Multiplexed activation of endogenous genes by CRISPRa elicits potent anti-tumor immunity (original) (raw)
. Author manuscript; available in PMC: 2020 Apr 14.
Published in final edited form as: Nat Immunol. 2019 Oct 14;20(11):1494–1505. doi: 10.1038/s41590-019-0500-4
Abstract
Immunotherapy has transformed cancer treatment. However, current immunotherapy modalities face various limitations. Here, we developed MAEGI, a new form of immunotherapy that elicits anti-tumor immunity through multiplexed activation of endogenous genes. We leveraged CRISPR activation (CRISPRa) to directly augment the in situ expression of endogenous genes, thereby the presentation of tumor antigens, leading to dramatic anti-tumor immune responses. Deploying this as a cell-based vaccination strategy showed efficacy in both prophylactic and therapeutic settings. Intratumoral adeno-associated virus delivery of CRISPRa libraries elicited strong anti-tumor immunity across multiple cancer types. Precision targeting of mutated gene sets eradicated a large fraction of established tumors at both local and distant sites. This treatment modality led to alterations of the tumor microenvironment, marked by enhanced T cell infiltration and anti-tumor immune signatures. Multiplexed endogenous gene activation is a versatile and highly scalable strategy to elicit potent immune responses against cancer, distinct from existing cancer therapies.
Introduction
Immunotherapy leverages the patient’s immune system against tumors, turning previously lethal cancers into manageable diseases for a subset of patients 1–5. Major types of immunotherapy include checkpoint blockade 6, adoptive cell transfer 7, human recombinant cytokines, and cancer vaccines 8. These regimens have transformed cancer treatment 9–11. In particular, checkpoint blockade immunotherapies targeting CTLA-4 and PD-1 pathways have yielded significant clinical benefits across a broad spectrum of cancer types, with durable responses even in late-stage, metastatic, and chemo-resistant tumors 12–15. However, only a fraction of patients show sustained clinical responses 5, urging for new types of immunotherapies.
As a consequence of cumulative genetic and epigenetic aberrations, cancers can be recognized and eliminated by the immune system if mutant or abnormally expressed antigens are adequately presented 16,17. Recognition of tumor-associated antigens (TAAs) formed by mutations and dysregulated gene expression programs is an essential step for cancer immunotherapy 17,18. However, the spontaneous immune recognition of tumor antigens is often insufficient to elicit effective immune responses, as the abnormal products may not be adequately presented 19. Moreover, neoantigen loss often occurs during malignancy 18. We reasoned that augmenting the expression and thus presentation of endogenous antigens in tumors could amplify the “non-self” identity of cancer cells, thereby flagging them for immune destruction 20.
Neoantigen-targeting approaches have demonstrated the concept of leveraging personalized neoantigens as cancer treatments, and are based on delivery of synthetic mutant peptides or transcripts 21–24. However, the efficacy and scalability of these approaches is limited. The CRISPR activation (CRISPRa) system uses a catalytically inactive Cas9 (dCas9) 25, enabling simple and flexible gene expression regulation through dCas9-transcriptional activators paired with single guide RNAs (sgRNAs) 26–29. Using CRISPRa, multiplexed augmentation of desired gene sets can be readily achieved by utilizing pools of guide RNAs27. Here, we developed CRISPRa-mediated Multiplexed Activation of Endogenous Genes as an Immunotherapy (MAEGI), which acts by directly augmenting the expression and presentation of endogenous genes that encode potentially immunogenic antigens. We demonstrate that MAEGI has therapeutic efficacy across three tumor types. Mechanistically, we show that MAEGI treatment elicits anti-tumor immune responses by recruiting effector T cells and remodeling the tumor microenvironment.
Results
CRISPRa enhances in situ antigen presentation and promotes T cell effector function
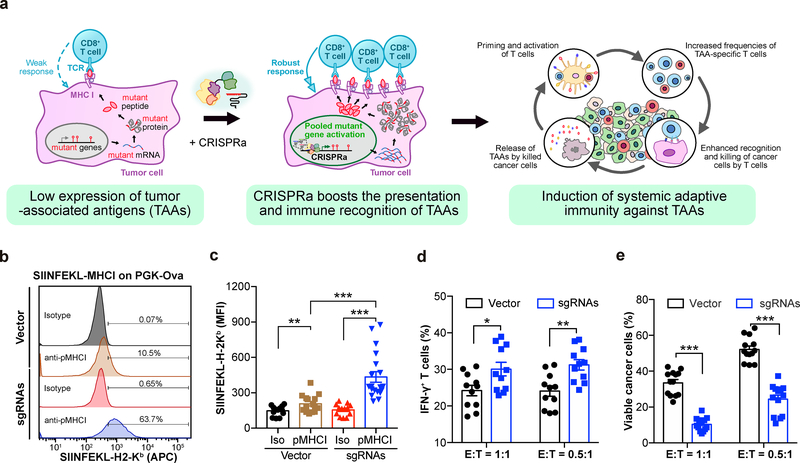
To investigate whether CRISPRa can elicit immune responses by enhancing the presentation of TAAs (Fig. 1a), we examined the effect of CRISPRa on the surface presentation of a target antigenic peptide. We transduced triple-negative breast cancer (TNBC) E0771 cells with CRISPRa lentiviral vectors expressing dCas9-VP64 and MS2-p65-HSF1 (E0771-dCas9-VP64-MPH) (Supplementary Fig. 1a). By introducing a model antigen transgene (chicken ovalbumin, OVA) driven by a phosphoglycerate kinase (PGK) promoter into E0771-dCas9-VP64-MPH cells (E0771-OVA cells), we found that PGK-targeting CRISPRa sgRNAs significantly enhanced the presentation of the target antigenic peptide (SIINFEKL) on the H-2Kb class I major histocompatibility complex (MHC-I) (Fig. 1b,c and Supplementary Table 1).
Figure 1: CRISPRa augments tumor antigen presentation to promote T cell effector function.
a, Schematic of the experimental design for using CRISPRa to enhance the immune recognition of tumor-associated antigens (TAAs), eliciting systemic immune responses. b, c, E0771-dCas9-VP64 cells were transduced with lentivirus to express ovalbumin (OVA) under a PGK promoter (E0771-OVA), and further transduced with either Vector or CRISPRa sgRNAs targeting the PGK promoter. (b), Representative flow cytometry analysis of surface staining for OVA-derived SIINFEKL-H-2Kb complex on cells transduced with Vector or sgRNAs. (c), Mean fluorescence intensity (MFI) of APC-SIINFEKL-H-2Kb on E0771-OVA cells transduced with Vector or sgRNAs. n = 15 cell replicates (SIINFEKL-H-2Kb staining in Vector), n = 13 (isotype in Vector), n = 19 (SIINFEKL-H-2Kb staining in CRISPRa sgRNAs), or n = 15 (isotype in CRISPRa sgRNAs) from four independent experiments. Two-sided Mann-Whitney test: SIINFEKL-H-2Kb staining vs. isotype in Vector, p = 0.0356; SIINFEKL-H-2Kb staining vs. isotype in CRISPRa sgRNAs, p < 0.0001; SIINFEKL-H-2Kb staining in CRISPRa sgRNAs vs. Vector, p < 0.0001. d, The percentage of IFN-γ-producing OT-I CD8+ T effector cells after co-culture with the indicated E0771-OVA cancer cells for 3h. n = 11 co-culture samples from three independent experiments. Welch’s two-tailed unpaired t-test: CRISPRa sgRNAs vs. vector in E:T = 1:1, p = 0.0218; CRISPRa sgRNAs vs. vector in E:T = 0.5:1, p = 0.002. e, The percentage of viable cancer cells (excluding dead cells and apoptotic cells) when co-cultured with OT-I CD8+ T effector cells for 24h. n = 11 co-culture samples from 3 independent experiments. Welch’s two-tailed unpaired t-test: CRISPRa sgRNAs vs. vector in E:T = 1:1, p < 0.0001; CRISPRa sgRNAs vs. vector in E:T = 0.5:1, p < 0.0001. Error bars: All data points in this figure are presented as mean ± s.e.m. Asterisks: * p < 0.05, ** p < 0.01, *** p < 0.001.
Additional supporting data: Supplementary Figure 1
We then utilized a co-culture assay to investigate the effect of CRISPRa-enhanced antigen presentation on cancer-immune cell interactions. To assess the effector function of T cells, we isolated the SIINFEKL-H-2Kb-specific CD8+ T cells from ovalbumin peptide-specific (OVA) OT-I mice, and then co-cultured them with E0771-OVA cells for 3h. E0771-OVA cells expressing PGK-targeting sgRNAs elicited a significantly higher percentage of interferon-γ (IFN-γ)-producing T cells (Fig. 1d and Supplementary Fig. 1b). To assess T cell-mediated killing of cancer cells, we co-cultured OT-I T cells with E0771-OVA cells for 24h and analyzed the viability and apoptosis of the cancer cells. Consistent with the increase in IFN-γ production, CRISPRa treatment led to a significantly lower percentage of viable cancer cells, with the majority of the cancer cells either killed or undergoing apoptosis (Fig. 1e and Supplementary Fig. 1c and Supplementary Table 1). These data demonstrate that CRISPRa enhanced the presentation of a target antigen in cancer cells, thereby eliciting a stronger T cell effector response.
CRISPRa-mediated gene activation leads to immune-mediated tumor clearance
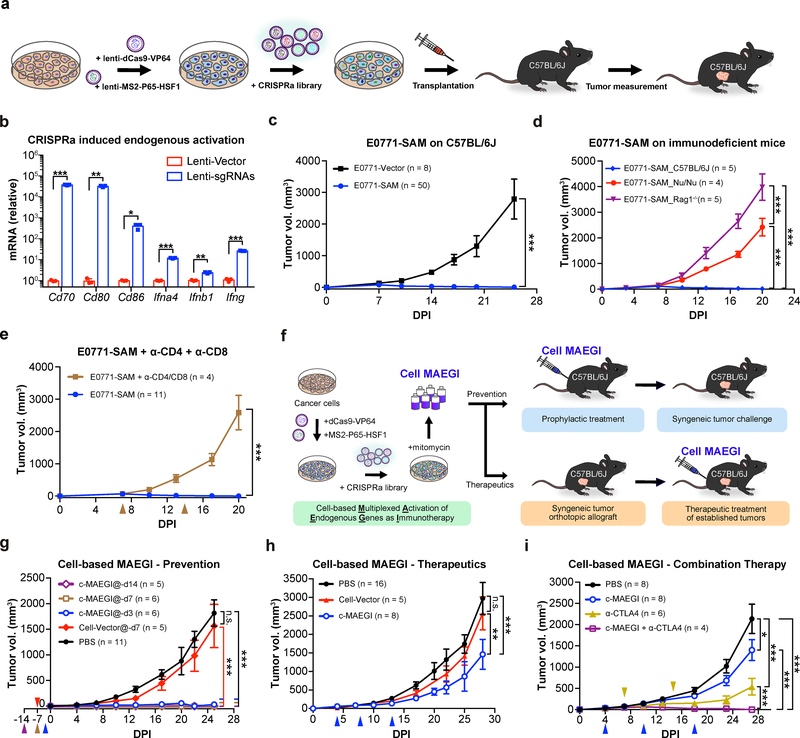
To test the effect of endogenous gene activation on the immunogenicity of cancer cells in vivo, we examined the tumorigenic ability of CRISPRa-transduced cancer cells in immunocompetent C57BL/6J mice (Fig. 2a and Methods). We transduced a mouse genome-scale lentiviral CRISPRa sgRNA library (mSAM) 27 into E0771-dCas9-VP64-MPH cells (E0771-SAM) (Supplementary Fig. 1d–e and Supplementary Table 2). After confirming the efficient activation of endogenous genes using this system (Fig. 2b), we transplanted the library-transduced E0771 cells into C57BL/6J mice. In sharp contrast to E0771-Vector cells (0/8 rejection, i.e. 8/8 engrafted, all with large tumors), the E0771-SAM cell pool was rejected in most (42/50, 84%) of the mice, with the remaining mice (8/50, 16%) developing small tumors (Fig. 2c). To examine the contribution of the immune system towards rejection of E0771-SAM cells, we transplanted E0771-SAM cells into immunodeficient Nude (_Foxn1_nu) and Rag1 −/− mice. Unlike immunocompetent C57BL/6J hosts (5/5 either completely rejected or with small modules), all immunodeficient Nude (_Foxn1_nu) (4/4) and Rag1 −/− (5/5) mice rapidly grew large tumors from E0771-SAM cells (Fig. 2d). When we depleted both CD4+ and CD8+ T cells in C57BL/6J mice using anti-CD4 and anti-CD8 monoclonal antibodies, all mice receiving E0771-SAM cells formed large tumors (4/4), in contrast to untreated mice (0/11 with tumors) (Fig. 2e). Together, these results indicate that pooled activation of endogenous genes induced potent tumor rejection in vivo only in immunocompetent hosts.
Figure 2. Multiplexed activation of endogenous genes as an immunotherapy (MAEGI) in a cellular formulation.
a, Schematics for generating genome-scale CRISPR activation (SAM) sgRNA library-transduced cells and assessment of tumorigenic capacity. b, Lentiviral CRISPRa-mediated transcriptional activation of Cd70, Cd80, Cd86, Ifnα4, Ifnβ1, and Ifnγ, normalized to vector-transduced controls. n = 3 infection replicates from one representative experiment. Two-sided unpaired t-test, sgRNAs vs. vector Cd70, p = 0.0006; Cd80, p = 0.0025; Cd86, p = 0.026; Ifnα4, p = 0.0007; Ifnβ1, p = 0.003; Ifnγ, p < 0.0001. c, Tumor growth curves of Vector (E0771-Vector, n = 8 mice) or SAM transduced E0771-dCas9-VP64-MPH cells (E0771-SAM; n = 50) transplanted in C57BL/6J mice. Two-way ANOVA: p < 0.0001. d, Tumor growth curves of E0771-SAM cells in syngeneic immunocompetent mice (C57BL/6J, n = 5 mice) and two strains of immunodeficient mice (Nude (_Foxn1_nu), n = 4; Rag1 −/−, n = 5). Two-way ANOVA: C57BL/6J vs. Nude (_Foxn1_nu), p < 0.0001; C57BL/6J vs. Rag1 −/−, p < 0.0001; Nude (_Foxn1_nu) vs. Rag1 −/−, p < 0.0001. e, Tumor growth curves of E0771-SAM cells, with (n = 4 mice) or without T cell depletion (n = 11). Two-way ANOVA, untreated vs. α-CD4 + α-CD8 treated, p < 0.0001. f, Schematics of the experimental design for harnessing multiplexed endogenous gene activation as immunotherapy (MAEGI) in a cellular vaccine formulation. The E0771 cell-based MAEGI (c-MAEGI) was generated by treating E0771-SAM cells (E0771 transduced with dCas9-VP64, MS2-P65-HSF1, and a genome-wide CRISPRa library) with mitomycin. The mock cell-vector treatment (Cell-Vector) was generated in parallel. g, Growth curves of orthotopic E0771 transplants in mice pre-inoculated with PBS, mock Cell-Vector vaccine, or c-MAEGI at various time points prior to tumor transplantation (PBS, n = 11 mice; E0771-Vector at dpi = −7 (abbreviated @-d7, n = 5); c-MAEGI@-d14, (n = 5); c-MAEGI@-d7, (n = 6); c-MAEGI@-d3, (n = 6)). Two-way ANOVA: Cell-Vector@-d7 vs. PBS, p = 0.2077; c-MAEGI@-d3 vs. PBS or Vector@-d7, p < 0.0001; c-MAEGI@-d7 vs. PBS or Vector@-d7, p < 0.0001; c-MAEGI@-d14 vs. PBS or Vector@-d7, p < 0.0001. h, E0771 tumor-bearing mice were treated with PBS (n = 16 mice), Cell-Vector (n = 5) or c-MAEGI (n = 8) at indicated times (blue arrows). Two-way ANOVA: PBS vs. Vector, p = 0.2955; c-MAEGI vs. PBS, p < 0.0001; c-MAEGI vs. Vector, p = 0.0005. i, Tumor growth curves in E0771 tumor-bearing mice that were treated by PBS (n = 8 mice), c-MAEGI (n = 8), α-CTLA4 (n = 6), or c-MAEGI + α-CTLA4 (n = 4). Blue arrows, c-MAEGI treatment; yellow arrows, α-CTLA4 treatment. Two-way ANOVA: c-MAEGI vs. PBS, p = 0.0105; α-CTLA4 vs. PBS, p < 0.0001; c-MAEGI + α-CTLA4 vs. α-CTLA4 alone, p = 0.0005; c-MAEGI + α-CTLA4 vs. c-MAEGI, p < 0.0001. Error bars: All data points in this figure are presented as mean ± s.e.m. Asterisks: * p < 0.05, ** p < 0.01, *** p < 0.001.
Additional supporting data: Supplementary Figure 1
Cell-based MAEGI has prophylactic and therapeutic efficacy in syngeneic tumor models
Intrigued by the immune rejection of E0771-SAM cells, we wondered whether CRISPRa-mediated activation of endogenous genes within tumor cells could be harnessed as a new approach for immunotherapy (MAEGI). We transduced the lentiviral SAM sgRNA library into E0771-dCas9-VP64-MPH cells to generate E0771-SAM cells, followed by mitomycin treatment to induce senescence while maintaining the integrity of peptide-MHC-I complexes (Fig. 2f), generating the cell-based MAEGI (c-MAEGI). We used E0771-Vector cells that were treated with mitomycin in parallel as the Cell-Vector control. We inoculated c-MAEGI or Cell-Vector into C57BL/6J mice prior to E0771 tumor induction (Fig. 2g and Methods). In sharp contrast to untreated mice or Cell-Vector control treated mice (both 0% tumor free), c-MAEGI treated mice had complete protection (100% tumor free) against subsequent tumor challenges with the unmodified parental E0771 cells (Fig. 2g). This held true across two vaccination regiments, i.e. 7 or 14 days prior to tumor challenge, while vaccination 3 days prior to tumor challenge granted near-complete protection (1/6 mice with a small nodule, 5/6 tumor-free) (Fig. 2g and Supplementary Table 1). The results demonstrate that genome-scale activation of endogenous genes using CRISPRa is an effective approach for prophylaxis against tumors with otherwise identical genetic background.
Given its efficacy as a prophylactic agent, we postulated that c-MAEGI could also potentially be used as a therapeutic intervention against established tumors. We implanted orthotopic E0771 tumors before treating the mice with c-MAEGI. Using a three-dose treatment scheme, we observed that tumors in c-MAEGI treated mice were significantly smaller than those in mice treated with Cell-Vector control or PBS (Fig. 2h and Supplementary Table 1). To examine the contribution of T cell-mediated immunity in the anti-tumor response, we depleted CD8+ T cells in C57BL/6J mice with anti-CD8 antibodies. We found that CD8+ T cell depletion abolished the therapeutic efficacy of c-MAEGI (Supplementary Fig. 1e–h), indicating that CD8+ T cells are essential for c-MAEGI-mediated anti-tumor responses. We then investigated whether c-MAEGI could be used in conjunction with other immunotherapies or immunomodulatory agents. The combination of c-MAEGI + anti-CTLA4 was significantly more effective than c-MAEGI alone or anti-CTLA4 alone, leading to complete regression of established tumors (Fig. 2i and Supplementary Table 1). Collectively, our data demonstrate that c-MAEGI has significant efficacy in both prophylactic and therapeutic settings.
AAV-mediated in situ gene activation as an immunotherapeutic modality (AAV-MAEGI)
Since enhanced antigen presentation by CRISPRa elicited strong immune responses, we reasoned that direct in vivo delivery of CRISPRa components into target tumors could boost the presentation level of TAAs, thereby serving as a therapeutic modality. Adeno-associated viruses (AAVs) are viral vectors capable of mediating efficient transgene delivery 30,31. To enable direct delivery of MAEGI to tumors, we devised an AAV version of MAEGI by generating an AAV-CRISPRa vector containing the CRISPRa modules (EF1α-MPH and U6-sgRNABackbone-MS2). The effectiveness of AAV-CRISPRa in activating endogenous genes was confirmed by infecting E0771-dCas9-VP64 cells with CRISPRa AAVs carrying a small pool of sgRNAs (Supplementary Fig. 2a). Using the E0771-OVA model, we confirmed that AAV-delivered sgRNAs targeting the PGK promoter significantly increased the presentation of antigenic peptide (Supplementary Fig. 2b).
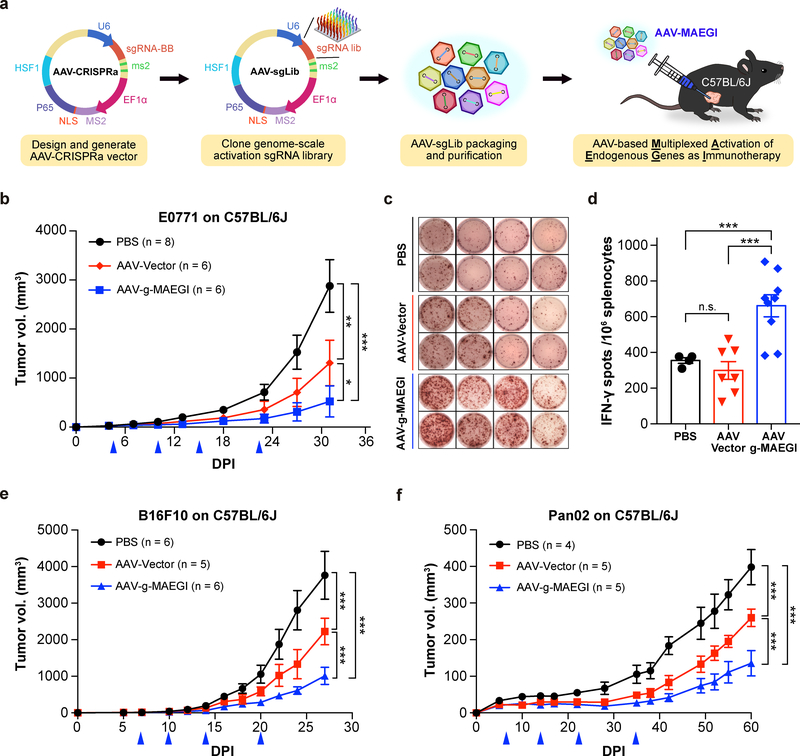
We then cloned the genome-scale SAM sgRNA library into the AAV-CRISPRa vector to produce AAV-g-MAEGI (Methods). We treated C57BL/6J mice bearing orthotopic E0771-dCas9-VP64 tumors with AAV-g-MAEGI by intratumoral administration (Fig. 3a). AAV-g-MAEGI treatment led to significantly reduced tumor burden compared to either AAV-Vector or PBS (Fig. 3b). AAV-Vector (EF1α-MPH and U6-sgRNABackbone-MS2) treatment also showed an anti-tumor effect, consistent with baseline immunogenicity of viral vectors carrying the MPH transgenes, but was nevertheless significantly weaker than AAV-g-MAEGI treatment containing the sgRNA library (Fig. 3b). Using ELISPOT, we found that AAV-g-MAEGI elicited higher frequencies of tumor-reactive immune cells (in terms of the frequencies of IFN-γ-producing splenocytes stimulated by tumor antigens) compared to AAV-Vector or PBS (Figs. 3c,d). No difference was observed in the frequencies of tumor-specific IFN-γ-secreting splenocytes between PBS and AAV-Vector (Fig. 3d). These data demonstrate that multiplexed in situ activation of endogenous genes by AAV-g-MAEGI elicits robust and specific anti-tumor immune responses in vivo.
Figure 3: Anti-tumor efficacy of MAEGI in an AAV-based formulation.
a, Schematics of the experimental design for intratumoral delivery of MAEGI in an AAV formulation. AAV-g-MAEGI was generated by cloning the genome-scale activation sgRNA SAM library into an AAV-CRISPRa vector, followed by pooled viral packaging into AAV9. b, Growth curves of orthotopic E0771-dCas9-VP64 tumor transplants in C57BL/6J mice treated with PBS, AAV-Vector, or AAV-g-MAEGI by intratumoral administration at indicated times (blue arrows) (PBS, n = 8 mice; AAV-Vector, n = 6; AAV-g-MAEGI, n = 6). Two-way ANOVA: PBS vs. AAV-Vector, p = 0.0013; AAV-g-MAEGI vs. PBS, p < 0.0001; AAV-g-MAEGI vs. AAV-Vector, p = 0.0303. c-d, Tumor-specific immune responses in tumor-bearing C57BL/6J mice, assessed by measuring the proportion of Ifn-γ-producing splenocytes with ELISPOT assays after stimulation with mitomycin-treated tumor cells. c, Representative images of spots obtained from four mice for each treatment group (PBS, AAV-Vector, and AAV-g-MAEGI). d, Quantification of ELISPOT assays from mice treated with PBS (n = 4 mice), AAV-Vector (n = 7) or AAV-g-MAEGI (n = 9). Two-sided unpaired t-test: AAV-Vector vs. PBS, p = 0.326; AAV-g-MAEGI vs. PBS, p = 0.0009; AAV-g-MAEGI vs. AAV-Vector, p = 0.0004. Results shown are from the aggregation of 2 independent experiments. e, Growth curves of orthotopic B16F10-dCas9-VP64 tumor transplants in C57BL/6J mice treated with PBS, AAV-Vector, or AAV-g-MAEGI at indicated times (blue arrows) (PBS, n = 6 mice; AAV-Vector, n = 5; AAV-g-MAEGI, n = 6). Two-way ANOVA: AAV-Vector vs. PBS, p < 0.0001; AAV-g-MAEGI vs. PBS, p < 0.0001; AAV-g-MAEGI vs. AAV-Vector, p < 0.0001. f, Growth curves of Pan02-dCas9-VP64 tumors in C57BL/6J mice treated with PBS (n = 4 mice), AAV-Vector (n = 5), or AAV-g-MAEGI (n = 5) at indicated times (blue arrows). Two-way ANOVA: AAV-Vector vs. PBS, p < 0.0001; AAV-g-MAEGI vs. PBS, p < 0.0001; AAV-g-MAEGI vs. AAV-Vector, p < 0.0001. Error bars: All data points in this figure are presented as mean ± s.e.m. Asterisks: * p < 0.05, ** p < 0.01, *** p < 0.001.
Additional supporting data: Supplementary Figure 2
To evaluate the broader utility of MAEGI, we tested the same treatment modality on other tumor types. On a syngeneic melanoma mouse model (B16F10, expressing dCas9-VP64), AAV-g-MAEGI again demonstrated significant efficacy (Fig. 3e). We further tested the efficacy of AAV-g-MAEGI in a syngeneic pancreatic cancer model using the Pan02 cell line. Against established Pan02-dCas9-VP64 tumors, we observed that while AAV-Vector had an anti-tumor effect compared to PBS, AAV-g-MAEGI has significantly stronger efficacy compared to both AAV-Vector and PBS (Fig. 3f and Supplementary Table 1). Thus, AAV-mediated delivery of genome-scale MAEGI elicits host immune responses against established tumors across multiple aggressive cancer types.
Multiplexed in situ activation of mutated gene sets as proof-of-concept precision immunotherapy
Since individual tumors have unique mutation profiles distinguishing them from normal tissues, we investigated whether MAEGI could be customized to tumor-specific mutated gene sets. We termed this approach precision MAEGI (p-MAEGI) (Fig. 4a). We hypothesized that the precision version could more specifically enhance the presentation of mutant peptides. To this end, we performed whole-exome sequencing of E0771 cells and called all genic SNPs, insertions and deletions (indels) by comparison to healthy mammary fat pad cells from wildtype C57BL/6J mice sequenced in parallel (Fig. 4a and Methods), revealing the E0771-specific mutation profile (Supplementary Fig. 3a,b). We then harnessed the differential mutation data for CRISPRa sgRNA library design, generating a library of 3,839 sgRNAs targeting 1,116 E0771-mutated genes (Supplementary Table 2). We synthesized and pool-cloned the library, verified successful cloning by sequencing, and produced the AAV pool as a precision MAEGI (AAV-p-MAEGI) (Supplementary Fig. 3c).
Figure 4. Exome-guided precision MAEGI eliminates established tumors.
a, Schematics of experimental design. Exome sequencing of cancer cells and normal tissues was performed to identify somatic mutations. A library of CRISPRa sgRNAs targeting the top mutant genes was synthesized and pool-cloned into the AAV-CRISPRa vector, then packaged into AAVs to generate AAV-p-MAEGI. b, Tumor growth curves of E0771-dCas9-VP64 syngeneic tumors in mice treated by PBS (n = 24 mice), AAV-Vector (n = 20), or AAV-p-MAEGI (n = 25) by intratumoral administration at indicated times (blue arrows). Two-way ANOVA: AAV-Vector vs. PBS, p < 0.0001; AAV-p-MAEGI vs. PBS, p < 0.0001; AAV-p-MAEGI vs. AAV-Vector, p < 0.0001. c, Response rates for each treatment, categorized by near-complete or complete response (nCR/CR), partial response (PR), or stable/progressive disease (SD/PD). d, Long-term spider growth plots of E0771-dCas9-VP64 tumors treated by PBS, AAV-Vector, or AAV-p-MAEGI from (b). Mice whose tumors had undergone complete response by AAV-p-MAEGI (n = 9) were subjected to tumor re-challenges (green arrows). No tumors grew with re-challenge. Treatment days are indicated by blue arrows. e, Schematic of experimental design for evaluating the induction of systemic anti-tumor immunity by AAV-p-MAEGI. 2*106 or 0.25*106 E0771-dCas9-VP64 tumor cells were respectively transplanted into the left or right flank of C57BL/6J mice to model local and distant tumors. AAV-p-MAEGI was administered only into the local tumors at the indicated times (blue arrows). f-g, Growth curves of E0771-dCas9-VP64 local (f) and distant (g) tumors in mice treated by AAV-Vector (n = 18), or AAV-p-MAEGI (n = 18). Inset, pie charts detailing the response rates for each treatment. (f) Two-way ANOVA: AAV-p-MAEGI vs. AAV-Vector, p < 0.0001. (g) Two-way ANOVA: AAV-p-MAEGI vs. AAV-Vector, p = 0.0003. h, Schematics of dual AAV CRISPRa system for MAEGI delivery into tumor-bearing mice. i, Growth curves of E0771 syngeneic tumors in mice treated by intratumoral injection of PBS (n = 10 mice), AAV-dCas9 + AAV-Vector (n = 10), or dual AAV-p-MAEGI (n = 10) at the indicated time points (blue arrows). Two-way ANOVA: AAV-dCas9+AAV-Vector vs. PBS, p = 0.0083; dual AAV-p-MAEGI vs. PBS, p < 0.0001; dual AAV-p-MAEGI vs. AAV-dCas9+AAV-Vector, p = 0.0002. Error bars: All data points in this figure are presented as mean ± s.e.m. Asterisks: * p < 0.05, ** p < 0.01, *** p < 0.001.
Additional supporting data: Supplementary Figures 3, 4, and 5
We treated C57BL/6J mice bearing orthotopic E0771 TNBC with AAV-p-MAEGI, along with AAV-Vector and PBS controls. While the AAV-Vector itself showed anti-tumor effects compared to PBS, AAV-p-MAEGI exhibited dramatic efficacy compared to both AAV-Vector and PBS (Fig. 4b and Supplementary Fig. 3d). We determined the complete response (CR) and near-complete response (nCR) rates for each treatment (Methods), which revealed a combined nCR and CR rate of 4% for PBS, 0% for AAV-Vector, and 44% for AAV-p-MAEGI (Fig. 4c). Three months after the initial tumor transplantation, we re-challenged 9 AAV-p-MAEGI treated mice that had undergone complete response. All (9/9) mice completely rejected the E0771 tumor re-challenges (Fig. 4d). The complete response of AAV-p-MAEGI treated mice retained a long-term complete remission for over 180 days (Fig. 4d), indicating that AAV-p-MAEGI had induced potent and durable anti-tumor responses.
The efficacy of AAV-p-MAEGI was abolished in immunodeficient Rag1 −/− mice (Supplementary Fig. 3e), indicating that the adaptive immune system is essential for the anti-tumor effect of AAV-p-MAEGI. Consistent with this, we observed CD8+ T cell infiltration into tumors by histology and immunohistochemistry (Supplementary Fig. 4a,b). AAV-p-MAEGI treated tumors also showed reduced proliferation (Supplementary Fig. 4c,d). Given the importance of adaptive immunity in driving these anti-tumor responses, we wondered whether intratumoral delivery of AAV-p-MAEGI could elicit systemic anti-tumor immunity and affect distant sites (Fig. 4e). We utilized a syngeneic model of E0771 TNBC in which mice bear both “local” and “distant” breast tumors. After administration of AAV-p-MAEGI only to local tumors, we observed significant anti-tumor effects against both local tumors and distant tumors that had not been directly infected by AAV-p-MAEGI (Fig. 4f, g and Supplementary Table 1). A 67% CR rate was observed at distant tumors following AAV-p-MAEGI treatment (Fig. 4g). The observation of an abscopal effect indicates that AAV-p-MAEGI had galvanized systemic anti-tumor immunity.
A critical step in demonstrating the potential of MAEGI as a therapeutic modality is to deliver all of the CRISPRa components in vivo. To assess the infection rate of intratumoral AAV delivery, we repeated the experiments using titer-matched AAVs expressing GFP. Four days following intratumoral AAV injection, cancer cells were successfully transduced in all tumors injected, with an infection rate of 4.2 ± 0.51% (mean ± s.e.m.) (Supplementary Fig. 4e). In contrast, pan-immune (CD45+) cells exhibited a lower infection rate of 0.74 ± 0.25% (Supplementary Fig. 4e). At day 12 following two AAV injections, the infection rate increased to 30.4 ± 5.1% for tumor cells (Supplementary Fig. 4f). The off-target infection rate for the spleen, liver, lung and heart in the same cohort of mice was 0 ± 0%, 0.04 ± 0.04%, 0.18 ± 0.06% and 0.11 ± 0.08%, respectively (Supplementary Fig. 4g). In addition, we did not observe any severe adverse effects with MAEGI treatment (Supplementary Table 1).
To enable delivery of all MAEGI components into tumors, we developed a dual AAV system in which one vector expresses dCas9 (AAV-dCas9) and a second vector expresses the sgRNA as well as MS2-p65-HSF1 (AAV-CRISPRa). To evaluate whether the dual AAV system can effectively activate target transcripts, we co-infected E0771 cells in vitro with both AAV-dCas9 and AAV-CRISPRa, and observed significantly increased expression of target transcripts 5–7 days post-infection (Supplementary Fig. 5a). We then examined the therapeutic efficacy of dual AAV-p-MAEGI by co-injection of AAV-dCas9 and AAV-p-MAEGI into unmodified E0771 tumors (Fig. 4h). Similar to the results obtained using tumor cells with lentivirally transduced dCas9-VP64, the co-injection of AAV-dCas9 and AAV-p-MAEGI showed significant anti-tumor efficacy compared to both AAV-dCas9+AAV-Vector and PBS, while the co-injection of AAV-dCas9 plus AAV-vector showed moderate anti-tumor effects compared to PBS (Fig. 4i and Supplementary Table 1). Taken together, these data demonstrate that by using a dual AAV system, all components of MAEGI can be efficiently delivered into tumors in vivo, resulting in anti-tumor responses.
AAV-p-MAEGI remodels the tumor immune microenvironment
To examine the adaptive immune responses over a time-course, we performed flow cytometry analysis of tumor-infiltrating T cell populations (Supplementary Fig. 5b). We found that AAV-p-MAEGI treatment significantly augmented CD4+ and CD8+ T cell infiltration in tumors from day 29 onwards, as compared to AAV-Vector or PBS treatment (Fig. 5a, b). We also examined other immune cell populations at day 19, as this is the time point with initial observation of tumor regression (Supplementary Fig. 5c). AAV-p-MAEGI treatment increased the fraction of MHC-II+ antigen-presenting cells over AAV-Vector treatment (Fig. 5c), with a trend of increased dendritic cells (Supplementary Fig. 5d), but no significant changes in macrophages, monocytes, or neutrophils (Supplementary Fig. 5e–g and Supplementary Table 1). These results indicate that AAV-p-MAEGI treatment increased T cell infiltration into tumors and altered the composition of tumor-infiltrating immune cells (TIICs), ultimately resulting in tumor regression.
Figure 5. AAV-p-MAEGI treatment promotes infiltration of tumor-reactive immune populations.
a, Flow cytometry time-course quantification of CD4+ T cells out of total tumor cells from mice treated with PBS, AAV-Vector, or AAV-p-MAEGI. Two-tailed Mann-Whitney test: AAV-p-MAEGI (n = 4 mice) vs. PBS (n = 4) at dpi 9 (p = 0.6857), AAV-p-MAEGI (n = 13) vs. PBS (n = 6) at dpi 19 (p = 0.0047), AAV-p-MAEGI (n = 14) vs. PBS (n = 10) at dpi 29 (p = 0.0048), AAV-p-MAEGI (n = 13) vs. PBS (n = 11) at dpi 36 (p = 0.0015); AAV-p-MAEGI (n = 4) vs. AAV-Vector (n = 4) at dpi 9 (p = 0.2000), AAV-p-MAEGI (n = 13) vs. AAV-Vector (n = 10) at dpi 19 (p = 0.2569), AAV-p-MAEGI (n = 14) vs. AAV-Vector (n = 8) at dpi 29 (p = 0.0159), AAV-p-MAEGI (n = 12) vs. AAV-Vector (n = 10) at dpi 36 (p = 0.0422). b, Flow cytometry time-course quantification of CD8+ T cells out of total tumor cells from mice treated with PBS, AAV-Vector, or AAV-p-MAEGI. Two-tailed Mann-Whitney test: AAV-p-MAEGI (n = 4 mice) vs. PBS (n = 4) at dpi 9 (p = 0.6857), AAV-p-MAEGI (n = 13) vs. PBS (n = 6) at dpi 19 (p = 0.0047), AAV-p-MAEGI (n = 14) vs. PBS (n = 10) at dpi 29 (p = 0.0005), AAV-p-MAEGI (n = 13) vs. PBS (n = 11) at dpi 36 (p = 0.0014); AAV-p-MAEGI (n = 4) vs. AAV-Vector (n = 4) at dpi 9 (p = 0.4857), AAV-p-MAEGI (n = 13) vs. AAV-Vector (n = 10) at dpi 19 (p = 0.2569), AAV-p-MAEGI (n = 13) vs. PBS (n = 11) at dpi 29 (p = 0.0064), AAV-p-MAEGI (n = 12) vs. AAV-Vector (n = 10) at dpi 36 (p = 0.0200). c, Percentage of CD45+MHC-II+ antigen presenting cells in tumors from mice treated with PBS (n = 6 mice), AAV-Vector (n = 10), or AAV-p-MAEGI (n = 12). Two-tailed Mann-Whitney test: AAV-Vector vs. PBS, p = 0.0225; AAV-p-MAEGI vs. PBS, p = 0.0001; AAV-p-MAEGI vs. AAV-Vector, p = 0.0147. d, Schematic of experimental design using ELISPOT to assess anti-tumor specificity of the immune response elicited by AAV-p-MAEGI or AAV-Vector. e,g Representative images of Ifn-γ ELISPOTs on splenocytes (e) or tumor-infiltrating immune cells (TIICs) (g) from AAV-Vector or AAV-p-MAEGI treated mice bearing E0771-dCas9-VP64 tumors, stimulated with either normal primary cells from wildtype C57BL/6J fat pad tissue (Normal) or E0771 cells (Cancer). f, Quantification of Ifn-γ ELISPOTs on splenocytes from AAV-Vector (n = 9 mice) or AAV-p-MAEGI (n = 9) treated mice bearing E0771-dCas9-VP64 tumors, stimulated with primary C57BL/6J normal cells or cancer cells. Two sided unpaired t-test: AAV-p-MAEGI vs. AAV-Vector stimulated by normal cells, p = 0.1667; AAV-p-MAEGI vs. AAV-Vector stimulated by E0771 cancer cells, p = 0.0057; Cancer vs. Normal cells stimulation within AAV-Vector group, p = 0.0004; Cancer vs. Normal cell stimulation within AAV-p-MAEGI group, p = 0.0002. Results shown are aggregated from 2 independent experiments. h, Quantification of Ifn-γ ELISPOTs for TIICs from AAV-Vector (n = 6) or AAV-p-MAEGI (n = 6) treated mice bearing E0771-dCas9-VP64 tumors that were stimulated with primary C57BL/6J normal cells or cancer cells. Two-sided unpaired t-test: AAV-p-MAEGI vs. AAV-Vector stimulated by Normal cells, p = 0.0577; AAV-p-MAEGI vs. AAV-Vector stimulated by cancer cells, p < 0.0001; Cancer vs. Normal cells stimulation for AAV-Vector group, p = 0.0175; Cancer vs. Normal cells stimulation for AAV-p-MAEGI group, p < 0.0001. Error bars: All data points in this figure are presented as mean ± s.e.m. Asterisks: * p < 0.05, ** p < 0.01, *** p < 0.001.
Additional supporting data: Supplementary Figure 5
Given these findings, we wondered if the adaptive immune responses induced by AAV-p-MAEGI were tumor-specific. To test this, we performed ELISPOT assays on both splenocytes and fluorescence activated cell sorting (FACS)-isolated TIICs, using either E0771 cancer cells or matched normal tissues (primary C57BL/6J wildtype cells isolated from mammary fat pad) as the antigen source (Fig. 5d). In both AAV-Vector and AAV-p-MAEGI conditions, the ELISPOT assays showed significantly more IFN-γ-producing splenocytes and TIICs after stimulation by E0771 tumor cells, as compared to stimulation with normal primary cells (Fig. 5e–h). Importantly, when stimulated with E0771 cancer cells, AAV-p-MAEGI treated mice had significantly higher frequencies of IFN-γ-secreting immune cells than AAV-Vector treated mice. This effect was particularly pronounced in TIICs, where AAV-p-MAEGI significantly augmented the frequencies of IFN-γ-secreting immune cells in response to E0771 stimulation, but not to matched normal cells (Fig. 5g,h). The anti-tumor specificity of AAV-p-MAEGI was similarly observed in splenocytes (Fig. 5e,f and Supplementary Table 1). The ELISPOT data therefore suggest that tumor-antigen specific immune responses were elicited by AAV-p-MAEGI.
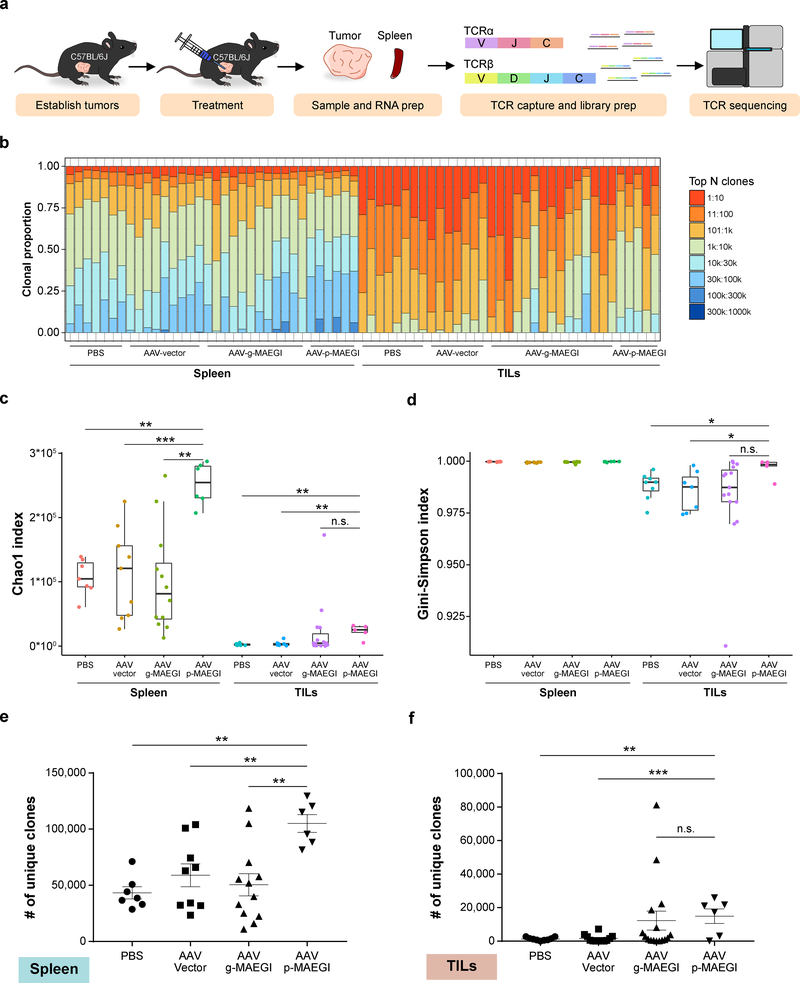
To understand the repertoire of T cells recruited by MAEGI, we performed T cell receptor sequencing (TCR-seq) to profile the TCR repertoire in mice treated with AAV-p-MAEGI, AAV-g-MAEGI, AAV-Vector or PBS (Fig. 6a and Methods). We assessed TCR clonal proportions and calculated various metrics (Fig. 6b–d). In comparison to PBS or AAV-Vector, AAV-p-MAEGI significantly increased TCR diversity in both spleens and TILs as measured by the Chao1 index 32 (Fig. 6c and Supplementary Table 2). In comparison to AAV-g-MAEGI treated mice (genome-wide AAV-MAEGI), splenocytes from AAV-p-MAEGI treated mice (precision AAV-MAEGI) had higher Chao1 indices, with no significant difference among TILs (Fig. 6c). Next, we calculated the Gini-Simpson indices, which quantify the evenness of TCR clonal abundances 32. We found that TILs from AAV-p-MAEGI treated mice had significantly higher Gini-Simpson indices compared to PBS or AAV-Vector treated mice, but not compared to AAV-g-MAEGI treated mice (Fig. 6d), indicating that MAEGI is associated with the recruitment of diverse T cell populations. Intrigued by the increased TCR diversity and clonal evenness among TILs from AAV-p-MAEGI treated mice, we wondered if these changes were due to differences in T cell infiltration, in accordance with the FACS data. We examined the number of unique CDR3 clonotypes identified in each sample (i.e. TCR richness) (Supplementary Fig. 6) 32, finding that AAV-p-MAEGI mice had significantly more unique clonotypes than PBS or AAV-Vector samples in both spleen and TIL samples (Fig. 6e,f). Collectively, the TCR-seq data affirmed our conclusion from the FACS analysis that AAV-p-MAEGI significantly enhanced T cell infiltration into tumors, while additionally revealing the richness and diversity of these tumor-infiltrating T cells.
Figure 6. Interrogation of the T cell repertoire in MAEGI-treated mice by TCR sequencing.
a, Schematics of TCR-seq experiment design. Mice bearing E0771-dCas9-VP64 tumors were treated with PBS, AAV-Vector, AAV-g-MAEGI (genome-wide), or AAV-p-MAEGI (exome-guided). Spleens and tumors were harvested and subjected to RT-based TCRα/TCRβ capture followed by Illumina sequencing. In spleen samples, n = 7 (PBS-treated mice), n = 9 (AAV-Vector treated mice), n = 12 (AAV-g-MAEGI treated mice), n = 6 (AAV-p-MAEGI treated mice) from two independent experiments. In the tumor-infiltrating lymphocytes (TILs) samples, n = 7 (PBS-treated mice), n = 7 (AAV-Vector treated mice), n = 15 (AAV-g-MAEGI treated mice), n = 5 (AAV-p-MAEGI treated mice) from two independent experiments. b, Global clonal proportion plot showing the relative frequencies of the top N clones. c, Boxplot of Chao1 indices (TCR diversity) for each group. Unpaired two-tailed Mann-Whitney test, spleens: AAV-p-MAEGI vs. PBS, p = 0.0012; AAV-p-MAEGI vs. AAV-Vector, p = 0.0008; AAV-p-MAEGI vs. AAV-g-MAEGI, p = 0.0013. Unpaired two-tailed Mann-Whitney test, TILs: AAV-p-MAEGI vs. PBS, p = 0.0031; AAV-p-MAEGI vs. AAV-Vector, p = 0.0051; AAV-p-MAEGI vs. AAV-g-MAEGI, p = 0.0983. d, Boxplot of Gini-Simpson indices (TCR evenness) for each group. Two-tailed Mann-Whitney test, TILs: AAV-p-MAEGI vs. PBS, p = 0.0295; AAV-p-MAEGI vs. AAV-Vector, p = 0.0303 AAV-p-MAEGI vs. AAV-g-MAEGI, p = 0.0526. e-f, Dot plots of the number of unique clonotypes identified (TCR richness) in each spleen sample (e) or TILs sample (f), compared across treatment conditions. Statistical significance was assessed by unpaired two-tailed t-test. Spleen: AAV-p-MAEGI vs. PBS, p = 0.002; AAV-p-MAEGI vs. AAV-Vector, p = 0.006 AAV-p-MAEGI vs. AAV-g-MAEGI, p = 0.0023. TILs: AAV-p-MAEGI vs. PBS, p = 0.002; AAV-p-MAEGI vs. AAV-Vector, p = 0.0006, AAV-p-MAEGI vs. AAV-g-MAEGI, p = 0.7875. All boxplots are Tukey boxplots (interquartile range (IQR) boxes with 1.5 × IQR whiskers). Error bars: Data points in this figure are presented as mean ± s.e.m. Asterisks: * p < 0.05, ** p < 0.01, *** p < 0.001.
Additional supporting data: Supplementary Figure 6
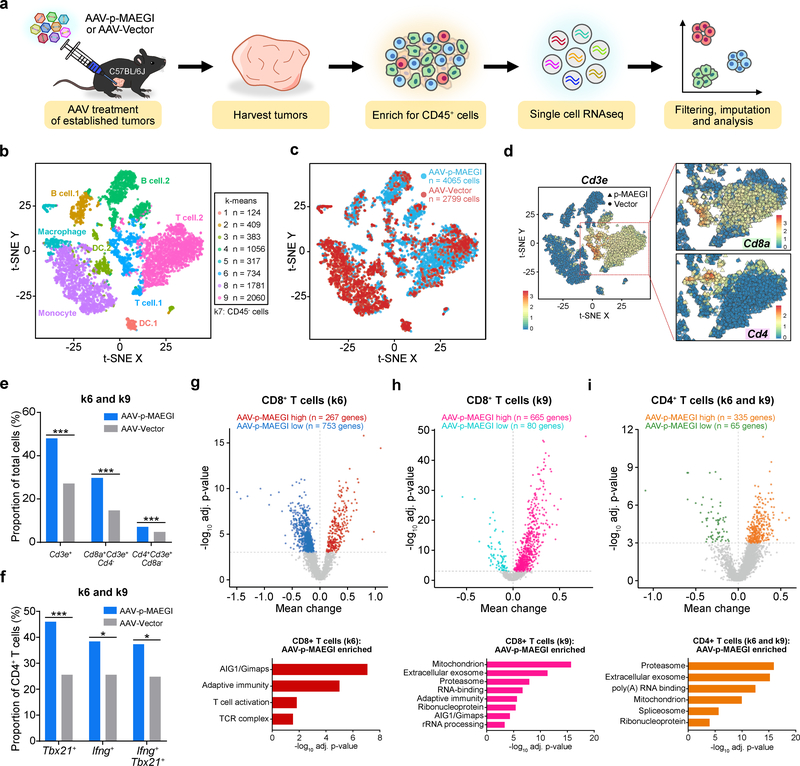
To further investigate the effect of AAV-p-MAEGI on the tumor microenvironment, we performed single cell RNA sequencing (scRNA-seq) to simultaneously profile the composition and transcriptome of TIICs (Fig. 7a and Methods). We isolated total TIICs by FACS of CD45+ cells and performed scRNA-seq, recovering a total of 4,381 cells from AAV-p-MAEGI and 3,482 cells from AAV-Vector treated mice (n = 3 mice pooled for each condition, Supplementary Table 2). We performed imputation and k-means clustering to identify a total of 9 clusters. Analysis of differentially expressed genes in each cluster revealed the cellular identities of each cluster (Fig. 7b and Supplementary Fig. 7a–f). After excluding a population of cells (k-means cluster 7, or k7) negative for CD45 gene expression (encoded by Ptprc) (Supplementary Fig. 7b), the final dataset comprised the transcriptomes of 4,065 TIICs from AAV-p-MAEGI and 2,799 TIICs from AAV-Vector treated mice (Fig. 7c).
Figure 7. Single cell RNA-seq profiling of immune populations in the tumor microenvironment.
a, Schematic of experimental design for single cell RNA-seq (scRNA-seq) analysis of immune populations. Cells from n = 3 independent mice (AAV-Vector) or n =3 mice (AAV-p-MAEGI) were pooled together for scRNA-seq library preparation. b, Scatter plot of t-SNE dimensional reduction, with cells colored by k-means cluster (cell numbers detailed on right). Putative cell types associated with each cluster are annotated. Cluster 7 (k7) is composed of CD45- cells and was excluded from further analyses (see Supplementary Fig. 7). c, Scatter plot of t-SNE dimensional reduction as in (b), colored by treatment group. scRNA-seq cell numbers after preprocessing: AAV-p-MAEGI, n = 4,065 cells; AAV-Vector, n = 2,799 cells. d, Identification of T cell populations from scRNA-seq. Left, t-SNE dimensional reduction, with cells colored by Cd3e expression. Right, zoomed-in view of clusters k6 and k9, colored by Cd8a expression (top) or Cd4 expression (bottom). e, Quantification of T cell populations in k6 and k9, out of total cells (AAV-p-MAEGI, n = 4,065 cells; AAV-Vector, n = 2,799). Two-sided Fisher’s exact test: p < 0.0001 for CD3+ T cells, CD8+ T cells, and CD4+ T cells. f, Quantification of putative CD4+ T cell subpopulations in k6 and k9 out of total CD4+ T cells (AAV-p-MAEGI, n = 289 cells; AAV-Vector, n = 133). Two-sided Fisher’s exact test: p < 0.0001 for Tbx21+ cells, p = 0.0111 for Ifng+ cells, and p = 0.0109 for Tbx21+Ifng+ cells. g-i, Top, differential expression volcano plots of CD8+ T cells in k6 (g), CD8+ T cells in k9 (h), and CD4+ T cells in k6 and k9 (i), comparing AAV-p-MAEGI to AAV-Vector. n = number of differentially expressed genes, assessed by two-sided Mann Whitney test with Benjamini-Hochberg multiple hypothesis correction. Bottom, gene ontology enrichment analysis of upregulated genes in T cells from AAV-p-MAEGI mice, determined by Fischer’s exact test with Benjamini-Hochberg multiple hypothesis correction. Error bars: All data points in this figure are presented as mean ± s.e.m. Asterisks: * p < 0.05, ** p < 0.01, *** p < 0.001.
Additional supporting data: Supplementary Figure 7
Given that the FACS and TCR-seq analyses had revealed increased T cell tumor infiltration with AAV-p-MAEGI compared to AAV-Vector, we sought to investigate whether these findings were recapitulated by scRNA-seq. To perform in silico analysis on T cell populations, we filtered by mRNA transcript expression levels (“mRNA-gated”) on cells in k6 or k9, as these clusters comprise the cells that robustly express Cd3e, Cd4, and Cd8a mRNA (Fig. 7d). Examining the Cd3e+ cells within k6 and k9, we then mRNA-gated these cells into Cd8a + Cd4 – and Cd4 + Cd8a – populations. Visualization of these cell groups revealed clear separation of putative CD8+ T cells (defined by Cd8a + Cd3e + Cd4 – mRNA gating) from CD4+ T cells (defined by Cd4 + Cd3e + Cd8a – mRNA gating) (Fig. 7d). We quantified the relative abundance of these different T cell populations in k6 and k9, finding that AAV-p-MAEGI mice had significantly more Cd3e + T cells, putative CD8+ T cells, and putative CD4+ T cells (Fig. 7e). Furthermore, among these putative CD4+ T cells, we found that AAV-p-MAEGI treated mice had increased abundance of Tbx21 +, Ifng +, and Tbx21 + Ifng + cells, indicative of TH1 cells (Fig. 7f and Supplementary Table 2). Thus, the single cell transcriptomic analyses on CD45+ TIICs reaffirmed our prior observations by FACS and TCR-seq that AAV-p-MAEGI promotes T cell infiltration into tumors.
To investigate the transcriptomic changes in tumor-infiltrating T cells following AAV-p-MAEGI, we performed differential expression analyses to compare T cells from AAV-p-MAEGI and AAV-Vector treated mice. Among the putative CD8+ T cells in k6 (characterized by higher Gzmb expression compared to k9) (Fig. 7g), we found 267 upregulated and 753 downregulated genes in AAV-p-MAEGI treated mice compared to AAV-Vector (Fig. 7g and Supplementary Table 2). Gene ontology analysis revealed enrichment of several T cell signatures within the upregulated gene set, including adaptive immunity, T cell activation, and the T cell receptor complex. The strongest enriched gene set was the AIG1 family of GTPases, particularly Gimap genes (GTPases of immunity-associated protein), whose associated proteins are involved in T cell activation and survival33. As for the putative CD8+ T cells in k9, 665 genes were upregulated and 80 genes downregulated with AAV-p-MAEGI treatment (Fig. 7h). Enriched gene ontologies in the upregulated gene set included mitochondrion, exosome, proteasome, RNA-binding, adaptive immunity, ribonucleoprotein, AIG1/Gimaps, and rRNA processing. The same analysis was performed on putative CD4+ T cells, revealing 335 upregulated and 65 downregulated genes with AAV-p-MAEGI treatment (Fig. 7i). Collectively, the scRNA-seq data reaffirmed that AAV-p-MAEGI treatment increases T cell infiltration into tumors, and further revealed significant transcriptomic changes in TILs following MAEGI treatment.
Discussion
Tumor cells harbor a multitude of mutated, partially truncated, or amplified genes that are potentially immunogenic 17,34,35. However, these mutant products might not be expressed at levels sufficient to elicit an effective T cell-mediated response, and cancer cells often downregulate antigen presentation to escape immune recognition 19. Several approaches have been described to increase the immunogenicity of tumor cells by in situ manipulation of the tumor microenvironment 36–43. A key advantage of MAEGI is its versatility, as CRISPR sgRNA libraries can be easily customized to augment the expression of any tumor antigens in a multiplexed manner 27,44,45, as well as alleged noncoding transcripts, immunomodulatory molecules, cytokines, chemokines, or transcription factors, either individually or in combinations.
With high-throughput exome sequencing, personalized immunotherapy targeting patient-specific mutations is now feasible 46. Synthesis of mutant peptides or transcripts has been used for personal neoantigen vaccine development 21–24, but the scalability of these approaches is limited. We reasoned that recently developed CRISPRa technologies could radically increase the scale of cancer antigen targeting. By targeting mutant genes for overexpression, MAEGI utilizes host cellular machinery to process and present these abnormal antigens in higher abundance. Our data demonstrates that MAEGI increases target gene expression, increases antigen presentation, increases T cell effector function, recruits CD4+ and CD8+ T cells to the tumor microenvironment, and promotes both local and systemic anti-tumor T cell responses, thereby eliciting potent anti-tumor immunity. Distinct from current neoantigen vaccines, AAV-MAEGI represents an orthogonal immunotherapy that directly targets the interplay between tumor cells and the immune microenvironment, inducing longer-term immune memory.
Of note, AAVs are now an FDA-approved therapeutic transgene vector (see voretigene neparvovec)47,48. As a potential viral gene therapy in oncology, administration of AAV-p-MAEGI could be performed to primary tumors, metastatic sites, or to residual disease as a surgery adjuvant. Importantly, since AAV-MAEGI activates endogenous genes, the acquisition of additional mutations by the tumor, which may have driven relapse by nullifying existing anti-tumor immunologic memory, can still be targeted by MAEGI, as long as the library contains sgRNAs targeting those genes. With a sufficiently diverse sgRNA library, AAV-g-MAEGI or AAV-p-MAEGI can potentially remain efficacious even in the face of continual tumor evolution and immunoediting. It is likely that neutralizing antibodies will be developed against AAV, dCas9, or other components of MAEGI. However not all the viruses are neutralized, as we found that it is important to have consecutive treatments to maintain efficacy. Local high dose administration may overcome this issue as long as a fraction of cells are transduced, thereby becoming hyper-presenters. Other AAV subtypes and other types of catalytically dead RNA-guided nucleases can also be used to reduce neutralization.
In summary, here we demonstrate that direct activation of endogenous mutant genes through CRISPRa amplifies the “non-self” signals of tumor cells, inducing potent anti-tumor adaptive immunity. The various forms of MAEGI, including AAV-g-MAEGI and AAV-p-MAEGI, as well as any future derivatives of endogenous gene activation-based therapies, offer an orthogonal modality of cancer immunotherapy that may serve either as single agents or in concert with other therapeutic modalities. Future clinical translation of MAEGI will require the exclusion of potentially harmful genes when overexpressed, optimization of composition and design, and toxicity evaluation in animal models, investigational new drug (IND)-enabling studies, before a Phase I clinical trial.
Online Methods
Institutional Approval
This study has received institutional regulatory approval. All recombinant DNA and biosafety work were performed under the guidelines of Yale Environment, Health and Safety (EHS) Committee with an approved protocol (Chen-rDNA-15–45). All animal work was performed under the guidelines of Yale University Institutional Animal Care and Use Committee (IACUC) with approved protocols (Chen-2015–20068, Chen-2018–20068), and was consistent with the Guide for Care and Use of Laboratory Animals, National Research Council, 1996 (Institutional Animal Welfare Assurance No. A-3125–01).
Cell lines
E0771 was from CH3. Pan02 and B16F10 were from ATCC. HEK293FT was from ThermoFisher. Various derivative lines were made in this study as described below. All cell lines tested negative for mycoplasma.
Lentivirus production
Mouse CRISPR activation plasmid libraries (lenti-U6-mSAM-EFS-Puro) were expanded by electroporation49. An estimated library coverage of >100× (>100 colonies per sgRNA) was achieved in electroporation, and the coverage of sgRNAs was subsequently sequence-verified by Illumina sequencing. For lentivirus production, 20 μg of plasmids of lenti-EF1a-NLS-dCas9-VP64-P2A-Blast, lenti-EF1a-MS2-p65-HSF1-P2A-Hygro, lenti-U6-sgRNA(ms2)-EFS-Puro vector or library, together with 10 μg of pMD2.G and 15 μg of psPAX2 were co-transfected into HEK293FT cells in a 150 mm-dish at 80–90% confluency using 130 μg polyethyleneimine (PEI) as the transfection reagent. 6–12 hours later, the media was replaced by fresh DMEM+10%FBS. Virus supernatant was collected 48 h and 72 h post-transfection, centrifuged at 1500 g for 10 min and passed through 0.45-μm filter to remove the cell debris; aliquoted and stored at −80°C. Library virus was titrated by infecting E0771 cells followed by the selection under 5 μg/ml puromycin.
Antigen presentation assay using an OVA-expressing cell line
A lentiviral mChOva-expressing vector (Lenti-pGK-mCherry-2A-Ova-WPRE; Addgene, #129600) was generated via Gibson Assembly. E0771 cells were then transduced with mChOva-expressing lentiviruses. 2–3 days post-transduction, mChOva-expressing cells (E0771-mChOva) were FACS sorted on a BD FACSAria. OVA expression was further confirmed by staining these cells with antibody specific to the complex of SIINFEKL peptide bound to class I major histocompatibility complex (APC anti-mouse H-2Kb bound to SIINFEKL Antibody). Then, E0771-mChOva cells were transduced lentiviral lenti-EF1a-NLS-dCas9-VP64–2A-Blast and lenti-EF1a-MS2–p65–HSF1–2A-Hygro, followed by 7 days of selection under the pressure of 10 μg/ml blasticidin and 500 μg/ml hygromycin to generate cells stably expressing dCas9-VP64 and MS2-P65-HSF1 (E0771-mChOva-dCa9-MPH).
To activate the expression and thus presentation of mChOva, PGK-promoter targeting sgRNAs were designed and cloned into Lenti: U6-sgRNA(ms2)-EFS-Puro and AAV:U6p-sgSapI-EF1a-MS2-p65-HSF1-WPRE; Addgene, #129599) vector. After lentivirus or AAV production, E0771-mChOva-dCa9-MPH were infected with these lentiviruses or AAVs to activate mChOva transcription. 3–7 days after viral transduction, cell surface SIINFEKL-H-2Kb was stained with PE anti-mouse H-2Kb bound to SIINFEKL Antibody (AAV experiments) or APC anti-mouse H-2Kb bound to SIINFEKL Antibody (lentiviral experiments). The genomic mean intensity of APC-SIINFEKL-H-2Kb/PE-SIINFEKL-H-2Kb were used to determine cell surface presentation of SIINFEKL.
Co-culture of MAEGI-treated cells and effector T cells
SIINFEKL-H-2Kb -specific naive CD8 T cells were isolated from OT-I mice using Naive mouse CD8 T cell kit (Miltenyi). Before use, the plate was coated with anti-CD3 (5 μg/ml in PBS) at 37°C for 1 h and then washed twice with PBS. Then naive T cells were cultured in the anti-CD3 coated plate using cRPMI (RPMI1640 + 10% FBS + 2 mM L- Glutamine + 49 nM b-mercaptoethanol + 100 U Pen/Strep) supplemented with 1 μg/ml anti-CD28, 2 ng/ml recombinant mouse IL-2, and 2 ng/ml recombinant mouse IL-12. 3 days later, the effector T cells were cultured in cRPMI supplemented with 2 ng/ml recombinant mouse IL-2 and 5 ng/ml recombinant mouse IL-7 for another 2 days. E0771-mChOva-dCa9-MPH cells transduced with vector or PGK-targeted sgRNAs were seeded in 24-well plates at concentration of 2×105 cells per well. 2–4 hours later, OT-1 T cells were counted, resuspended in cRPMI supplemented with IL-2, and added to tumor cells seeded plate at ratios of Effector : Tumor (E:T) = 0.5, or 1. For killing and apoptosis analysis, the effector T cells and E0771 cells were co-cultured for 24 hrs. All the samples were collected and stained with anti-CD8a Apc/Cy7 to exclude the T cells. The dead and apoptotic cancer cells were analyzed using PE Annexin V Apoptosis Detection Kit with 7-AAD. For interferon detection, 5 μg/ml Brefeldin A was added and co-cultured for 3 h. Then, samples were collected, and stained with anti-CD45 PerCP/Cy5.5 and anti-CD8a Apc/Cy7 for 30 min. After washing twice, the cells were fixed with BD Cytofix buffer for 10 minutes on ice and washed with Cytoperm/Wash Buffer. Cells were then subjected to intracellular staining with anti-IFNγ APC for 30 minutes on ice. After washing once with Cytoperm/Wash Buffer, the samples were resuspended in MACS Buffer and analyzed via flow cytometry.
Generation of mSAM library- and vector- transduced cells
E0771 cells stably expressing dCas9-VP64 and MS2-P65-HSF1 (E0771-dCas9-VP64-MPH) were generated by transducing lentiviral lenti-EF1a-NLS-dCas9-VP64–2A-Blast and lenti-EF1a-MS2–p65–HSF1–2A-Hygro into E0771 cells, followed by 7 days of selection under the pressure of 10 μg/ml blasticidin and 500 μg/ml hygromycin. E0771-dCas9-VP64-MPH cells were then infected with lentiviral mSAM-library or vector to obtain mSAM-transduced E0771 (E0771-SAM) or vector-transduced E0771 (E0771-Vector). 1 × 108 cells were initially infected by lentiviral mSAM sgRNAs library at a calculated MOI of 0.2 with a minimal representation of 200× transduced cells per sgRNA, as described previously 49. Lentivirus-infected cells were cultured at 37°C more than 1 day before replacing with 5 μg/ml puromycin containing media, and the transduced cells were drug-selected for 7 days before use.
Sequencing confirmation of SAM sgRNA library representation
Library transduced cells were subjected to genomic DNA (gDNA) extraction using standard molecular biology protocols. The sgRNA library readout was performed using a two-step PCR strategy, where the first PCR includes enough genomic DNA to preserve full library complexity and the second PCR adds appropriate sequencing adapters to the products from the first PCR.
PCR1 primers:
Forward AATGGACTATCATATGCTTACCGTAACTTGAAAGTATTTCG
Reverse CTTTAGTTTGTATGTCTGTTGCTATTATGTCTACTATTCTTTCCC
PCR was performed using Phusion Flash High Fidelity Master Mix (PF) (ThermoFisher). For reactions using PF, in PCR#1, the thermocycling parameters were: 98 °C for 1 min, 16–20 cycles of (98 °C for 1s, 62 °C for 5s, 72 °C for 30 s), and 72 °C for 2 min. In each PCR#1 reaction, we used 3 μg of total gDNA. A total of 8 to 12 of PCR#1 reactions was used to capture the full representation of the library in the cells. PCR#1 products for each biological sample were pooled and used for amplification with barcoded second PCR primers.
Barcoding second PCR primers used:
SF5 AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTTCGATCGTTACCATCTTGTGGAAAGGACGAAACACCG
SF6 AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTATCGATTCCTTGGTTCTTGTGGAAAGGACGAAACACCG
SR1 CAAGCAGAAGACGGCATACGAGATAAGTAGAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTTCTACTATTCTTTCCCCTGCACTGT
SR2 CAAGCAGAAGACGGCATACGAGATACACGATCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTATTCTACTATTCTTTCCCCTGCACTGT
SR3 CAAGCAGAAGACGGCATACGAGATCGCGCGGTGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTGATTCTACTATTCTTTCCCCTGCACTGT
SR4 CAAGCAGAAGACGGCATACGAGATCATGATCGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTCGATTCTACTATTCTTTCCCCTGCACTGT
SR5 CAAGCAGAAGACGGCATACGAGATCGTTACCAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTCGATCTCTACTATTCTTTCCCCTGCACTGT
SR6 CAAGCAGAAGACGGCATACGAGATTCCTTGGTGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTTCTACTATTCTTTCCCCTGCACTGT
For reactions using Phusion Flash, in PCR#2, the thermocycling parameters were: 98 °C for 1 min, 18–24 cycles of (98 °C for 1s, 60 °C for 5s, 72 °C for 30s), and 72 °C for 2 min. Second PCR products were pooled and then normalized for each biological sample before combining uniquely barcoded separate biological samples. The pooled product was then gel purified from a 2% E-gel EX (Life Technologies) using the QIAquick Gel Extraction Kit (Qiagen). The purified pooled library was then quantified with a gel-based method using the Low-Range Quantitative Ladder Life Technologies, dsDNA High-Sensitivity Qubit (Life Technologies), BioAnalyzer (Agilent) and/or qPCR. Diluted libraries with 5–20% PhiX were sequenced with MiSeq, HiSeq 2500 or HiSeq 4000 systems (Illumina).
Raw single-end fastq read files were filtered and demultiplexed using Cutadapt 50. To remove sgRNA scaffold sequences downstream (i.e. 3’ end) of the sgRNA spacer sequences, we used the following command: cutadapt --discard-untrimmed -a GTTTTAGAGCTAGGCCAAC. As the forward PCR primers used to readout sgRNA representation were designed to have a variety of barcodes to facilitate multiplexed sequencing, we then demultiplexed these filtered reads with the following settings: cutadapt -g file:fbc.fasta --no-trim, where fbc.fasta contained the possible barcode sequences within the forward primers. Finally, to remove non-sgRNA sequences upstream (i.e. 5’ end) of the sgRNA spacers, we used the following command: cutadapt --discard-untrimmed –g GTGGAAAGGACGAAACACCG. Through this procedure, the raw fastq read files were trimmed to the 20 bp sgRNA spacer sequences. The 20 bp sgRNA spacer sequences from each demulitplexed sample were mapped the sgRNA spacers to the mSAM library using Bowtie 1.1.2 51: bowtie -v 2 --suppress 4,5,6,7 --chunkmbs 2000 –best. Using the resultant mapping output, we quantitated the number of reads that had mapped to each sgRNA within the library.
Mice
Mice of both sexes, between the ages of 5–12 weeks of age were used for the study. Various animals were used in this study. 5–10 week old C57BL/6J mice were used for experiments unless otherwise specified. Female mice were used for breast cancer (E0771) models. Male mice were used for pancreatic adenocarcinoma (Pan02) models. A mix of both male and female mice were used for melanoma (B16F10) models. All animals were housed in standard individually ventilated, pathogen-free conditions, with 12h:12h or 13h:11h light cycle, room temperature (21–23°C) and 40–60% relative humidity. Sample size determination (i.e. number of mice per treatment group) was based on similar tumor models in the field. When a cohort of animals was to receive different treatments, animals were randomized by 1) randomly assigning littermates to different groups prior to treatment, maximizing the evenness or representation of mice from different cages in each group, 2) group animals prior to treatment so that each group has even distribution of initial tumor sizes; and/or 3) random assignment of mice to minimize the effect of litter, and small differences in age, cage, or housing position, where applicable.
Tumorigenesis of E0771-SAM in C57BL/6J, Nude (_Foxn1_nu), or Rag1 −/− mice
5 × 106 of E0771-SAM or E0771-Vector tumor cells were injected into the orthotopic mammary fat pad of syngeneic 5–8 weeks old female C57BL/6J mice, Nude (_Foxn1_nu), or Rag1 −/− mice. Tumor sizes were measured every 3–4 days by caliper on the three diameters, and sizes were calculated with the formula: Vol = π/6 *x*y*z. Statistical significance of all tumor growth curves in the study was assessed by analysis of variance (two-way ANOVA), jointly considering the effect of treatment and the passage of time on tumor growth. For CD4 and CD8 T cell depletion, 200 ug anti-CD4 (GK1.5, BioXcell) and 200 ug anti-CD8 (YTS 169.4) were intraperitoneally injected into tumor-bearing C57BL/6J mice at dpi 7 and dpi 14. The successful depletion of CD8 T cells was confirmed by isolating PBMCs from mice at dpi 14 and 21 and analyzing the CD8+ T cell population with flow cytometry.
ELISPOT assay
Ifn-γ ELISPOT mouse kits (BD Biosciences) were used according to the manufacturer’s instructions. Briefly, 96-well filtration plates were coated overnight at 4°C with Ifn-γ capturing monoclonal antibody; the plates were then washed and blocked with RPMI-1640 medium containing 10% FBS for 2h at room temperature. Splenocytes and tumor-infiltrating immune cells (TIICs) isolated from different treatment groups were counted and resuspended in 10% FBS supplemented RPMI-1640. Splenocytes and TIICs were seeded into the 96-well filtration plate at concentrations of 1 × 106 cells/well and 2.5 × 104 cells/well, respectively. After mitomycin treatment, 5 × 104 C57BL/6J fat pad derived primary cells or E0771 tumor cells were added as different stimulators to stimulate the Ifn-γ secretion. The plates were cultured for approximately 45 hours at 37°C and 5% CO2. The co-culture were stopped by soaking in DI water, and washing three times with PBST; the plates were incubated with biotinylated detection antibody at room temperature for 2 h. Then, plates were incubated with HRP-conjugated streptavidin at room temperature for 1 h after washing three times with PBST. Spots were revealed using an AEC substrate reagent kit (BD Bioscience) at room temperature and counted using an Immunospot Reader (Cellular Technology). Statistical significance was assessed by two-tailed unpaired Welch’s t-test.
Generation of AAV-MAEGI
An AAV version of the CRISPR activation vector (AAV-CRISPRa vector, i.e. U6p-sgSapI-EF1a-MS2-p65-HSF1-sPA; Addgene, #129602), was generated by restriction cloning and Gibson assembly. The sgRNA libraries of mSAM were cloned into the above CRISPRa plasmid by linearization with SapI digestion and Gibson assembly. The purification and electroporation of Gibson products into Endura electrocompetent cells were performed as previously described 52, with at least 100x coverage of colonies represented per sgRNA. AAV was produced by co-transfecting HEK293FT cells with above AAV plasmids of CRISPR activation vector or library (AAV-mSAM), AAV2, AAV9, or AAV-DJ serotype plasmid and helper plasmid PDF6 to generate AAV-g-MAEGI. Briefly, 5.2 μg of AAV-vector, AAV-mSAM or other AAV-sgLib plasmid, 8.7 μg of plasmid AAV9 or AAV-DJ serotype, and 10.4 μg of pDF6 were mixed with PEI, room temperature 10–15 min before drop-wise adding them into HEK293FT cells in 150mm-dish at 80–90% confluency. Replicates collected from multiple dishes were pooled to enhance production yield. Cells were collected 72 h post transfection. For AAV purification, 1/10 volume of chloroform was added and the mixture was vigorously shaken for 1 h at 37°C. NaCl was added to a final concentration of 1 M and the mixture was shaken until dissolved and then pelleted at 20,000 g at 4°C for 15 min. The aqueous layer was discarded while the chloroform layer was transferred to another tube. PEG8000 was added to 10% (w/v) and shaken until dissolved. The mixture was incubated at 4°C for 1 h and then spun at 20,000 g at 4°C for 15 min. The supernatant was discarded and the pellet was resuspended in DPBS plus MgCl2 and treated with Benzonase (Sigma) and incubated at 37°C for 30 min. Chloroform (1:1 volume) was then added, shaken, and spun down at 12,000 g at 4°C for 15 min. The aqueous layer was isolated and passed through a 100 kDa MWCO (Millipore). The concentrated solution was washed with PBS and the filtration process was repeated. Genomic copy number (GC) of AAV was determined by real-time quantitative PCR using custom Taqman assays (ThermoFisher) targeted to the engineered U6 promoter.
Therapeutic testing of AAV-g-MAEGI in syngeneic tumor models
Syngeneic orthotopic breast tumors were established by transplanting 2 × 106 E0771-dCas9-VP64 cells into the mammary fat pad of 5–8 weeks old female C57BL/6J mice. 4, 10, 14, and 21 days after transplantation, 5–10 × 1010 GCs of AAV-g-MAEGI, AAV-Vector, or PBS were injected intratumorally to tumor-bearing mice. For the B16F10 melanoma model, 5 × 105 dCas9-VP64 expressing cancer cells were injected subcutaneously into the flank of C57BL/6J mice. 7, 10, 14, and 20 days after transplantation, 5–10 × 1010 GCs of AAV-g-MAEGI, AAV-Vector, or PBS were intratumorally administrated into tumor-bearing mice. For the pancreatic tumor model, 2 × 106 Pan02-dCas9-VP64 cancer cells were injected subcutaneously into the flank of C57BL/6J mice. 5, 13, 21, and 35 days after transplantation, 5–10 × 1010 GCs of AAV-g-MAEGI, AAV-Vector, or PBS were intratumorally administrated into tumor-bearing mice. Tumors were measured every 3–4 days using caliper and sizes were calculated with the formula: Vol = π/6 *x*y*z. Two-way ANOVA was used to compare growth curves between treatment groups.
Isolation of splenocytes and tumor infiltrating lymphocytes
Syngeneic breast tumors were established by orthotopically transplanting 2 × 106 E0771-dCas9-VP64 cells into the mammary fat pad of 5–8 weeks old female C57BL/6J mice. Tumor-bearing mice were randomly assembled into different treatment groups and treated with various reagents where applicable as described in the legends. Mice were euthanized at 9, 19, 29, or 36 days post-transplantation as indicated, tumors and spleens were collected and kept in ice-cold 2% FBS. For spleens, they were placed in ice-cold 2% FBS and mashed through a 100 μm filter. Splenocytes were washed once with 2% FBS. Tumors were minced into 1–3 mm size pieces using a scalper and then digested using 100 U/mL Collagenase IV for 30–60 min while stirred at 37°C. Tumor suspensions were filtered twice through 100 μm cell strainer, and once through 40 μm cell strainer to remove large bulk masses. RBCs were lysed with 1ml of ACK Lysis Buffer (Lonza) per spleen by incubating 2–5 mins at room temperature, which was followed by the dilution with 10 ml 2% FBS and pass through a 40 μm filter. Splenocytes were resuspended in 2% FBS buffer, counted for flow cytometry staining or RNA isolation. Single cell suspensions of tumors were used for flow cytometry staining, further FACS sorting, or Ficoll purification to obtain tumor-infiltrating lymphocytes. For Ficoll-Paque purification, single cell suspensions at density of (~107 cells/ml) were carefully layered onto Ficoll-Paque media (GE Healthcare) and centrifuged at 400g for 30 min. Cells at the interface were carefully collected, and washed twice with 2% FBS, counted, and used for RNA isolation or flow cytometry staining.
Flow cytometry
All antibodies for flow cytometry were purchased from Biolegend or eBiosciences. Single cell suspensions from tumors or spleens were prepared using a gentleMACS tissue dissociation system. The panels of antibodies used in the flow cytometry staining as follows: Panel 1: anti-CD45.2-APC/Cy7, anti-CD3-PE, anti-CD4-FITC, anti-CD8a-APC; Panel 2: anti-CD45-PerCP/Cy5.5, anti-CD3-FITC, anti-CD4-PE/Cy7; anti-CD8-APC/Cy7; or Panel 3: anti-CD45-APC/Cy7, anti-I-A/I-K-PerCP/Cy5.5, anti-CD11b-FITC, anti-CD11c-PE/Dazzle594; anti-Ly6c-APC; anti-F4/80-PE, anti-CD24-PE/Cy7. All flow antibodies were used at 1:100 dilutions for staining unless otherwise noted. For surface staining, cells were blocked with anti-Fc receptor anti-CD16/CD32, and then stained with surface marker antibodies in the staining buffer of 2% FBS in PBS on ice for 30 min. Samples were washed twice with 2% FBS in PBS before analysis. For intracellular staining, eBioscience™ Intracellular Fixation & Permeabilization Buffer Set was used to fix and permeabilize cells by following manufacturer’s instructions. Briefly, after the staining of surface makers, cells were resuspended in 100 μl Fixation/Permeabilization working solution, and incubated on ice for 10 min before washing with 1× Permeabilization buffer by centrifugation at 600 g for 5 min. Then cell pellet was resuspended in 50 μl 1× Permeabilization buffer with anti-Fc receptor anti-CD16/CD32, and incubated on ice for 10 min, before adding 50 μl 2× intracellular staining antibodies and incubated on ice for 30 min. After staining, cells were centrifuged at 600 g for 5 min, and washed twice with staining buffer before being analyzed or sorted on a BD FACSAria. The data was analyzed using FlowJo software (v9.9.4 or v10.3). A previously reported strategy was used to define the populations of monocytes, neutrophils, dendritic cell, and macrophages in tumor 53. Antibodies used for flow cytometry are listed in the Supplementary Table 1. Statistical significance was assessed by two-tailed Mann-Whitney test.
Fluorescence-activated cell sorting (FACS) isolation of CD45+ cells from tumors
Single tumor cell suspensions were prepared using the gentleMACS system with the method described above. Tumor cells were blocked using anti-Fc receptor anti-CD16/CD32. Live cells were distinguished from dead cells in flow cytometry by staining with LIVE/DEAD™ Fixable Near-IR Dead Cell Stain Kit following manufacturer’s instructions. Cells at a density of 107/ml were stained with DMSO dissolved live/dead staining dye and PerCP/Cy5.5 or APC conjugated CD45 antibody in PBS + 2% FBS, and incubated on ice for 30 min. Stained cells were washed three times before being analyzed and FACS isolated on a BD FACSAria. Tumor-infiltrating CD45+ pan-immune cells (TIICs) were isolated by gating on PerCP/Cy7+APC/Cy7- or APC+APC/Cy7- populations. Sorted TIICs were counted, and used for scRNA-seq and ELISpot analysis.
Immune cell profiling by single-cell RNA-seq
FACS sorted cells were collected in PBS+2%FBS and concentrated by centrifugation at 400 g for 5 min. Cell numbers and viabilities were assessed by trypan blue staining and analysis in Countess II FL Automated Cell Counter (Thermo). 10,000 CD45+ TIICs isolated from tumors were used for single-cell RNA-seq library prep by following the protocol from 10x Genomics.
Single cell RNA-seq data processing
Pan-immune cell scRNA-seq data were pre-processed using both established pipelines and custom scripts. Briefly, raw Illumina data files were processed by Cell Ranger 1.3 (10x Genomics) 54, using cellranger mkfastq to wrap Illumina’s bcl2fastq to correctly demultiplex sequencing samples and to convert barcode and read data to FASTQ files. Then, cellranger count was used to take FASTQ files and perform alignment to the mouse genome (mm10) 55, filtering, and UMI counting. Cells with fewer than 2,000 UMIs were subsequently filtered from analysis, leaving a final set of 7,863 cells for further analysis. We next performed imputation using ALRA 56, which is a conservative strategy for distinguishing technical “dropout” zeros from true biological zeros. After log normalization, we additionally filtered the 27,998 genes/features using a flat cutoff metric such that genes with variance < 0.1 were excluded, leaving 7,632 genes. Using the final normalized and processed dataset above, we performed t-SNE dimension reduction 57,58. Individual data points were colored based on the treatment condition for each cell, or based on unbiased k-means clustering performed on the normalized dataset. Differential expression analyses between clusters and/or treatment conditions was performed by nonparametric two-tailed Mann Whitney test, filtering for genes with variance > 0.1 across the cell population of interest. Multiple hypothesis correction was performed by the Benjamini-Hochberg method. Significantly differentially expressed genes were defined as having a Benjamini-Hochberg adjusted p < 0.001. A cutoff of 0 was used for determining whether cells expressed a particular gene of interest. The statistical significance of differences in cell type proportions between AAV-p-MAEGI and AAV-Vector conditions was assessed by two-tailed Fisher’s exact test. Gene ontology analysis was performed with DAVID 59.
RNA extraction, reverse transcription, and quantitative PCR
RNA in cells, splenocytes, and tumor infiltrating lymphocytes were extracted using TRIzol Reagent (Invitrogen) by following standard RNA extraction protocols. The first-strand cDNA of RNA was synthesized using SuperScript™ IV Reverse Transcriptase (Invitrogen). After normalizing the concentrations of cDNA with nuclease-free water, quantitative PCR (qPCR) was performed by adding designated Taqman probe of interested genes, and GAPDH was used as an internal positive control.
Western Blot
E0771, E0771-dCas9-VP64, and E0771-dCas9-VP64-MPH cells in 6-well plates were washed twice with ice cold PBS before lysis with 1× RIPA buffer which was kept on ice for 15 min. Cell lysates were centrifuged at 12,000 g for 15 min at 4°C and protein-containing supernatant was collected. Protein concentration was measured using a standard Bradford assay (BioRad) and 20 μg of protein in each sample were loaded into SDS-PAGE gel. After electrophoresis, proteins separated in gel were transferred into nitrocellulose membranes. Membranes were blocked at room temperature for 1 h using 5% skim-milk in TBST, followed by the incubation with primary antibody in 4°C overnight. After washing three times with TBST, horseradish peroxidase (HRP)-conjugated secondary antibody was added and incubated at room temperature for 30 – 60 min. The chemiluminescent substrate (Clarity Western ECL Substrate, Bio-Rad) was added on top of blot membrane according to the manufacturer’s instructions. The signals were captured using a CCD camera-based imager (GE Healthcare).
AAV infectivity assay
AAVs carrying GFP reporter (AAV-GFP) were used to assess the infection rate of intratumoral AAV delivery. Orthotopic breast tumors were established by transplanting 2 × 106 E0771-dCas9-VP64 cells into the mammary fat pad of 5–8 weeks old female C57BL/6J mice. 3 and 10 days after transplantation, 1 × 1011 GCs of AAV-GFP were injected intratumorally to tumor-bearing mice. Four days following intratumoral AAV injection, we harvested tumor cells, tumor-infiltrating immune cells (TIICs, CD45+), and cells from non-tumor organs. To analyze the persistent expression of AAV delivered transgene, 12 days after initial AAV injection (AAV-GFP were injected at day 3 and day 10), we harvested tumor cells, tumor-infiltrating immune cells (TIICs, CD45+). Single tumor cell suspensions were prepared. LIVE/DEAD™ Fixable Near-IR Dead Cell Stain Kit was used to distinguish live cells from dead cells. Cells at a density of 107/ml were stained with DMSO dissolved live/dead staining dye and PerCP/Cy5.5 conjugated CD45 antibody in PBS + 2% FBS, and incubated on ice for 30 min. Stained cells were washed three times before being analyzed and FACS isolated on a BD FACSAria.
Histology and immunohistochemistry
At matched time points, tumors from different treatment group were collected and fixed in 10% neutral formalin for 2–5 days. Tissues were transferred into 70% ethanol for long-term storage. Haematoxylin and eosin (H&E) staining or antibody staining were performed on 3–5 μm tissue sections using standard procedures at Yale Pathology Core Facility.
T cell receptor sequencing (TCR-seq)
Mice in different treatment groups were euthanized at 30–40 days post-transplantation. Tumors and spleens were collected and kept in ice-cold 2% FBS before use. Single cell suspensions from tumors or spleens were prepared as previous described. The RNA from collected splenocytes and TILs were extracted using Trizol and an Ambion RNA extraction kit. TCR library prep was performed using the SMARTer® Mouse TCR a/b Profiling Kit (Takara) according to manufacturer’s instructions.
TCR-seq data analysis
Raw fastq files from TCR-seq were processed to clonotypes using MiXCR following the author recommendations 60. Specifically, the mixcr align function was first used to align to the V segment transcript, followed by clonotype assembly using mixcr assemble and mixcr exportClones. Samples with fewer than 1,000 UMIs were removed from further analysis. Subsequently, TCR-seq data were analyzed using the tcR package61 to determine clonal proportions and occupied clonal homeostasis in each sample. Various metrics such as Gini-Simpson and Chao1 indices were calculated.
Exome sequencing of E0771 and characterization of mutational spectrum
E0771 cells and freshly-dissected mammary fat pads from healthy, untreated 6–8wk old female C57BL/6J mice were subjected to gDNA extraction following standard protocols. A total of 2 μg of gDNA per sample were subjected to exome capture using mouse exome probes (Roche) and then Illumina library preparation following manufacturer’s protocols. Exome capture paired-end fastq files were mapped to the mm10 genome using Bowtie v2.2.9 62, and sorted by Samtools 63. VarScan v2.3.9 64 was then used in somatic mode to call indels and SNPs that were specific to E0771 cells and not found in wildtype C57BL/6J mammary fat pad tissue. Germline calls were subsequently filtered out. The remaining variants were collapsed to the gene level, and the number of indels or SNPs for each gene was tabulated.
Generation of exome-guided precision AAV-MAEGI (AAV-p-MAEGI)
Genes were ranked by their number of somatic variants present in E0771 cells but not in healthy mammary fat pad. Both intronic and exonic variants were included. A set of 1,116 top mutated genes were chosen as differentially mutated genes in E0771. Up to 3 sgRNAs per gene (3 for the majority of genes) targeting promoter regions were chosen. A CRISPRa library consisting of 3,839 sgRNAs was designed for the E0771 top mutated gene set (Addgene, #129601), pool-synthesized (CustomArray), and cloned into the AAV-CRISPRa vector described above. The production of library AAV was done as described above, generating AAV-p-MAEGI for E0771.
Therapeutic testing of AAV-p-MAEGI
Syngeneic TNBC were established by orthotopic transplantation of 2 × 106 E0771-dCas9-VP64 cells into the mammary fat pad of 5–8 weeks old female C57BL/6J mice. 5, 12 and 18 days after tumor induction, 5 – 10 × 1010 GCs of AAV-p-MAEGI, AAV-Vector, or PBS were intratumorally administrated into tumor-bearing mice. For testing of the abscopal effect, syngeneic TNBC were established by orthotopic transplantation of 2 × 106 E0771 cells into an ipsilateral mammary fat pad, and 2.5 × 105 E0771 cells into the contralateral mammary fat pad. 4, 11, 18 and 24 days after tumor induction, 5 – 10 × 1010 GCs of AAV-p-MAEGI or AAV-Vector were intratumorally administrated into the ipsilateral tumor. Tumors were measured every 3–4 days using caliper and sizes were calculated with the formula: Vol = π/6 *x*y*z. Two-way ANOVA was used to compare growth curves between treatment groups. Response to therapy was defined by a method similar to RECIST 1.1 criteria (CR, complete response, where endpoint tumor size = 0; Near-CR, near complete response, where tumor size < 50 mm3 for the last 2 measurements; PR, partial response, where tumor sizes decreased for at least 2 consecutive measurements; SD, stable disease, where tumor size is between 70~100% of initial tumor size at the first treatment; PD, progressive disease, where tumor size > initial tumor size at the first treatment).
A dual AAV system for in vitro and in vivo delivery of MAEGI
A dual AAV vector system of the CRISPR activation (AAV-dCas9, i.e. EFs-dSpCas9-spA (Addgene, #129598); and AAV-CRISPRa vector, i.e. U6p-sgSapI-EF1a-MS2-p65-HSF1-sPA (Addgene, #129602)), was generated by restriction cloning and Gibson assembly. Individual sgRNA or sgRNA libraries were cloned into the AAV-CRISPRa plasmid. dCas9 or U6-sgRNA-EF1a-MS2-p65-HSF1 expressing AAVs were produced using the method as described above in “Generation of AAV-based MAEGI”. For in vitro and in vivo delivery, AAV-dCas9 and AAV-CRISPRa were simultaneously added into cultured cells or co-injected into tumors at a ratio of 1:1.
Therapeutic effects of dual AAV system delivered p-MAEGI
Syngeneic TNBC were established by orthotopic transplantation of 2 × 106 E0771 cells into the mammary fat pad of 5–8 weeks old female C57BL/6J mice. 4, 9 (11) and 18 days after tumor induction, 1–5 × 1010 GCs of AAV-dCas9, together with same titers of AAV-p-MAEGI or AAV-Vector, or PBS were intratumorally administrated into tumor-bearing mice. Tumors were measured every 3–4 days using caliper and sizes were calculated with the formula: Vol = π/6 *x*y*z. Two-way ANOVA was used to compare growth curves between treatment groups.
Statistical analysis
Data analysis was performed using GraphPad Prism7 and R 3.0. Unpaired two-sided Mann-Whitney test was used to compare two groups unless indicated otherwise. Two-way ANOVA was used to compare multiple groups in the tumor growth curves with two independent variables. P values < 0.05 were considered statistically significant.
Data and resource availability
Source data and statistics for non-NGS experiments such as tumor studies, flow cytometry, ELISPOT, and qPCR are provided in an excel table (Supplementary Table 1). Processed genomic sequencing data are provided in an excel table (Supplementary Table 2). Original exome sequencing data are available via NCBI SRA (PRJNA553203). Single cell transcriptome sequencing data are available via GEO (GSE133983). Other data, reagents, methods, computational codes and materials that support the findings of this research are available from the corresponding author upon reasonable request.
Code availability
Custom codes used to support the findings of this research are available from the corresponding author upon reasonable request.
Supplementary Material
1
Supp_Tab1
Supplementary Table 1: Raw data and statistics of mouse and cellular experiments
Supp_Tab2
Supplementary Table 2: Processed data and analysis of high-throughput experiments
4
Acknowledgments
We thank all members in the Chen laboratory, as well as various colleagues in Department of Genetics, Systems Biology Institute, Immunobiology Program, BBS Program, Comprehensive Cancer Center and Stem Cell Center at Yale, for their assistance and/or scientific discussion. We thank the Center for Genome Analysis, Center for Molecular Discovery, Pathology Tissue Services, Histology Services, High Performance Computing Center, West Campus Analytical Chemistry Core and West Campus Imaging Core and Keck Biotechnology Resource Laboratory at Yale, for technical support. We thank P. Cresswell for assistance on ELISPOT.
S.C. is supported by Yale SBI/Genetics Startup Fund, NIH/NCI (DP2CA238295, R01CA231112, R33CA225498, U54CA209992-8697, RF1DA048811, P50CA196530-A10805, P50CA121974-A08306), Damon Runyon Dale Frey Award (DFS-13-15), Melanoma Research Alliance (412806, 16-003524), St-Baldrick’s Foundation (426685), Breast Cancer Alliance, Cancer Research Institute (CLIP), AACR (499395, 17-20-01-CHEN), The Mary Kay Foundation (017-81), The V Foundation (V2017-022), Ludwig Family Foundation, DoD (W81XWH-17-1-0235), Sontag Foundation, and Chenevert Family Foundation. CSF is supported by NIH/NCI Cancer Center Support Grant (3P30CA016359). GW is supported by CRI Irvington and RJ Anderson Postdoctoral Fellowships. RDC, JP and MBD are supported by the Yale MSTP training grant from NIH (T32GM007205). PR is supported by Yale PhD training grant from NIH (T32GM007499). XD is supported by C Revson Postdoctoral Fellowship.
Footnotes
Competing Interests Declaration
A patent application has been filed by Yale University related to data in this study.
References
- 1.Sharma P, Hu-Lieskovan S, Wargo JA & Ribas A Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 168, 707–723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herbst RS, Morgensztern D & Boshoff C The biology and management of non-small cell lung cancer. Nature 553, 446–454 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg SA & Restifo NP Adoptive cell transfer as personalized immunotherapy for human cancer. Science 348, 62–68 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robert C et al. Pembrolizumab versus Ipilimumab in Advanced Melanoma. N Engl J Med 372, 2521–2532 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Sharma P & Allison JP The future of immune checkpoint therapy. Science 348, 56–61 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Ribas A & Wolchok JD Cancer immunotherapy using checkpoint blockade. Science 359, 1350–1355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.June CH, O’Connor RS, Kawalekar OU, Ghassemi S & Milone MC CAR T cell immunotherapy for human cancer. Science 359, 1361–1365 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Sahin U & Tureci O Personalized vaccines for cancer immunotherapy. Science 359, 1355–1360 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Chen DS & Mellman I Oncology meets immunology: the cancer-immunity cycle. Immunity 39, 1–10 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Chen DS & Mellman I Elements of cancer immunity and the cancer-immune set point. Nature 541, 321–330 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Mellman I, Coukos G & Dranoff G Cancer immunotherapy comes of age. Nature 480, 480–489 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamid O et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med 369, 134–144 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodi FS et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 363, 711–723 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolchok JD et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med 369, 122–133 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Topalian SL et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 366, 2443–2454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn GP, Bruce AT, Ikeda H, Old LJ & Schreiber RD Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol 3, 991–998 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Schumacher TN & Schreiber RD Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Verdegaal EM et al. Neoantigen landscape dynamics during human melanoma-T cell interactions. Nature 536, 91–95 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Khong HT & Restifo NP Natural selection of tumor variants in the generation of “tumor escape” phenotypes. Nat Immunol 3, 999–1005 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spiotto MT et al. Increasing tumor antigen expression overcomes “ignorance” to solid tumors via crosspresentation by bone marrow-derived stromal cells. Immunity 17, 737–747 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Ott PA et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sahin U et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Hilf N et al. Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature 565, 240–245 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Keskin DB et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature 565, 234–239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi LS et al. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 152, 1173–1183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert LA et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konermann S et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chavez A et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods 12, 326–328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS & Vale RD A Protein-Tagging System for Signal Amplification in Gene Expression and Fluorescence Imaging. Cell 159, 635–646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zincarelli C, Soltys S, Rengo G & Rabinowitz JE Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol Ther 16, 1073–1080 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Mingozzi F & High KA Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat Rev Genet 12, 341–355 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Kirsch I, Vignali M & Robins H T-cell receptor profiling in cancer. Mol Oncol 9, 2063–2070 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciucci T & Bosselut R Gimap and T cells: a matter of life or death. Eur J Immunol 44, 348–351 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martincorena I & Campbell PJ Somatic mutation in cancer and normal cells. Science 349, 1483–1489 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Liu XS & Mardis ER Applications of Immunogenomics to Cancer. Cell 168, 600–612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van den Boorn JG & Hartmann G Turning Tumors into Vaccines: Co-opting the Innate Immune System. Immunity 39, 27–37 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Marabelle A, Tselikas L, de Baere T & Houot R Intratumoral immunotherapy: using the tumor as the remedy. Ann Oncol 28, 33–43 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Townsend SE & Allison JP Tumor rejection after direct costimulation of CD8+ T cells by B7-transfected melanoma cells. Science 259, 368–370 (1993). [DOI] [PubMed] [Google Scholar]
- 39.Chen L et al. Costimulation of antitumor immunity by the B7 counterreceptor for the T lymphocyte molecules CD28 and CTLA-4. Cell 71, 1093–1102 (1992). [DOI] [PubMed] [Google Scholar]
- 40.Ribas A et al. Oncolytic Virotherapy Promotes Intratumoral T Cell Infiltration and Improves Anti-PD-1 Immunotherapy. Cell 170, 1109–1119 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zamarin D et al. Localized oncolytic virotherapy overcomes systemic tumor resistance to immune checkpoint blockade immunotherapy. Sci Transl Med 6, 226ra232 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suva ML et al. Reconstructing and Reprogramming the Tumor-Propagating Potential of Glioblastoma Stem-like Cells. Cell 157, 580–594 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roulois D et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell 162, 961–973 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shalem O et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang T, Wei JJ, Sabatini DM & Lander ES Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robbins PF et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med 19, 747–752 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russell S et al. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet 390, 849–860 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bainbridge JW et al. Long-term effect of gene therapy on Leber’s congenital amaurosis. N Engl J Med 372, 1887–1897 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
Supp_Tab1
Supplementary Table 1: Raw data and statistics of mouse and cellular experiments
Supp_Tab2
Supplementary Table 2: Processed data and analysis of high-throughput experiments
4