shown), and in association with a few scattered cells in the more ventral caudate putamen, but it was low throughout the majority of caudate putamen and lateral septum (Fig. 2B,C). Within the hippocampal formation, cells in the hilus and stratum pyramidale were labeled moderately densely (Fig. 2A). In stratum pyramidale the labeling densities declined around the curve of the cell layer from CA3c to CA1 and along the temporal-to-septal axis. Stratum granulosum was not labeled (Fig.2A).
Fig. 2.
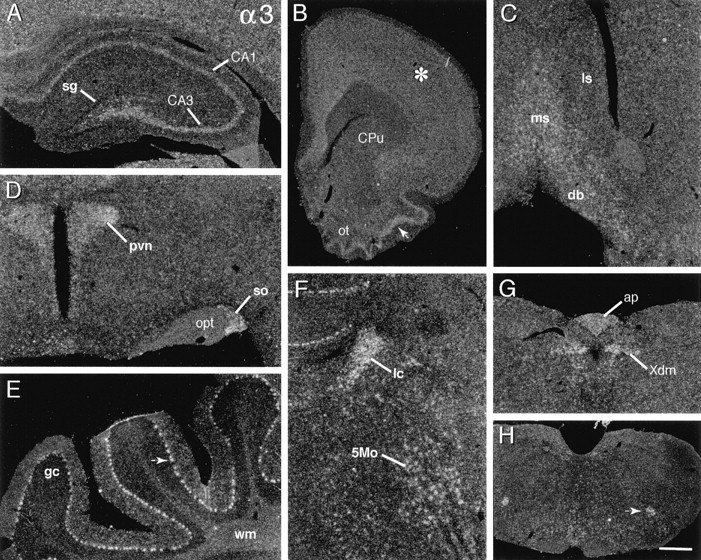
Integrin α3 mRNA is broadly distributed. Dark-field photomicrographs show the autoradiographic localization of α3 cRNA labeling in select brain regions. A, Rostral hippocampal section showing that labeling is distributed within the pyramidal cell layer (CA3, CA1), but not within stratum granulosum (sg). B, Rostral forebrain section shows that labeling is broadly distributed within neocortex (asterisk), moderately dense within superficial piriform cortex (arrow) and olfactory tubercle (ot), but low within caudate putamen (CPu). C, Within the septal region, hybridization is moderately dense in the medial septal nucleus (ms) and the diagonal bands of Broca (db) but is at low neuropil levels within lateral septum (ls). D, Section through hypothalamus showing relatively dense labeling in the paraventricular (pvn) and supraoptic (so) nuclei (opt, optic tract). E, Photomicrograph showing dense labeling of the cerebellar Purkinje cells (arrow) (gc, granule cell layer;wm, deep cerebellar white matter). F, Section showing dense hybridization in the locus coeruleus (lc) and in association with the large neurons of the motor trigeminal nucleus (5Mo). G,H, Sections through lower brainstem showing labeling within the dorsal motor nucleus of the vagus (G;Xdm), area postrema (G;ap), and nucleus ambiguus (H; arrow). Scale bar (shown in H): A, 400 μm; B, 1.2 mm; C, H, 900 μm; D, 320 μm; E, F, 250 μm; G, 450 μm.
In diencephalon the labeling was greatest in the magnocellular hypothalamic nuclei (i.e., paraventricular, supraoptic; Fig.2D) and was moderately dense in the anterodorsal, midline, intralaminar, and medial geniculate thalamic nuclei, in the subthalamic nucleus, and in association with scattered cells in the lateral hypothalamus. Labeling was low in the ventral and lateral thalamic nuclei. Within mesencephalon, cells in the substantia nigra/ventral tegmental area and medial aspects of the oculomotor nuclear complex were labeled the most densely.
At more caudal levels there was a scattering of densely labeled cells, including neurons in the reticular formation, superior olivary complex, motor trigeminal nucleus (Fig. 2F), dorsal motor nucleus of the vagus (Fig. 2G), nucleus ambiguus (Fig.2H), and ventral medullary reticular nucleus. Although cells were labeled in other cranial nerve nuclei (e.g., abducens, facial motor, and hypoglossal), these regions were not labeled as densely or completely as those seen with other α-integrin cRNAs (e.g., α7, vida infra). In contrast, as shown in Figure2F, the locus coeruleus was labeled very densely with α3 cRNA; this was the only α integrin prominently expressed by this noradrenergic cell group. In cerebellum the Purkinje cells were well labeled, but the granule cell layer was not (Fig.2E); indeed, the granule cell layer was labeled less densely than the underlying cerebellar white matter. Finally, there was labeling in the subfornical organ and area postrema (Fig.2G), but not in association with blood vessels, the ventricular ependyma, or the pia mater.
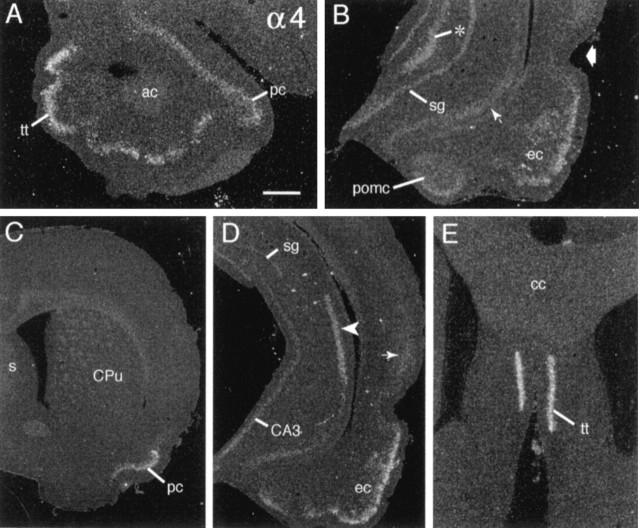
α4 mRNA
Of the various α transcripts, α4 mRNA was distributed the most narrowly, being limited almost entirely to discrete regions of the limbic telencephalon. Hybridization was greatest in rostral planes, with dense labeling of the tenia tecta (Fig.3A,E) and moderate labeling of rostral piriform cortex (Fig. 3A). Labeling continued caudally through piriform (Fig. 3C) and entorhinal (Fig.3B,D) cortex. In the hippocampal formation the labeling was not dense but was greatest in region CA2 stratum pyramidale (Fig.3D, arrowhead), the fasciola cinerium, and stratum granulosum and was quite light in CA3 stratum pyramidale. Hybridization densities increased along a septotemporal gradient in both CA3 and stratum granulosum (Fig. 3B,D). CA1 stratum pyramidale did not appear to be labeled although there was low-density hybridization in the temporal subiculum (Fig. 3B, arrow) and, as seen in the same sections, the posterior corticomedial amygdala (Fig. 3B). Finally, at caudal levels only, labeling extended from layers 2/3 of olfactory cortex and into the superficial neuronal layers of neocortex; this neocortical labeling was sparse and dissipated with distance from the rhinal fissure (Fig. 3D, arrow). With the exception of sparse labeling lateral to the third ventricle, further hybridization was not detected in diencephalon, mesencephalon, cerebellum, or lower brainstem (Table 3). In distinction with most other α-integrin cRNAs, patchy labeling of the neuropil, as might signify expression by glial cells, was not in evidence.
Fig. 3.
α4 mRNA is distributed discretely in limbic forebrain. A, Section through the rostral olfactory peduncle showing hybridization in tenia tecta (tt) and rostral piriform cortex (pc) (ac, anterior commissure).B, Section through temporal hippocampal formation showing labeling in the entorhinal cortex (ec), posterior corticomedial amygdala (pomc), temporal stratum granulosum (sg), CA3c (asterisk), and subiculum (small arrow); the broad arrow indicates the rhinal fissure. C, Low-magnification photomicrograph showing exclusive labeling of the piriform cortex (pc) in a section through the level of the septum (s) and caudate putamen (CPu). D, Section through the midcaudal hippocampal formation shows prominent labeling of CA2 stratum pyramidale (arrowhead) and entorhinal cortex (ec) as well as faint labeling in superficial layers of temporal cortex (thin arrow). In a comparison of_D_ and B one can see evidence for the increasing septotemporal labeling gradient within stratum granulosum (sg), with just faint labeling in the mid/dorsal dentate gyrus (D) as compared with greater labeling in the temporal field (B). E, Section through rostral septum shows dense labeling of tenia tecta (tt) (cc, corpus callosum). Scale bar (shown in A): A, E, 500 μm; B, 720 μm; C, 1.6 mm;D, 1 mm.
α5 mRNA
Like α3 cRNA, the α5 cRNA generated low-density, diffuse labeling throughout gray and white matter. In addition, variable density labeling was associated with neuronal cytoarchitectonics. As exemplified by the section through the olfactory peduncle shown in Figure 4A, labeling was distributed evenly through the neuronal layers of the anterior olfactory nucleus and tenia tecta. Labeling similarly was distributed broadly in neuronal fields of neocortex and amygdala (data not shown) and in the major neuronal cell layers of the hippocampal formation (Fig. 4C). Just a few areas stood out as being more densely labeled. In forebrain the most densely labeled cells were distributed above the deep white matter of medial allocortex and neocortex (Fig.4B); these cells scattered more broadly away from the white matter in lateral, as compared with dorsal, fields and did not extend all the way down to the rhinal fissure. In addition, labeling was denser in hippocampal stratum pyramidale than in stratum granulosum and was denser in the horizontal limb of the diagonal bands/magnocellular preoptic area and the paratenial thalamic nuclei than in surrounding fields. Low-density labeling was distributed broadly to neuronal fields in midbrain and lower brainstem but was conspicuously laminated in cerebellum. As shown in Figure4D, the Purkinje cells were well labeled, and α5 cRNA labeling was lower in the internal granule cell layer than in either the cerebellar molecular layer or the deep white matter.
Fig. 4.
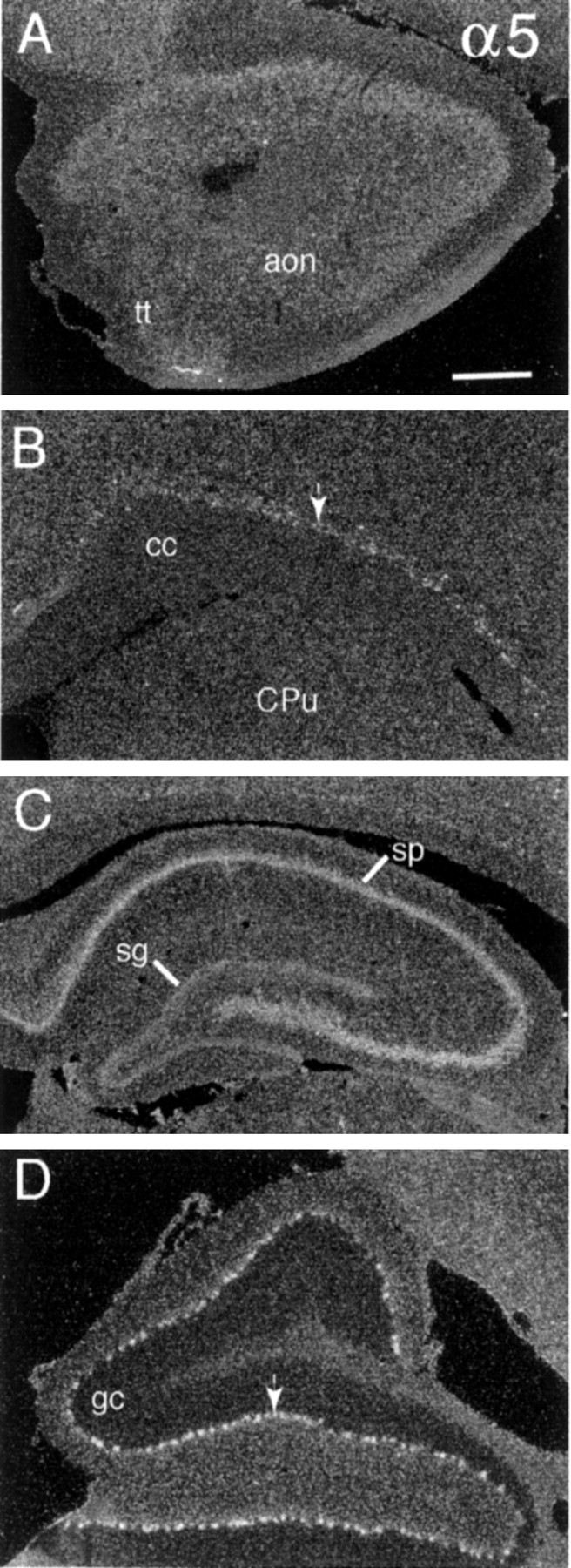
Relatively low-density α5 cRNA labeling is distributed across most neuronal fields. Dark-field photomicrographs show α5 cRNA labeling in (A) olfactory peduncle (tt, tenia tecta; aon, anterior olfactory nucleus), (B) deep neocortex (cc, corpus callosum; CPu, caudate putamen), (C) rostral hippocampus (sg, stratum granulosum; sp, stratum pyramidale), and (D) floccular lobe of cerebellum (arrow indicates Purkinje cell layer; gc, granule cell layer). Individual neurons were labeled densely in only two areas: deep neocortex (C, arrow) and the Purkinje cell layer of cerebellum (D, arrow). Scale bar (shown in_A_): A, B, 500 μm;C, 625 μm; D, 350 μm.
α6 mRNA
α6 cRNA labeling was diffuse over the neuropil of gray and white matter, giving rise to patchy labeling within the hippocampal and cortical molecular layers, but was associated most prominently with sharply defined cortical laminae. As summarized in Table 2 and illustrated in Figure 5, moderately dense hybridization was associated with neurons in the deeper aspect of the anterior olfactory nucleus (Fig. 5A) and formed an extremely thin layer just above the subcortical white matter (Fig.5D–G). Cells in the latter field spanned the full cortical mantle, at some levels merging with broader fields of labeled cells in the endopiriform nuclei (Fig. 5D). The piriform cortex was not well labeled at rostral planes (Fig. 5A,D), but in the caudal piriform and rostral entorhinal cortices the labeled cells were scattered in deeper layers and were laminated sharply in layer 2 near the rhinal fissure (Fig. 5B). At progressively more caudal planes the latter field of cells broadened to encompass, eventually, the full extent of entorhinal cortex (Fig. 5E,F). Very little labeling was evident in the hippocampal formation (Fig.5E,G). In rostral sections only CA2 stratum pyramidale was labeled clearly, although at very low densities (Fig. 5G,arrow). More caudally, labeling was associated with a few scattered hilar neurons and was evident at low levels in stratum granulosum (Fig. 5E). In contrast, cells in the caudal subicular pyramidal cell layer were well labeled (Fig.5F).
Fig. 5.
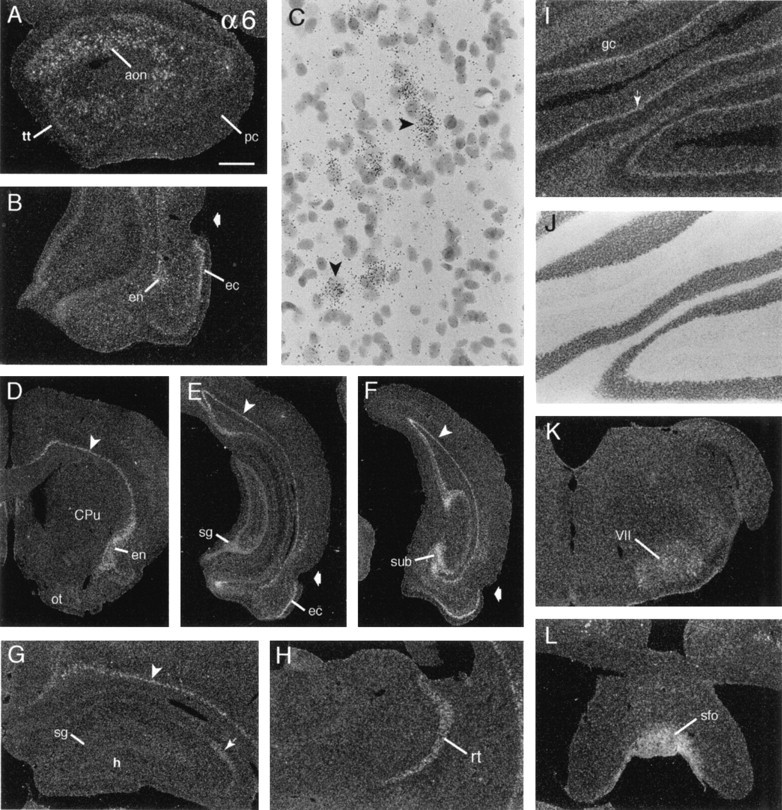
α6 mRNA is distributed most prominently within deep cortex. Dark-field photomicrographs show the distribution of α6 cRNA labeling in select brain regions. A, Labeled cells are distributed in the anterior olfactory nucleus (aon), but not rostral piriform cortex (pc) or tenia tecta (tt). B, Shown are labeled cells in layer 2 of entorhinal cortex (ec) and the ventral endopiriform nucleus (en) (broad arrow in_B_, E, and F indicates the rhinal fissure). C, High-magnification photomicrograph showing labeled cells (arrowheads) just above the white matter in retrosplenial cortex; note that smaller, more densely Nissl-stained cells are not autoradiographically labeled.D, Section through rostral forebrain showing the distribution of labeled cells above the cortical white matter (arrowhead) extending ventrolaterally into the endopiriform nucleus (en) and the absence of clear labeling in the olfactory tubercle (ot) and caudate putamen (CPu). E, F, Photomicrographs of sections through caudal hippocampus (E) and the more caudal subiculum (F) showing labeled cells in (1) the deepest layers of neocortex (arrowhead) and allocortex, (2) entorhinal cortex (ec), (3) caudal hippocampal stratum granulosum (sg), and (4) subiculum (sub).G, Section through rostral hippocampus showing faint labeling in CA2 stratum pyramidale (arrow) and denser labeling in the overlying deep neocortex (arrowhead;sg, stratum granulosum; h, hilus).H, Section showing labeling of the reticular thalamic nucleus (rt). I, J, The same field of paravermal cerebellum is shown with dark-field (I) and bright-field (J) illumination to show labeling of the Purkinje cell layer (arrow in I) relative to the Nissl-stained cytoarchitectonics (J) (gc, granule cell layer in_I_). K, L, Sections showing labeling of the facial motor nucleus (VII) and the subfornical organ (sfo). Scale bar (shown in A):A, 480 μm; B, 685 μm;C, 60 μm; D–F, 1.25 mm;G, H, L, 600 μm;I, J, 400 μm.
α6 mRNA was detected in a few subcortical regions, but labeling in these fields was generally not dense. In particular, the reticular thalamic nucleus (Fig. 5H), the paraventricular, dorsal ventromedial, and arcuate hypothalamic nuclei and median eminence (data not shown) were labeled. Beyond this, there was low-density labeling in the oculomotor, facial motor (Fig.5K), and hypoglossal nuclei, the mesencephalic, principal, and motor trigeminal nuclei, and the Purkinje cell layer (Fig. 5I,J). Finally, labeling was moderately dense in the subfornical organ (Fig. 5L) and area postrema and was low overlying the pia mater and ventricle epithelium.
α7 mRNA
Hybridization of α7 cRNA was distributed broadly across the neuraxis (Tables 2, 3), including particularly dense labeling in the hippocampal formation and brainstem cranial nerve nuclei. As described for other transcripts above, there was very low-density, irregularly distributed labeling across the neuropil and far higher density labeling that was aligned with neuronal cytoarchitectonics. In rostral sections (Fig. 6A,D), scattered cells in the anterior olfactory nucleus, tenia tecta, and the extreme rostral piriform cortex were lightly to moderately well labeled. At all levels there was fairly low-density but clearly laminated labeling of the neuronal layers of neocortex (Fig.6A,B), with labeling of individual neurons being greatest in upper layer 5 (Fig. 6B). In contrast to the distribution of other α transcripts, α7 mRNA expression was not laminated in the majority of olfactory cortex (Fig.6F) or superficial olfactory tubercle (Fig.6A). Like neocortex, labeling in the amygdala was generally low and not sharply differentiated, although grain densities were higher in the posterodorsal medial and central nuclei as compared with the cortical and basolateral nuclei (data not shown).
Fig. 6.
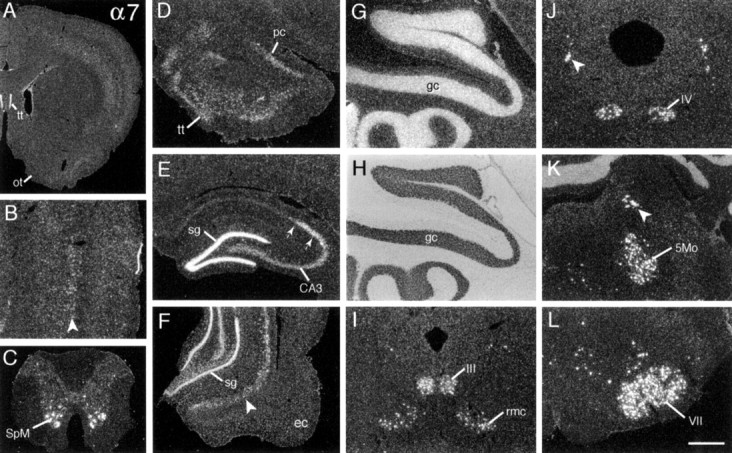
α7 mRNA is expressed at high levels by neuronal groups distributed across the neuraxis. Photomicrographs show the distribution of α7 cRNA labeling in (A) rostral forebrain with labeling evident in the tenia tecta (tt) and neocortex, but not olfactory tubercle (ot); (B) parietal cortex with most prominently labeled cells evident in deep layer 5 (arrowhead; superficial to the right); (C) spinal cord with dense labeling of the spinal motor neurons (SpM); (D) olfactory peduncle showing labeling of the tenia tecta (tt) and the extreme rostrodorsal piriform cortex (pc); (E) rostral hippocampus showing labeling in stratum granulosum (sg) and stratum pyramidale of regions CA3 and CA2 (between arrows); and (F) temporal hippocampus and entorhinal cortex (ec) showing labeling of scattered cells in the subiculum (arrowhead). The same field of cerebellar cortex in dark-field (G) and bright-field (H) illumination is shown and demonstrates α7 mRNA expression in the granule cell layer (gc). I–L show extremely dense labeling of neurons in motor cranial nerve nuclei and other discrete nuclei of mesencephalon and lower brainstem, including the red (rmc) and oculomotor (III) nuclei (in I), the mesencephalic trigeminal (arrowhead) and trochlear (IV) nuclei (in J), the mesencephalic (arrowhead) and motor (5Mo) trigeminal nuclei (in K), and the facial motor nucleus (VII; in L). Scale bar (shown in_L_): A, 1.7 mm; B, G, H, 400 μm; C, D, K, 650 μm; E, F, 800 μm; I, J, L, 500 μm.
α7 cRNA labeling differentiated the major hippocampal subfields. As shown for the rostral hippocampal formation in Figure6E, hybridization was dense in stratum granulosum, CA2 stratum pyramidale, and the fasciola cinerium, moderately dense in CA3 stratum pyramidale, and at general neuropil levels in CA1. The density of CA3 labeling and the incidence of labeled hilar neurons increased across the septotemporal axis of hippocampus. Moreover, in temporal fields one could see relatively dense labeling of scattered cells in the prosubiculum and subiculum (Fig. 6F, arrowhead).
With the exception of moderately dense labeling in the supraoptic nucleus, hybridization densities were low in the diencephalon, with diffuse but greater than neuropil levels in the anteroventral, supramamillary, and medial geniculate nuclei. In sharp contrast, α7 mRNA levels were extremely high in discrete brainstem regions and, most particularly, within all motor and several sensory cranial nerve nuclei (Table 3). Figure 6 shows the dense labeling of neurons in the oculomotor nuclei and the red nucleus (Fig. 6I), the trochlear and mesencephalic trigeminal nuclei (Fig.6J), the motor trigeminal nucleus (Fig.6K), and the facial motor nucleus (Fig.6L). In addition, α7 cRNA labeling was evident at lower levels or in association with fewer cells in the vestibular, cochlear, spinal trigeminal, and inferior olivary nuclei and in the reticular formation (Fig. 6L, Table 3). High α7 mRNA expression by the many cholinergic efferent nuclei of brainstem suggested that this transcript might be localized in spinal motor neurons. As shown for the section of spinal cord illustrated in Figure6C, the α7 cRNA labeled these motor neurons at extremely high density.
Finally, α7 mRNA levels were high in the cerebellum. In distinction from other α transcripts, labeling was most prominent in the granule cell layer (Fig. 6G,H), although at higher magnification the labeling of Purkinje cells was evident as well.
α8 mRNA
Prominent α8 cRNA labeling was entirely telencephalic (Tables 2,3; Fig. 7). Labeled cells were distributed within the anterior olfactory nucleus (Fig. 7A) and adjacent to the deep cortical white matter (Fig. 7B), extending into a broader, well labeled claustrum rostrally (Fig.7D) and endopiriform nucleus caudally (Fig.7G,H). Although deep cortical labeling was evident at all levels, it was greatest in medial prelimbic and limbic cortex (Fig.7C, arrow) and scattered away from the white matter to a variable degree among regions. For example, labeled cells were scattered well away from the white matter in the cingulate (Fig.7B), insular, and auditory (Fig. 7G, arrow) cortices but were limited to fewer cells near the white matter in parietal, somatosensory cortex (Fig. 7D, arrowhead). The distribution of these cells corresponded with that of large neocortical layer 6 neurons, as seen in hematoxylin-stained tissue sections. Labeling was generally low in superficial olfactory cortex (Fig.7C,D), although labeled cells were scattered in deeper layers. A narrow field of rostral lateral septum was labeled densely (Fig. 7D), but hybridization was at background levels throughout most of medial and lateral septum and in the diagonal bands of Broca. The α8 cRNA also labeled caudate putamen, but not the globus pallidus or nucleus accumbens. As shown in Figure 7D, striatal labeling was patchy and increased along a dorsomedial-to-ventrolateral gradient, being densest and extending furthest caudal within the fundis of the striatum (Swanson, 1992).
Fig. 7.
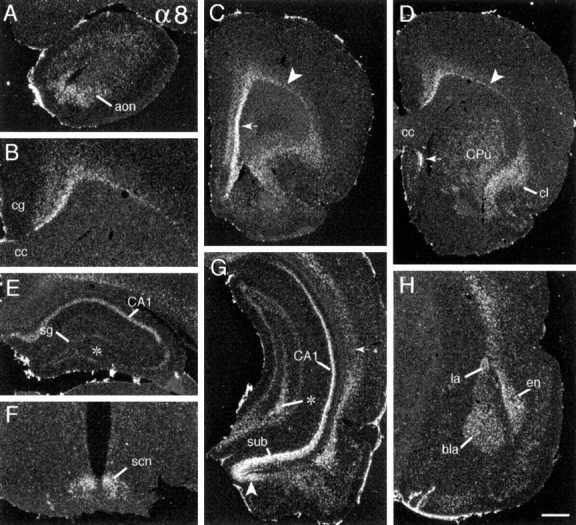
α8 mRNA is expressed predominantly in forebrain. Dark-field photomicrographs showing the distribution of α8 cRNA labeling in (A) the olfactory peduncle (aon, anterior olfactory nucleus); (B) cortex overlying the corpus callosum (cc) (cg, cingulate cortex; medial is to the left); (C) rostral forebrain in which there is dense labeling of neurons in deep prelimbic cortex (arrow) and lesser labeling of cells above the remainder of the subcortical white matter (arrowhead); (D) the rostral caudate putamen (CPu) and, in the same plane, cells above the cortical white matter (arrowhead), within claustrum (cl), and in a discrete field of lateral septum (small arrow). Photomicrographs show labeling in (E) field CA1 of rostral hippocampus (sg, stratum granulosum), (F) the hypothalamic suprachiasmatic nucleus (scn), (G) temporal hippocampus (sub, subiculum) and the amygdalohippocampal transition area (arrowhead), and (H) amygdala (bla, basolateral amygdala; la, lateral amygdala dorsal part; en, endopiriform nucleus). Note the regional differences in cortical lamination, with labeled cells abutting the subcortical white matter in frontal and rostral parietal cortices (arrowheads in C_and D, respectively) but being distributed more diffusely in primary auditory cortex (arrow in_G). In a comparison of E and_G_ one can see that labeling in hippocampal fields CA1 and CA3 increases along a septotemporal gradient (septal pole is shown in E; temporal pole is represented in the lower part of_G_; asterisk indicates CA3c in both). Scale bar (shown in H): A,E, 640 μm; B, 500 μm;C, D, 1.0 mm; F, 430 μm;G, 800 μm; H, 575 μm.
Dense regionally differentiated labeling was observed in the hippocampal formation and amygdala. As shown in Figure 7, _E_and G, for rostral and caudal hippocampus, respectively, CA1 stratum pyramidale and the subiculum were well labeled, and hybridization densities increased markedly across the septotemporal axis. CA3c stratum pyramidale and hilar neurons did not appear to be labeled, and the remainder of CA3 and the stratum granulosum were labeled very lightly at rostral planes, but hybridization increased to moderate levels in each of these fields across the septotemporal axis. CA2 stratum pyramidale was not labeled. As shown in Figure 7,G and H, in amygdala the hybridization was dense in the basolateral and dorsolateral nucleus, moderately dense in the lateral nuclei and the amygdalohippocampal transition area, low and irregularly distributed within the corticomedial nuclei, and at neuropil levels in the central nucleus (Fig. 7G).
With the exception of moderately dense labeling of cells in the suprachiasmatic (Fig. 7F) and arcuate nuclei, α8 cRNA labeling was low in diencephalon and at lower levels of the neuraxis (Table 3).
αV mRNA
As shown in Figure 8, among the transcripts evaluated here αV mRNA was the most abundant and broadly distributed in neocortex. Labeling spanned the neuronal layers but was relatively denser in layers 2/3 in rostral planes (Fig.8B,H, Table 2). There was some diffuse and patchy hybridization in cortical layer 1 and in molecular layers of other regions, suggesting expression by glial cells. Expression was relatively high in the anterior olfactory nucleus (Fig.8A) and the superficial, but not deep, layers of piriform (Fig. 8B,C) and entorhinal (Fig.8E,G) cortex. The piriform cortical labeling extended into the superficial olfactory tubercle (Fig. 8C); in the latter region the islands of Calleja and striatal bridge regions were labeled at lower densities. Within the amygdala, labeling densities were low and relatively undifferentiated, with the exception of moderately well labeled cells in the posterior cortical nuclei (Fig.8E). Within the hippocampal formation, expression was moderate to low in stratum granulosum, low in CA1 stratum pyramidale, and at neuropil levels within CA3 stratum pyramidale (Fig.8G, Table 2). For this and all transcripts previously described, there was no clear evidence for the labeling of neurons within the hippocampal molecular layers.
Fig. 8.
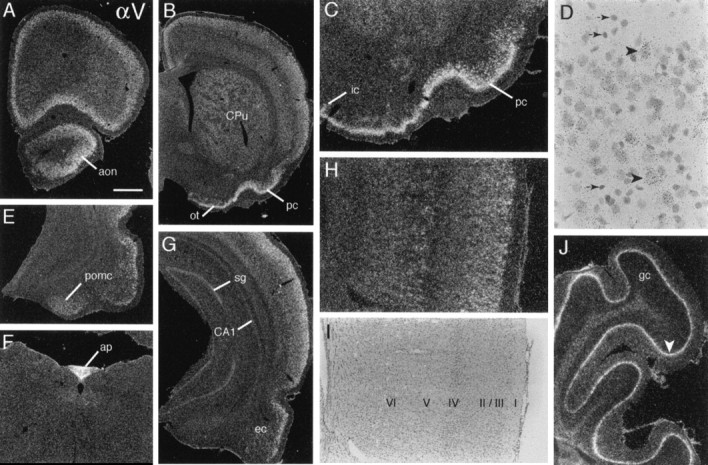
αV mRNA is distributed broadly in cortex and striatum. A, B, Sections through extreme rostral (A) and somewhat more caudal (B) forebrain show broadly distributed αV cRNA labeling in neocortex, anterior olfactory nucleus (aon), caudate putamen (CPu), piriform cortex (pc), and olfactory tubercle (ot).C, Higher magnification view of basal forebrain shows labeling is dense within piriform layer II (pc) and scattered deeper cells and is light in the islands of Calleja (ic). D, High-magnification bright-field photomicrograph of superficial neocortex shows that labeling is associated with larger cells with the Nissl-staining characteristics of neurons (arrowheads) and not with smaller, more densely stained glial-like cells (small arrows).E–G, Sections show hybridization within posteromedial cortical amygdala (pomc; E), area postrema (ap; F), and caudal hippocampus (G). G, Shown is light labeling in hippocampal CA1 stratum pyramidale and stratum granulosum (sg) and labeling in entorhinal cortex (ec). H, I, Photomicrographs of the same field of visual cortex, using dark-field (H) and bright-field (I) illumination to visualize the distribution of αV cRNA labeling (H) relative to cytoarchitectonic lamination (I); labeling is most prominent in layers 2/3 and in association with large cells in deep layer 5, with diffuse and lower density labeling in the nonpyramidal layer 4.J, Field of cerebellar cortex showing rather continuous labeling of the Purkinje cell layer (arrowhead;gc, granule cell layer). Scale bar (shown in_A_): A, B, 1.0 mm;C, 50 μm; D, 40 μm; E,G, 820 μm; F, 585 μm;H, I, 410 μm; J, 340 μm.
As shown in Figure 8B, αV mRNA was the most abundant of the α transcripts in striatum. Labeling was distributed rather evenly across the full caudate putamen, with the exception of distributed patches of lower hybridization densities, giving the impression of expression by matrix neurons (Gerfen, 1992). In contrast, labeling was at neuropil levels in nucleus accumbens and globus pallidus.
With the exception of dense to moderately dense labeling in the area postrema (Fig. 8F), median eminence, and tuberomamillary nuclei, αV mRNA levels were low (e.g., medial geniculate, pararubral field) or at neuropil levels throughout diencephalon, mesencephalon, and lower brainstem (Table 3). However, within cerebellum the Purkinje cell layer was labeled densely (Fig.8J). In contrast to the very punctate cellular distribution of the α5 and α3 mRNAs within this lamina, αV cRNA labeling appeared more diffuse, suggesting that the mRNA either was distributed partly into neuronal processes (thereby blurring perikaryal boundaries) or was localized within glial cells.
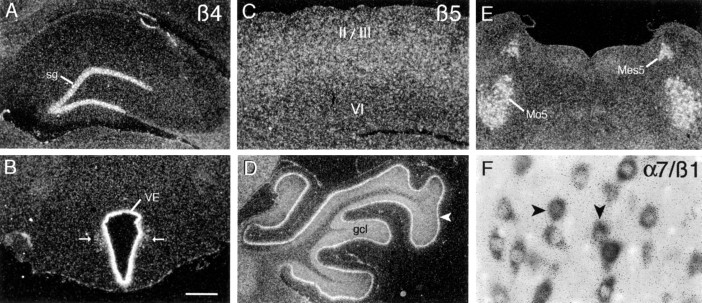
β4 mRNA
Labeling with β4 cRNA was extremely rare. Neuronal labeling was limited to the granule cell layer of the dentate gyrus (Fig.9A) and to a few cells scattered within the dorsal motor nucleus of the vagus. In addition, the circumventricular organs, ventricular ependyma (Fig.9B), and pia mater were clearly labeled.
Fig. 9.
Localization of transcripts for integrin subunits β4, β5, and β1. A, B, Shown is the autoradiographic localization of β4 cRNA labeling in the dentate gyrus stratum granulosum (sg; A) and the epithelium of the third ventricle (VE;B); arrows indicate labeled cells scattered near the ventricle in B. Shown is β5 cRNA labeling in (C) parietal cortex (layers_II/III_ and VI are indicated), (D) lateral cerebellar cortex (arrowhead indicates labeled Purkinje cell layer;gcl, granule cell layer), and (E) the motor (Mo5) and mesencephalic (Mes5) trigeminal nuclei. The scatter of β5 cRNA labeling into the cerebellar molecular layer (D) and surrounding the well labeled perikarya in the trigeminal nuclei (E) suggests that mRNAs may be localized within proximal dendrites. F, Shown are double-labeled cells in the hypoglossal nucleus of a tissue section processed for the simultaneous colorimetric and autoradiographic localization of α7 and β1 mRNAs, respectively. As in this example, all labeled neurons within the hypoglossal and other efferent cranial nerve nuclei were double-labeled, indicating total α7β1 colocalization in these fields. Scale bar (shown in B): A, B, D, E, 1 mm; C, 720 μm; F, 72 μm.
β5 mRNA
Similar to the distribution of α3 cRNA labeling, the β5 cRNA generated both diffuse, low-density labeling throughout most regions of neuropil and greater hybridization densities in specific neuronal fields distributed across the neuraxis (Tables 2, 3). In telencephalon the labeling was most prominent within cortex and basal forebrain. In the neocortex low-to-moderate hybridization densities spanned layers 2–5, with densities being greatest in layers 4/5 (Fig. 9C). In allocortex the labeling was distributed in the deeper layers, being conspicuously low in layer 2 of the granular cingulate and retrosplenial cortices. Labeling was diffuse and at low densities in olfactory cortical regions and amygdala, with the exception of the posterodorsal medial amygdala, which stood out as being moderately densely labeled. Within the hippocampal formation there was diffuse and somewhat patchy labeling over much of the neuropil and a few well labeled cells scattered within the hilus and hippocampal molecular layers. At high magnification the latter cells were Nissl-pale and appeared neuron-like. In addition, labeling increased across the septotemporal axis of CA1 and CA3 stratum pyramidale and was moderately dense in the temporal subicular cell layer. In diencephalon there was low to moderately dense labeling in the paratenial, anteroventral, and ventroposterior thalamic nuclei, the ventral thalamus, and much of hypothalamus (Table 3).
Labeling with the β5 cRNA was denser and regionally differentiated at lower levels of the neuraxis. As summarized in Table 3, in addition to low-density hybridization over much of the reticular formation, hybridization was dense to moderately dense in association with the efferent cranial nerve nuclei; this is illustrated for the mesencephalic and motor trigeminal nuclei in Figure 9E. In addition, ventral horn neurons evident at the lowest levels of brainstem were labeled very densely. Within cerebellum, hybridization was dense in both the Purkinje and granule cell layers and low to moderately dense in the deep cerebellar nuclei. Labeling of the Purkinje cell layer appeared to dissipate into the overlying molecular layer, suggesting that transcripts were, in part, distributed into dendrites.
Colocalization of α7 and β1 mRNAs
As summarized in Table 3, the distribution of α7 mRNA within lower brain centers and, in particular, within efferent cranial nerve nuclei overlaps the brainstem distribution of β1 integrin mRNA described in an earlier study from this laboratory (Pinkstaff et al., 1998). To determine whether α7 and β1 mRNAs are coexpressed by individual brainstem neurons, we processed sections through cerebellum, lower brainstem, and spinal cord for the simultaneous colorimetric and autoradiographic localization of α7 and β1 mRNAs, respectively. Labeling with the two cRNAs was fully colocalized in the efferent cranial nerve nuclei; i.e., all labeled cells were double-labeled. This colocalization is illustrated for neurons of the hypoglossal nucleus in Figure 9F. In addition, vestibular, reticular formation, and ventral horn spinal neurons were double-labeled. The relative density of labeling did vary between regions. A majority of labeled cells in the efferent cranial nerve nuclei and spinal ventral horn were labeled densely by both the α7 and β1 cRNAs. In contrast, neurons of the vestibular nuclei were labeled only moderately densely with the two probes, and cells in the dorsal motor nucleus of the vagus were densely labeled with β1 cRNA and lightly labeled with α7 cRNA. The cerebellar granule cells and some scattered cells in the reticular formation were labeled with α7 cRNA alone.
DISCUSSION
The present results demonstrate that mRNA expression for eight α and two β integrin subunits is regionally differentiated in the adult brain. Only transcripts for α2, β2, and β3 were not detected. Dense neuronal labeling was evident at all levels of the neuraxis. Some transcripts also were expressed at low levels within molecular layers and white matter. Although the latter labeling was never densely clustered over individual cells, it is typical of astroglial labeling obtained with isotopic in situ hybridization (Pinkstaff et al., 1998). Expression by both neurons and glia was expected from studies of mRNA and protein content in dissociated cells (Neugebauer and Reichardt, 1991; Tomaselli et al., 1993; Tawil et al., 1994;DeFreitas et al., 1995; Shaw et al., 1996) and brain (Grooms et al., 1993; Einheber et al., 1996; Cousin et al., 1997; Nishimura et al., 1998), but the distributions and levels of gene expression for subunits that were evaluated here were not anticipated in the literature. A previous study demonstrated that integrin β1 mRNA is expressed at high levels by select neuronal groups in adult brain (Pinkstaff et al., 1998). The present findings show that the general features of β1 expression (prominently neuronal and region- and cell-specific) are indeed typical of other integrin subunits and that neurons containing moderate-to-high levels of each of the integrin subunit mRNAs are distributed differentially.
Both α and β integrin subunits contribute to ligand recognition (Aplin et al., 1998) and to the specificity of integrin/matrix interactions that are considered critical for neuronal migration, process outgrowth, and synaptogenesis (Dodd and Jessell, 1988; Sanes, 1989; Reichardt and Tomaselli, 1991; Zhang and Galileo, 1998). In regard to this, it is intriguing that subunit expression profiles differentiated brain systems and levels of the neuraxis. For example, α4 mRNA expression was limited to defined regions of limbic forebrain. The α8 and αV mRNAs were also predominantly, although more broadly, expressed in forebrain: α8 mRNA was localized within discrete neuronal groups in limbic telencephalon (hippocampus, amygdala), whereas αV mRNA levels were greatest in neocortex. In contrast, α3 and α5 mRNAs were distributed broadly and diffusely across the neuraxis.
Within some regions the subunit expression patterns were complementary. For example, neocortical layers 2/3, 5, and deep 6 most prominently expressed different α subunits (αV, α1, and α8, respectively). Within the hippocampal formation the principal neurons of the dentate gyrus, CA3, CA2, and CA1 were distinguished by higher mRNA levels for the α7, α1, α4, and α8 subunits, respectively. In cerebellum, α7 and β5 mRNAs were abundant in the granule cells, whereas α3 and β1 mRNAs were highly expressed by the Purkinje cells but were not detected in the granule cells. Together with the results described above, these observations indicate that integrins provide functional cell surface markers that discriminate morphologically distinct populations of neurons between and within regions of adult brain.
Although there are many potential αβ integrin combinations, the true diversity of integrin receptors appears to be much more limited. As reviewed elsewhere (Hynes, 1992; Schnapp et al., 1995), all of the α subunits that were studied here can associate with β1. αV also forms heterodimers with β3, β5, β6, and β8, and subunits α4 and α6 can associate with β7 and β4, respectively. In some instances, subunits compete for association with limiting concentrations of the appropriate dimer partner (Moyle et al., 1991). With these constraints in mind, the mRNA distributions reported here and for β1 by Pinkstaff et al. (1998) (summarized on Tables 2 and 3for comparison purposes) give rise to specific predictions about the integrin receptors likely to be expressed by different neuronal groups in adult brain (see Table 4). For example, the colocalization of α7 and β1 mRNAs in the efferent cranial nerve nuclei and spinal ventral horn suggests that the α7β1 laminin receptor is highly expressed by these groups of motor neurons. Overlapping mRNA distributions suggest that within the hippocampal formation the α7β1 and α1β1 (laminin, collagen) receptors are abundant in CA3 stratum pyramidale, but not in CA1 stratum pyramidale or stratum granulosum, whereas α8β1 (tenascin, fibronectin, vitronectin) is abundant in CA1. Similarly, the αVβ5 vitronectin receptor would be predicted to be expressed by Purkinje cells, but not by granule cells within cerebellum. Nishimura et al. (1998) recently demonstrated that β8 and αV immunoreactivities are similarly localized to synapse-like puncta throughout the neuropil of telencephalon and in cerebellum. Thus, the αVβ8 vitronectin receptor appears to be expressed by αV mRNA-positive hippocampal, striatal, and cortical neurons.
Table 4.
Potential integrin heterodimer combinations in select brain regions: predictions from regional mRNA distributions and known pairings in other systems
| α1β1 | α3β1 | α4β1 | α5β1 | α6β1 | α7β1 | α8β1 | αVβ1 | α6β4 | αVβ5 |
|---|---|---|---|---|---|---|---|---|---|
| Neocortex 2/3 | • | • | • | • | • | • | |||
| Neocortex 4 | • | • | • | • | • | ||||
| Neocortex 5 | • | • | • | • | • | • | |||
| Neocortex 2 | • | • | • | • | • | • | |||
| Entorhinal 2 | • | • | • | • | • | • | • | • | |
| CA1, sp | • | • | • | • | • | • | |||
| CA3, sp | • | • | • | • | • | • | |||
| SG | • | • | |||||||
| PVN | • | • | • | • | • | • | |||
| SFO | • | • | • | • | • | ||||
| C. Purkinje | • | • | • | • | • | • | |||
| C. granule | |||||||||
| III (M) | • | • | • | • | |||||
| VI (M) | • | ||||||||
| XII (M) | • | • | • | • | • | • | • | ||
| Motor V (M) | • | • | • | ||||||
| Facial (M) | • | • | • | • | • | • | |||
| Ambiguus (SE) | • | • | • | ||||||
| DM Vagus (SE) | • | • | • | ||||||
| Mes. V (S) | • | • | • | ||||||
| Spinal V (S) | • | • | |||||||
| Vestibular (S) | • | • | • | • | |||||
| RF | • | • | • |
The nearly exclusive localization of β4 mRNA to the ventricular epithelium and the dentate gyrus granule cells is intriguing in light of the proposed role of the α6β4 laminin receptor in maintaining the integrity of cytoplasmic plaques associated with hemidesmosomes (Spinardi et al., 1995). Although α6 mRNA was not detected in the granule cells in untreated rats, it is upregulated in these neurons by seizures (J. Pinkstaff and C. Gall, unpublished observations). The granule cells form puncta adherens (Ribak and Anderson, 1980), and ventricular epithelial cells form macula adherens (Peters et al., 1976). Thus, α6β4 expression may be linked to the organization of tight junctions with submembranous plaques in brain as in other tissues.
Aspects of the present results are surprising and raise issues that will require further information for resolution. Prominent examples here include the absence of obvious dimer partners for α7 and β5 in the dentate gyrus and cerebellar granule cells, respectively. The α7 subunit reportedly associates exclusively with β1 (Hynes, 1992), but β1 mRNA levels are negligible in dentate granule cells (Pinkstaff et al., 1998). Similarly, β5 is thought to associate with αV alone (Hynes, 1992), but αV mRNA was not detected in cerebellar granule cells. It is possible that heterodimer partners are lacking in these instances and that the unpaired subunits are degraded. However, it is also possible that novel integrin proteins are expressed and form previously uncharacterized receptors in these regions. Alternatively, the conventional heterodimer partners may be present but are transcribed at such low levels that mRNAs were not detected. This interpretation also could account for discrepancies between the brain distributions of mRNAs and immunoreactivities for α8 (Einheber et al., 1996) and α1 (Murase and Hayashi, 1998). For these subunits the immunostaining patterns overlap but extend beyond the mRNA distributions reported here. These region-specific inconsistencies suggest that brain subdivisions and neuronal populations differ not only in the types of integrins they express but also in the rates at which these receptors are metabolized: slow turnover would be associated with low and potentially undetectable mRNA levels. This further implies that neurons differ in the rates at which adhesive contacts are modified or replaced. Possibly relevant to the idea that integrin mRNA levels reflect structural plasticity are results showing that seizures, which induce synaptic remodeling (Gall et al., 1997), increase β1 (Pinkstaff et al., 1998) and α1 (J. Pinkstaff and C. Gall, unpublished observations) gene expression in hippocampus and cortex and, in the latter instance, reveal synthetic capacity that accords with immunocytochemical results.
Integrins are not catalytically active but are linked to the cytoskeleton and to cytoplasmic signaling cascades by association with other molecules (Clark and Brugge, 1995; Katz and Yamada, 1997); the specific molecular interactions are determined mainly by the subunit composition of the receptor (Huhtala et al., 1995; Jewell et al., 1995;Giancotti, 1996; Sastry et al., 1996; Pfaff et al., 1998). These integrin-driven cascades have profound subunit-specific effects on proliferation, differentiation, gene expression, and cell survival in a variety of tissues (Clark and Brugge, 1995; Lafrenie and Yamada, 1996), and recent studies suggest that integrins mediate similar processes in the adult brain. For example, depending on the integrin expression profile, disruption of integrin/matrix binding can lead to apoptosis (Frisch and Francis, 1994; Judware et al., 1998). Chen and Strickland (1997) recently reported that disruption of laminin adhesion within hippocampus exacerbates the excitotoxic effects of kainic acid on CA1 and CA3 stratum pyramidale, but not on stratum granulosum. Subunit expression profiles reported here suggest that β1-containing laminin receptors (e.g., α1β1, α3β1, α7β1; Table 4) expressed within stratum pyramidale, but not stratum granulosum, mediate this effect. The integrins regulate the responsiveness of peripheral cells to cytokines and growth factors (Miyamoto et al., 1996; Sastry et al., 1996; Katz and Yamada, 1997). Recent studies point to the conclusion that the integrins also influence trophic factor signaling in the adult brain, albeit via a novel mechanism. Specifically, integrin antagonists selectively increase gene expression for brain-derived neurotrophic factor and its receptor _trk_B in mature cultured hippocampal slices, thereby indicating that integrins tonically suppress these growth-related genes in differentiated central neurons (J. Pinkstaff, A. Yong, C. Gall, G. Lynch, unpublished data).
On matrix binding the integrin receptor cytoplasmic domains nucleate the formation of elaborate protein complexes, with strong influences on the actin-based cytoskeleton (Clark and Brugge, 1995; Katz and Yamada, 1997). Precisely how these aggregates vary is not known, but, because of differences in subunit-associated proteins, it reasonably can be assumed that different integrins will favor different cytoskeletal arrangements. If so, the class or classes of integrin receptors inserted into individual synapses could shape the morphology of postsynaptic densities and dendritic spines. Subunit composition is also likely to affect the ease with which synaptic integrins are activated, broken down, and replaced, variables that may be of primary importance with regard to modifying synaptic structure. Related to this, integrin antagonists block the consolidation of LTP; i.e., potentiation develops normally but decays gradually toward baseline (Xiao et al., 1991). LTP is accompanied by changes in synaptic morphology (Lee et al., 1980; Desmond and Levy, 1983; Buchs and Muller, 1996), and modifications to integrin-dependent adhesive relationships are an obvious route for adjusting and then stabilizing local anatomy. From this it follows that the types of integrins and other adhesion molecules (Lüthi et al., 1994; Tang et al., 1998) expressed by a neuron could be responsible for the different forms of plasticity exhibited by the diverse populations of synapses in adult brain.
Footnotes
This work was supported by National Institute of Neurological and Communicative Disorders and Stroke Grants NS26748 and NS37799 to C.M.G. and Air Force Office of Scientific Research Grant F49620-J-0304 to G.L. We thank Dr. Julie Lauterborn for comments on this manuscript and the following persons who generously provided cDNAs used in this project: Drs. L. Reichardt (University of California, San Francisco; α1 and α8), S. Santoro (Washington University, St. Louis, MO; α2), J.-J. Cassiman (University of Leuven, Belgium; α4), R. Hynes (Massachusetts Institute of Technology, Cambridge, MA; α5), V. Quaranta (The Scripps Research Institute, La Jolla, CA; α6), S. Kaufman (University of Illinois; α7), L. Rome (University of California, Los Angeles; β1), S. Teitelbaum (Washington University, St. Louis, MO; β3), and S. Kennel (Oak Ridge National Laboratory, TN; β4).
Correspondence should be addressed to Christine M. Gall, Ph.D., Department of Anatomy and Neurobiology, University of California at Irvine, Irvine, CA 92697-1275.
REFERENCES
- 1.Aplin A, Howe A, Alahari S, Juliano R. Signal transduction and signal modulation by cell adhesion receptors: the role of integrins, cadherins, immunoglobulin-cell adhesion molecules, and selectins. Pharmacol Rev. 1998;50:197–263. [PubMed] [Google Scholar]
- 2.Bahr B, Lynch G. Purification of an Arg-Gly-Asp selective matrix receptor from brain synaptic plasma membranes. Biochem J. 1992;281:137–142. doi: 10.1042/bj2810137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahr B, Staubli U, Xiao P, Chun D, Ji Z, Esteban E, Lynch G. Arg-Gly-Asp-Ser-selective adhesion and the stabilization of long-term potentiation: pharmacological studies and the characterization of a candidate matrix receptor. J Neurosci. 1997;17:1320–1329. doi: 10.1523/JNEUROSCI.17-04-01320.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bizon JM, Lauterborn JC, Isackson PJ, Gall CM. Acidic fibroblast growth factor mRNA is expressed by basal forebrain and striatal cholinergic neurons. J Comp Neurol. 1995;366:379–389. doi: 10.1002/(SICI)1096-9861(19960311)366:3<379::AID-CNE1>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 5.Buchs PA, Muller D. Induction of long-term potentiation is associated with major ultrastructural changes of activated synapses. Proc Natl Acad Sci USA. 1996;93:8040–8045. doi: 10.1073/pnas.93.15.8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capaldi D, Rosario R, Esteban E, Bahr B. A 27-kDa matrix receptor from rat brain synaptosomes: selective recognition of the Arg-Gly-Asp-Ser domain and unique resistance to calcium-dependent proteolysis. Neurosci Res. 1997;28:275–279. doi: 10.1016/s0168-0102(97)00049-7. [DOI] [PubMed] [Google Scholar]
- 7.Chiba M, Teitelbaum S, Cao X, Ross F. Retinoic acid stimulates expression of the functional osteoclast integrin αV β3: transcriptional activation of the β3 but not the αV gene. J Cell Biochem. 1996;62:467–475. doi: 10.1002/(SICI)1097-4644(19960915)62:4%3C467::AID-JCB4%3E3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 8.Clark E, Brugge J. Integrins and signal transduction pathways. Science. 1995;268:233–239. doi: 10.1126/science.7716514. [DOI] [PubMed] [Google Scholar]
- 9.Cooper H, Tamura R, Quaranta V. The major laminin receptor of mouse embryonic stem cells is a novel isoform of the α6 β1 integrin. J Cell Biol. 1991;115:843–850. doi: 10.1083/jcb.115.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cousin B, Leloup C, Penicaud L, Price J. Developmental changes in integrin β-subunits in rat cerebral cortex. Neurosci Lett. 1997;234:161–165. doi: 10.1016/s0304-3940(97)00683-6. [DOI] [PubMed] [Google Scholar]
- 11.Damsky C, Werb Z. Signal transduction by integrin receptors for extracellular matrix: cooperative processing of extracellular information. Curr Opin Cell Biol. 1992;4:772–781. doi: 10.1016/0955-0674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- 12.DeFreitas M, Yoshida C, Frazier W, Mendrick D, Kypta R, Reichardt L. Identification of integrin α3 β1 as a neuronal thrombospondin receptor mediating neurite outgrowth. Neuron. 1995;15:333–343. doi: 10.1016/0896-6273(95)90038-1. [DOI] [PubMed] [Google Scholar]
- 13.De Meirsman C, Schollen E, Jaspers M, Ongena K, Matthijs G, Marynen P, Cassiman J. Cloning and characterization of the promoter region of the murine α-4 integrin subunit. DNA Cell Biol. 1994;13:743–754. doi: 10.1089/dna.1994.13.743. [DOI] [PubMed] [Google Scholar]
- 14.Denda S, Muller U, Crossin KL, Erickson HL, Reichardt LF. Utilization of a soluble integrin–alkaline phosphatase chimera to characterize integrin α8 β1 receptor interactions with tenascin: murine α8 β1 binds to the RGD site in tenascin-C fragments, but not to native tenascin-C. Biochemistry. 1998;37:5464–5474. doi: 10.1021/bi9727489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desmond NL, Levy WB. Synaptic correlates of associative potentiation/depression: an ultrastructural study in the hippocampus. Brain Res. 1983;265:21–30. doi: 10.1016/0006-8993(83)91329-x. [DOI] [PubMed] [Google Scholar]
- 16.Dodd J, Jessell T. Axon guidance and the patterning of neuronal projections in vertebrates. Science. 1988;242:692–699. doi: 10.1126/science.3055291. [DOI] [PubMed] [Google Scholar]
- 17.Einheber S, Schnapp L, Salzer J, Cappiello Z, Milner T. The regional and ultrastructural distribution of the α8 integrin subunit in the developing and adult rat brain suggests a role in synaptic plasticity. J Comp Neurol. 1996;370:105–134. doi: 10.1002/(SICI)1096-9861(19960617)370:1<105::AID-CNE10>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 18.Falcioni R, Cimino L, Gentileschi M, D’Agnano I, Zupi G, Kennel S, Sacchi A. Expression of β1, β3, β4, and β5 integrins by human lung carcinoma cells of different histotypes. Exp Cell Res. 1994;210:113–122. doi: 10.1006/excr.1994.1017. [DOI] [PubMed] [Google Scholar]
- 19.Frisch SM, Francis H. Disruption of epithelial cell–matrix interactions induces apoptosis. J Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gall C, Lauterborn J, Guthrie K, Stinis C. Seizures and the regulation of neurotrophic factor expression: associations with structural plasticity in epilepsy. Adv Neurol. 1997;72:9–23. [PubMed] [Google Scholar]
- 21.Gerfen C. The neostriatal mosaic: multiple levels of compartmental organization. Trends Neurosci. 1992;15:133–139. doi: 10.1016/0166-2236(92)90355-c. [DOI] [PubMed] [Google Scholar]
- 22.Giancotti FG. Signal transduction by the α6β4 integrin: charting the path between laminin binding and nuclear events. J Cell Sci. 1996;109:1165–1172. doi: 10.1242/jcs.109.6.1165. [DOI] [PubMed] [Google Scholar]
- 23.Gong J-G, Ko TC, Brattain MG. Disruption of fibronectin binding to the α5β1 integrin stimulates the expression of cyclin-dependent kinases and DNA synthesis through activation of extracellular signal-regulated kinase. J Biol Chem. 1998;273:1662–1669. doi: 10.1074/jbc.273.3.1662. [DOI] [PubMed] [Google Scholar]
- 24.Grooms S, Jones L. RGDS tetrapeptide and hippocampal in vitro kindling in rats: evidence for integrin-mediated physiological stability. Neurosci Lett. 1997;231:139–142. doi: 10.1016/s0304-3940(97)00524-7. [DOI] [PubMed] [Google Scholar]
- 25.Grooms S, Terracio L, Jones L. Anatomical localization of β1 integrin-like immunoreactivity in rat brain. Exp Neurol. 1993;122:253–259. doi: 10.1006/exnr.1993.1125. [DOI] [PubMed] [Google Scholar]
- 26.Grotewiel M, Beck C, Wu K, Zhu X, Davis R. Integrin-mediated short-term memory in Drosophila. Nature. 1998;391:455–460. doi: 10.1038/35079. [DOI] [PubMed] [Google Scholar]
- 27.Guthrie K, Nguyen T, Gall C. Insulin-like growth factor-1 mRNA is increased in deafferented hippocampus: spatiotemporal correspondence of a trophic event with axon sprouting. J Comp Neurol. 1995;352:147–160. doi: 10.1002/cne.903520111. [DOI] [PubMed] [Google Scholar]
- 28.Huhtala P, Humphries M, McCarthy J, Tremble P, Werb Z, Damsky C. Cooperative signaling by α5 β1 and α4 β1 integrins regulates metalloproteinase gene expression in fibroblasts adhering to fibronectin. J Cell Biol. 1995;129:867–879. doi: 10.1083/jcb.129.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hynes R. Integrins, versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 30.Ignatius M, Large T, Houde M, Tawil J, Barton A, Esch F, Carbonetto S, Reichardt L. Molecular cloning of the rat integrin α1-subunit: a receptor for laminin and collagen. J Cell Biol. 1990;111:709–720. doi: 10.1083/jcb.111.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jewell K, Kapron-Bras C, Jeevaratnam P, Dedhar S. Stimulation of tyrosine phosphorylation of distinct proteins in response to antibody-mediated ligation and clustering of α3 and α6 integrins. J Cell Sci. 1995;108:1165–1174. doi: 10.1242/jcs.108.3.1165. [DOI] [PubMed] [Google Scholar]
- 32.Judware R, McCormick TS, Mohr S, Yun JK, Lapetina EG. Propensity for macrophage apoptosis is related to the pattern of expression and function of integrin extracellular matrix receptors. Biochem Biophys Res Commun. 1998;246:507–512. doi: 10.1006/bbrc.1998.8650. [DOI] [PubMed] [Google Scholar]
- 33.Katz BZ, Yamada KM. Integrins in morphogenesis and signaling. Biochimie. 1997;79:467–476. doi: 10.1016/s0300-9084(97)82738-1. [DOI] [PubMed] [Google Scholar]
- 34.Kil S, Lallier T, Bronner-Fraser M. Inhibition of cranial neural crest adhesion in vitro and migration in vivo using integrin antisense oligonucleotides. Dev Biol. 1996;179:91–101. doi: 10.1006/dbio.1996.0243. [DOI] [PubMed] [Google Scholar]
- 35.Lafrenie R, Yamada K. Integrin-dependent signal transduction. J Cell Biochem. 1996;61:543–553. doi: 10.1002/(sici)1097-4644(19960616)61:4<543::aid-jcb7>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 36.Lee K, Schottler F, Oliver M, Lynch G. Brief bursts of high-frequency stimulation produce two types of structural change in rat hippocampus. J Neurophysiol. 1980;44:247–258. doi: 10.1152/jn.1980.44.2.247. [DOI] [PubMed] [Google Scholar]
- 37.Lüthi A, Laurent J, Figurov A, Muller D, Schachner M. Hippocampal long-term potentiation and neural cell adhesion molecules L1 and NCAM. Nature. 1994;372:777–779. doi: 10.1038/372777a0. [DOI] [PubMed] [Google Scholar]
- 38.Malek-Hedayat S, Rome L. Expression of a β1-related integrin by oligodendroglia in primary culture: evidence for a functional role in myelination. J Cell Biol. 1994;124:1039–1046. doi: 10.1083/jcb.124.6.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maren MS, Baudry M. Properties and mechanisms of long-term synaptic plasticity in the mammalian brain: relationships to learning and memory. Neurobiol Learn Mem. 1995;63:1–18. doi: 10.1006/nlme.1995.1001. [DOI] [PubMed] [Google Scholar]
- 40.Miyamoto S, Teramoto H, Gutkind J, Yamada K. Integrins can collaborate with growth factors for phosphorylation of receptor tyrosine kinases and MAP kinase activation: roles of integrin aggregation and occupancy of receptors. J Cell Biol. 1996;135:1633–1642. doi: 10.1083/jcb.135.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moyle M, Napier MA, McLean JW. Cloning and expression of a divergent integrin subunit β8. J Biol Chem. 1991;266:19650–19658. [PubMed] [Google Scholar]
- 42.Murase S, Hayashi Y. Integrin α1 localization in murine central and peripheral nervous system. J Comp Neurol. 1998;395:161–176. doi: 10.1002/(sici)1096-9861(19980601)395:2<161::aid-cne2>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 43.Neugebauer K, Reichardt L. Cell-surface regulation of β1-integrin activity on developing retinal neurons. Nature. 1991;350:68–71. doi: 10.1038/350068a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishimura SL, Boylen KP, Einheber S, Milner TA, Ramos DM, Pytela R. Synaptic and glial localization of the integrin αVβ8 in mouse and rat brain. Brain Res. 1998;791:271–282. doi: 10.1016/s0006-8993(98)00118-8. [DOI] [PubMed] [Google Scholar]
- 45.Paxinos G, Watson C. The rat brain in stereotaxic coordinates. Academic; New York: 1986. [DOI] [PubMed] [Google Scholar]
- 46.Peters A, Palay SL, Webster H. The neurons and supporting cells. Saunders; Philadelphia: 1976. The fine structure of the nervous system. [Google Scholar]
- 47.Pfaff M, Liu S, Erle D, Ginsberg M. Integrin β-cytoplasmic domains differentially bind to cytoskeletal proteins. J Biol Chem. 1998;273:6104–6109. doi: 10.1074/jbc.273.11.6104. [DOI] [PubMed] [Google Scholar]
- 48.Pinkstaff J, Lynch G, Gall C. Localization and seizure-regulation of integrin β1 mRNA in the adult rat brain. Mol Brain Res. 1998;55:265–276. doi: 10.1016/s0169-328x(98)00007-2. [DOI] [PubMed] [Google Scholar]
- 49.Reichardt L, Tomaselli K. Extracellular matrix molecules and their receptors: functions in neural development. Annu Rev Neurosci. 1991;14:531–570. doi: 10.1146/annurev.ne.14.030191.002531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ribak CE, Anderson L. Ultrastructure of the pyramidal basket cells in the dentate gyrus of the rat. J Comp Neurol. 1980;192:903–916. doi: 10.1002/cne.901920416. [DOI] [PubMed] [Google Scholar]
- 51.Ruoslahti E. Integrin signaling and matrix assembly. Tumour Biol. 1996;17:117–124. doi: 10.1159/000217975. [DOI] [PubMed] [Google Scholar]
- 52.Sanes J. Extracellular matrix molecules that influence neural development. Annu Rev Neurosci. 1989;12:491–516. doi: 10.1146/annurev.ne.12.030189.002423. [DOI] [PubMed] [Google Scholar]
- 53.Sastry S, Lakonishok M, Thomas D, Muschler J, Horwitz A. Integrin α-subunit ratios, cytoplasmic domains, and growth factor synergy regulate muscle proliferation and differentiation. J Cell Biol. 1996;133:169–184. doi: 10.1083/jcb.133.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schnapp LM, Hatch N, Ramos DM, Klimanskaya IV, Scheppard D, Pytela R. The human integrin α8 β1 functions as a receptor for tenascin, fibronectin, and vitronectin. J Biol Chem. 1995;270:23196–23202. doi: 10.1074/jbc.270.39.23196. [DOI] [PubMed] [Google Scholar]
- 55.Shaw C, Milner R, Compston A, ffrench-Constant C. Analysis of integrin expression on oligodendrocytes during axo–glial interaction by using rat–mouse xenocultures. J Neurosci. 1996;16:1163–1172. doi: 10.1523/JNEUROSCI.16-03-01163.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shinar DM, Schmidt A, Halperin D, Rodan GA, Weinreb M. Expression of αV and β3 integrin subunits in rat osteoclasts in situ. J Bone Miner Res. 1993;8:403–414. doi: 10.1002/jbmr.5650080404. [DOI] [PubMed] [Google Scholar]
- 57.Song W, Wang W, Foster R, Bielser D, Kaufman S. H36–α7 is a novel integrin alpha chain that is developmentally regulated during skeletal myogenesis. J Cell Biol. 1992;117:643–657. doi: 10.1083/jcb.117.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spinardi L, Einheber S, Cullen T, Milner TA, Giancotti FG. A recombinant tail-less integrin β4 subunit disrupts hemidesmosomes but does not suppress α6β4-mediated cell adhesion to laminins. J Cell Biol. 1995;129:473–487. doi: 10.1083/jcb.129.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Staubli U, Vanderklish P, Lynch G. An inhibitor of integrin receptors blocks long-term potentiation. Behav Neural Biol. 1990;53:1–5. doi: 10.1016/0163-1047(90)90712-f. [DOI] [PubMed] [Google Scholar]
- 60.Staubli U, Chun D, Lynch G. Time-dependent reversal of long-term potentiation by an integrin antagonist. J Neurosci. 1998;18:3460–3469. doi: 10.1523/JNEUROSCI.18-09-03460.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Swanson L. Brain maps: structure of the rat brain. Elsevier; Amsterdam: 1992. [Google Scholar]
- 62.Tamura R, Cooper H, Collo G, Quaranta V. Cell type-specific integrin variants with alternative alpha chain cytoplasmic domains. Proc Natl Acad Sci USA. 1991;88:10183–10187. doi: 10.1073/pnas.88.22.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang L, Hung CP, Schuman EM. A role for the cadherin family of cell adhesion molecules in hippocampal tong-term potentiation. Neuron. 1998;20:1165–1175. doi: 10.1016/s0896-6273(00)80497-3. [DOI] [PubMed] [Google Scholar]
- 64.Tawil N, Wilson P, Carbonetto S. Expression and distribution of functional integrins in rat CNS glia. J Neurosci Res. 1994;39:436–447. doi: 10.1002/jnr.490390411. [DOI] [PubMed] [Google Scholar]
- 65.Tomaselli K, Doherty P, Emmett C, Damsky C, Walsh F, Reichardt L. Expression of β1 integrins in sensory neurons of the dorsal root ganglion and their functions in neurite outgrowth on two laminin isoforms. J Neurosci. 1993;13:4880–4888. doi: 10.1523/JNEUROSCI.13-11-04880.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilson R, O’Brien W, Beaudet A. Nucleotide sequence of the cDNA from the mouse leukocyte adhesion protein CD18. Nucleic Acids Res. 1989;17:5397. doi: 10.1093/nar/17.13.5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu J, Santoro S. Differential expression of integrin α-subunits supports distinct roles during lung branching morphogenesis. Dev Dyn. 1996;206:169–181. doi: 10.1002/(SICI)1097-0177(199606)206:2<169::AID-AJA6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 68.Xiao P, Bahr B, Staubli U, Vanderklish P, Lynch G. Evidence that matrix recognition contributes to stabilization but not induction of LTP. NeuroReport. 1991;2:461–464. doi: 10.1097/00001756-199108000-00013. [DOI] [PubMed] [Google Scholar]
- 69.Yamada KM, Miyamoto S. Integrin transmembrane signaling and cytoskeletal control. Curr Opin Cell Biol. 1995;7:681–689. doi: 10.1016/0955-0674(95)80110-3. [DOI] [PubMed] [Google Scholar]
- 70.Yang J, Rayburn H, Hynes R. Embryonic mesodermal defects in α5 integrin-deficient mice. Development. 1993;119:1093–1105. doi: 10.1242/dev.119.4.1093. [DOI] [PubMed] [Google Scholar]
- 71.Zhang A, Galileo DS. Retroviral transfer of antisense integrin α6 or α8 sequences results in laminar redistribution or clonal cell death in developing brain. J Neurosci. 1998;18:6928–6938. doi: 10.1523/JNEUROSCI.18-17-06928.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]