Ribosomopathies: old concepts, new controversies (original) (raw)
. Author manuscript; available in PMC: 2020 Oct 1.
Published in final edited form as: Trends Genet. 2019 Jul 31;35(10):754–767. doi: 10.1016/j.tig.2019.07.004
Abstract
Ribosomopathies are a diverse subset of diseases caused by reduced expression of or mutations in factors necessary for making ribosomes, the protein translation machinery in the cell. Despite a ubiquitous need for ribosomes in all cell types, ribosomopathies manifest with tissue specific defects and sometimes increased cancer susceptibility, with few treatments that target the underlying cause. Here, by highlighting new research in the field, we review current hypotheses for the basis of this tissue specificity. Based on new work, we broaden our understanding of the role of ribosome biogenesis in diverse tissue types throughout embryonic development. We also pose the question of whether previously described human diseases, like aging, can be at least partially attributed to defects in making ribosomes.
Keywords: Ribosomopathy, cancer, specialized ribosome, ribosome concentration hypothesis, neurodegeneration, aging
Human diseases of making ribosomes, the ribosomopathies
Ribosomopathies are a diverse subset of largely developmental disorders that result from aberrant ribosome production. Ribosome synthesis is an essential and energy intensive cellular process that requires coordination of all three RNA polymerases, approximately 200 accessory factors and 80 ribosomal proteins (r-proteins) to process and assemble the mature ribosomal RNAs (rRNAs) [1]. In humans, the bulk of ribosome biogenesis initiates in the nucleolus with transcription of the 47S pre-rRNA (pre-rRNA) from ribosomal DNA (rDNA) loci by RNA polymerase I (RNAPI). Following transcription, the pre-rRNA is processed by a series of endo- and exo- nucleolytic steps to yield three of the four mature rRNAs (18S, 5.8S, and 28S) [2]. The rRNA is also modified by box C/D and H/ACA small nucleolar ribonucleoproteins (snoRNPs), and assembled with r-proteins and the fourth rRNA (5S), transcribed by RNA polymerase III from an extranucleolar locus [2]. Together, these steps yield the mature small subunit (SSU; 40S) and large subunit (LSU; 60S) of the ribosome that come together in the cytoplasm to translate messenger RNA (mRNA) into proteins. At the time of the first discovery that difficulties in making ribosomes could lead to the bone marrow failure syndrome, Diamond Blackfan Anemia (DBA [3]), it was unclear how a defect in the ubiquitous process of making ribosomes could lead to a tissue-specific disorder. Since then, new ribosomopathies have been identified, each with tissue-specific manifestations.
There have been several recent comprehensive reviews on ribosomopathies [4–8], so here we discuss only the new and controversial aspects of their pathogenesis. First, we highlight new ribosomopathies based on the step in which ribosome biogenesis is impacted. Second, we discuss the tissue-specific manifestations of the ribosomopathies and present one hypothesis defining how defects in the global process of making ribosomes may only affect some tissues. Finally, we pose key open questions and discuss current controversies surrounding ribosomopathies, such as: (1) Do specialized ribosomes influence the pathogenesis of ribosomopathies? (2) How can defects in cell growth cause cancer? (3) Are aging and neurodegenerative diseases also ribosomopathies? While there is currently no single unifying principle that explains the pathogenesis of all of the known ribosomopathies, substantial progress has been made in recognizing the source of these diseases and in gaining mechanistic insights into their pathogenesis. The hope is that in the future we may see the development of novel therapeutic options.
Highlighting the molecular pathology of ribosomopathies
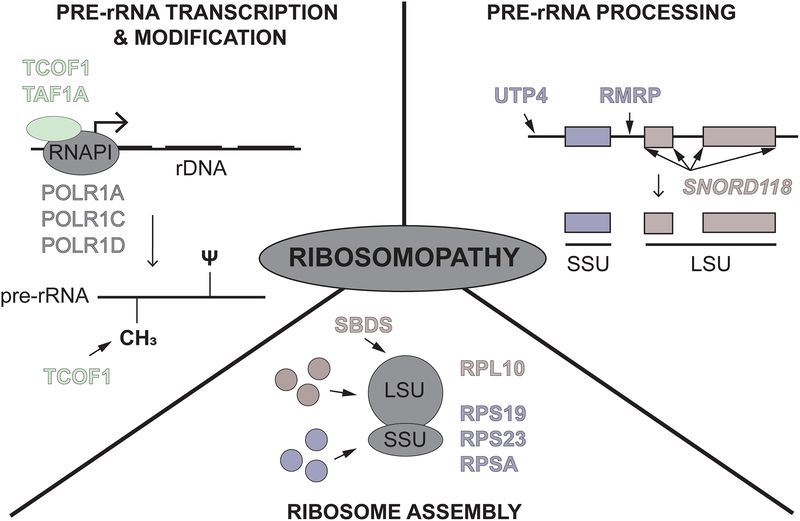
The diversity of clinical presentations of ribosomopathies makes them difficult to unify. While all ribosomopathies share defects in ribosome production, not all are caused by defects at the same step in the process. Here, we classify ribosomopathies by the step at which ribosome production is impacted. Based on our current knowledge, there are ribosomopathies that result from defects in (1) pre-rRNA transcription and modification, (2) pre-rRNA processing, and (3) ribosome assembly (Fig 1). Highlights from the last five years will be discussed.
Figure 1.
How ribosomopathies impact ribosome production. Ribosomopathies can affect: (top left) pre-ribosomal RNA (pre-rRNA) transcription and modification including pseudouridylations (ψ) and 2’-O-methylations (CH3), (top right) pre-rRNA processing, and (bottom) ribosome assembly. Proteins named in each section are highlighted in this review, including RNAPI subunits (grey), RNAPI-associated factors (green), small subunit (SSU) components and factors (purple), and large subunit (LSU) components and factors (red).
Pre-rRNA transcription and modification
Multiple ribosomopathies are caused by proteins implicated in pre-rRNA transcription and modification, including the well-studied mandibulofacial dysostosis Treacher-Collins Syndrome (TCS). TCS is caused by mutations in treacle (TCOF1) and the RNAPI subunits, POLR1C and POLR1D [9–12]. In depletion and knock-out models of TCOF1, a nearly 50% reduction in transcription of the 47S transcript was observed, suggesting a subsequent reduction in overall ribosome numbers [13]. TCOF1 also interacts with a core component of the box C/D snoRNP, NOP56, and studies in animal models reveal an effect of TCOF1 on 2’-O-methylation of the pre-18S rRNA [14].
In addition to TCS, two newly described ribosomopathies also result from defects in RNAPI transcription of the rDNA. First, compound heterozygous mutations in the TAF1A RNAPI-associated factor were reported in two sisters that presented with end-stage dilated cardiomyopathy. The cardiac phenotype was recapitulated in a zebrafish taf1a knockout model [15]. However, biochemical follow-up was not performed [15]. Second, mutations in the largest RNAPI subunit, POLR1A, cause acrofacial dysostosis, Cincinnati type, resulting in craniofacial abnormalities in both humans and zebrafish [16]. Here, levels of the pre-rRNA were tested by qPCR, which revealed a significant reduction in the pre-rRNA in polr1a mutants, again suggesting an associated reduction in overall ribosome numbers and a role for ribosome biogenesis in the pathogenesis of this disease.
Pre-rRNA processing
Ribosomopathies can also arise from defects in processing of the 47S pre-rRNA into the mature rRNAs. Such disorders include North American Indian childhood cirrhosis caused by a mutation in the SSU processome component Cirhin (UTP4 [17, 18]) and cartilage hair hypoplasia (CHH) caused by mutations in RMRP, which encodes the RNA component of the MRP endoribonuclease complex [19]. In addition, a new disorder caused by mutations in the U8 snoRNA has recently been described. While typically involved in rRNA modification, a small number of snoRNPs are required for pre-rRNA cleavage events. Among these is the U8 snoRNP, required for maturation of the 5.8S and 28S LSU rRNAs, likely by guiding proper pre-rRNA folding and recruiting the necessary cleavage factors [20, 21]. Mutations in the U8 snoRNA (transcribed from the SNORD118 locus) were originally described in 40 patients who presented with leukoencephalopathy with calcifications and cysts (LCC or Labrune Syndrome; [22]) in the brain. Since then, there have been several reports of additional patients with LCC who have mutations in the U8 snoRNA [23–26], providing additional support that mutations in U8 are the root cause of LCC pathogenesis. Biochemical experiments support an underlying defect in the function of the mutated U8 snoRNAs [22], but these mutations have yet to be introduced into an animal model for direct verification of their centrality.
Ribosome assembly
Finally, there are also several ribosomopathies that result from mutations in large and small subunit r-proteins and assembly factors that impact the number or proportion of functional ribosomes. Examples of these include Diamond Blackfan anemia (DBA) and Shwachman Diamond syndrome (SDS). In DBA, mutations in one of 18 r-proteins that result in haploinsufficiency have been implicated in causing the disease. While ribosome composition remains unaffected, overall ribosome levels are reduced [27]. In SDS, on the other hand, disease is caused by mutations in SBDS in a majority of patients, which leads to a failure in the cytoplasmic maturation of the LSU and prevents LSU and SSU joining, reducing the overall number of translationally competent ribosomes [4].
Mutations in the r-protein, RPL10 (uL16), were first discovered to be associated with autism [28, 29]. More recently, however, mutations in RPL10 (K78E) have also been shown to cause X-linked microcephaly, intellectual disability, and seizures [30, 31]. When the latter was modeled in zebrafish, Rpl10 depletion caused a reduction in head size and increased apoptosis as assayed by TUNEL staining - defects that could be rescued by expression of the human mRNA and autism-associated mutations, but not with mRNA that carried the K78E mutation, highlighting the important of studying these ortholgous mutations in animal models [30].
Additionally, two autosomal dominant ribosomopathies were recently described that are likely caused by r-protein haploinsufficiency. Two unrelated children presented with overlapping symptomatology including brachycephaly, trichomegaly, and development delay (BTDD). Whole exome sequencing revealed amino acid changes in RPS23 (uS12; [32]). Each child bears a variant of RPS23 (R67K or F120I) in one RPS23 allele. When tested for function in Saccharomyces cerevisiae, the orthologous R67K mutation resulted in slower growth, smaller colony size, and decreased translational fidelity, indicating that the mutation is deleterious. Studies in patient fibroblasts also revealed reduced translational fidelity. Likewise, mutations in RPSA (uS2) cause isolated congenital asplenia (ICA; [33, 34]). Knockdown of Rpsa in Xenopus tropicalis revealed impaired spleen development and pre-rRNA processing defects, and the ICA-causing mutation was unable to rescue either defect [35]. Interestingly, however, the heterozygous null mouse (Rpsa +/−) did not demonstrate ICA [33]. The plethora of these newly identified ribosomopathies, with more likely to come, will continue to spark insight into the role of ribosome biogenesis in embryonic development and disease.
It is important to note, however, that while most of the ribosomopathies discussed here may be traced to a defect in a particular aspect of ribosome biogenesis, the disease process may also include additional defective steps in making ribosomes. Furthermore, it is possible that the implicated ribosome biogenesis factors may also influence extraribosomal cellular functions [36, 37]. The potential contributions of these alternative functions to disease pathogenesis remain to be elucidated. Thus, to define ribosomopathies based solely on the step at which ribosome biogenesis is affected, while convenient for this discussion, may not fully encapsulate the complexity of ribosomopathies as we currently understand them. Further studies, particularly in animal models, will be necessary to gain a more comprehensive understanding of how failure in ribosome production contributes to the natural history of each condition.
Hints at the basis for the tissue specificity of ribosomopathies
The ribosomopathies manifest as diverse disorders, each with a tissue-specific clinical presentation. Despite the requirement for ribosome function in all cell types, disruptions in the process of making ribosomes often affect the development of certain tissues. Here we attempt to synthesize the expansive list of affected tissues by dividing them into ribosomopathies that affect the tissues derived from the neural crest and ribosomopathies that affect non-neural crest derived tissues. The mechanisms dictating how a specific mutation in a protein required for this ubiquitous process can affect only particular tissues is an active area of investigation.
Ribosomopathies derived from hindered development of neural crest cells
It has been known for some time that there is a strong link between neural crest development and ribosome biogenesis [38]. Neural crest cells in the developing embryo are multipotent migratory cells that differentiate into numerous cell and tissue types including the teeth, peripheral nervous system and glia, heart, pigment cells of the skin, and head skeletal structures. Multiple ribosomopathies present with defective tissues derived from neural crest cells during embryogenesis (Fig 2). For example, heart defects can be found in multiple ribosomopathies including DBA [39], the RPL10 ribosomopathy [31], SDS ([40]), and non-ischemic dilated cardiomyopathy caused by TAF1A mutations [15]. Also, skin and pigmentation defects can be found in aplasia cutis congenita [41], dyskeratosis congenita [42–45], DBA (mouse model) [46], ANE syndrome [47], a ribosomopathy arising from mutations in DNAJC21 similar to SDS [48, 49], and the uS12 ribosomopathy [32].
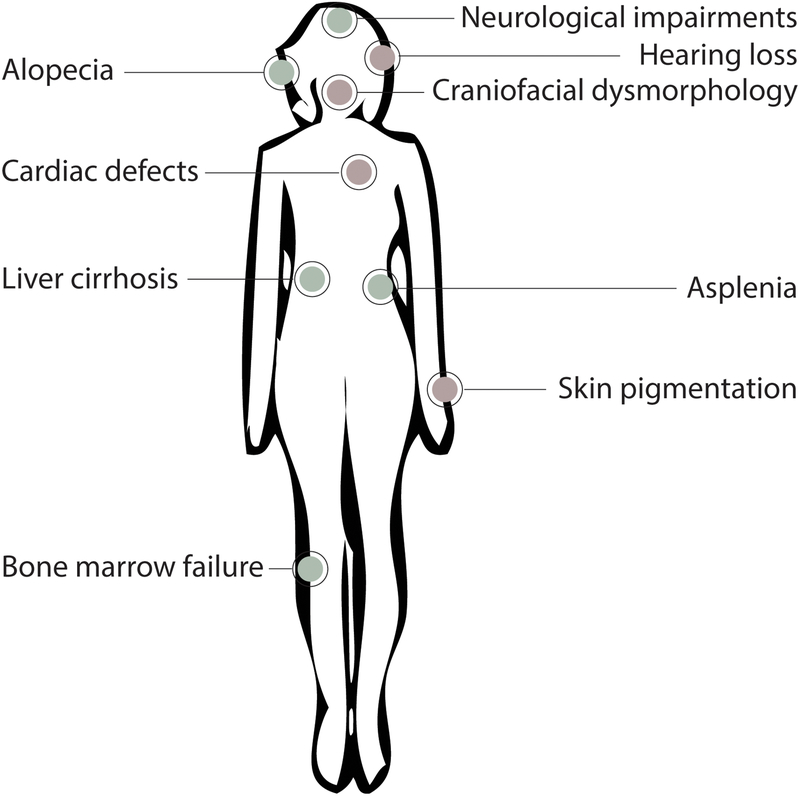
Figure 2.
The clinical manifestations of ribosomopathies. The diverse pathologies associated with ribosomopathies are depicted. These pathologies arise from neural crest cell (red) and non-neural crest cell lineages (green).
The best studied neural crest-derived ribosomopathies affect development of the face. Craniofacial development is affected in a number of ribosomopathies, including TCS (reviewed in [50], DBA [51], and acrofacial dysostosis, Cincinnati type [16] (Table 1). Further linking development of the face and ribosomopathies, several ribosome biogenesis factors that have not been associated with human disease cause craniofacial defects in model organisms (Table 1). Additionally, ear development is influenced by neural crest cells [52], and multiple ribosomopathies manifest in hearing loss. Hearing loss is usually seen in ribosomopathies that affect development of the face and head as a whole, such as TCS [53], the RPS23 ribosomopathy [32], the RPS10 ribosomopathy [30], DBA [54], a ribosomopathy resulting from mutations in DNAJC21 similar to SDS [48], and acrofacial dysostosis, Cincinnati type [16]. Therefore, aberrant ribosome biogenesis often results in defects manifested through neural crest-derived tissues.
Table 1.
Genes with roles in both ribosome biogenesis and craniofacial development.
| Gene Name | Function in ribosome biogenesis | Defects in craniofacial development | Name of human disease | Reference |
|---|---|---|---|---|
| DDX11 | rDNA transcription | Long faces, narrow eyes, low mouths | Warsaw Breakage syndrome (WABS) | [126, 135, 136] |
| DDX21 | rDNA binding, transcription of r-proteins | Hypoplasia of mandible/zygomatic complex | n/a but may be linked to Treacher Collins syndrome | [57] |
| ESF1 | Pre-rRNA processing | Jaw malformations, microcephaly | n/a | [137] |
| NOL11 | rDNA transcription and pre-rRNA processing | Microcephaly, reduced size of pharyngeal cartilages | n/a | [58] |
| PAK1IP1 | Pre-rRNA processing | Midline facial cleft | n/a | [138, 139] |
| POLR1A | rDNA transcription | Range of mandibulofacial dystoses including downslanting palpebral fissures, eyelid clefts, and micrognathia | acrofacial dystosis, Cincinnati type | [16] |
| RPL38/eL38 | Large subunit ribosomal protein | midline facial cleft, cleft palate | n/a | [78] |
| RPS19/eS19, RPL5/uL18, RPL11/uL5, RPL35a/eL33, RPS26/eS26, RPS24/eS24, RPS17/eS17, RPS7/eS7, RPS10/eS10, RPL19/eL19, RPL26/uL24, RPS29/uS14, RPL31/eL31, RPS28/eS28, RPS20/uS10, RPL15/eL15, RPL17/uL22, GATA1, TSR2 | Mainly ribosomal proteins | Cleft lip, cleft palate, flat nasal bridge, hypertelorism | Diamond-Blackfan anemia | [51] |
| TCOF1, POLR1C, POLR1D | rDNA transcription | Hypoplasia of mandible/zygomatic complex, some dental anomalies, cleft palate | Treacher Collins syndrome | [13, 140, 141] |
| WDR43 | rDNA transcription | Reduced size of pharyngeal cartilages, hydrocephaly | Linked to 3-M syndrome | [59] |
P53-mediated nucleolar stress response
Some, but not all, of the disease manifestations of ribosomopathies have been attributed to the increased sensitivity of the affected neural crest progenitor cells to the stabilization of the pro-apoptotic factor p53. This occurs in response to dysfunctional ribosome biogenesis during a window of development [7]. Supporting this idea, the clinical manifestations of several ribosomopathies can be rescued by co-depletion of p53 [55–59]. However, not all phenotypes can be rescued this way, suggesting that not all of the signs and symptoms associated with ribosomopathies are mediated by p53 [4, 56, 60–64]. Additionally, the examination of other stress response pathways in various affected and unaffected tissues have yet to be fully explored [65]. In the future, it would be pertinent to examine the sensitivity of various cell types to p53 throughout development in order to untangle some of the questions surrounding the cell type specificity of ribosomopathies.
Ribosomopathies derived from hindered development of non-neural crest cells
Some ribosomopathies affect tissues that are not derived from the neural crest (Fig 2). For example, bone marrow failure and anemia are present in SDS (also characterized by neutropenia [66]), DBA [51], 5q- syndrome [67], dyskeratosis congenita [68, 69], and a newly described ribosomopathy arising from mutations in DNAJC21 [49]. Additionally, hair development is affected in ANE syndrome [47], CHH [19], the RPS23 ribosomopathy [32], and a hair loss disorder (hereditary hypotrichosis simplex) caused by mutations in RPL21 (eL21) [70]. As both the bone marrow and hair develop from continuously dividing cells, it is possible that these tissues would have an increased reliance on making ribosomes for their growth. However, neither North American Indian childhood cirrhosis, which affects liver function [71], nor ICA, which affects spleen development [33], fit this model. Also, ribosomopathies often present with neurodevelopmental defects like microcephaly and intellectual disability [22, 25, 30–32, 47, 72]. Rescue experiments via co-knockout of p53 in animal models have succeeded for some phenotypes in a subset of these disorders, suggesting that some non-neural crest derived cell types also have an increased susceptibility to p53 [61–64]. Additional studies of animal models of these developmental disorders will therefore be needed to parse out the mechanisms of these tissue-specific clinical manifestations.
Do specialized ribosomes influence the pathogenesis of ribosomopathies?
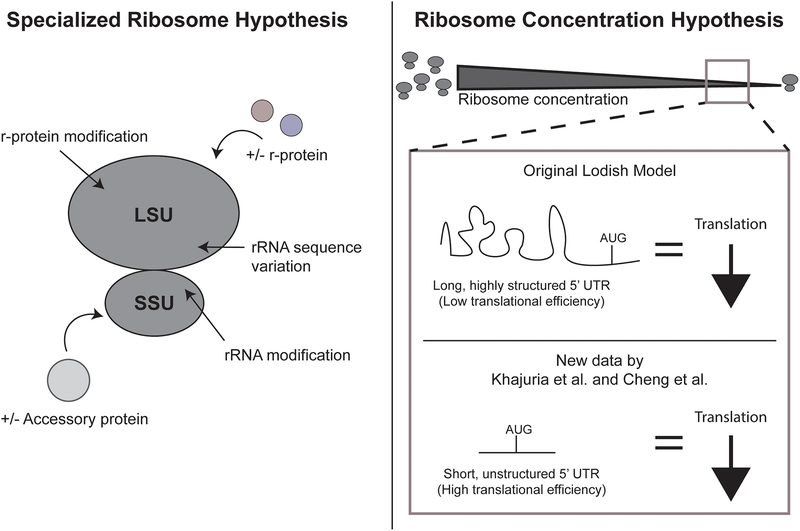
One explanation for how ribosomopathies can manifest in tissue-specific defects is that different tissues are comprised of ribosomes with varied compositions that endow them with “specialized” functions in translating certain mRNAs. The specialized ribosome hypothesis is as old as the discovery of ribosomes themselves; Francis Crick postulated that each individual gene had a single ribosome responsible for its translation (often referred to as the one gene - one ribosome – one protein hypothesis) [73]. Ribosome heterogeneity would be predicted to occur through a multitude of mechanisms including through changes in rRNA sequence, core r-protein composition, rRNA or protein modifications, or binding of accessory proteins (Fig 3; reviewed in [74]). These subtle changes could thus lead to the altered translation of mRNAs required for the proper development of some cell types but not others.
Figure 3.
Two hypotheses for the tissue-specific effects of ribosomopathies. (Left) The specialized ribosome hypothesis states that modification or changes in rRNA sequence, core r-protein composition, or accessory proteins in either the large subunit (LSU) or small subunit (SSU) of the ribosome can result in heterogeneous ribosomes with differential translation abilities in diverse tissues. (Right) The ribosome concentration hypothesis states that decreased ribosome concentration results in decreased protein levels for specific mRNAs. At low ribosome concentrations, the original Lodish model [81] proposed that mRNAs with long, highly structured 5’ UTRs would be most affected by changes in ribosome concentration (above), whereas new data by Khajuria et al. [27] and Cheng et al. [84] propose the opposite (below).
Despite enthusiasm for finding specialized ribosomes, their existence remains controversial. This is because a high standard must be applied to demonstrate not only that there is variation in ribosome composition but also that such variation results in functional differences in protein synthesis. Evidence supporting the specialized ribosome hypothesis has been thoroughly reviewed elsewhere [74–76], so here we highlight a few of the most controversial and recent developments in the field.
Ribosomal proteins as the basis for ribosome heterogeneity
Since the ribosome is made of rRNAs and r-proteins, one place to search for specialization is in the r-protein composition of the ribosomes themselves. A seminal work in support of the specialized ribosome hypothesis comes from investigating the role of RPL38 (eL38) in murine development [77, 78]. It has previously been shown that the skeletal patterning defects observed in tail-short (Ts) mice are due to mutations in RPL38 that affect its role in the translation of certain Hox mRNAs [77, 78]. However, ribosomes lacking RPL38 in different tissues were not directly measured [77, 78]. Another recent example in support of specialized ribosomes examined the role of RPL10A (uL1) in binding various mRNAs [79]. While the authors did find that ribosomes containing RPL10A bound a different set of mRNAs that those without [79], these studies utilized tagged r-proteins, which can affect their function (reviewed in [74]). Especially notable for these experiments is that control experiments employ a different tag altogether (HA-RPL22/eL22) than the experimental group (FLAG-RPL10A) [79]. Finally, recent work in yeast has shown that differential mRNA translation occurs in ribosomes with and without Rps26 (eS26), an essential r-protein mutated in DBA [80]. Because Rps26 is essential, it is puzzling that ribosomes could be made at all when Rps26 was depleted. Perhaps its essential role is not in ribosome assembly but only in ribosome function. Clearly, ribosomes appear to be more heterogeneous than was previously thought.
That there are specialized ribosomes is both an old and new concept brought recently to attention by new technological developments. While we embrace these studies that have been on the vanguard of discovery, we caution that finding specialized ribosomes requires additional supporting evidence with special care taken to have stringent controls. One problem with examining a heterogenous ribosome population is that it may represent failed synthesis intermediates or defective ribosomes on their way to being degraded. Current methods aggregate results because they rely on pools of cells (e.g. sucrose gradient sedimentation) and therefore have limited resolution. Additional orthogonal approaches such as single-cell mass spectrometry and cryo-electron microscopy may help to validate recent discoveries. Nevertheless, to answer the question of whether there are specialized ribosomes, it is sure to be a multi-pronged approach that combines these new techniques with the old.
Alternative view: the ribosome concentration hypothesis
A second hypothesis put forth to explain the tissue-specificity of ribosomopathies is the ribosome concentration hypothesis (Fig 3). Like the specialized ribosome hypothesis, it was originally proposed several years ago by Lodish [81] but was recently re-popularized by Mills and Green [6]. The ribosome concentration hypothesis postulates that the number of available cytoplasmic ribosomes per volume results in mRNAs that are translated differently based on structural features such as 5’ untranslated region (UTR) length or structural elements, open reading frame (ORF) length, and Kozak context. Linking back to the problem of tissue specificity of ribosomopathies, the expression of specific proteins may be sensitive to decreased ribosome concentration in that particular cell type.
While the ribosome concentration hypothesis might at first glance seem more straightforward than the specialized ribosome hypothesis, experimental evidence supporting this theory has also been hard to generate. It has been difficult to directly measure ribosome concentration on a per cell basis, with most studies only inferring reduced ribosome number.
The most well-studied examples in support of the ribosome concentration hypothesis have focused on the protein GATA1, a lineage-determining hematopoietic transcription factor that is implicated in the pathogenesis of DBA. Two recent papers with overlapping groups of authors argue that decreased ribosome concentration leads to reductions in GATA1 protein levels, resulting in the erythropoiesis-specific defects observed in patients with DBA [27, 82]. However, the mechanistic explanation for how reductions in GATA1 protein levels occur is inconsistent between the two studies, making it difficult to reconcile the underlying mechanism. In the first, it is because GATA1 has a highly structured, long 5’ UTR; while in the second, it is because GATA1 has a short, unstructured 5’ UTR [27, 82]. This discrepancy may partially arise from methodological differences: older studies utilized the technique rapid amplification of 5’ cDNA ends (5’ RACE) [82] while more recent studies utilize cape analysis gene expression (CAGE) [27] to define the GATA1 5’ UTR. The GATA1 NCBI reference sequence, NM_002049.3, reveals a 5’ UTR of 91 nt, which supports a shorter 5’ UTR for GATA1 than the average length of 210.2 nt for human 5’ UTRs [83]. Additional new ribosome profiling results in yeast support the idea that more efficiently translated mRNAs are more affected by reduced ribosome concentration [84]. In contrast, the original mathematical modeling by Lodish [81] and echoed by more recent reviews [6, 74] postulated that poorly translated mRNAs (like GATA1 if it had a highly structured, long 5’ UTR) would be dependent on the cell having a high available ribosome concentration while well-translated mRNAs would not have such dependency. These apparently contradictory results emphasize the degree of our current lack of understanding of how the structural elements of mRNAs impact their translation. In addition, examination of the molecular basis of ribosomopathies should continue to probe the effects of compensatory processes such as ribosome recycling [6].
It is important to note that these two hypotheses - specialized ribosome and ribosome concentration - are not mutually exclusive and that both may contribute to the clinical manifestations observed in patients with ribosomopathies. In addition, the role of p53 stabilization in tissue specificity cannot be overlooked. It is possible that either or both of the proposed mechanisms could result in reduced translation of a protein that leads to p53 stabilization and apoptosis, ultimately causing the tissue-specific defects. In the end, it is likely that no single mechanism will be able to fully explain all of the signs and symptoms of these disorders, which are complicated not only by their tissue specificity, but also by their developmental timing.
How can defects in cell growth cause cancer?
Several ribosomopathies, disorders of hypoproliferation, often coincide with a predisposition to cancer, a disease of hyperproliferation. This juxtaposition is referred to as Dameshek’s riddle [85]. Although cancer predisposition has not been studied for every ribosomopathy, this paradox has been studied in the context of SDS, caused by mutations in SBDS [86, 87]. The neutropenia and bone marrow abnormalities characteristic of SDS often progress to myelodysplastic syndromes (MDS) and acute myelogenous leukemia (AML) [88, 89]. Reinforcing this link to cancer, SBDS physically interacts with the r-protein uL16, mutated in pediatric T-cell leukemia (T-ALL, [90, 91]). Perhaps the hyperproliferation results as a consequence of cellular compensation for the dysfunctional ribosome biogenesis. Supporting this, some cells compensate for SBDS mutations by deleting the EIF6 gene (del(20)q) to upregulate protein synthesis [92, 93]. This has been associated with a lower risk of developing MDS and AML, although more longitudinal studies with larger patient cohorts are needed to confirm this [94–96]. In contrast, SDS patients often also acquire p53 mutations [97]. It is possible that cells sense disruptions in making ribosomes by upregulating p53 levels, with cancer resulting when the cells acquire additional mutations to bypass this effect. Further insight into Dameshek’s riddle may be found in studies showing that niche-derived inflammatory signaling may facilitate or even drive malignant progression in MDS in an effort to compensate for perturbations in hematopoietic stem cells [5, 98, 99]. In the future, we hope to be able to evaluate the natural progression of all of the compensatory mutations in patients with diverse ribosomopathies.
Are aging and neurodegenerative diseases also ribosomopathies?
Recent studies on premature aging diseases, models of longevity, and neurodegeneration continue to support a connection between aging and nucleolar morphology and activity. This connection is consistent with the longstanding proposal that rDNA instability underlies aging phenotypes [100–104]. Hutchinson-Gilford progeria syndrome (HGPS), caused by mutated lamin A/C, results in increased nucleolar function through increased rDNA transcription and translation [105]. In contrast, both Werner syndrome (WS) and Cockayne syndrome (CS), canonical disorders of DNA repair [106, 107], result in decreased nucleolar activity through reduced rDNA transcription [108]. Furthermore, WS has been shown to disrupt nuclear pores and lamin B1 [109], and lamin B2 has been shown to regulate nucleolar morphology and function [110], suggesting a broader link among the nuclear membrane, the nucleolus, and aging.
Intriguingly, neurodegenerative diseases share similar nucleolar phenotypes to WS and CS. While this has been reviewed in greater detail previously [111–116], in brief, observations in Alzheimer’s disease (AD) support reduced nucleolar size, activity and translation [117, 118]. Likewise, in Parkinson’s disease (PD) nucleolar disruption has been observed [114], and when the RNAPI transcription factor RRN3 (TIF-1A) is depleted specifically in adult mouse dopaminergic neurons, p53-dependent apoptosis and PD-like symptoms are observed, supporting a link between nucleolar stress and neurodegeneration [119]. However, recent work studying children heterozygous for a gain-of-function mutation in the RNAPI transcription factor UBTF (E210K) also have neurodegeneration suggesting that our understanding of the association between nucleolar function and neurodegeneration is still incomplete [120].
Finally, recently it has been discerned that small nucleolar size predicts a longer lifespan not only in C. elegans, but also in fly, mouse, and humans [121]. Despite contrasting with the above observations on neurodegeneration, reduced nucleolar size and activity as a hallmark of longevity is consistent with the longstanding research on mTOR (mechanistic/mammalian target of rapamycin) and the use of inhibitors to treat age-related diseases [122]. Further confounding is a new study that identified increased CpG methylation in the rDNA of aged mice (a pattern conserved to canids and human models) [123]. This would suggest decreased rDNA transcription and greater genome stability in aged individuals, which is contrary to much of the existing literature. Perhaps increased methylation is a compensatory mechanism to counteract cellular processes that are no longer maintaining genome stability. Furthermore, defects in ribosome recycling have also recently been implicated in aging and neurodegeneration [124, 125]. Thus, additional studies are required to gain a more comprehensive understanding of the role of ribosome biogenesis in aging and neurodegenerative disease.
Concluding remarks and future perspectives
Can we classify old diseases as new ribosomopathies (see outstanding questions)? As we continue to understand more about embryonic development and its relationship to making ribosomes, we will be compelled to re-examine old disorders for new links to ribosome biogenesis. Along these lines, recent research indicates that the microcephaly, small forehead, elongated face, clinodactyly of the fifth fingers, and intellectual disability seen in the cohesinopathy Warsaw Breakage syndrome (WABS) may be due in part to a newfound role for the protein DDX11 in RNAPI transcription [126]. Two proteins in another bone marrow failure syndrome, Fanconi Anemia (FA), have recently been found to have a role outside of DNA repair in RNAPI transcription, suggesting that there may be a dual pathogenesis for FA [127, 128]. In addition, recent results have shown differential expression of r-proteins in response to ethanol exposure, highlighting a potential link between the craniofacial defects observed in fetal alcohol syndrome and making ribosomes [129]. Discovery-based approaches for factors that influence human ribosome biogenesis [130–134] may be critical in identifying these new links. Moving forward, by defining the mechanisms underlying the pathogenesis of ribosomopathies, we hope to eventually be able to alleviate the human suffering that they cause.
Acknowledgements
Special thanks to Dr. Kathleen L. McCann for inspiration for figure 2. Thanks also to the members of the Baserga laboratory for careful reading and insightful questions. SJB acknowledges the support of 1R35GM131687, 1R01GM115710, and 1R01GM122926 from the NIH and the Breast Cancer Alliance for support of her laboratory. KIF acknowledges T32GM007223 and F31DE026946 from the NIH for funding. LMO acknowledges T32GM007223 and F31AG058405 from the NIH for funding.
References
- 1.Woolford JL and Baserga SJ (2013) Ribosome Biogenesis in the Yeast Saccaromyces cerevisiae. Genetics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henras AK, et al. (2015) An overview of pre-ribosomal RNA processing in eukaryotes. Wiley interdisciplinary reviews. RNA 6, 225–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Draptchinskaia N, et al. (1999) The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet 21, 169–175 [DOI] [PubMed] [Google Scholar]
- 4.Warren AJ (2018) Molecular basis of the human ribosomopathy Shwachman-Diamond syndrome. Advances in biological regulation 67, 109–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sulima SO, et al. (2019) Cancer Biogenesis in Ribosomopathies. Cells 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mills EW and Green R (2017) Ribosomopathies: There’s strength in numbers. Science 358 [DOI] [PubMed] [Google Scholar]
- 7.Danilova N and Gazda HT (2015) Ribosomopathies: how a common root can cause a tree of pathologies. Disease models & mechanisms 8, 1013–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aspesi A and Ellis SR (2019) Rare ribosomopathies: insights into mechanisms of cancer. Nat Rev Cancer 19, 228–238 [DOI] [PubMed] [Google Scholar]
- 9.Dauwerse JG, et al. (2011) Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat Genet 43, 20–22 [DOI] [PubMed] [Google Scholar]
- 10.Splendore A, et al. (2000) High mutation detection rate in TCOF1 among Treacher Collins syndrome patients reveals clustering of mutations and 16 novel pathogenic changes. Hum Mutat 16, 315–322 [DOI] [PubMed] [Google Scholar]
- 11.Altug Teber O, et al. (2004) Genotyping in 46 patients with tentative diagnosis of Treacher Collins syndrome revealed unexpected phenotypic variation. Eur J Hum Genet 12, 879–890 [DOI] [PubMed] [Google Scholar]
- 12.Bowman M, et al. (2012) Gross deletions in TCOF1 are a cause of Treacher-Collins-Franceschetti syndrome. Eur J Hum Genet 20, 769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valdez BC, et al. (2004) The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proceedings of the National Academy of Sciences of the United States of America 101, 10709–10714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzales B, et al. (2005) The Treacher Collins syndrome (TCOF1) gene product is involved in pre-rRNA methylation. Human Molecular Genetics 14, 2035–2043 [DOI] [PubMed] [Google Scholar]
- 15.Long PA, et al. (2017) Recessive TAF1A mutations reveal ribosomopathy in siblings with end-stage pediatric dilated cardiomyopathy. Human Molecular Genetics 26, 2874–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weaver KN, et al. (2015) Acrofacial Dysostosis, Cincinnati Type, a Mandibulofacial Dysostosis Syndrome with Limb Anomalies, Is Caused by POLR1A Dysfunction. Am J Hum Genet 96, 765–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chagnon P, et al. (2002) A missense mutation (R565W) in cirhin (FLJ14728) in North American Indian childhood cirrhosis. Am J Hum Genet 71, 1443–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freed EF and Baserga SJ (2010) The C-terminus of Utp4, mutated in childhood cirrhosis, is essential for ribosome biogenesis. Nucleic Acids Res 38, 4798–4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kostjukovits S, et al. (2017) Analysis of clinical and immunologic phenotype in a large cohort of children and adults with cartilage-hair hypoplasia. J Allergy Clin Immunol 140, 612–614 e615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peculis BA and Steitz JA (1993) Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell 73, 1233–1245 [DOI] [PubMed] [Google Scholar]
- 21.Peculis BA (1997) The sequence of the 5’ end of the U8 small nucleolar RNA is critical for 5.8S and 28S rRNA maturation. Molecular and cellular biology 17, 3702–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenkinson EM, et al. (2016) Mutations in SNORD118 cause the cerebral microangiopathy leukoencephalopathy with calcifications and cysts. Nat Genet 48, 1185–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pahuja L, et al. (2017) Labrune syndrome: A unique leukoencephalopathy. Ann Indian Acad Neurol 20, 59–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwasaki Y, et al. (2017) Longitudinal clinical and neuro-radiological findings in a patient with leukoencephalopathy with brain calcifications and cysts (Labrune syndrome). eNeurologicalSci 8, 28–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hermens M, et al. (2018) A brother and sister with intellectual disability and characteristic neuroimaging findings. Eur J Paediatr Neurol 22, 866–869 [DOI] [PubMed] [Google Scholar]
- 26.Shtaya A, et al. (2019) Leukoencephalopathy, intracranial calcifications, cysts and SNORD118 mutation (Labrune Syndrome) with obstructive hydrocephalus. World Neurosurg [DOI] [PubMed] [Google Scholar]
- 27.Khajuria RK, et al. (2018) Ribosome Levels Selectively Regulate Translation and Lineage Commitment in Human Hematopoiesis. Cell 173, 90–103 e119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klauck SM, et al. (2006) Mutations in the ribosomal protein gene RPL10 suggest a novel modulating disease mechanism for autism. Mol Psychiatry 11, 1073–1084 [DOI] [PubMed] [Google Scholar]
- 29.Chiocchetti A, et al. (2011) Mutation and expression analyses of the ribosomal protein gene RPL10 in an extended German sample of patients with autism spectrum disorder. Am J Med Genet A 155A, 1472–1475 [DOI] [PubMed] [Google Scholar]
- 30.Brooks SS, et al. (2014) A Novel Ribosomopathy Caused by Dysfunction of RPL10 Disrupts Neurodevelopment and Causes X-Linked Microcephaly in Humans. Genetics 198, 723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bourque DK, et al. (2018) A de novo mutation in RPL10 causes a rare X-linked ribosomopathy characterized by syndromic intellectual disability and epilepsy: A new case and review of the literature. European Journal of Medical Genetics 61, 89–93 [DOI] [PubMed] [Google Scholar]
- 32.Paolini NA, et al. (2017) A Ribosomopathy Reveals Decoding Defective Ribosomes Driving Human Dysmorphism. American journal of human genetics 100, 506–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolze A, et al. (2013) Ribosomal Protein SA Haploinsufficiency in Humans with Isolated Congenital Asplenia. Science 340, 976–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolze A, et al. (2018) Incomplete penetrance for isolated congenital asplenia in humans with mutations in translated and untranslated RPSA exons. Proceedings of the National Academy of Sciences of the United States of America 115, E8007–E8016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffin JN, et al. (2018) RPSA, a candidate gene for isolated congenital asplenia, is required for pre-rRNA processing and spleen formation in Xenopus. Development 145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pederson T (1998) The plurifunctional nucleolus. Nucleic Acids Research 26, 3871–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warner JR and McIntosh KB (2009) How common are extraribosomal functions of ribosomal proteins? Mol Cell 34, 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watt KEN and Trainor PA (2014) Chapter 17 - Neurocristopathies: The Etiology and Pathogenesis of Disorders Arising from Defects in Neural Crest Cell Development In Neural Crest Cells (Trainor PA, ed), pp. 361–394, Academic Press [Google Scholar]
- 39.Vlachos A, et al. (2018) Increased Prevalence of Congenital Heart Disease in Children With Diamond Blackfan Anemia Suggests Unrecognized Diamond Blackfan Anemia as a Cause of Congenital Heart Disease in the General Population: A Report of the Diamond Blackfan Anemia Registry. Circulation. Genomic and precision medicine 11, e002044–e002044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myers KC, et al. (2014) Variable clinical presentation of Shwachman-Diamond syndrome: update from the North American Shwachman-Diamond Syndrome Registry. J Pediatr 164, 866–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marneros AG (2013) BMS1 is mutated in aplasia cutis congenita. PLoS genetics 9, e1003573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiferaw B, et al. (2015) A case report on a rare disease: dyskeratosis congenita. Journal of clinical medicine research 7, 361–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marrone A, et al. (2007) Telomerase reverse-transcriptase homozygous mutations in autosomal recessive dyskeratosis congenita and Hoyeraal-Hreidarsson syndrome. Blood 110, 4198–4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dokal I (2000) Dyskeratosis congenita in all its forms. Br J Haematol 110, 768–779 [DOI] [PubMed] [Google Scholar]
- 45.Ayas M and Ahmed SO (2017) Chapter 18 - Dyskeratosis Congenita In Congenital and Acquired Bone Marrow Failure (Aljurf MD, et al. , eds), pp. 225–233, Elsevier [Google Scholar]
- 46.McGowan KA, et al. (2008) Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet 40, 963–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nousbeck J, et al. (2008) Alopecia, neurological defects, and endocrinopathy syndrome caused by decreased expression of RBM28, a nucleolar protein associated with ribosome biogenesis. Am J Hum Genet 82, 1114–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.D’Amours G, et al. (2018) Refining the phenotype associated with biallelic DNAJC21 mutations. Clinical Genetics 94, 252–258 [DOI] [PubMed] [Google Scholar]
- 49.Tummala H, et al. (2016) DNAJC21 Mutations Link a Cancer-Prone Bone Marrow Failure Syndrome to Corruption in 60S Ribosome Subunit Maturation. The American Journal of Human Genetics 99, 115–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kadakia S, et al. (2014) Treacher Collins Syndrome: The genetics of a craniofacial disease. International Journal of Pediatric Otorhinolaryngology 78, 893–898 [DOI] [PubMed] [Google Scholar]
- 51.Lipton JM, et al. (2006) Improving clinical care and elucidating the pathophysiology of Diamond Blackfan anemia: An update from the Diamond Blackfan Anemia Registry. Pediatric Blood & Cancer 46, 558–564 [DOI] [PubMed] [Google Scholar]
- 52.Sandell L (2014) Chapter 9 - Neural Crest Cells in Ear Development In Neural Crest Cells (Trainor PA, ed), pp. 167–187, Academic Press [Google Scholar]
- 53.Rosa F, et al. (2016) Ear malformations, hearing loss and hearing rehabilitation in children with Treacher Collins syndrome. Acta Otorrinolaringologica (English Edition) 67, 142–147 [DOI] [PubMed] [Google Scholar]
- 54.Gripp KW, et al. (2014) Diamond–Blackfan anemia with mandibulofacial dystostosis is heterogeneous, including the novel DBA genes TSR2 and RPS28. American Journal of Medical Genetics Part A 164, 2240–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones NC, et al. (2008) Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nature medicine 14, 125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watt KEN, et al. (2018) tp53-dependent and independent signaling underlies the pathogenesis and possible prevention of Acrofacial Dysostosis–Cincinnati type. Human Molecular Genetics 27, 2628–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Calo E, et al. (2018) Tissue-selective effects of nucleolar stress and rDNA damage in developmental disorders. Nature 554, 112–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Griffin JN, et al. (2015) The Ribosome Biogenesis Factor Nol11 Is Required for Optimal rDNA Transcription and Craniofacial Development in Xenopus. PLoS genetics 11, e1005018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao C, et al. (2014) Tissue Specific Roles for the Ribosome Biogenesis Factor Wdr43 in Zebrafish Development. PLoS genetics 10, e1004074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosas MG, et al. (2019) Proteasomal inhibition attenuates craniofacial malformations in a zebrafish model of Treacher Collins Syndrome. Biochem Pharmacol 163, 362–370 [DOI] [PubMed] [Google Scholar]
- 61.Tourlakis ME, et al. (2015) In Vivo Senescence in the Sbds-Deficient Murine Pancreas: Cell-Type Specific Consequences of Translation Insufficiency. PLoS genetics 11, e1005288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Provost E, et al. (2012) Ribosomal biogenesis genes play an essential and p53-independent role in zebrafish pancreas development. Development 139, 3232–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wan Y, et al. (2016) Transcriptome analysis reveals a ribosome constituents disorder involved in the RPL5 downregulated zebrafish model of Diamond-Blackfan anemia. BMC Med Genomics 9, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aspesi A, et al. (2014) Dissecting the transcriptional phenotype of ribosomal protein deficiency: implications for Diamond-Blackfan Anemia. Gene 545, 282–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.James A, et al. (2014) Nucleolar stress with and without p53. Nucleus 5, 402–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shwachman H, et al. (1964) THE SYNDROME OF PANCREATIC INSUFFICIENCY AND BONE MARROW DYSFUNCTION. J Pediatr 65, 645–663 [DOI] [PubMed] [Google Scholar]
- 67.Van den Berghe H, et al. (1974) Distinct haematological disorder with deletion of long arm of no. 5 chromosome. Nature 251, 437–438 [DOI] [PubMed] [Google Scholar]
- 68.Li F, et al. (2019) Clinical features of dyskeratosis congenita in mainland China: case reports and literature review. International Journal of Hematology 109, 328–335 [DOI] [PubMed] [Google Scholar]
- 69.Agarwal S (2018) Evaluation and Management of Hematopoietic Failure in Dyskeratosis Congenita. Hematol Oncol Clin North Am 32, 669–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou C, et al. (2011) Mutation in ribosomal protein L21 underlies hereditary hypotrichosis simplex. Human Mutation 32, 710–714 [DOI] [PubMed] [Google Scholar]
- 71.Weber SC and Brangwynne CP (2015) Inverse size scaling of the nucleolus by a concentration-dependent phase transition. Curr Biol 25, 641–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vincent M, et al. (2016) Treacher Collins syndrome: a clinical and molecular study based on a large series of patients. Genet Med 18, 49–56 [DOI] [PubMed] [Google Scholar]
- 73.Crick FH (1958) On protein synthesis. Symp Soc Exp Biol 12, 138–163 [PubMed] [Google Scholar]
- 74.Ferretti MB and Karbstein K (2019) Does functional specialization of ribosomes really exist? RNA 25, 521–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Emmott E, et al. (2019) Ribosome Stoichiometry: From Form to Function. Trends Biochem Sci 44, 95–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Genuth NR and Barna M (2018) The Discovery of Ribosome Heterogeneity and Its Implications for Gene Regulation and Organismal Life. Mol Cell 71, 364–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xue S, et al. (2015) RNA regulons in Hox 5’ UTRs confer ribosome specificity to gene regulation. Nature 517, 33–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kondrashov N, et al. (2011) Ribosome mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145, 383–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shi Z, et al. (2017) Heterogeneous Ribosomes Preferentially Translate Distinct Subpools of mRNAs Genome-wide. Molecular Cell 67, 71–83.e77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ferretti MB, et al. (2017) Rps26 directs mRNA-specific translation by recognition of Kozak sequence elements. Nature Structural &Amp; Molecular Biology 24, 700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lodish HF (1974) Model for the regulation of mRNA translation applied to haemoglobin synthesis. Nature 251, 385–388 [DOI] [PubMed] [Google Scholar]
- 82.Ludwig LS, et al. (2014) Altered translation of GATA1 in Diamond-Blackfan anemia. Nature medicine 20, 748–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mignone F, et al. (2002) Untranslated regions of mRNAs. Genome Biol 3, REVIEWS0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheng Z, et al. (2019) Small and Large Ribosomal Subunit Deficiencies Lead to Distinct Gene Expression Signatures that Reflect Cellular Growth Rate. Mol Cell 73, 36–47 e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dameshek W (1967) Riddle: what do aplastic anemia, paroxysmal nocturnal hemoglobinuria (PNH) and “hypoplastic” leukemia have in common? Blood 30, 251–254 [PubMed] [Google Scholar]
- 86.Alter BP, et al. (2018) Cancer in the National Cancer Institute inherited bone marrow failure syndrome cohort after fifteen years of follow-up. Haematologica 103, 30–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boocock GR, et al. (2003) Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet 33, 97–101 [DOI] [PubMed] [Google Scholar]
- 88.Savage SA and Dufour C (2017) Classical inherited bone marrow failure syndromes with high risk for myelodysplastic syndrome and acute myelogenous leukemia. Seminars in hematology 54, 105–114 [DOI] [PubMed] [Google Scholar]
- 89.Donadieu J, et al. (2012) Classification of and risk factors for hematologic complications in a French national cohort of 102 patients with Shwachman-Diamond syndrome. Haematologica 97, 1312–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.De Keersmaecker K, et al. (2013) Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet 45, 186–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weis F, et al. (2015) Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat Struct Mol Biol 22, 914–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pressato B, et al. (2012) Deletion of chromosome 20 in bone marrow of patients with Shwachman-Diamond syndrome, loss of the EIF6 gene and benign prognosis. Br J Haematol 157, 503–505 [DOI] [PubMed] [Google Scholar]
- 93.Valli R, et al. (2013) Different loss of material in recurrent chromosome 20 interstitial deletions in Shwachman-Diamond syndrome and in myeloid neoplasms. Mol Cytogenet 6, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pressato B, et al. (2015) Cytogenetic monitoring in Shwachman-Diamond syndrome: a note on clonal progression and a practical warning. J Pediatr Hematol Oncol 37, 307–310 [DOI] [PubMed] [Google Scholar]
- 95.Valli R, et al. (2019) Shwachman-Diamond syndrome with clonal interstitial deletion of the long arm of chromosome 20 in bone marrow: haematological features, prognosis and genomic instability. British Journal of Haematology 184, 974–981 [DOI] [PubMed] [Google Scholar]
- 96.Valli R, et al. (2017) Novel recurrent chromosome anomalies in Shwachman–Diamond syndrome. Pediatric Blood & Cancer 64, e26454. [DOI] [PubMed] [Google Scholar]
- 97.Lindsley RC, et al. (2017) Prognostic Mutations in Myelodysplastic Syndrome after Stem-Cell Transplantation. The New England journal of medicine 376, 536–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zambetti NA, et al. (2016) Mesenchymal Inflammation Drives Genotoxic Stress in Hematopoietic Stem Cells and Predicts Disease Evolution in Human Pre-leukemia. Cell stem cell 19, 613–627 [DOI] [PubMed] [Google Scholar]
- 99.Pronk E and Raaijmakers M (2019) The mesenchymal niche in MDS. Blood 133, 1031–1038 [DOI] [PubMed] [Google Scholar]
- 100.Guarente L (1997) Link between aging and the nucleolus. Genes Dev 11, 2449–2455 [DOI] [PubMed] [Google Scholar]
- 101.Sinclair DA and Guarente L (1997) Extrachromosomal rDNA circles--a cause of aging in yeast. Cell 91, 1033–1042 [DOI] [PubMed] [Google Scholar]
- 102.Ganley AR and Kobayashi T (2014) Ribosomal DNA and cellular senescence: new evidence supporting the connection between rDNA and aging. FEMS Yeast Res 14, 49–59 [DOI] [PubMed] [Google Scholar]
- 103.Tiku V and Antebi A (2018) Nucleolar Function in Lifespan Regulation. Trends Cell Biol 28, 662–672 [DOI] [PubMed] [Google Scholar]
- 104.Turi Z, et al. (2019) Impaired ribosome biogenesis: mechanisms and relevance to cancer and aging. Aging (Albany NY) 11, 2512–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Buchwalter A and Hetzer MW (2017) Nucleolar expansion and elevated protein translation in premature aging. Nature communications 8, 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mukherjee S, et al. (2018) Werner Syndrome Protein and DNA Replication. International journal of molecular sciences 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Karikkineth AC, et al. (2017) Cockayne syndrome: Clinical features, model systems and pathways. Ageing Res Rev 33, 3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hannan KM, et al. (2013) Dysregulation of RNA polymerase I transcription during disease. Biochim Biophys Acta 1829, 342–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li Z, et al. (2013) Werner complex deficiency in cells disrupts the Nuclear Pore Complex and the distribution of lamin B1. Biochim Biophys Acta 1833, 3338–3345 [DOI] [PubMed] [Google Scholar]
- 110.Sen Gupta A and Sengupta K (2017) Lamin B2 Modulates Nucleolar Morphology, Dynamics, and Function. Molecular and cellular biology 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Parlato R and Kreiner G (2013) Nucleolar activity in neurodegenerative diseases: a missing piece of the puzzle? J Mol Med (Berl) 91, 541–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hetman M and Pietrzak M (2012) Emerging roles of the neuronal nucleolus. Trends Neurosci 35, 305–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sia PI, et al. (2016) Role of the nucleolus in neurodegenerative diseases with particular reference to the retina: a review. Clin Exp Ophthalmol 44, 188–195 [DOI] [PubMed] [Google Scholar]
- 114.Parlato R and Liss B (2014) How Parkinson’s disease meets nucleolar stress. Biochim Biophys Acta 1842, 791–797 [DOI] [PubMed] [Google Scholar]
- 115.Herrmann D and Parlato R (2018) C9orf72-associated neurodegeneration in ALS-FTD: breaking new ground in ribosomal RNA and nucleolar dysfunction. Cell and tissue research 373, 351–360 [DOI] [PubMed] [Google Scholar]
- 116.Parlato R and Bierhoff H (2015) Role of nucleolar dysfunction in neurodegenerative disorders: a game of genes? AIMS Molecular Science 2, 211–224 [Google Scholar]
- 117.Hernandez-Ortega K, et al. (2016) Altered Machinery of Protein Synthesis in Alzheimer’s: From the Nucleolus to the Ribosome. Brain Pathol 26, 593–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ding Q, et al. (2005) Ribosome dysfunction is an early event in Alzheimer’s disease. The Journal of neuroscience : the official journal of the Society for Neuroscience 25, 9171–9175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rieker C, et al. (2011) Nucleolar disruption in dopaminergic neurons leads to oxidative damage and parkinsonism through repression of mammalian target of rapamycin signaling. The Journal of neuroscience : the official journal of the Society for Neuroscience 31, 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Edvardson S, et al. (2017) Heterozygous De Novo UBTF Gain-of-Function Variant Is Associated with Neurodegeneration in Childhood. Am J Hum Genet 101, 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tiku V, et al. (2017) Small nucleoli are a cellular hallmark of longevity. Nature communications 8, 16083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Walters HE and Cox LS (2018) mTORC Inhibitors as Broad-Spectrum Therapeutics for Age-Related Diseases. International journal of molecular sciences 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang M and Lemos B (2017) Ribosomal DNA copy number amplification and loss in human cancers is linked to tumor genetic context, nucleolus activity, and proliferation. PLoS genetics 13, e1006994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sudmant PH, et al. (2018) Widespread Accumulation of Ribosome-Associated Isolated 3’ UTRs in Neuronal Cell Populations of the Aging Brain. Cell Rep 25, 2447–2456 e2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ishimura R, et al. (2014) RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science 345, 455–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sun X, et al. (2015) The Warsaw breakage syndrome-related protein DDX11 is required for ribosomal RNA synthesis and embryonic development. Human Molecular Genetics 24, 4901–4915 [DOI] [PubMed] [Google Scholar]
- 127.Sondalle SB, et al. (2019) Fanconi anemia protein FANCI functions in ribosome biogenesis. Proceedings of the National Academy of Sciences 116, 2561–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Johnston GC and Singer RA (1978) RNA synthesis and control of cell division in the yeast S.cerevisiae. Cell 14:951–958, 951–958 [DOI] [PubMed] [Google Scholar]
- 129.Berres ME, et al. (2017) Transcriptome Profiling Identifies Ribosome Biogenesis as a Target of Alcohol Teratogenicity and Vulnerability during Early Embryogenesis. PLoS One 12, e0169351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Farley-Barnes KI, et al. (2018) Diverse Regulators of Human Ribosome Biogenesis Discovered by Changes in Nucleolar Number. Cell Rep 22, 1923–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Badertscher L, et al. (2015) Genome-wide RNAi Screening Identifies Protein Modules Required for 40S Subunit Synthesis in Human Cells. Cell Rep 13, 2879–2891 [DOI] [PubMed] [Google Scholar]
- 132.Wild T, et al. (2010) A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS Biol 8, e1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tafforeau L, et al. (2013) The Complexity of Human Ribosome Biogenesis Revealed by Systematic Nucleolar Screening of Pre-rRNA Processing Factors. Mol Cell 51, 539–551 [DOI] [PubMed] [Google Scholar]
- 134.Stamatopoulou V, et al. (2018) Use of the iNo score to discriminate normal from altered nucleolar morphology, with applications in basic cell biology and potential in human disease diagnostics. Nature protocols 13, 2387–2406 [DOI] [PubMed] [Google Scholar]
- 135.van der Lelij P, et al. (2010) Warsaw Breakage Syndrome, a Cohesinopathy Associated with Mutations in the XPD Helicase Family Member DDX11/ChlR1. The American Journal of Human Genetics 86, 262–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Capo-Chichi J-M, et al. (2013) Identification and biochemical characterization of a novel mutation in DDX11 causing Warsaw breakage syndrome. Human mutation 34, 103–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen JY, et al. (2018) The ribosome biogenesis protein Esf1 is essential for pharyngeal cartilage formation in zebrafish. FEBS J 285, 3464–3483 [DOI] [PubMed] [Google Scholar]
- 138.Yu W, et al. (2011) PAK1IP1, a ribosomal stress-induced nucleolar protein, regulates cell proliferation via the p53–MDM2 loop. Nucleic Acids Research 39, 2234–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ross AP, et al. (2013) A Mutation in Mouse Pak1ip1 Causes Orofacial Clefting while Human PAK1IP1 Maps to 6p24 Translocation Breaking Points Associated with Orofacial Clefting. PLoS ONE 8, e69333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Noack Watt KE, et al. (2016) The Roles of RNA Polymerase I and III Subunits Polr1c and Polr1d in Craniofacial Development and in Zebrafish Models of Treacher Collins Syndrome. PLoS genetics 12, e1006187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.da Silva Dalben G, et al. (2006) Prevalence of dental anomalies, ectopic eruption and associated oral malformations in subjects with Treacher Collins syndrome. Oral Surgery, Oral Medicine, Oral Pathology, Oral Radiology, and Endodontology 101, 588–592 [DOI] [PubMed] [Google Scholar]