Retrospective motion correction protocol for high‐resolution anatomical MRI (original) (raw)
Abstract
Modern computational brain morphology methods require that anatomical images be acquired at high resolution and with a high signal‐to‐noise ratio. This often translates into long acquisition times (>20 minutes) and images susceptible to head motion. In this study we tested retrospective motion correction (RMC), common for functional MRI (fMRI) and PET image motion correction, as a means to improve the quality of high‐resolution 3‐D anatomical MR images. RMC methods are known to be effective for correcting interscan motion; therefore, a single high‐resolution 3‐D MRI brain study was divided into six shorter acquisition segments to help shift intrascan motion into interscan motion. To help reduce intrascan head motion, each segment image was reviewed for motion artifacts and repeated if necessary. Interscan motion correction was done by spatially registering images to the third image and forming a single average motion‐corrected image. RMC was tested on 35 subjects who were considered at high risk for head motion. Our results show that RMC provided better contrast‐to‐noise ratio and boundary detail when compared to nonmotion‐corrected averaged images. Hum Brain Mapp, 2006. © 2006 Wiley‐Liss, Inc.
Keywords: brain, structural anatomy, high‐resolution imaging
INTRODUCTION
Computational morphometry methods use high‐resolution anatomical images to assess regional cortical atrophy [Kochunov et al.,2005; Magnotta et al.,1999; Sowell et al.,2003; Thompson et al.,2003], measure cortical gray matter thickness [Fischl and Dale,2000; Lerch and Evans,2005], and study patterns of cortical folding [Van Essen,1997]. These methods analyze fine details of cortical anatomy using high‐quality 3‐D MR anatomical images and improve with increasing spatial‐resolution and contrast‐to‐noise ratio (CNR). Spatial resolution under 1 mm with good gray matter (GM) to white matter (WM) contrast are now possible with 3 T MR imaging systems. However, this translates into long scan times, requiring subjects to remain motionless for periods of 20 to 30 minutes. Unfortunately, long scan times increase the likelihood of head motion and, in our experience, lead to intolerable head motion artifacts in more than 25% of studies.
Head motion is a well‐known problem in positron emission tomography (PET) studies of cerebral blood flow (CBF) and functional MRI (fMRI) studies due to long imaging times. A number of motion correction approaches have been proposed, with retrospective motion correction (RMC) techniques being ubiquitous. Most RMC approaches postulate two categories of motion: intra‐ and interscan. Intrascan head motion during fMRI studies leads to phase‐related artifacts. Such artifacts cannot be adequately corrected using RMC, but they are generally assumed to be minimal for fMRI experiments where acquisition times for individual image volumes are very short (a few seconds). A similar relationship between intra‐ and interscan motion is seen for PET O‐15 water CBF studies where imaging times are several minutes, with 10‐minute interscan intervals. The predominant source of head motion is assumed to be interscan for such studies. Interscan head movement can be modeled as a 3‐D rigid‐body motion and corrected using spatial registration algorithms to realign images. Rigid‐body registration algorithms calculate three position and three orientation parameters that best register each image in the group to a single reference image within the study [Fox,1995]. A number of registration algorithms have been introduced and studied extensively for RMC applications [Ciulla and Deek,2002; Morgan et al.,2001].
We investigated the utility of RMC with motion artifacts encountered in the acquisition of high‐quality high‐resolution anatomical MR images in patients at high risk for movement. We felt that it would be better to break up long‐duration high‐resolution 3‐D brain studies into multiple smaller time segments with interscan breaks. The premise was that realignment and averaging of multiple images would provide a better final image than a single scan of the same overall duration for studies when head motion is a problem. The time‐segmented images help to cast head movements into the interscan category where RMC methods are known to be effective. To test this approach we devised a high‐resolution anatomical MRI time‐segmented protocol consisting of six sequential 4‐minute full‐resolution image volumes with interscan breaks of approximately 10 seconds.
SUBJECTS AND METHODS
All experiments were performed on a Siemens Trio, 3T scanner (Siemens, Erlangen) at the Research Imaging Center, University Health Science Center at San Antonio. Heads were stabilized using the lateral clamp motion suppression system provided by Siemens for head imaging. Imaging was done using an 8‐channel Siemens head coil and a parallel imaging technique to shorten acquisition time to approximately 4 minutes per image. The FLIRT registration application, provided in the FSL package (http://www.fmrib.ox.ac.uk/), was used to correct for interscan motion in the six images. Motion‐corrected images were averaged to form a single high‐resolution 3‐D image volume. FLIRT has been extensively validated for spatial registration between pairs of high‐resolution anatomical images, not just for fMRI and PET studies [Jenkinson et al.,2002].
Subjects
The six‐segment RMC method was tested in a group of 35 volunteers enrolled in four existing research projects at the Research Imaging Center, all of whom were considered high risk for head motion. This group included five children diagnosed with ADHD, nine adolescents diagnosed with bipolar II depression, seven young adults with genetic predisposition to alcohol abuse, and 14 adults with developmental stuttering. The average subject age was 23.3 ± 9.5 years and ranged from 8 to 49 years. Based on previous experience with long‐duration 3‐D MRI, the subjects enrolled in these protocols were considered to be at high risk for intrascan motion. In fact, more than 25% of high‐resolution 3‐D MR images from similar patient studies were not acceptable for analysis because of motion artifacts. All protocols were reviewed and approved by the UTHSCSA Institutional Review Board and all subjects signed an informed consent.
Imaging Procedure
MRI sequence
Subjects were imaged using a high‐resolution (0.8 mm isotropic, axial orientation, 320 × 224 matrix, field of view (FoV) = 256 × 180 mm, 208 slices) T1‐weighted, 3‐D TurboFLASH sequence with an adiabatic inversion contrast pulse with the following scan parameters: TE/TR/TI = 3.04/2100/785 ms, flip angle = 13°. This sequence was optimized to provide an average white matter (WM) to gray matter (GM) contrast of approximately 23%, while maintaining a signal‐to‐noise ratio (SNR) of approximately 11:1 when acquired with the GeneRalized Autocalibrating Partially Parallel Acquisitions (GRAPPA) parallel imaging method with an acceleration factor of two along the phase encoding direction and 12 phase encoding reference lines. Six image volumes were acquired per subject (Fig. 1A). The acquisition time for single volume was 260 seconds and the total acquisition time for six volumes was approximately 25 minutes. Each segment image was acquired using the standard tune‐up procedure, which performed calibration for central frequency.
Figure 1.
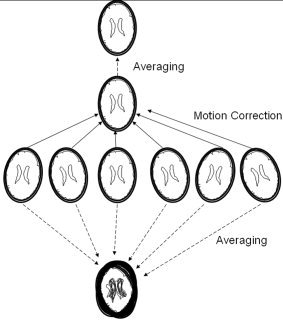
Six high‐resolution images acquired and averaged with and without motion correction.
MRI protocol
Subjects were instructed not to move their head during each of the six image acquisitions and informed that if they moved the acquisition would be repeated. Subjects were notified of the beginning and end of each imaging segment. Between acquisitions subjects were allowed to slightly adjust their posture in an attempt to maintain a comfortable position for the duration of the study. Between acquisitions the scanner operator screened the image using a 3‐D viewer for motion artifacts observable as “ghosting/blurring/striping” in the phase‐encoding directions (∼20 s). If artifacts were present, the subject was informed and the segment repeated. If motion continued the study would be stopped and rescheduled, although this was not necessary for any subjects in our investigation.
Motion correction
For each subject, the six time‐segmented 3‐D raw images were motion‐corrected by registering each to the approximate mid‐time image of the study (third image). FLIRT [Jenkinson et al.,2002] image registration software was used with the following settings: six parameters (three translations and three rotations); normalized spatial correlation as the cost function; and 17 × 17 × 17‐voxel sinc function as the interpolation kernel. FLIRT takes approximately 15 minutes to perform registration between two 3‐D brain images on a single Pentium 4 processor system (2.5 GHz), leading to a total processing time of approximately 1.5 hours per study to align the six images. FLIRT processing was done in parallel on a cluster of 30 computers improving throughput to approximately 15 minutes per study.
Motion study
The magnitude of x, y, and z translations and rotations about the x, y, and z axes were calculated from the 4 × 4 matrices used for registration. Additionally, a software application (RMSDIFF, distributed as part of FSL) was used to estimate the root mean square (RMS) movement distance. The RMSDIFF program calculates the RMS difference in transformed locations for two transforms applied to a simple brain model. We compared the individual 4 × 4 alignment matrices to a 4 × 4 identity matrix which served as the no‐movement reference. The brain model was a sphere spanning approximately 25% of the image volume, with the origin at the center of the volume. RMS distance includes both translation and rotation effects, providing an index of motion.
Image quality
When images with interscan motion are averaged the result is a spatially blurred average image. For translation‐only motions this blurring can be modeled using a single blurring point spread function (PSF). However, head movement during imaging is both translational and rotational, and the rotational component of motion leads to PSFs which broaden with distance from the axis of rotation. Rotation‐induced blurring must be modeled using spatially varying PSFs. For supine head imaging the axis of rotation is likely near the posterior surface of the head, leading to the largest rotation type blurring toward the anterior surface of the head. Overall blurring is a combination of these two rigid body motions with a net effect to broaden the imaging system PSF. As the system PSF increases, partial volume averaging increases, which alters tissue signal distributions in histograms of the averaged images (Fig. 2). We analyzed GM and WM distributions to assess image quality difference for averaged images with and without motion correction.
Figure 2.
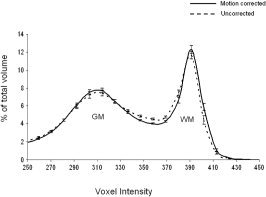
Average voxel intensity histograms for motion‐corrected and uncorrected images.
In order to properly calculate histograms, the effects of RF inhomogeneity in the raw 3‐D brain images must be corrected. The RF inhomogeneity correction program supplied with the FSL toolkit (FAST) was used to make corrections on raw images prior to averaging. To use FAST each image must first be deskulled. We performed this using the FSL Brain Extraction Tool (BET) [Smith,2002]. A 3 × 3 × 3 median filter was used during calculations of RF‐bias fields to improve SNR in raw while preserving GM/WM borders.
RESULTS
Intrascan Motion
Subject cooperation was rated high for the RMC acquisition protocol. The operator found significant intrascan motion artifacts in only 5 of 35 studies (3 ADHD and 2 stuttering subjects). In 4 of these subjects (2 ADHD and 2 stutterers), a single 4.2‐minute scan was repeated, and in 1 case (ADHD) two acquisitions were repeated, thus extending the scan time by 4.2 and 8.4 minutes, respectively. Subjects usually attributed intrascan head motion to a respiratory distress such as sneezing or coughing. Using the review and repeat approach and allowing small head movements during breaks between imaging segments resulted in minimal intrascan motion artifacts and acceptable scan times.
Interscan Motion
Translation and rotation
Translation and rotation values used in motion correction were estimated from the transform matrices for Images 1–6 in the 35 subjects (Table I). Head motions were smallest for left–right movement (x‐translation) and largest for inferior–superior movement (z‐translation). The smallest orientation change was for rotation about the _z_‐axis and largest for rotation about the _x_‐axis. These interscan motion parameter estimates follow the expected head movement pattern for the physical motion suppression system supplied by Siemens, where two lateral clamps fix the head at the temples. This system best suppresses left–right head motion and rotation about the _z_‐axis and is far less effective in suppressing superior–inferior motion and rotations about the _x_‐axis, i.e., “nodding‐like” motions.
Table I.
Analysis of motion
| Measurement | Image 1 | Image 2 | Image 3 | Image 4 | Image 5 | Image 6 | Average |
|---|---|---|---|---|---|---|---|
| Root mean square distance (mm) | 1.24 ± 0.16 | 0.85 ± 0.15 | 0.00 ± 0.001 | 0.72 ± 0.11 | 1.23 ± 0.18 | 2.26 ± 0.54 | 1.05 ± 0.12 |
| Translation X (mm) | 0.46 ± 0.35 | 0.21 ± 0.12 | 0.02 ± 0.001 | 0.31 ± 0.16 | 0.48 ± 0.38 | 0.76 ± 0.35 | 0.24 ± 0.10 |
| Translation Y (mm) | 0.80 ± 0.57 | 0.50 ± 0.34 | 0.01 ± 0.002 | 0.48 ± 0.21 | 0.73 ± 0.48 | 1.00 ± 0.72 | 0.58 ± 0.24 |
| Translation Z (mm) | 1.41 ± 1.20 | 0.87 ± 0.52 | 0.02 ± 0.004 | 0.85 ± 0.41 | 1.33 ± 1.20 | 2.21 ± 1.38 | 1.21 ± 0.38 |
| Rotation X (degrees) | 0.81 ± 0.81 | 0.60 ± 0.54 | 0.01 ± 0.008 | 0.54 ± 0.74 | 0.82 ± 0.82 | 1.10 ± 1.21 | 0.88 ± 0.93 |
| Rotation Y (degrees) | 0.37 ± 0.33 | 0.26 ± 0.15 | 0.00 ± 0.005 | 0.23 ± 0.11 | 0.26 ± 0.18 | 0.39 ± 0.30 | 0.25 ± 0.20 |
| Rotation Z (degrees) | 0.20 ± 0.21 | 0.11 ± 0.97 | 0.00 ± 0.004 | 0.09 ± 0.04 | 0.15 ± 0.12 | 0.25 ± 0.23 | 0.13 ± 0.12 |
RMS distance
The average RMS motion distance averaged for all six scans was 1.05 ± 0.12 mm, surprisingly small. Inspection of RMS distances for Images 1–6 indicates that motion increases with time relative to reference Image 3. The small RMS distance for Image 3 provides an estimate of the baseline error since it was registered to itself. Images 2 and 4 had similar RMS motion, as did Images 1 and 5, each grouping having similar time differences from Image 3. Image 6, which had the greatest time difference, showed the largest RMS motion distance. Standard deviations also generally increased with time relative to Image 3.
Image quality
Voxel intensity histograms were calculated for uncorrected and motion‐corrected images, following RF‐homogeneity correction (Fig. 2). The location of the GM peak is similar in histograms from both uncorrected and motion‐corrected images. The WM peak location was shifted slightly lower for the uncorrected study. The number of voxels with values near the WM and GM peak diminished with motion, while the number off‐peak increased. These observations are as predicted and motion increases the system PSF and partial volume averaging between cerebrospinal fluid (CSF), WM, and GM. Since WM has the highest signal, assessment of loss from WM is a measure of the effect of partial volume averaging. The fraction of WM voxels above the valley point between WM and GM decreased from 31% to 28% due to motion, while the fraction of valley voxels increased from 17.5% to 20% (Fig. 2). While this histogram analysis indicates adverse effects from a larger system PSF, the spatial impact of this loss is probably best illustrated by improvements in clarity of the cortical ribbon (Fig. 3).
Figure 3.
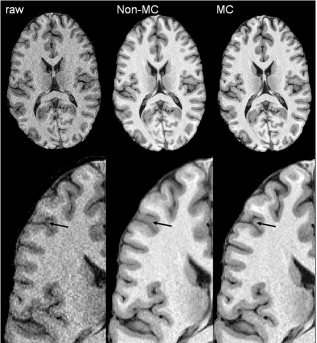
Patient with minimal motion during study (asleep). The third raw image in the study was used as a target for motion correction (left), uncorrected average image (middle), and motion‐corrected average image (right). The arrow shows an anastomotic sulcus that was blurred out by subject's motion, but recovered following motion correction,
DISCUSSION
MRI Protocol
Retrospective motion correction (RMC) based on averaging of multiple sequential high‐resolution 3‐D images reduced the incidence of motion artifacts compared with prior studies in our lab that used a single high‐resolution 3‐D image. Others have proposed prospective methods that are acquisition‐based techniques. Such methods include PROPELLER MRI [Pipe,1999], orbital navigator echoes [Fu et al.,1995], radial imaging [McLeish et al.,2004], or navigator volumes (PACE) techniques [Thesen et al.,2000]. The prospective motion‐correction methods are conceptually attractive; unfortunately, their general use is limited by difficulty of implementing these sequences for rotation and translations not coincident with acquisition defined axes, e.g., PROPELLER and radial imaging will not correct for out‐of‐plane motion. Also, these methods are specific to pulse sequence and/or scanner platforms. The proposed RMC method is capable of correcting for full 3‐D motion and not specific to the pulse sequences and/or scanner platforms. The proposed method is computationally intensive, but modern computers can reduce this to acceptable times for research sites, and the entire processing stream can be fully automated.
The RMC method is most effective for high‐resolution anatomical acquisitions if intrascan motion is minimized. We targeted having individual scans with imaging times of approximately 4 minutes under the assumption that most subjects could be motionless for this time period. We used parallel imaging with an acceleration factor of two and the GRAPPA imaging algorithm [Griswold et al.,2002]. This accelerated imaging approach supported both the desired 0.8‐mm sample spacing and 4‐minute acquisition times without excessive loss in SNR. Further improvement in motion correction quality could potentially be obtained by shortening the acquisition time by using a higher acceleration factor.
Image comparison
Our comparison of average 3‐D images with and without motion correction is potentially biased, since subjects were allowed to move during interscan intervals to adjust their posture and take a more comfortable position. Such movements can drastically reduce the quality of the nonmotion‐corrected average image. To test for improvement by RMC where interscan motion was expected to be small, we studied an ADHD child who was asleep throughout the study. The average RMS distance was 0.56 mm, or about half of the group‐average RMS motion distance. Visually, the corrected image (Fig. 3, right) showed a modest improvement in cortical ribbon clarity over the uncorrected image (Fig. 3, middle), with a more pronounced GM/WM interface and less blurring in the intrasulcal spaces.
Limitations
The proposed motion correction method averages magnitude images. Complex domain averaging can have an advantage over magnitude domain averaging, since it can reduce noise level in the regions with zero mean signal, e.g., background [Henkelman,1985]. However, complex domain averaging was not practical in our study since the k‐space for multichannel coils was not accessible as a single k‐space, but rather as a collection of k‐spaces for individual coils. Obtaining the combined k‐space would require off‐scanner reconstruction, and would only be available for centers with a research agreement with the scanner vendor, thus limiting the applicability of the proposed approach. To deal with this issue, we used an adaptive threshold to zero out pixel values below the median background value in individual images. This approach forced a large number of background pixels to zero, leading to reduced spatial coherence in image noise when averaging and a reduction in the background signal (Fig. 3).
CONCLUSION
Retrospective motion correction can be reliably applied to improve image quality in high‐resolution 3‐D MR images in a population of patients with a high incidence of motion. This method could potentially provide further improvement in motion correction quality by shortening the acquisition time by using higher acceleration factor.
REFERENCES
- Ciulla C, Deek FP (2002): Performance assessment of an algorithm for the alignment of fMRI time series. Brain Topogr 14: 313–332. [DOI] [PubMed] [Google Scholar]
- Fischl B, Dale AM (2000): Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A 97: 11050–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox P (1995): Spatial normalization origins: objectives, applications, and alternatives. Hum Brain Mapp 3: 161–164. [Google Scholar]
- Fu ZW, Wang Y, Grimm RC, Rossman PJ, Felmlee JP, Riederer SJ, Ehman RL (1995): Orbital navigator echoes for motion measurements in magnetic resonance imaging. Magn Reson Med 34: 746–753. [DOI] [PubMed] [Google Scholar]
- Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A (2002): Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med 47: 1202–1210. [DOI] [PubMed] [Google Scholar]
- Henkelman RM (1985): Measurement of signal intensities in the presence of noise in MR images. Med Phys 12: 232–233. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Bannister PR, Brady JM, Smith SM (2002): Improved optimisation for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17: 825–841. [DOI] [PubMed] [Google Scholar]
- Kochunov P, Mangin JF, Coyle T, Lancaster J, Thompson P, Riviere D, Cointepas Y, Regis J, Schlosser A, Royall DR, Zilles K, Mazziotta J, Toga A, Fox PT (2005): Age‐related morphology trends of cortical sulci. Hum Brain Mapp 26: 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch J, Evans A (2005): Cortical thickness analysis examined through power analysis and a population simulation. Neuroimage 24: 163–173. [DOI] [PubMed] [Google Scholar]
- Magnotta VA, Andreasen NC, Schultz SK, Harris G, Cizadlo T, Heckel D, Nopoulos P, Flaum M (1999): Quantitative in vivo measurement of gyrification in the human brain: changes associated with aging. Cereb Cortex 9: 151–160. [DOI] [PubMed] [Google Scholar]
- McLeish K, Kozerke S, Crum WR, Hill DL (2004): Free‐breathing radial acquisitions of the heart. Magn Reson Med 52: 1127–1135. [DOI] [PubMed] [Google Scholar]
- Morgan VL, Pickens DR, Hartmann SL (2001): Comparison of functional MRI image realignment tools using a computer‐generated phantom. Magn Reson Med 46: 510–514. [DOI] [PubMed] [Google Scholar]
- Pipe JG (1999): Motion correction with PROPELLER MRI: application to head motion and free‐breathing cardiac imaging. Magn Reson Med 42: 963–969. [DOI] [PubMed] [Google Scholar]
- Thesen S, Heid O, Mueller E, Schad LR (2000): Prospective acquisition correction for head motion with image‐based tracking for real‐time fMRI. Magn Reson Med 44: 457–465. [DOI] [PubMed] [Google Scholar]
- Smith SM (2002): Fast robust automated brain extraction. Hum Brain Mapp 17: 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowell ER, Peterson BS, Thompson PM, Welcome SE, Henkenius AL, Toga AW (2003): Mapping cortical change across the human life span. Nat Neurosci 6: 309–315. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, de Zubicaray G, Janke AL, Rose SE, Semple J, Herman D, Hong MS, Dittmer SS, Doddrell DM, Toga AW (2003): Dynamics of gray matter loss in Alzheimer's disease. J Neurosci 23: 994–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC (1997): A tension‐based theory of morphogenesis and compact wiring in the central nervous system. Nature 385: 313–318. [DOI] [PubMed] [Google Scholar]