Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner (original) (raw)
. Author manuscript; available in PMC: 2020 Jul 25.
SUMMARY
Immune cells residing in white adipose tissue have been highlighted as important factors contributing to the pathogenesis of metabolic diseases, but the molecular regulators that drive adipose tissue immune cell remodeling during obesity remain largely unknown. Using index and transcriptional single-cell sorting, we comprehensively map all adipose tissue immune populations in both mice and humans during obesity. We describe a novel and conserved Trem2+ lipid-associated macrophage (LAM) subset and identify markers, spatial localization, origin, and functional pathways associated with these cells. Genetic ablation of Trem2 in mice globally inhibits the downstream molecular LAM program, leading to adipocyte hypertrophy as well as systemic hypercholesterolemia, body fat accumulation, and glucose intolerance. These findings identify Trem2 signaling as a major pathway by which macrophages respond to loss of tissue-level lipid homeostasis, highlighting Trem2 as a key sensor of metabolic pathologies across multiple tissues and a potential therapeutic target in metabolic diseases.
INTRODUCTION
The obesity pandemic has reached alarming magnitudes, with over 44% of the adult world population estimated to be overweight. In addition to its widespread prevalence, obesity is considered a major risk factor for a number of metabolic diseases, including type II diabetes mellitus, non-alcoholic fatty liver disease, atherosclerosis, and ischemic cardiovascular disease. Furthermore, obesity is a predisposing factor for numerous other diseases that are not typically classified as metabolic/endocrine, such as cancer and neurodegeneration. Collectively, the global obesity pandemic has far-reaching consequences on life expectancy, quality of life, and healthcare costs (Stevens et al., 2015). The human tissue most strongly involved in the pathogenesis of obesity and its metabolic complications is the adipose tissue. Adipose tissue undergoes marked morphological changes during the development of obesity, including adipocyte hypertrophy and extensive vascularization (Sung et al., 2013). In addition, obese adipose tissue is characterized by a distinct repertoire of soluble mediators, typically referred to as adipokines, that influence both the tissue itself and several distal organ sites (Rosen and Spiegelman, 2014).
Major contributors to the adipose tissue secretome during obesity are tissue-resident immune cells (Mathis, 2013). Under both homeostatic and pathological conditions, adipose tissue is interspersed by a large range of immune cells, which dramatically increase in total abundance with greater adiposity. Adipose-resident immune cells display markedly distinct molecular characteristics compared to their circulating counterparts. Recently identified examples include regulatory T and B cells that interact with adipocytes (Feuerer et al., 2009; Nishimura et al., 2013), γδ T and NKT cells that drive thermogenesis (Kohlgruber et al., 2018; Lynch et al., 2016), memory T cells protective against infection (Han et al., 2017), cytotoxic innate lymphoid cells (Boulenouar et al., 2017), dendritic cells and macrophages expressing the transcription factor PPARγ (Cipolletta et al., 2012; Macdougall et al., 2018; Odegaard et al., 2007), as well as macrophages forming “crown-like” structures around large adipocytes that are suggested to preserve tissue integrity in face of massive adipocyte cell death (McNelis and Olefsky, 2014).
While inflammation is generally considered a driver of the metabolic derangements that accompany obesity, including insulin resistance and dyslipidemia (Winer et al., 2016), the molecular triggers, sensory receptors and signaling pathways of immune cell accumulation in adipose tissue remain incompletely understood. There is thus an urgent need to identify the regulatory mechanisms driving disease-associated immune cell behavior in obese adipose tissue and to understand their function in driving or protecting from obesity-related metabolic derangements. Here, we performed extensive time-resolved single-cell RNA-sequencing of the entire immune cell compartment in adipose tissue of obese mice. We identified the lipid receptor Trem2 as a major driver of tissue-level immune cell remodeling. In a subset of lipid-associated macrophages (LAM) prominently arising under obese conditions from circulating monocytes and positioned around enlarged adipocytes, Trem2 drives a gene expression program involved in phagocytosis, lipid catabolism and energy metabolism. Single-cell RNA-seq of human adipose tissue shows that LAM cells, as well as the Trem2 pathway, are highly conserved. The absence of Trem2 prevents downstream activation of the LAM molecular program including the adiposity-associated pathways. Consequently, Trem2 deletion abrogates the recruitment of macrophages to enlarged adipocytes and causes massive adipocyte hypertrophy, systemic hypercholesterolemia, inflammation, and glucose intolerance. Integration of these results with recent findings on the role of Trem2 in neurodegeneration indicates that Trem2 is a major macrophage sensor of extracellular lipids that drives a conserved and protective tissue immune cell response to loss of metabolic homeostasis. These findings may have important implications for the development of Trem2-targeted immunotherapies for a wide range of metabolic diseases.
RESULTS
Time-resolved single-cell characterization of adipose tissue immune cells in obesity
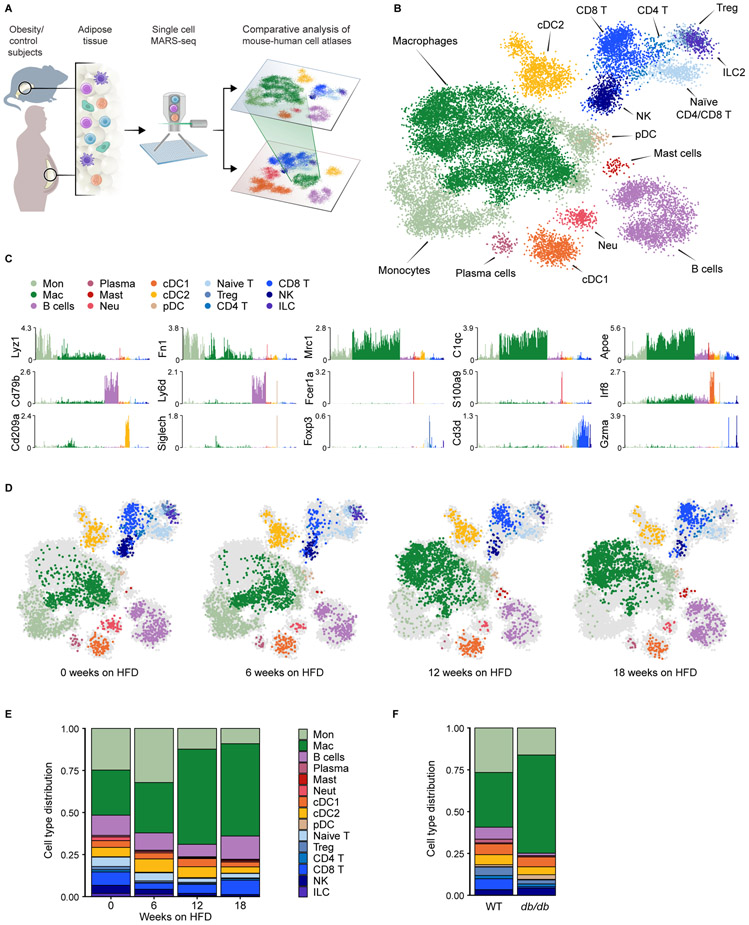
To identify key factors in adipose tissue immune cell remodeling during metabolic disease, we first studied a well-established diet-induced obesity model using mice fed a high-fat diet (HFD) to trigger weight gain (STAR Methods). We isolated epididymal visceral adipose tissue (EAT) at regular intervals after 6, 12, and 18 weeks of HFD feeding, to cover the entire timespan from the lean state to morbid adiposity, followed by massively-parallel single-cell RNA-sequencing (MARS-seq) of tissue-resident CD45+ immune cells (Figure 1A) (Jaitin et al., 2014). At each time point, we compared all animals to age-matched littermates fed a normal chow (NC) diet. Expectedly, HFD feeding induced weight gain and adiposity over the course of the experiment (Figures S1A and S1B). We profiled a total of 21,210 quality control (QC)-positive cells sampled from 20 mice at different time points. We applied the MetaCell algorithm (Baran et al., 2018) to identify homogeneous and robust groups of cells (“metacells”; STAR Methods) from single-cell RNA-seq data, resulting in a detailed map of 244 metacells organized into 15 broad immune cell types (Figure 1B and Table S1). We classified the different groups based on expression levels of the most variable genes and used the differentially expressed genes to annotate broad myeloid and lymphoid cell types (Figures 1B, 1C, S1C and Table S1). Following this approach, we detected a massive reorganization of the immune cell population in visceral adipose tissue between 6 and 12 weeks on HFD (Figure 1D), while NC diet did not induce alterations in the immune cell compartment over time (Figure S1D). The most prominent changes induced by HFD included an expansion of adipose tissue macrophages and a reduction of regulatory T cells and type 2 innate lymphoid cells (ILCs) (Figures 1E, S1E and S1F), in line with previous reports (Biswas and Mantovani, 2012; Cipolletta et al., 2012; Feuerer et al., 2009; Kanneganti and Dixit, 2012; Mathis, 2013; McNelis and Olefsky, 2014).
Figure 1. Single-cell characterization of the adipose tissue immune niche during obesity progression.
A. Schematic of experimental approach: single-cell RNA-seq pipeline of mouse and human adipose tissue immune cells during obesity. B. kNN graph of 21,210 QC-positive immune cells (244 metacells) from EAT of 20 mice fed a normal chow (NC) or high-fat diet (HFD). C. Log2 of average unique molecular identifier (UMI) count of selected genes across metacells. D. kNN graph of EAT immune cells of WT mice on HFD, down-sampled to 2,283 cells (in each condition), annotated as in (B). Each time-point is contributed by either 2 (6 weeks) or 4 mice (12 or 18 weeks) on HFD. E, F. Immune cell type distribution of WT mice on HFD (E), and 7-week old db/db mice and WT littermates (F) of a total of 8,372 QC-positive single cells (75 metacells).
See also Figure S1 and Tables S1 and S2.
To determine whether this global rearrangement of the immune cell population was induced by the dietary conditions or associated with obesity per se, we used a genetic model of obesity, the db/db mouse (Bahary et al., 1990), which harbors a mutation in the leptin receptor gene that causes hyperphagia and massive adiposity even on NC diet (Figure S1G). We profiled a total of 8,372 QC-positive cells sampled from two 7 week-old, two 15 week-old db/db, and four wild-type littermate control mice (Table S2). The db/db mice underwent similar remodeling of the immune cell compartment as observed in mice fed on HFD (Figures 1F and S1H), indicating that these changes occur across different dietary conditions.
Macrophage and monocyte compartments undergo dramatic remodeling during obesity
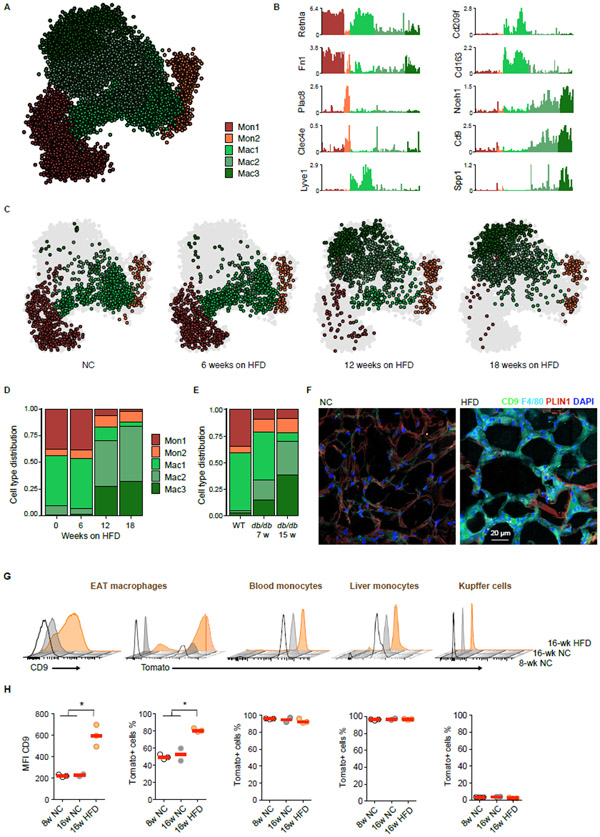
By far the most significant cell population changes during obesity development occurred in the monocyte/macrophage compartment, characterized by two monocyte and three macrophage subgroups comprising of 136 metacells (Figure 2A). The two monocyte subsets, Mon1 and Mon2, were distinguished by the expression of Retnla, Fn1 (Mon1), Plac8, and Clec4e (Mon2), among other differentially expressed genes (Figures 2B and S2A). One macrophage population (Mac1) highly expressed Retnla, Cd163, Lyve1, and Cd209f, a signature that was recently associated with perivascular macrophages (Chakarov et al., 2019) (Figures 2B and S2A). Two additional macrophage populations (Mac2 and Mac3) were characterized by gradual expression of genes, such as Cd9 and Nceh1, in line with the recently described CD9+ subset of adipose tissue macrophages (Hill et al., 2018). Among the differentially expressed genes between these macrophage subsets was the osteopontin-encoding gene Spp1 (Figures 2B and S2A). Both Mac2 and Mac3 emerged only under obese conditions and represented more than 75% of the myeloid compartment after 18 weeks of HFD (Figures 2C and 2D). Similar dynamic changes in the monocyte/macrophage compartment were observed in db/db mice, with the Mac2 and Mac3 subsets being strongly expanded in obese mice compared to littermate controls (Figures 2E and S2B-S2D). We validated these findings on the protein level by flow cytometry, which confirmed the emergence of CD9+CD63+ macrophages during obesity, as predicted by single-cell RNA-seq (Figure S2E). In addition, immunofluorescence staining revealed that these CD9-expressing macrophages accumulated in “crown-like” structures surrounding adipocytes of HFD fed mice (Figure 2F) (Hill et al., 2018).
Figure 2. Large changes during obesity in monocyte and macrophage subtypes are mainly characterized by the expansion of a distinct macrophage subset.
A. kNN graph of 11,241 QC-positive immune cells (136 metacells) of the monocyte/macrophage compartment from Figure 1B. B. Shown are Log2 average UMI count of selected genes across metacells of the Monocyte/Macrophage compartment. C. kNN graph of the monocyte/macrophage compartment of WT mice on HFD, downsampled to 1,327 cells (in each condition), annotated as in (A). D. Cell type distribution within the monocyte/macrophage compartment of WT mice on HFD. E. Cell type distribution of 4,988 QC-positive single cells (45 metacells) within the monocyte/macrophage compartment of 7 and 15 week-old db/db mice and WT littermates. F. Representative immunofluorescence images of CD9 (green), F4/80 (cyan) and perilipin-1 (red) in EAT sections of 16-week old WT mice on NC (left), or after 12 weeks on HFD (right). Cell nuclei are stained with DAPI (blue). Scale bar, 20 μm. G. Representative flow cytometry 3D histograms of the CD9 and of the Tomato (Ms4a3 lineage) signals among adipose tissue macrophages, Ly6Chi blood monocytes, Ly6Chi liver monocytes and Kupffer cells from 8 week-old or 16 week-old Ms4a3Cre-Rosa26tdTomato mice fed with NC or HFD for 8 weeks. H. Quantification of the different populations presented in (G). Each dot represents an independent mouse. * p < 0.05 using Student’s t-test.
See also Figures S2 and S3.
We next asked whether the expanded CD9+CD63+ population was derived from proliferating tissue-resident macrophages or recruited from circulating monocyte precursors. We used a newly developed lineage tracing system (Ms4a3Cre-Rosa26tdTomato), whereby fate mapping for Ms4a3 expression labels all bone marrow-derived monocytes, but not mature tissue-resident macrophages, enabling to disentangle embryonic-derived tissue macrophages from monocyte-derived macrophages (Zhaoyuan Liu, 2019). We validated this system by measuring recombination/labeling in monocytes from the blood and liver, which were almost exclusively Ms4a3 lineage-positive, while tissue-resident Kupffer cells that are of embryonic origin were not derived from an Ms4a3+ monocyte precursor (Figures 2G, 2H, and S3A). In 8- and 16-weeks old mice, ~50% of the adipose tissue macrophage population was Ms4a3 linage-positive under lean conditions (Figures 2G, 2H, and S3A). In contrast, under HFD 80% of CD9+CD63+ macrophages were Ms4a3 lineage-positive (Figures 2G, 2H, and S3A), supporting the notion that most of the Mac3 cells originate from recruited circulating monocytes. To infer the most probable molecular differentiation trajectory from monocytes towards the Mac3 subset, we used Slingshot, a recently published method for pseudo-time inference (Street et al., 2018), and characterized a gradual acquisition of Mac3 genes from HFD monocytes. Slingshot suggested a linear transition of adipose tissue HFD monocytes to Mac3 along a clear differentiation trajectory, characterized by loss of expression of Ly6c2, Il1b, Ccr2, Lyz2 and S100a10, and by acquisition of C1qa, Cd9, Cd63 and Cd68 expression (Figure S3B). Together, these data provide a time-resolved single-cell map of the immune cell compartment in visceral adipose tissue and highlight a stereotypical change in specific monocyte and macrophage subsets during obesity.
Lipid-associated macrophages are characterized by Trem2 expression in mice and humans
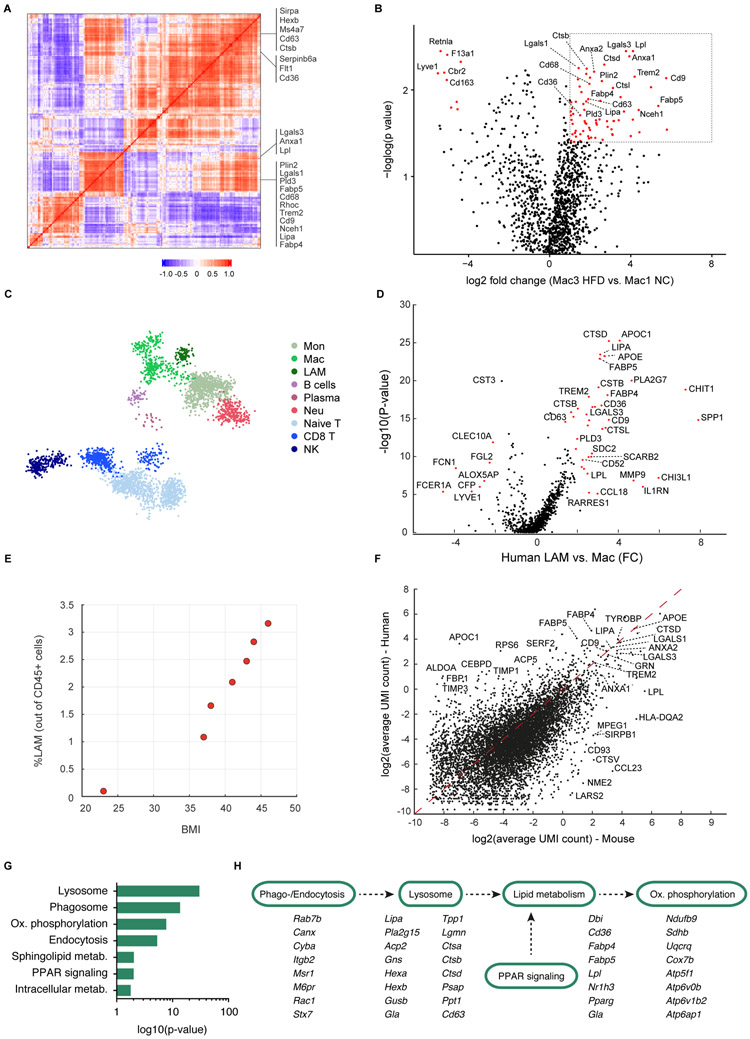
Given its strong expansion under obese conditions, we next sought to functionally characterize the Mac3 subset. Analysis of gene modules most highly associated with this subset revealed a transcriptional signature of Trem2, Lipa, Lpl, Ctsb, Ctsl, Fabp4, Fabp5, Lgals1, Lgals3, Cd9 and Cd36 (Figures 3A and 3B), signatures associated with lipid metabolism and phagocytosis. Hence we named these cells “lipid-associated macrophages” (LAM). In contrast, the Mac1 and Mac2 populations did not express these pathways (Figures 3B and S4A). This signature and specifically Trem2 expression was not found in any other adipose tissue immune cell type (Figures S4B and S4C). Immunofluorescence staining further verified the specificity of Trem2-expression by LAMs on the protein level and confirmed co-expression of Trem2 with CD9 (Figure S4D). LAM cells were also present in EAT harvested from 15-week-old db/db mice (Figure S4E).
Figure 3. A conserved Trem2 signature characterizes the obesity-related adipose tissue macrophages in mice and humans.
A. Gene-gene Pearson correlation heatmap of 200 most variable genes within the monocyte/macrophage compartment. B. Volcano plot showing the fold change of genes (log2 scale) between the HFD Mac3 to NC Mac1 (x-axis) and their p-value significance (y-axis, −loglog scale). Highly significant genes are indicated by a red dot. p-values were determined by Mann-Whitney U test with FDR correction. See Table S5. C. kNN graph of 15,150 QC-positive single cells (172 metacells) of human omental adipose tissue (OAT). See Table S3. D. Volcano plot of LAM vs Mac1 macrophages fold change in the human OAT (x-axis) and their p-value significance (y-axis). Highly significant genes are indicated by a red dot. p-values were determined by Mann-Whitney U test with FDR correction. See Table S6. E. Percentage of LAM cells in human adipose tissue donors as a function of the donors’ body mass index (BMI). F. Scatterplot showing the average UMI counts (log2 scale) of human LAM (y-axis) compared with the mouse LAM cells (x-axis). G. KEGG pathway analysis of LAM genes shared between mouse and human. H. Pathway visualization for selected genes contributing to the KEGG annotation in (G).
See also Figures S4 and S5.
We next sought to explore whether LAM cells can be found outside of adipose tissue, as this lipid metabolism signature is reminiscent of “disease-associated microglia” (DAM) found in the context of Alzheimer’s disease (Keren-Shaul et al., 2017). With the exception of a few tissue specific genes (e.g. Cd36, Fabp4, P2ry13 and Cst7) LAM cells in adipose tissue indeed expressed a highly similar gene profile as DAM cells (Figure S4F). In order to evaluate the conservation of the Trem2 pathway to other tissues we extended our analysis to the liver, given the strong accumulation of lipids in this organ during obesity and the close association between adiposity and non-alcoholic fatty liver disease in humans. Indeed, when examining hepatic CD45+ cells by single-cell RNA-sequencing, we detected _Trem2_-expressing macrophages featuring the LAM gene signature arising under HFD conditions (Figure S4G). These cells demonstrated a remarkable similarity with both adipose tissue LAMs and brain DAMs (Figure S4H), suggesting that the module of _Trem2_-associated genes is characteristic of a conserved macrophage response signature that is present across different tissues.
Given the unexpected finding of LAM cells expressing the Trem2 signature, we sought to confirm their existence in the context of human obesity. To this end, we analyzed the stromal-vascular fraction of visceral adipose tissue from an obese human donor by single-cell RNA-seq of 15,150 QC-positive cells, resulting in 172 metacells (Figures 3C, S5A, S5B, and Table S3). Indeed, _TREM2_-expressing human LAM cells constituted a defined cluster (Figure S5B), which was characterized by a highly conserved gene signature compared to what we had observed in mice, including LIPA, CTSB, CTSL, FABP4, FABP5, LGALS3, CD9 and CD36 (Figure 3D). To further explore the connection of LAM cells and obesity, we analyzed by MARS-seq 4,694 CD45+ cells from visceral adipose tissue of 5 additional obese donors with a BMI range from 36 to 46, and one lean donor with BMI of 23. LAM cells were only observed in obese individuals, and the proportion of LAM cells strongly correlated with BMI (Figure 3E). As in mice, TREM2 was not found in any other adipose tissue cell type (Figure S5B). In addition to the conserved gene signature, human LAM cells expressed a small number of unique genes, including the metallopeptidase inhibitors TIMP1 and TIMP3 as well as the aldolase A gene ALDOA and the apolipoprotein C1 gene APOC1 (Figures 3F).
Next, we investigated the pathways activated in human and mouse LAM cells. We ranked all genes by their relative enrichment in the Trem2-expressing subsets, followed by functional classification of the most characteristic genes according to KEGG pathways. This approach revealed a strong enrichment of LAM-expressed genes in pathways related to phago- and endocytosis as well as lipid metabolism (Figures 3G, 3H, S5C, and S5D), many of which were shared between mouse and human (Figures 3G and 3H), in line with previous reports on the function of Trem2 as a lipid receptor (Coats et al., 2017; Ulland et al., 2017; Wang et al., 2015; Xu et al., 2013), and with the notion of “metabolically-activated” adipose tissue macrophages (Hill et al., 2018; Kratz et al., 2014; Xu et al., 2013). The enriched gene signature was indicative of a highly active pathway initiated by phago- and endocytosis, coupled with lipid metabolism and oxidative phosphorylation (Figure 3H). To functionally validate these findings, we isolated mouse LAM cells by flow cytometry based on the expression of CD9 and CD63 (Figure S2E). We first confirmed the presence of the LAM gene signature by qPCR in sorted CD9+CD63+ macrophages (Figures S5E and S5F). Indeed, this subset was characterized by the expression of Trem2 and the same gene module we had identified for Trem2+ cells (Figure S5F). Bodipy staining confirmed the presence of intracellular lipids specifically in the CD9+CD63+ subset (Figure S5G), further emphasizing the functional role of LAM cells in lipid metabolism (Hill et al., 2018). These data identify Trem2+ LAM cells as a conserved cell type in both mouse and human visceral adipose tissue as well as mouse liver under obese conditions.
Trem2 is essential for adipose tissue macrophage remodeling during obesity
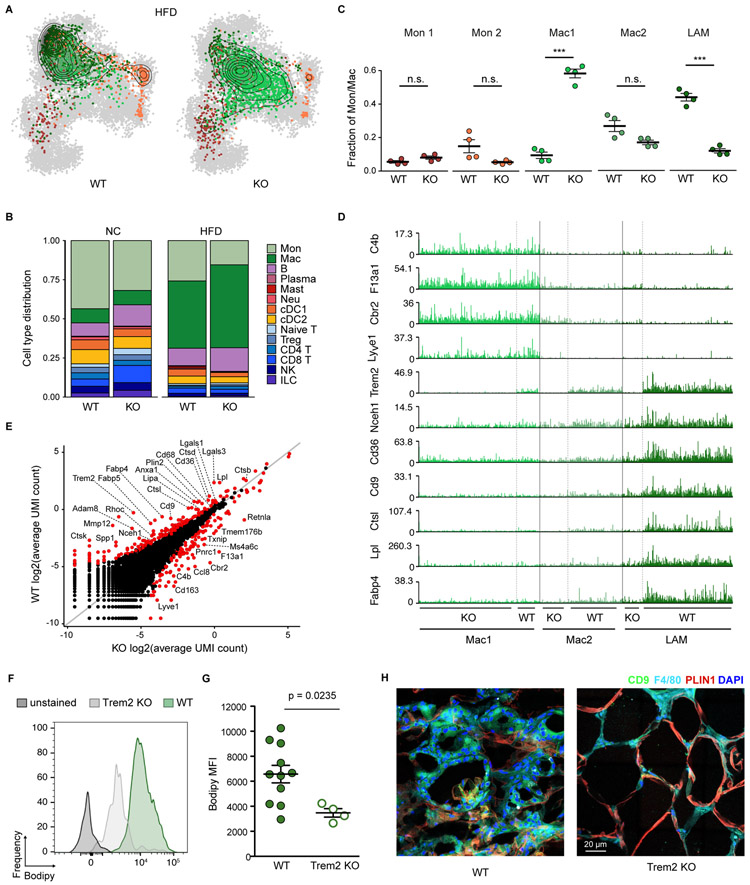
We next determined the functional importance of Trem2 for macrophage remodeling in obese adipose tissue. To this end, we used _Trem2_-deficient mice and littermate controls and analyzed the adipose tissue immune cell compartment after 12 weeks of HFD feeding. We profiled a total of 10,042 QC-positive cells by single-cell RNA-seq, resulting in 133 metacells (Table S4). Remarkably, the monocyte/macrophage compartment of Trem2−/− mice did not fully progress towards the HFD-associated state observed in wild-type littermates. Instead, these cells retained multiple features of NC controls (Figure 4A). While the strong recruitment/expansion of the monocyte-macrophage compartment upon HFD feeding still took place in the absence of Trem2, macrophages in _Trem2_-deficient mice retained the Mac1 and partially Mac2 gene expression signature instead of accumulating LAM cells (Figures 4B, 4C and S4A). This indicates that this surface receptor is not merely a characteristic marker, but an essential driver of the LAM cell molecular program. Trem2−/− macrophages lacked the majority of the LAM gene signature, featuring markedly reduced levels of Lipa, Lpl, Ctsb, Ctsl, Fabp4, Fabp5, Lgals1/3 and Cd36 (Figures 4D and 4E). Consequently, the lipid uptake and storage function of LAMs was abrogated in the absence of Trem2 (Figures 4F and 4G). Trem2 was also required for the formation of LAM cell-rich crown-like structures in obese adipose tissue, as accumulation of LAM cells surrounding adipocytes under HFD conditions was substantially reduced in Trem2−/− (Figure 4H). This function of Trem2 was specific to the metabolically-challenged condition, since Trem2-deficient mice on NC did not show any abnormalities in their adipose tissue immune cell population (Figures S6B and S6C). Other HFD-associated changes in the immune cell compartment were not affected by Trem2 deficiency, with the exception of a modest reduction in number of cDC1 and mast cells (Figures S6D and S6E). Together, these data indicate that Trem2 is a critical checkpoint for the response of adipose tissue myeloid cells to obese conditions and highlight its importance for macrophage remodeling, LAM cell formation, and the assembly of crown-like structures during obesity.
Figure 4. Trem2 is essential for adipose tissue macrophage remodeling during obesity.
A. Projection of the monocyte/macrophage compartment onto the kNN graph of Figure 2A for a total of 10,042 QC-positive immune cells (133 metacells) from EAT of Trem2 knock-out(KO) mice or WT littermate controls on HFD. Contour lines indicate the 2D density of projected cells, down-sampled to 2,289 cells (in each condition). See Table S4. B. Immune cell type distribution of WT and Trem2 KO littermates in NC or HFD. C. Frequency of the EAT monocyte/macrophage subsets as defined in Figure 2 in four KO and four WT mice on HFD. D. Expression level (UMI counts) per cell of selected marker genes in the Mac1, Mac2 and LAM populations found in EAT of WT and KO mice. E. Scatterplot showing the average molecule (UMI) count (log2 scale) of KO (x-axis) compared to WT macrophages (y-axis) in 732 cells from each group, randomly down-sampled from the area of highest density in (A) for equal cell numbers. F. Bodipy signal histograms from CD9+CD63+ adipose tissue macrophages of representative Trem2 WT (green) and Trem2 KO (light gray) mice. G. Analysis of intracellular neutral lipid accumulation by Bodipy MFI in adipose tissue macrophages of Trem2 WT and KO mice. Data are presented as mean ± SEM. P-value indicated was obtained using the Student’s t-test. H. Representative immunofluorescence images of CD9 (green), F4/80 (cyan) and perilipin-1 (red) in EAT sections from Trem2 WT mice (left) and Trem2 KO (right) 12 weeks on HFD. Cell nuclei are shown in blue (DAPI). Scale bar, 20 μm.
See also Figure S6.
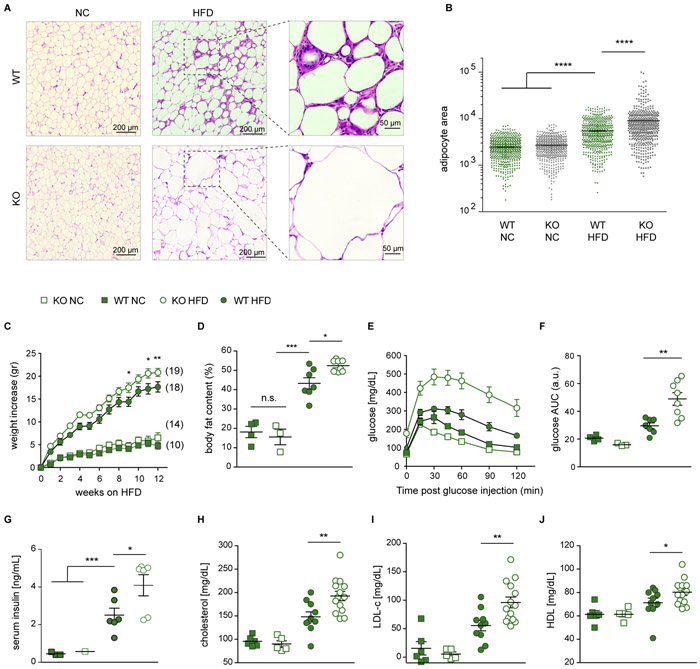
Trem2 prevents adipocyte hypertrophy and loss of systemic metabolic homeostasis
Finally, we sought to determine the physiological importance of LAM cells. Histological analysis of visceral adipose tissue revealed massive adipocyte hypertrophy in the absence of Trem2 (Figures 5A and 5B). This inability to control adipocyte size in the absence of Trem2 was not observed under NC conditions, indicating that LAM cells are required to prevent adipocyte expansion upon loss of adipose tissue homeostasis. Additionally, upon HFD feeding, _Trem2_-deficient mice featured accelerated weight gain (Figure 5C), enhanced body fat accumulation (Figure 5D), marked glucose intolerance (Figures 5E and 5F), elevated serum insulin levels (Figure 5G), hypercholesterolemia (Figure 5H) and increased levels of LDL and HDL cholesterol (Figures 5I and 5J). To ascertain immune cell-specificity for Trem2 deficiency, we generated bone marrow chimeras, in which both wild-type and _Trem2_-deficient mice served as donors of bone marrow transplantation after irradiation. The efficiency of immune cell reconstitution in adipose tissue was over 90% (Figure S7A). Upon HFD feeding, the bone marrow chimeras presented the same metabolic phenotype as global Trem2 knockout mice, including body fat accumulation, hypercholesterolemia, weight gain, and glucose intolerance (Figure S7B-S7H). These data demonstrate that immune cell-intrinsic Trem2 expression is critical for the maintenance of metabolic health. Taken together, these findings suggest a new role for _Trem2_-expressing lipid-metabolizing LAM cells in the local containment of adipocytes that is critical for the prevention of metabolic derangements upon loss of adipose tissue homeostasis.
Figure 5. Trem2 prevents adipocyte hypertrophy and loss of systemic metabolic homeostasis.
A. Representative images of hematoxylin and eosin (H&E) stain of fixated EAT sections. Scale bars, 200 μm, and 50 μm for HFD sections zoomed in areas. B. Area quantification of 500 adipocytes per genotype/diet tissue sections from photos taken to H&E sections. Bars indicate mean ± SEM. **** p < 0.0001 by one-way ANOVA. C. Weight gain over time on HFD. Number of mice in each group is indicated next to each curve. Filled symbols, Trem2 WT; open symbols, Trem2 KO; squares, mice on normal chow (NC); circles, mice on HFD. Data are presented as mean ± SEM. * p < 0.05; ** p < 0.01 by two-way ANOVA. D. Percentage of body fat content. Bars indicate mean ± SEM. * p < 0.05; *** p < 0.001; n.s., non-significant. E. Glucose tolerance test was performed at fasted mice on week 11 of HFD. F. Area under the curve (AUC) as a measure of glucose intolerance, calculated for each individual mouse in (E). Bars indicate mean ± SEM. ** p < 0.01. G. Fasting insulin concentration in the blood of Trem2 cohorts at week 15 on HFD or NC control. Bars indicate mean ± SEM. * p < 0.05; *** p < 0.001. H-J. Total cholesterol (H), LDL (I), and HDL (J) levels in mouse serum from Trem2 cohorts at week 12 on HFD or NC control. Bars indicate mean ± SEM. * p < 0.05; ** p < 0.01.
See also Figure S7.
DISCUSSION
In this study, we identified the lipid receptor Trem2 as a major driver of adipose tissue macrophage responses during obesity. Using single-cell genomic analysis, we explored the immune niche of visceral adipose tissue and found that Trem2+ LAM cells are the most strongly expanded immune cell subset in adipose tissue in multiple mouse models of obesity. The signature of Trem2 and associated genes involved in phagocytosis and lipid catabolism is found in mouse and human macrophages across multiple different organs that undergo loss of metabolic homeostasis, suggesting that this pathway is characteristic of a conserved macrophage response triggered by aberrations in lipid composition, levels, and distribution. In addition, we found that Trem2 signaling drives the formation of LAM cells in crown-like structures in adipose tissue, preventing adipocyte hypertrophy and loss of systemic lipid homeostasis under obese conditions.
These findings may have multiple important implications for our understanding of tissue macrophage biology and Trem2 signaling. First, macrophages are critically involved in the progression of obesity and its metabolic complications (Hotamisligil, 2017). The number of macrophages in adipose tissue positively correlates with the exacerbation of metabolic syndrome and the secretion of proinflammatory mediators from adipose tissue, which has been linked to the development of insulin resistance (Okin and Medzhitov, 2016). Consequently, strategies are being developed to block the infiltration of immune cells into adipose tissue (Kraakman et al., 2015; Sullivan et al., 2013). Our findings highlight a new beneficial function of a subset of adipose tissue macrophages during obesity. Trem2 expression on macrophages prevents the exacerbation of metabolic derangements, by promoting the formation of crown-like structures surrounding lipid-laden and cell death-prone adipocytes. While the formation of these micro-anatomical structures has primarily been correlated with the severity of obesity and metabolic syndrome, the primordial function of macrophage formation around large adipocytes might be driven by Trem2 to locally contain lipid plaques. As such, these findings provide a new perspective on the teleology of immune cell infiltration into adipose tissue during obesity and suggest that interventional strategies targeting immune cells in adipose tissue need to carefully differentiate between beneficial versus pathological immune cell subsets and their relevant pathways. In addition, LAM cells expressed many genes associated with immune suppression (e.g. Lgals1 and Lgals3), suggesting they may be involved in modulating inflammatory responses in the vicinity of cell death and lipid accumulation. Future investigations of the function of the Trem2 pathway and LAM cells in the context of obesity and inflammation will likely yield important insights into their amenability to pharmacologic intervention.
Furthermore, while the gene expression programs of tissue macrophages are generally highly divergent and strongly shaped by the tissue environment, with distinct gene modules driving the functional characteristics of locally resident macrophages (Lavin et al., 2014; Okabe and Medzhitov, 2016), our findings suggest that the module of the Trem2-pathway may represent a conserved and general macrophage response for detection of extracellular pathogenic lipids across multiple tissues (Deczkowska et al., 2018). The gene signature we discovered in adipose tissue macrophages is highly similar to that found in DAM cells in the brains of mice and humans with neurodegenerative disease (Keren-Shaul et al., 2017), hepatic macrophages during obesity (Figures S4G and S4H), and in aortic macrophages during atherosclerosis (Cochain et al., 2018). Accumulation of extracellular lipids and inflammation, as occurring upon excessive cell death, are common features of adiposity, progressive neurodegeneration, and atherosclerosis. As such, Trem2 might function as a pattern-recognition receptor for common signals indicative of loss of tissue homeostasis, and may be found in additional tissues and contexts characterized by extracellular lipid accumulation. As obesity, vascular plaque formation and neurodegeneration share numerous common elements in their pathophysiology (Cai, 2013; Khera et al., 2016; Lee, 2011; Lovren et al., 2015; Spielman et al., 2014), a deeper understanding of the biology of Trem2-expressing macrophages may yield fundamental insights into the common molecular etiology of these diseases and identify novel therapeutic handles to target neurodegeneration and metabolic disorders.
Finally, our study provides a new perspective on the function of Trem2 in metabolic disease. An initial report using global overexpression of Trem2 had documented a role for this receptor in adipogenesis and weight gain (Park et al., 2015). In contrast, a recent human study found a positive correlation between the levels of circulating soluble TREM2 and insulin resistance, but not with body mass index (Tanaka et al., 2018). Our results underline the potential importance of Trem2 in adipose tissue macrophages for the prevention of adipocyte hypertrophy and the regulation of systemic cholesterol levels. Given its recently appreciated centrality in Alzheimer’s disease (Guerreiro et al., 2013; Jonsson et al., 2013; Keren-Shaul et al., 2017; Ulland et al., 2017; Wang et al., 2015), numerous efforts are underway to stimulate TREM2 signaling pharmacologically. Our study highlights the possibility that harnessing TREM2 function might have beneficial effects in multiple disease contexts beyond neurodegeneration.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents should be directed to and will be fulfilled by lead author Ido Amit (ido.amit@weizmann.ac.il).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Wild-type (WT) mice (C57Bl/6) were purchased from Harlan and housed in the Weizmann Institute animal facility. Only male mice were used. All mice were provided with normal chow and water ad libitum, and housed under a strict 12-hour light-dark cycle. At age 8-9 weeks, for some mice the normal chow was replaced with a high-fat diet (HFD; irradiated Rodent Diet with 60 kcal% fat, D12492i Research Diets Inc., New Brunswick, NJ). Trem2−/− knock-out (KO) mice were provided by Prof. Marco Colonna (Turnbull et al., 2006). Founding KO breeders were crossed with WT mice at the Weizmann Institute animal facility to produce second-generation cohorts of WT and KO littermates. F1 offspring was bred to produce homozygous WT or KO. Heterozygous F2 mice were not used for experiments. For bone marrow chimera experiments, mice were given a sublethal dose of total body irradiation (900 Gy). 16 hours later mice were transplanted with 3x106 bone marrow cells. Mice were allowed 8 weeks for reconstitution before HFD treatment. Reconstitution efficiency was tracked using bone marrow with the congenic marker CD45.1. All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC).
Ms4a3 reporter experimental model
The Ms4a3Cre mouse model was generated at the Shanghai Model Organisms Center, Inc. Briefly, an Ires-Cre gene fusion was inserted into the 3' untranslated region of the Ms4a3 gene by CRISPR-Cas9 technique in C57BL/6 zygotes. To eliminate off-target effects, knock-in mice were then backcrossed onto a C57BL/6 background for three generations. This strain was genotyped by PCR using the following primers: Common forward primer 5’- AGAGAAATCATCAGGGCAGAAAT -3’ Mutant reverse primer 5’- TTGGCGAGAGGGGAAAGAC -3’(412 bp fragment) Wild-type reverse primer 5’-GAAAGGGGAACAAGCGAAGAT-3’ (517 bp fragment). Rosa26tdTomato reporter mice have been previously described (Madisen et al., 2010). All mice were bred in a specific pathogen-free animal facility at the Shanghai Jiao Tong University School of Medicine. All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Shanghai Jiao Tong University School of Medicine and were performed in compliance with the University’s guidelines for the care and use of laboratory animals.
Human samples
Biopsies from visceral adipose tissue from the omental depot (OAT) were obtained from six obese individuals and one lean donor with participant informed consent obtained after the nature and possible consequences of the studies were explained under protocols approved by the Institutional Review Boards of the Perelman School of Medicine at the University of Pennsylvania, the Children’s Hospital of Philadelphia, or the Tel Aviv Sourasky Medical Center. The obese donors underwent bariatric surgery, the lean donor underwent cholecystectomy. OAT samples were placed in 1 mL of DMEM, and finely minced under sterile conditions before digestion in 50 mL of DMEM with 3 mg/1 mL collagenase IV (Gibco). Samples were incubated at 37°C in a rotating oven for 20-60 min. Adipocyte and stromal vascular fractions (SVF) were separated by centrifugation, and red blood cells (RBCs) were removed from the SVF by histopaque gradient (Sigma). Single-cell RNA-sequencing libraries were prepared using the MARS-seq pipeline or Chromium platform (10x genomics), and sequenced on the MiSeq 500 or HiSeq 2500 Sequencing System (Illumina). Participants were between 31-73 years of age, with a BMI range of 23-46.
METHOD DETAILS
Isolation of adipose tissue-derived leukocytes
Mice were sacrificed by cervical dislocation and perfused immediately through the left ventricle of the heart with 20 ml of phosphate-buffered saline (PBS) to remove circulating leukocytes from the tissue. The epididymal (visceral) adipose tissue (EAT) was readily located and excised right above the epididymis. The tissue was placed in a 50 ml tube with 10 ml DMEM without phenol red at room temperature, cut into tiny bits with scissors, incubated with collagenase II at 37°C for 20 min. while gently agitating, filtered through a 100-μm cell strainer, and spun down at 500g for 10 min with low acceleration/brake, beginning at room temperature and cooling to 4°C. Cells were resuspended in 500 μl RBC lysis solution (Sigma) and incubated on ice for 2-5 min. (depending on the initial amount of tissue) and washed. The resulting cell suspension was incubated with anti-CD45 or a cocktail of antibodies for selected markers.
Flow cytometry and single-cell capture
After staining, cells were washed and resuspended in cold FACS buffer (0.5% BSA and 2 mM EDTA in PBS), stained with fluorophore-conjugated anti-mouse CD45 antibody, and filtered through a 40-μm strainer. Right before sorting, cells were stained with propidium iodide to exclude dead/dying cells. Cell sorting was performed using a BD FACSAria Fusion flow cytometer (BD Biosciences), gating for CD45+ cells (leukocytes) after exclusion of dead cells and doublets. Single cells were sorted into 384-well capture plates containing 2 μl of lysis solution and barcoded poly(T) reverse-transcription (RT) primers for scRNA-seq as described previously (Jaitin et al., 2014). Immediately after sorting, plates were spun down to ensure cell immersion into the lysis solution, snap-frozen on dry ice and stored at −80°C until further processing.
For marker-based validations, samples were stained using the following antibodies: APC-conjugated CD45 (leukocytes), eFluor450-conjugated CD3, CD19, NK1.1, and Ly6G (Lineage negative to exclude T, B, NK and neutrophils), PE-conjugated CD11b (myeloid cells), APC/Cy7-conjugated Ly6C (monocytes), PerCP/Cy5.5-conjugated F4/80 (macrophages), FITC-conjugated CD9 and PE/Cy7-conjugated CD63, purchased from eBioscience or Biolegend, and DAPI (in the same channel with Lineage staining), for live/dead cell detection. Cell populations from selected gates (Figures S2E and S3A) were sorted for RT-qPCR validation (Figures S5E and S5F). Cells were analyzed using BD FACSDIVA software (BD Bioscience) and FlowJo software (FlowJo, LLC).
Single cell library preparation
Single cell libraries were prepared with Massively Parallel Single-Cell RNA-seq method (MARS-seq) (Jaitin et al., 2014). In brief, mRNA from single cells sorted into cell capture plates was barcoded, converted into cDNA and pooled using an automated pipeline. Subsequently, the pooled sample was linearly amplified by T7 in vitro transcription, and the resulting RNA was fragmented and converted into a sequencing-ready library by tagging the samples with pool barcodes and Illumina sequences during ligation, reverse transcription, and PCR. Each pool of cells was tested for library quality and library concentration was assessed.
Immunofluorescence
Immunostaining of frozen adipose tissue were performed as described previously with some modifications (Honvo-Houeto and Truchet, 2015). Mice were sacrificed and epididymal white adipose tissues harvested and chopped into small pieces, fixed in 4% Paraformaldehyde solution in PBS (Santa Cruz Biotechnology) overnight at 4°C, transferred to 10-30% sucrose in PBS at 4°C for 3 days, then embedded in OCT (optimal cutting temperature compound), frozen on dry ice and stored at −80°C until further processing. Immunofluorescent staining was carried out on 35-μm thick sections. Samples were permeabilized with 0.3% Triton X-100 for 15 min. After washing three times with PBS, sections were blocked with a solution containing 5% FBS and 1% BSA in PBS pH 7.4 for 2 hours at room temperature. The blocking solution was then replaced with a cocktail of primary antibodies, including rabbit anti-CD9 (1:200, Abcam), rat anti-F4/80 (1:200, Abcam), goat anti-perilipin-1 (1:200, Abcam) diluted in blocking solution, and incubated overnight at 4°C. The secondary antibody mixture included 1:500 dilutions of the following antibodies: DyLight550 donkey anti-Goat IgG-heavy and light (Bethyl), donkey anti-Rat IgG-heavy and light chain cross-adsorbed Antibody DyLight594 Conjugated (Bethyl), and Alexa Fluor 647-AffiniPure donkey anti-rabbit IgG (H+L; Jackson ImmunoResearch Labs) and was diluted in 1% BSA in PBS and incubated for 1 hour at room temperature and followed DAPI staining. Trem2 protein was visualized using rat anti-Trem2 antibody (1:50, a gift from Prof. Christian Haass, Ludwig-Maximilians University Munich, Germany;(Xiang et al., 2016)) together with CD9, perilipin-1 and DAPI. All sections were imaged on a Leica STED confocal microscope with 63X objective (Leica Microsystems, Germany). Images were analyzed using the Fiji software (Schindelin et al., 2012). Single adipocytes were manually demarcated using the selection tool and these areas were tracked using the ROI Manager. Only complete adipocytes in the field of view were considered. The relative area of the demarcated adipocytes was then quantified (Figure 5B).
Histology
Adipose tissues were fixed in in a 4% paraformaldehyde solution in PBS for two days and embedded in paraffin (Leica Biosystems). Four μm-thick sections were stained with hematoxylin and eosin. All sections were imaged on a Leica DMi8 widefield microscope with 20X objective (Leica Microsystems, Germany), and processed with Fiji.
Intracellular neutral lipid accumulation
Bodipy powder was resuspended in DMSO and aliquoted at 1mg/ml. The staining was performed at a dilution of 1/200. Single cell suspensions of adipose tissue immune cells were stained for Bodipy along with surface markers for 20 min at 4°C. Mean fluorescence intensity (MFI) of each gated population was used to quantify the Bodipy signal by flow cytometry.
Glucose tolerance test
Mice were fasted for 14 h and subsequently given 200 μl of a 0.1 g/ml (10%) glucose solution (JT Baker) by intraperitoneal injection. Blood glucose was determined at 0, 15, 45, 30, 60, 90 and 120 min post injection (Contour blood glucose meter, Bayer).
Body composition measurements
Lean, fluid and fat mass of of individual mice was measured by live non-invasive magnet resonance at week eleven on high-fat diet (HFD) with a Minispec LF50 body com position device (Bruker).
Lipid profiling
Mice were fasted for 14h and subsequently anesthetized by intraperitoneal injection of 200 microliter of 10 vol. % ketamine. Blood was obtained from the eye. Measurements of concentrations of triglycerides, HDL, and cholesterol from blood plasma were performed with the Kenshin-2 kit (Medtechnica) at a SPOTCHEM EZ sp-4430 (Arkray).
Concentration of insulin in the blood serum
Concentrations of insulin (Ultra-sensitive mouse insulin ELISA kit, Crystal Chem,#90080) in the serum were measured using ELISA according to the manufacturer’s instructions.
SINGLE-CELL RNA-SEQUENCING ANALYSES
MARS-seq processing
scRNA-seq libraries (pooled at equimolar concentration) were sequenced on an Illumina NextSeq 500 at a median sequencing depth of ~40,000 reads per cell. Sequences were mapped to the mouse (mm10) and human (hg38) genome, respectively. Demultiplexing and filtering was performed as previously described (Jaitin et al., 2014), with the following adaptations: Mapping of reads was performed using HISAT (version 0.1.6); reads with multiple mapping positions were excluded. Reads were associated with genes if they were mapped to an exon, using the ensembl gene annotation database (embl release 90). Exons of different genes that shared a genomic position on the same strand were considered as a single gene with a concatenated gene symbol. The level of spurious unique molecular identifiers (UMIs) in the data were estimated by using statistics on empty MARS-seq wells, and excluded rare cases with estimated noise > 5% (median estimated noise over all experiments was 2%).
Metacell modeling
We used the R package “MetaCell” (Baran et al., 2018) with the following specific parameters (complete script reproducing all analyses from raw data will be available). We removed specific mitochondrial genes, immunoglobulin genes, and genes linked with poorly supported transcriptional models (annotated with the prefix “Rp-”). We then filtered cells with less than 400 UMIs. Gene features were selected using the parameter Tvm=0.3 and a minimum total UMI count > 50. See Table S7 for list of genes selected as features in each analysis. We subsequently performed hierarchical clustering of the correlation matrix between those genes (filtering genes with low coverage and computing correlation using a down-sampled UMI matrix) and selected the gene clusters that contained anchor genes.
The gene selection strategy discussed above retained a total of 425 marker gene features for the computation of the MetaCell balanced similarity graph. We used K = 150, 500 bootstrap iterations and otherwise standard parameters. Metacells were annotated as Monocytes /Macrophages or others, by applying a straightforward analysis of known cell type marker genes (e.g. Ear2, Fn1, Ccr2, Mrc1, Cd3d, Cd79b, and more). Subsets of Monocytes and Macrophages were obtained by hierarchical clustering of the confusion matrix (Figure S1C) and supervised analysis of enriched genes in homogeneous groups of metacells.
In Tables S1-S4, we refer to the log2 of a metacell’s footprint, which is the gene-wise average UMI count per metacell normalized by the median UMI count across all metacells (Baran et al., 2018).
kNN projection of cells on the graph of Figure 2A
To project the adipose tissue Monocytes/Macrophages obtained from Trem2 knock-out mice and their littermate wild-type controls onto the kNN graph of Figure 2A, we generated a UMI matrix of the 200 most variable genes of the Monocytes/Macrophages from Figure 2A. As query data, we utilized the UMI matrix of those genes of the Monocytes/Macrophages from the Trem2 knock-out experiment. To obtain the K=6 nearest neighbors of Pearson correlation to every cell in the query data, we applied the knn.index.dist function of the R package “KernelKnn” (version 1.0.8). We averaged the x- and y-coordinates of the nearest neighbors to calculate the position of every cell from the Trem2 knock-out experiment on the graph of Figure 2A.
Defining a module gene signature
To define a module gene signature for Monocytes/Macrophages (Figure 3A), we identified a group of 33 genes (incl. Trem2, Cd9, Lpl) that exhibited a strong Pearson correlation across the metacells’ log2 footprint expression of the 200 most variable genes excluding genes associated with cell cycle and stress that were filtered from these lists in advance.
Trajectory analysis
To infer trajectories and align cells along a differentiation pseudo-time, we used the published package Slingshot (Street et al., 2018). In short, Slingshot is a tool that uses pre-existing clusters to infer lineage hierarchies (based on minimal spanning tree, MST) and align cells in each cluster on a pseudo-time trajectory. Heatmaps presented in Figure S3B show smoothed normalized expression of selected genes across slingshot trajectory.
Human-mouse gene comparison
To compare the gene expression of LAM and other macrophages between human and mouse (Figure 3F), we used the orthologous gene annotation from ensembl 95 of the BioMart browser. We only plotted genes for which an orthologue between mouse and human existed. KEGG assignments were done using DAVID (Dennis et al., 2003).
Statistical Analyses
Differential gene expression analysis was performed upon down-sampling of the UMI matrix as part of the MetaCell package on molecules/1,000 UMIs by Mann Whitney U test with false-discovery rate (FDR) correction. Metabolic parameters in Figure 5 were compared by one- or two-way ANOVA. Relative frequencies of cell types in Figures S1 and S6 were analyzed by Mann Whitney U test.
DATA AND SOFTWARE AVAILABILITY
The accession number for the raw sequence reported in this paper is EGA: . The accession number for the processed data reported in this paper is NCBI GEO: GSE . Scripts reproducing the analysis will be available at: https://bitbucket.org/amitlab/.
Supplementary Material
Supplemental
Figure S1. Single-cell characterization of the adipose tissue immune niche during obesity progression. Related to Figure 1. A, B. Weight gain (A) and body composition (B) of mice on high-fat diet (HFD) or normal chow (NC). Error bars indicate standard deviation. N=8. C. Heatmap of differential gene expression for all cells colored by the assigned metacells, as in Figure 1B. D. kNN graph of epididymal adipose tissue immune cells of wild-type mice on NC, down-sampled to 2,187 cells each time-point, annotated as in Figure 1B. Each time-point is contributed by either 2 (6 weeks) or 4 mice (12 or 18 weeks). E, F. Relative frequencies of higher abundant (E) and less abundant (F) immune cell types of mice on HFD. Every dot represents a mouse. * p < 0.05; ** p < 0.01; *** p < 0.001, NS. not significant by Mann Whitney U-test. G. Weight gain of mice on db/db mice and littermate wild-type controls with age. Error bars indicate SD. N=6. H. kNN graph of epididymal adipose tissue immune cells of db/db mice and littermate wild-type controls with age, down-sampled to 1,985 cells each.
Figure S2. Analysis of monocyte and macrophage subsets. Related to Figure 2. A. Log2 of unique molecular identifier (UMI) count of selected genes in individual cells on the kNN graph of Figure 2A. B. kNN graph of monocytes and macrophages from epididymal adipose tissue of db/db mice. C. Log2 of average UMI count of selected genes across metacells within the monocyte/macrophage compartment. D. Log2 of UMI count of selected genes in individual metacells from the kNN graph in (B). E. Representative pseudocolor cell density FACS plots showing the strategy to enrich for macrophage (CD11b+F4/80+) subpopulations using the CD9 and CD63 markers. Numbers indicate percentage of gated cells. Macrophages from obese mice are predominantly CD9+CD63+. Upper row, 16-week old WT mouse on NC; bottom row, WT mouse on HFD for 12 weeks.
Figure S3. Adipose tissue macrophages during obesity are derived from monocytes. Related to Figure 2. A. Flow cytometric analysis of the frequency of Tomato+ cells (Ms4a3 lineage) among adipose tissue macrophages, Ly6Chi blood monocytes, Ly6Chi liver monocytes and Kupffer cells from 8 week-old or 16 week-old Ms4a3Cre-Rosa26tdTomato mice fed with NC or HFD during 8 weeks. Numbers indicate percentage of gated cells, used for the analysis shown in Figure 2H. B. Gene expression profiles driving the molecular differentiation trajectory from monocyte towards the Mac3 macrophage subset, ordered according to Slingshot pseudo-time trajectory (STAR Methods).
Figure S4. Conserved Trem2 signature characterizes obesity-related macrophages in mice and humans. Related to Figure 3. A. Volcano plot showing the fold change of genes (log2 scale) between Mac2 from wild-type mice on HFD to Mac1 from wild-type mice on NC (x-axis) and their significance (y-axis, −loglog scale). Highly significant genes are indicated by a red dot. P-values were determined by Mann-Whitney U-test with FDR correction. B. Log2 of average unique molecular identifier (UMI) count of Trem2 across all metacells as defined in Figure 1. C. Log2 of unique molecular identifier (UMI) count of Trem2 gene expression in individual cells on the kNN graph of Figure 1B. D. Representative immunofluorescence images of Trem2 (Cyan), CD9 (green), and perilipin-1 (red) in EAT sections of 16-week old WT mice after 12 weeks on HFD (above) or on NC (below). Cell nuclei are stained with DAPI (blue). Scale bar, 50 μm. Shown are the individual channels and their composite image on the right. E. Scatterplot comparing Z-scores of log2 fold changes of the Trem2 module genes between LAM versus Mac1 in wild-type mice (x-axis) versus db/db (y-axis). R indicates the Pearson correlation coefficient. F. Scatterplot showing the average UMI counts (log2 scale) of mouse LAM (y-axis) compared with the mouse DAM from the brain of Alzheimer’s disease mice (Keren-Shaul et al., 2017) (x-axis). G. Detection of LAM cells in the liver of mice feeding on HFD. Shown are percentages of immune cells expressing a LAM signature above a LAM score threshold of 10 in the liver of two mice fed a HFD for 27 weeks and two mice on NC. H. Scatterplot showing the average UMI counts (log2 scale) of LAM detected in the liver (y-axis) compared with LAM isolated from EAT (x-axis) in mice fed a HFD.
Figure S5. Characteristics of LAM cells in mice and humans. Related to Figure 3. A. kNN graph of 15,150 cells of human adipose tissue cells. See Table S3. B. Log2 of UMI count of selected genes in individual cells on the kNN graph from (A). C, D. KEGG pathway analysis for top-expressed genes of mouse (C) and human (D) LAM. E, F. qPCR of bulk-sorted subsets of adipose tissue macrophages for discovered LAM marker genes (E) and Trem2 module genes (F). G. Bodipy staining of bulk-sorted subsets of adipose tissue macrophages.
Figure S6. Characterization of adipose tissue immune cells in Trem2 knock-out mice. Related to Figure 4. A. Cell type distribution within the monocyte/macrophage compartment of KO and WT mice on HFD and NC. B. Projection of the monocyte/macrophage compartment onto the kNN graph of Figure 2A from a total of 10,042 QC-positive immune cells (133 metacells; Table S4) from EAT of Trem2 knock-out (KO) mice or WT littermate controls on NC. Contour lines indicate the 2D density of projected cells, down-sampled to 453 cells. C. Frequency of the EAT Mon/Mac subsets as defined in Figure 2 in each of two KO and two WT mice on NC. D, E. Relative frequencies of abundant (D) and less abundant (E) immune cell types. Each symbol represents a mouse. * p < 0.05; ** p < 0.01; *** p < 0.001, NS. not significant by a Mann Whitney U-test.
Figure S7. Immune cell specificity of Trem2 in metabolic homeostasis. Related to Figure 5. A. Representative FACS plots showing the degree of reconstitution eight weeks post bone marrow transplantation from CD45.2 donors into CD45.1 recipient mice, at the time of setting ten mice in each group, WT and Trem2 KO (20 mice in total), to feed a HFD. B. Percentage body fat content. Fat mass, lean mass and liquid mass of individual animals was measured by live non-invasive magnet resonance on week 18 of HFD. Filled circles, Trem2 WT; open circles, Trem2 KO. C-E. Total cholesterol (C), LDL (D), and HDL (E) levels in mouse serum from Trem2 cohorts at week 12 on HFD or NC control. F. Weight gain over time on HFD. G. Glucose tolerance test was performed on fasted mice at week 18 of HFD. H. Area under the curve (AUC) as a measure of glucose intolerance, calculated for each individual mouse in (G). Bars indicate mean ± SEM. * p < 0.05; n.s., non-significant.
Table S1
Table S1. Mouse epididymal adipose tissue metacell data. Related to Figure 1. Footprint gene expression of 244 metacells obtained from scRNA-seq of 21,210 visceral adipose tissue immune cells from wild-type mice.
Table S2
Table S2. Leptin receptor db/db mutant mouse epididymal adipose tissue metacell data. Related to Figures 1 and 2. Footprint gene expression of 75 metacells obtained from scRNA-seq of 8,372 visceral adipose tissue immune cells from db/db mice and wild-type littermate controls.
Table S3
Table S3. Human omental adipose tissue metacell data. Related to Figure 3. Footprint gene expression of 172 metacells obtained from scRNA-seq of 15,150 whole omental adipose tissue cells from an obese human subject.
Table S4
Table S4. Trem2 knockout mouse epididymal adipose tissue metacell data. Related to Figure 4. Footprint gene expression of 133 metacells obtained from scRNA-seq of 10,042 visceral adipose tissue immune cells from Trem2 knock-out mice and wild-type littermate controls.
Table S5
Table S5. Differential gene expression analysis between mouse macrophage subsets. Related to Figure 3. log2 fold change and respective p value of a Mann Whitney U test with false discovery rate (FDR) correction per gene comparing Mac3 HFD and Mac1 NC (Figure 3B), Mac2 HFD and Mac1 NC (Figure S3A), or Mac3 HFD and Mac2 HFD (Figure S3B), respectively.
Table S6
Table S6. Differential gene expression analysis between human macrophage subsets. Related to Figure 3. Fold change and respective p value of a Mann Whitney U test with false discovery rate (FDR) correction per gene comparing LAM and Mac in the human data.
Table S7
Table S7. Gene groups used as features for metacells clustering. List of genes used for clustering in the various metacells analysis throughout the paper, as well as genes used for gene-gene correlation analysis in figure 3A.
ACKNOWLEDGMENTS
We thank Genia Brodsky from the Scientific Illustration unit of the Weizmann Institute for artwork, Tomer Meir Salame from the Cell Cytometry unit, Calanit Raanan and Marina Cohen for histology, and members of the Amit and Elinav laboratories for discussions. We thank Prof. Christian Haass from the LMU Munich for providing the anti Trem2 antibody (Xiang et al., 2016). I.A. is supported by the Chan Zuckerberg Initiative (CZI), the HHMI International Scholar award, the European Research Council Consolidator Grant (ERC-COG) 724471-HemTree2.0, the Thompson Family Foundation, an MRA Established Investigator Award (509044), the Israel Science Foundation (703/15), the Ernest and Bonnie Beutler Research Program for Excellence in Genomic Medicine, the Helen and Martin Kimmel award for innovative investigation, the NeuroMac DFG/Transregional Collaborative Research Center Grant, an International Progressive MS Alliance/NMSS PA-1604-08459 and an Adelis Foundation grant. I.A. is the incumbent of the Alan and Laraine Fischer Career Development Chair. D.A.H. is supported by NIH DK116668 and a CHOP Junior Faculty Grant. M.A.L. is supported by NIH DK49780. D.A.H. and M.A.L. acknowledge The Center for Applied Genomics at the Children's Hospital of Philadelphia, and the Human Metabolic Tissue Bank of the Penn Diabetes Research Center (P30 NIH DK19525). C.A.T. is supported by the Edward Mallinckrodt, Jr. Foundation, the Agilent Early Career Professor Award, the Global Probiotics Council, and grants by the PennCHOP Microbiome Program, the Penn Institute for Immunology, the Penn Center for Molecular Studies in Digestive and Liver Diseases (P30-DK-050306), the Penn Skin Biology and Diseases Resource-based Center (P30-AR-069589), and the Penn Diabetes Research Center (P30-DK-019525). N.Z. is supported by the Gilead Sciences International Research Scholars Program in Liver Disease. H.S. is supported by The V. R. Schwartz Research Fellow Chair. E.E. is supported by: Y. and R. Ungar; Leona M. and Harry B. Helmsley Charitable Trust; the Adelis Foundation; J. L. and V. Schwartz; D. L. Schwarz; and by grants funded by the European Research Council; the Israel Science Foundation; the Helmholtz Foundation; and the Bill & Melinda Gates Foundation. E.E. is a senior fellow, Canadian Institute of Advanced Research (CIFAR), and an international scholar, Bill & Melinda Gates Foundation and Howard Hughes Medical Institute (HHMI). Raw and processed single-cell RNA-sequencing data can be downloaded from NCBI GEO with accession number GSE128518.
Footnotes
Declaration of Interests
A patent application has been filed related to this work.
REFERENCES
- Bahary N, Leibel RL, Joseph L, and Friedman JM (1990). Molecular mapping of the mouse db mutation. Proc Natl Acad Sci U S A 87, 8642–8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran Y, Sebe-Pedros A, Lubling Y, Giladi A, Chomsky E, Meir Z, Hoichman M, Lifshitz A, and Tanay A (2018). MetaCell: analysis of single cell RNA-seq data using k-NN graph partitions. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas SK, and Mantovani A (2012). Orchestration of metabolism by macrophages. Cell metabolism 15, 432–437. [DOI] [PubMed] [Google Scholar]
- Boulenouar S, Michelet X, Duquette D, Alvarez D, Hogan AE, Dold C, O'Connor D, Stutte S, Tavakkoli A, Winters D, et al. (2017). Adipose Type One Innate Lymphoid Cells Regulate Macrophage Homeostasis through Targeted Cytotoxicity. Immunity 46, 273–286. [DOI] [PubMed] [Google Scholar]
- Cai D (2013). Neuroinflammation and neurodegeneration in overnutrition-induced diseases. Trends Endocrinol Metab 24, 40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakarov S, Lim HY, Tan L, Lim SY, See P, Lum J, Zhang XM, Foo S, Nakamizo S, Duan K, et al. (2019). Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science 363. [DOI] [PubMed] [Google Scholar]
- Cipolletta D, Feuerer M, Li A, Kamei N, Lee J, Shoelson SE, Benoist C, and Mathis D (2012). PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature 486, 549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coats BR, Schoenfelt KQ, Barbosa-Lorenzi VC, Peris E, Cui C, Hoffman A, Zhou G, Fernandez S, Zhai L, Hall BA, et al. (2017). Metabolically Activated Adipose Tissue Macrophages Perform Detrimental and Beneficial Functions during Diet-Induced Obesity. Cell Rep 20, 3149–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochain C, Vafadarnejad E, Arampatzi P, Pelisek J, Winkels H, Ley K, Wolf D, Saliba AE, and Zernecke A (2018). Single-Cell RNA-Seq Reveals the Transcriptional Landscape and Heterogeneity of Aortic Macrophages in Murine Atherosclerosis. Circ Res 122, 1661–1674. [DOI] [PubMed] [Google Scholar]
- Deczkowska A, Keren-Shaul H, Weiner A, Colonna M, Schwartz M, and Amit I (2018). Disease-Associated Microglia: A Universal Immune Sensor of Neurodegeneration. Cell 173, 1073–1081. [DOI] [PubMed] [Google Scholar]
- Dennis G Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, and Lempicki RA (2003). DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4, P3. [PubMed] [Google Scholar]
- Feuerer M, Herrero L, Cipolletta D, Naaz A, Wong J, Nayer A, Lee J, Goldfine AB, Benoist C, Shoelson S, et al. (2009). Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med 15, 930–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JS, Younkin S, et al. (2013). TREM2 variants in Alzheimer's disease. The New England journal of medicine 368, 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SJ, Glatman Zaretsky A, Andrade-Oliveira V, Collins N, Dzutsev A, Shaik J, Morais da Fonseca D, Harrison OJ, Tamoutounour S, Byrd AL, et al. (2017). White Adipose Tissue Is a Reservoir for Memory T Cells and Promotes Protective Memory Responses to Infection. Immunity 47, 1154–1168 e1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DA, Lim HW, Kim YH, Ho WY, Foong YH, Nelson VL, Nguyen HCB, Chegireddy K, Kim J, Habertheuer A, et al. (2018). Distinct macrophage populations direct inflammatory versus physiological changes in adipose tissue. Proc Natl Acad Sci U S A 115, E5096–E5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honvo-Houeto E, and Truchet S (2015). Indirect Immunofluorescence on Frozen Sections of Mouse Mammary Gland. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS (2017). Inflammation, metaflammation and immunometabolic disorders. Nature 542, 177–185. [DOI] [PubMed] [Google Scholar]
- Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, et al. (2014). Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, et al. (2013). Variant of TREM2 associated with the risk of Alzheimer's disease. The New England journal of medicine 368, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti TD, and Dixit VD (2012). Immunological complications of obesity. Nature immunology 13, 707–712. [DOI] [PubMed] [Google Scholar]
- Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, et al. (2017). A Unique Microglia Type Associated with Restricting Development of Alzheimer's Disease. Cell 169, 1276–1290.e1217. [DOI] [PubMed] [Google Scholar]
- Khera AV, Emdin CA, Drake I, Natarajan P, Bick AG, Cook NR, Chasman DI, Baber U, Mehran R, Rader DJ, et al. (2016). Genetic Risk, Adherence to a Healthy Lifestyle, and Coronary Disease. The New England journal of medicine 375, 2349–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlgruber AC, Gal-Oz ST, LaMarche NM, Shimazaki M, Duquette D, Nguyen HN, Mina AI, Paras T, Tavakkoli A, von Andrian U, et al. (2018). gammadelta T cells producing interleukin-17A regulate adipose regulatory T cell homeostasis and thermogenesis. Nat Immunol 19, 464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraakman MJ, Kammoun HL, Allen TL, Deswaerte V, Henstridge DC, Estevez E, Matthews VB, Neill B, White DA, Murphy AJ, et al. (2015). Blocking IL-6 trans-signaling prevents high-fat diet-induced adipose tissue macrophage recruitment but does not improve insulin resistance. Cell metabolism 21, 403–416. [DOI] [PubMed] [Google Scholar]
- Kratz M, Coats BR, Hisert KB, Hagman D, Mutskov V, Peris E, Schoenfelt KQ, Kuzma JN, Larson I, Billing PS, et al. (2014). Metabolic dysfunction drives a mechanistically distinct proinflammatory phenotype in adipose tissue macrophages. Cell Metab 20, 614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, and Amit I (2014). Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EB (2011). Obesity, leptin, and Alzheimer's disease. Ann N Y Acad Sci 1243, 15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovren F, Teoh H, and Verma S (2015). Obesity and atherosclerosis: mechanistic insights. Can J Cardiol 31, 177–183. [DOI] [PubMed] [Google Scholar]
- Lynch L, Hogan AE, Duquette D, Lester C, Banks A, LeClair K, Cohen DE, Ghosh A, Lu B, Corrigan M, et al. (2016). iNKT Cells Induce FGF21 for Thermogenesis and Are Required for Maximal Weight Loss in GLP1 Therapy. Cell Metab 24, 510–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdougall CE, Wood EG, Loschko J, Scagliotti V, Cassidy FC, Robinson ME, Feldhahn N, Castellano L, Voisin MB, Marelli-Berg F, et al. (2018). Visceral Adipose Tissue Immune Homeostasis Is Regulated by the Crosstalk between Adipocytes and Dendritic Cell Subsets. Cell Metab 27, 588–601 e584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis D (2013). Immunological goings-on in visceral adipose tissue. Cell Metab 17, 851–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNelis JC, and Olefsky JM (2014). Macrophages, immunity, and metabolic disease. Immunity 41, 36–48. [DOI] [PubMed] [Google Scholar]
- Nishimura S, Manabe I, Takaki S, Nagasaki M, Otsu M, Yamashita H, Sugita J, Yoshimura K, Eto K, Komuro I, et al. (2013). Adipose Natural Regulatory B Cells Negatively Control Adipose Tissue Inflammation. Cell Metab. [DOI] [PubMed] [Google Scholar]
- Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, Red Eagle A, Vats D, Brombacher F, Ferrante AW, et al. (2007). Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature 447, 1116–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe Y, and Medzhitov R (2016). Tissue biology perspective on macrophages. Nature immunology 17, 9–17. [DOI] [PubMed] [Google Scholar]
- Okin D, and Medzhitov R (2016). The Effect of Sustained Inflammation on Hepatic Mevalonate Pathway Results in Hyperglycemia. Cell 165, 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Yi JW, Kim EM, Yoon IJ, Lee EH, Lee HY, Ji KY, Lee KH, Jang JH, Oh SS, et al. (2015). Triggering receptor expressed on myeloid cells 2 (TREM2) promotes adipogenesis and diet-induced obesity. Diabetes 64, 117–127. [DOI] [PubMed] [Google Scholar]
- Rosen ED, and Spiegelman BM (2014). What we talk about when we talk about fat. Cell 156, 20–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman LJ, Little JP, and Klegeris A (2014). Inflammation and insulin/IGF-1 resistance as the possible link between obesity and neurodegeneration. Journal of neuroimmunology 273, 8–21. [DOI] [PubMed] [Google Scholar]
- Stevens J, Oakkar EE, Cui Z, Cai J, and Truesdale KP (2015). US adults recommended for weight reduction by 1998 and 2013 obesity guidelines, NHANES 2007-2012. Obesity (Silver Spring) 23, 527–531. [DOI] [PubMed] [Google Scholar]
- Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, and Dudoit S (2018). Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan TJ, Miao Z, Zhao BN, Ertl LS, Wang Y, Krasinski A, Walters MJ, Powers JP, Dairaghi DJ, Baumgart T, et al. (2013). Experimental evidence for the use of CCR2 antagonists in the treatment of type 2 diabetes. Metabolism 62, 1623–1632. [DOI] [PubMed] [Google Scholar]
- Sung HK, Doh KO, Son JE, Park JG, Bae Y, Choi S, Nelson SM, Cowling R, Nagy K, Michael IP, et al. (2013). Adipose vascular endothelial growth factor regulates metabolic homeostasis through angiogenesis. Cell Metab 17, 61–72. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Honda T, Yamakage H, Hata J, Yoshida D, Hirakawa Y, Shibata M, Inoue T, Kusakabe T, Satoh-Asahara N, et al. (2018). A potential novel pathological implication of serum soluble triggering receptor expressed on myeloid cell 2 in insulin resistance in a general Japanese population: The Hisayama study. Diabetes Res Clin Pract 146, 225–232. [DOI] [PubMed] [Google Scholar]
- Turnbull IR, Gilfillan S, Cella M, Aoshi T, Miller M, Piccio L, Hernandez M, and Colonna M (2006). Cutting edge: TREM-2 attenuates macrophage activation. J Immunol 177, 3520–3524. [DOI] [PubMed] [Google Scholar]
- Ulland TK, Song WM, Huang SC, Ulrich JD, Sergushichev A, Beatty WL, Loboda AA, Zhou Y, Cairns NJ, Kambal A, et al. (2017). TREM2 Maintains Microglial Metabolic Fitness in Alzheimer's Disease. Cell 170, 649–663 e613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cella M, Mallinson K, Ulrich JD, Young KL, Robinette ML, Gilfillan S, Krishnan GM, Sudhakar S, Zinselmeyer BH, et al. (2015). TREM2 lipid sensing sustains the microglial response in an Alzheimer's disease model. Cell 160, 1061–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer DA, Luck H, Tsai S, and Winer S (2016). The Intestinal Immune System in Obesity and Insulin Resistance. Cell Metab 23, 413–426. [DOI] [PubMed] [Google Scholar]
- Xiang X, Werner G, Bohrmann B, Liesz A, Mazaheri F, Capell A, Feederle R, Knuesel I, Kleinberger G, and Haass C (2016). TREM2 deficiency reduces the efficacy of immunotherapeutic amyloid clearance. EMBO Mol Med 8, 992–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Grijalva A, Skowronski A, van Eijk M, Serlie MJ, and Ferrante AW Jr. (2013). Obesity activates a program of lysosomal-dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab 18, 816–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhaoyuan Liu, Y.G., Svetoslav Chakarov, Camille Bleriot, Xin Chen, Amanda Shin, Dress Regine J., Charles-Antoine Dutertre, Andreas Schlitzer, Jinmiao Chen, Honglin Wang, Zhiduo Liu, Bing Su and Florent Ginhoux (2019). Fate mapping via Ms4a3 expression history traces monocyte-derived cells. Cell, under revision. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental
Figure S1. Single-cell characterization of the adipose tissue immune niche during obesity progression. Related to Figure 1. A, B. Weight gain (A) and body composition (B) of mice on high-fat diet (HFD) or normal chow (NC). Error bars indicate standard deviation. N=8. C. Heatmap of differential gene expression for all cells colored by the assigned metacells, as in Figure 1B. D. kNN graph of epididymal adipose tissue immune cells of wild-type mice on NC, down-sampled to 2,187 cells each time-point, annotated as in Figure 1B. Each time-point is contributed by either 2 (6 weeks) or 4 mice (12 or 18 weeks). E, F. Relative frequencies of higher abundant (E) and less abundant (F) immune cell types of mice on HFD. Every dot represents a mouse. * p < 0.05; ** p < 0.01; *** p < 0.001, NS. not significant by Mann Whitney U-test. G. Weight gain of mice on db/db mice and littermate wild-type controls with age. Error bars indicate SD. N=6. H. kNN graph of epididymal adipose tissue immune cells of db/db mice and littermate wild-type controls with age, down-sampled to 1,985 cells each.
Figure S2. Analysis of monocyte and macrophage subsets. Related to Figure 2. A. Log2 of unique molecular identifier (UMI) count of selected genes in individual cells on the kNN graph of Figure 2A. B. kNN graph of monocytes and macrophages from epididymal adipose tissue of db/db mice. C. Log2 of average UMI count of selected genes across metacells within the monocyte/macrophage compartment. D. Log2 of UMI count of selected genes in individual metacells from the kNN graph in (B). E. Representative pseudocolor cell density FACS plots showing the strategy to enrich for macrophage (CD11b+F4/80+) subpopulations using the CD9 and CD63 markers. Numbers indicate percentage of gated cells. Macrophages from obese mice are predominantly CD9+CD63+. Upper row, 16-week old WT mouse on NC; bottom row, WT mouse on HFD for 12 weeks.
Figure S3. Adipose tissue macrophages during obesity are derived from monocytes. Related to Figure 2. A. Flow cytometric analysis of the frequency of Tomato+ cells (Ms4a3 lineage) among adipose tissue macrophages, Ly6Chi blood monocytes, Ly6Chi liver monocytes and Kupffer cells from 8 week-old or 16 week-old Ms4a3Cre-Rosa26tdTomato mice fed with NC or HFD during 8 weeks. Numbers indicate percentage of gated cells, used for the analysis shown in Figure 2H. B. Gene expression profiles driving the molecular differentiation trajectory from monocyte towards the Mac3 macrophage subset, ordered according to Slingshot pseudo-time trajectory (STAR Methods).
Figure S4. Conserved Trem2 signature characterizes obesity-related macrophages in mice and humans. Related to Figure 3. A. Volcano plot showing the fold change of genes (log2 scale) between Mac2 from wild-type mice on HFD to Mac1 from wild-type mice on NC (x-axis) and their significance (y-axis, −loglog scale). Highly significant genes are indicated by a red dot. P-values were determined by Mann-Whitney U-test with FDR correction. B. Log2 of average unique molecular identifier (UMI) count of Trem2 across all metacells as defined in Figure 1. C. Log2 of unique molecular identifier (UMI) count of Trem2 gene expression in individual cells on the kNN graph of Figure 1B. D. Representative immunofluorescence images of Trem2 (Cyan), CD9 (green), and perilipin-1 (red) in EAT sections of 16-week old WT mice after 12 weeks on HFD (above) or on NC (below). Cell nuclei are stained with DAPI (blue). Scale bar, 50 μm. Shown are the individual channels and their composite image on the right. E. Scatterplot comparing Z-scores of log2 fold changes of the Trem2 module genes between LAM versus Mac1 in wild-type mice (x-axis) versus db/db (y-axis). R indicates the Pearson correlation coefficient. F. Scatterplot showing the average UMI counts (log2 scale) of mouse LAM (y-axis) compared with the mouse DAM from the brain of Alzheimer’s disease mice (Keren-Shaul et al., 2017) (x-axis). G. Detection of LAM cells in the liver of mice feeding on HFD. Shown are percentages of immune cells expressing a LAM signature above a LAM score threshold of 10 in the liver of two mice fed a HFD for 27 weeks and two mice on NC. H. Scatterplot showing the average UMI counts (log2 scale) of LAM detected in the liver (y-axis) compared with LAM isolated from EAT (x-axis) in mice fed a HFD.
Figure S5. Characteristics of LAM cells in mice and humans. Related to Figure 3. A. kNN graph of 15,150 cells of human adipose tissue cells. See Table S3. B. Log2 of UMI count of selected genes in individual cells on the kNN graph from (A). C, D. KEGG pathway analysis for top-expressed genes of mouse (C) and human (D) LAM. E, F. qPCR of bulk-sorted subsets of adipose tissue macrophages for discovered LAM marker genes (E) and Trem2 module genes (F). G. Bodipy staining of bulk-sorted subsets of adipose tissue macrophages.
Figure S6. Characterization of adipose tissue immune cells in Trem2 knock-out mice. Related to Figure 4. A. Cell type distribution within the monocyte/macrophage compartment of KO and WT mice on HFD and NC. B. Projection of the monocyte/macrophage compartment onto the kNN graph of Figure 2A from a total of 10,042 QC-positive immune cells (133 metacells; Table S4) from EAT of Trem2 knock-out (KO) mice or WT littermate controls on NC. Contour lines indicate the 2D density of projected cells, down-sampled to 453 cells. C. Frequency of the EAT Mon/Mac subsets as defined in Figure 2 in each of two KO and two WT mice on NC. D, E. Relative frequencies of abundant (D) and less abundant (E) immune cell types. Each symbol represents a mouse. * p < 0.05; ** p < 0.01; *** p < 0.001, NS. not significant by a Mann Whitney U-test.
Figure S7. Immune cell specificity of Trem2 in metabolic homeostasis. Related to Figure 5. A. Representative FACS plots showing the degree of reconstitution eight weeks post bone marrow transplantation from CD45.2 donors into CD45.1 recipient mice, at the time of setting ten mice in each group, WT and Trem2 KO (20 mice in total), to feed a HFD. B. Percentage body fat content. Fat mass, lean mass and liquid mass of individual animals was measured by live non-invasive magnet resonance on week 18 of HFD. Filled circles, Trem2 WT; open circles, Trem2 KO. C-E. Total cholesterol (C), LDL (D), and HDL (E) levels in mouse serum from Trem2 cohorts at week 12 on HFD or NC control. F. Weight gain over time on HFD. G. Glucose tolerance test was performed on fasted mice at week 18 of HFD. H. Area under the curve (AUC) as a measure of glucose intolerance, calculated for each individual mouse in (G). Bars indicate mean ± SEM. * p < 0.05; n.s., non-significant.
Table S1
Table S1. Mouse epididymal adipose tissue metacell data. Related to Figure 1. Footprint gene expression of 244 metacells obtained from scRNA-seq of 21,210 visceral adipose tissue immune cells from wild-type mice.
Table S2
Table S2. Leptin receptor db/db mutant mouse epididymal adipose tissue metacell data. Related to Figures 1 and 2. Footprint gene expression of 75 metacells obtained from scRNA-seq of 8,372 visceral adipose tissue immune cells from db/db mice and wild-type littermate controls.
Table S3
Table S3. Human omental adipose tissue metacell data. Related to Figure 3. Footprint gene expression of 172 metacells obtained from scRNA-seq of 15,150 whole omental adipose tissue cells from an obese human subject.
Table S4
Table S4. Trem2 knockout mouse epididymal adipose tissue metacell data. Related to Figure 4. Footprint gene expression of 133 metacells obtained from scRNA-seq of 10,042 visceral adipose tissue immune cells from Trem2 knock-out mice and wild-type littermate controls.
Table S5
Table S5. Differential gene expression analysis between mouse macrophage subsets. Related to Figure 3. log2 fold change and respective p value of a Mann Whitney U test with false discovery rate (FDR) correction per gene comparing Mac3 HFD and Mac1 NC (Figure 3B), Mac2 HFD and Mac1 NC (Figure S3A), or Mac3 HFD and Mac2 HFD (Figure S3B), respectively.
Table S6
Table S6. Differential gene expression analysis between human macrophage subsets. Related to Figure 3. Fold change and respective p value of a Mann Whitney U test with false discovery rate (FDR) correction per gene comparing LAM and Mac in the human data.
Table S7
Table S7. Gene groups used as features for metacells clustering. List of genes used for clustering in the various metacells analysis throughout the paper, as well as genes used for gene-gene correlation analysis in figure 3A.
Data Availability Statement
The accession number for the raw sequence reported in this paper is EGA: . The accession number for the processed data reported in this paper is NCBI GEO: GSE . Scripts reproducing the analysis will be available at: https://bitbucket.org/amitlab/.