SARS-CoV-2 productively infects human gut enterocytes (original) (raw)
Abstract
The virus severe acute respiratory syndrome–coronavirus 2 (SARS-CoV-2) can cause coronavirus disease 2019 (COVID-19), an influenza-like disease that is primarily thought to infect the lungs with transmission via the respiratory route. However, clinical evidence suggests that the intestine may present another viral target organ. Indeed, the SARS-CoV-2 receptor angiotensin converting enzyme 2 (ACE2) is highly expressed on differentiated enterocytes. In human small intestinal organoids (hSIOs), enterocytes were readily infected by SARS-CoV and SARS-CoV-2 as demonstrated by confocal- and electron-microscopy. Consequently, significant titers of infectious viral particles were detected. mRNA expression analysis revealed strong induction of a generic viral response program. Hence, intestinal epithelium supports SARS-CoV-2 replication, and hSIOs serve as an experimental model for coronavirus infection and biology
Severe acute respiratory syndrome (SARS), caused by coronavirus SARS-CoV, emerged in 2003 (1). In late 2019, a novel transmissible coronavirus (SARS-CoV-2) was noted to cause an influenza-like disease ranging from mild respiratory symptoms to severe lung injury, multi-organ failure, and death (2–4). SARS-CoV and SARS-CoV-2 belong to the Sarbecovirus subgenus (genus Betacoronavirus, family Coronaviridae) (5–7). The SARS-CoV receptor is the angiotensin-converting enzyme 2 (ACE2) (8, 9). The spike proteins of both viruses bind to ACE2, whereas soluble ACE2 blocks infection by SARS-CoV as well as by SARS-CoV-2 (10–13). Transmission of SARS-CoV-2 is thought to occur through respiratory droplets and fomites. The virus can be detected in upper respiratory tract samples, implicating the nasopharynx as site of replication. In human lung, ACE2 is expressed mainly in alveolar epithelial type II cells and ciliated cells (14–16). However, highest expression of ACE2 in the human body occurs in the brush border of intestinal enterocytes (14, 17). Even though respiratory symptoms dominate the clinical presentation of COVID-19, gastrointestinal symptoms are observed in a subset of patients (18, 19). Moreover, viral RNA can be found in rectal swabs, even after nasopharyngeal testing has turned negative, implying gastro-intestinal infection and a fecal–oral transmission route (20–22).
SARS-CoV-2 infects airway and gut organoids
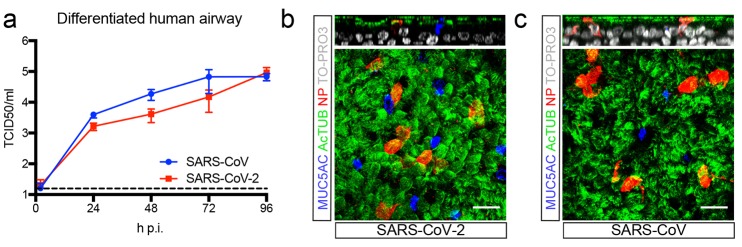
Organoids are 3D structures, that can be grown from adult stem cells (ASCs) and recapitulate key aspects of the organ from which the ASCs derive. Since SARS-CoV and SARS-CoV-2 target the lung, we added virus to organoid-derived human airway epithelium cultured in 2D and observed that SARS-CoV and SARS-CoV2 readily infected differentiated airway cultures. (Fig. 1A). Immunostaining reveal that the viruses targeted ciliated cells, but not goblet cells (Fig. 1, B and C).
Fig. 1. SARS-CoV and SARS-CoV-2 infect 2D human airway cultures.
(a) Live virus titers can be observed by virus titrations on VeroE6 cells of apical washes at 2, 24, 48, 72 and 96h after infection with SARS-CoV (blue) and SARS-CoV-2 (red). The dotted line indicates the lower limit of detection. Error bars represent SEM. N=4. *P<0.05, **P<0.01, ***P<0.001. (b and c) Immunofluorescent staining of SARS-CoV-2 (b) and SARS-CoV (c) infected differentiated airway cultures. Nucleoprotein (NP) stains viral nucleocapsid (red), which colocalized with the ciliated cell marker AcTUB (green). Goblet cells are identified by MUC5AC (blue). Nuclei are stained with TO-PRO3 (white). Scale bars indicate 20μM. Top panels are side-view while bottom panels are top-view.
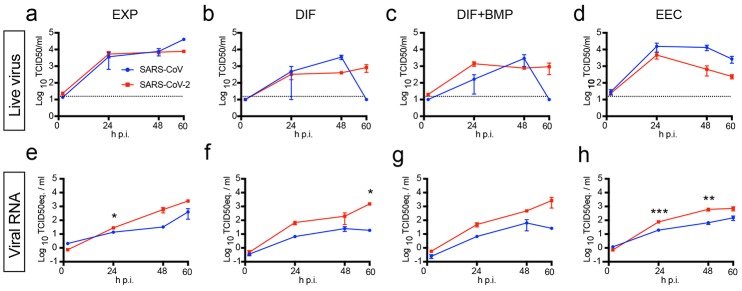
Human small intestinal organoids (hSIOs) are established from primary gut epithelial stem cells, can be expanded indefinitely in 3D culture and contain all proliferative and differentiated cell types of the in vivo epithelium (23). Of note, hSIOs have allowed the first in vitro culturing of Norovirus (24). We exposed ileal hSIOs grown under four different culture conditions (‘EXP’, ‘DIF’, ‘DIF-BMP’ and ‘EEC’) to SARS-CoV and SARS-CoV-2 at a multiplicity of infection (MOI) of 1. hSIOs grown in Wnt-high expansion medium (EXP) overwhelmingly consist of stem cells and enterocyte progenitors. Organoids grown in differentiation medium (DIF) contain enterocytes, goblet cells, and low numbers of enteroendocrine cells (EECs). Addition of BMP2/4 to DIF (DIF-BMP) leads to further maturation (25). In the final condition, we induced expression of NeuroG3 from a stably transfected vector with doxycycline to raise EECs numbers (fig. S3D). Samples were harvested at multiple timepoints post infection and processed for the analyses given in Figs. 2 to 5. Both SARS-CoV and SARS-CoV-2 productively infected hSIOs as assessed by qRT-PCR for viral sequences and by live virus titrations on VeroE6 cells (see Fig. 2 for lysed organoids and fig. S1 for organoid supernatant). Infectious virus particles and viral RNA increased to significant titers for both viruses in all conditions. Since EXP medium supported virus replication (Fig. 2, A and E), enterocyte progenitors appeared to be a primary viral target. Differentiated organoids (DIF; DIF-BMP) produced slightly (non-statistically significant) lower levels of infectious virus (Fig. 2 and fig. S1). In organoids induced to generate EECs, virus yields were similar to those in EXP medium (Fig. 2, D and H). In differentiated hSIOs, SARS-CoV-2 titers remained stable at 60 hours post infection, whereas SARS-CoV titers dropped 1-2 log (Fig. 2, B, C, F, and G). The latter decline was not observed in infected hSIOs grown in EXP. Culture supernatants across culture conditions contained lower levels of infectious virus compared to lysed hSIOs, implying that virus was primarily secreted apically (fig. S1, A to D). Despite this, viral RNA was detected readily in culture supernatants correlating with the infectious virus levels within hSIOs (Fig. 2, E to H, and fig. S1, E to H).
Fig. 2. SARS-CoV and SARS-CoV-2 replicate in hSIOs.
(a to d) Live virus titers can be observed by virus titrations on VeroE6 cells of lysed organoids at 2, 24, 48 and 60h after infection with SARS-CoV (blue) and SARS-CoV-2 (red). Different medium compositions show similar results. (e to h) qPCR analysis targeting the E gene of similar timepoints and medium compositions as (a) to (d). The dotted line indicates the lower limit of detection. Error bars represent SEM. N=3. *P<0.05, **P<0.01, ***P<0.001.
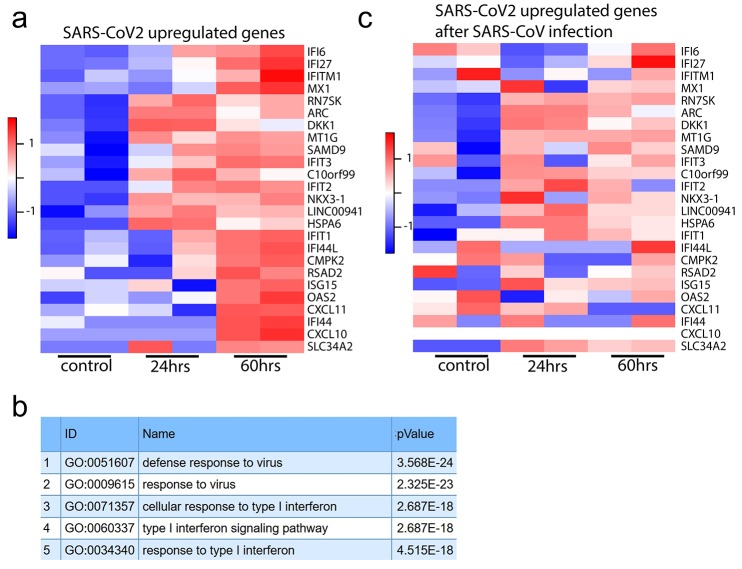
Fig. 5. Transcriptomic analysis of SARS-CoV-2 infected intestinal organoids.
(a) Heatmaps depicting the 25 most significantly enriched genes upon SARS-CoV-2 infection in expanding intestinal organoids. (b) Colored bar represents Z-score of log2 transformed values. GO term enrichment analysis for biological processes of the 50 most significantly up-regulated genes upon SARS-CoV-2 infected in intestinal organoids. (c) Heatmaps depicting the genes from (a) in SARS-CoV infected expanding organoids. Colored bar represents Z-score of log2 transformed values.
ACE2 mRNA expression differed greatly between the four conditions. EXP-hSIOs express 300-fold less ACE2 mRNA compared to DIF-hSIOs, when analyzed in bulk (fig. S2). BMP treatment induced 6.5-fold up-regulation of ACE2 mRNA compared to DIF treatment alone. Since this did not yield infection rate differences, the DIF-BMP condition was not analyzed further.
SARS-CoV-2 infects enterocyte lineage cells
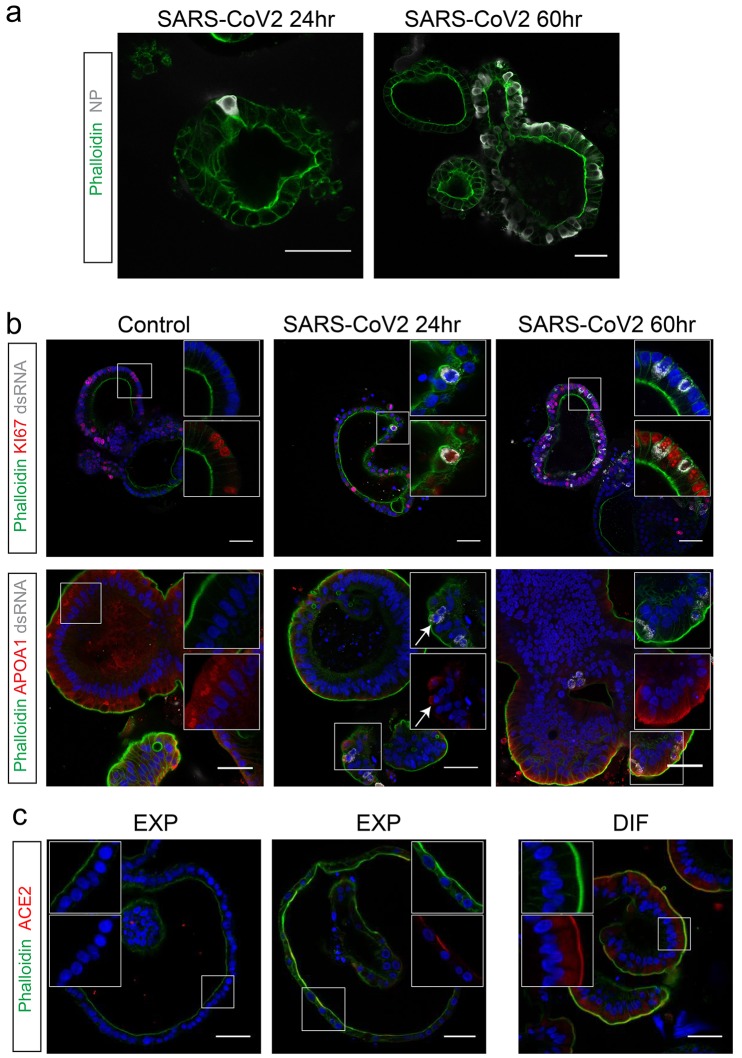
To determine the target cell type, we then performed confocal analysis on hSIOs cultured in EXP, DIF, or EEC conditions. We stained for viral dsRNA, viral nucleocapsid protein, KI67 to visualize proliferative cells, actin (using phalloidin) to visualize enterocyte brush borders, DNA (DAPI) and cleaved caspase 3 to visualize apoptotic cells. Generally, comparable rates of viral infections were observed in the organoids growing in all three conditions. We typically noted staining for viral components (white) in rare, single cells at 24 hours. At 60 hours, the number of infected cells had dramatically increased (Fig. 3A). Infected cells invariably displayed proliferative enterocyte progenitor-phenotypes (EXP; Fig. 3B, top) or ApoA1+ enterocyte-phenotypes (DIF; Fig. 3B, bottom). Of note, SARS-CoV also readily infected enterocyte lineage cells (fig. S3, A and B) as shown previously (26, 27). Some infected enterocyte progenitors were in mitosis (fig. S3C). Whereas EEC-organoids produced appreciable titers, we never observed infection of Chromogranin-A+ EECs (fig. S3, D and E). We also did not notice infection of Goblet cells across culture conditions. At 60 hours, apoptosis became prominent in both SARS-CoV and SARS-CoV-2 infected enterocytes (fig. S5). ACE2 protein was readily revealed as a bright and ubiquitous brush border marker in hSIOs in DIF medium (Fig. 3C). In hSIOs in EXP medium, ACE2 staining was much lower -yet still apical- in occasional cells in a subset of organoids that displayed a more mature morphology (Fig. 3C). In immature (cystic) organoids within the same cultures, the ACE2 signal was below the detection threshold. The percentage of infected organoids under EXP and DIF conditions are given in fig. S4. Figure S5 shows images and quantification of apoptotic cells upon infection.
Fig. 3. SARS-CoV-2 infects proliferating cells and enterocytes.
(a) Immunofluorescent staining of SARS-CoV-2-infected intestinal organoids. Nucleoprotein (NP) stains viral capsid. After 24 hours, single virus-infected cells are generally observed in organoids. These small infection clusters spread through the whole organoid after 60 hours. (b) SARS-CoV-2 infects both post-mitotic enterocytes identified by Apolipoprotein A1 (APOA1) and dividing cells that are KI67-positive. Infected cells are visualized by dsRNA staining. Enterocytes are shown in differentiated organoids, and proliferating cells in expanding organoids. Arrows point to APOA1-positive cells. (c) Immunofluorescent staining of ACE2 in intestinal organoids in expansion and differentiation condition. All scale bars are 50 μm.
Ultrastructural analysis of the viral life cycle in enterocytes
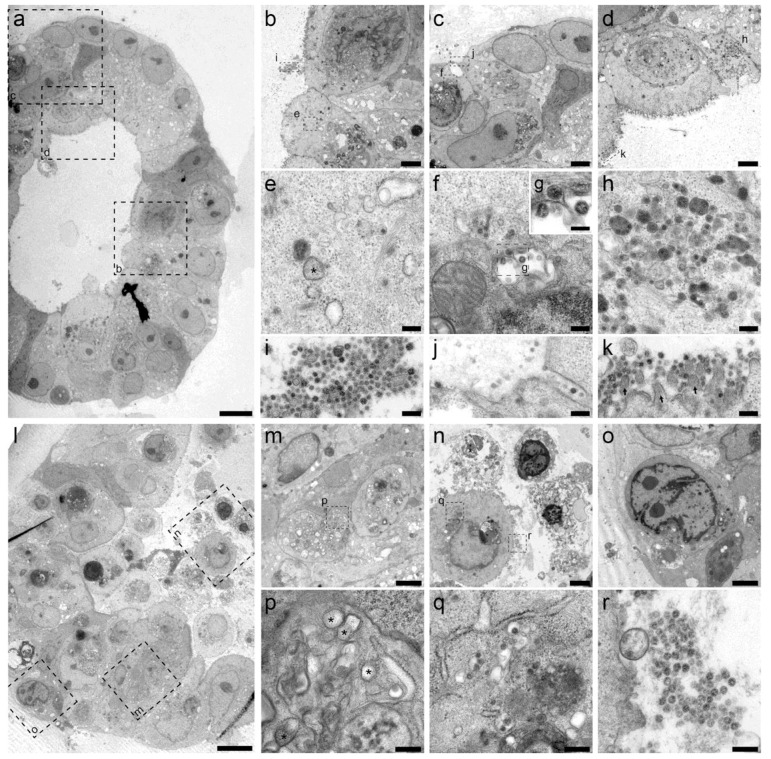
Unsupervised transmission electron microscopy (TEM) (28) was performed on selected highly infected samples. Figure 4 shows two hSIOs, selected from 42 imaged hSIOs at 60 hours post SARS-CoV-2 infection. These differ in the state of infection: whereas the cellular organization within organoid 1 was still intact (Fig. 4A, entire organoid; B to D, intermediate magnification; E to K, high magnification), many disintegrated cells can be seen in organoid 2 (Fig. 4, bottom; L, entire organoid; M to O, intermediate magnification; P to R, high magnification). Viral particles of 80-120 nm occurred in the lumen of the organoid (Fig. 4I), at the basolateral (Fig. 4J) and apical side (Fig. 4K) of enterocytes. The double-membrane vesicles which are the subcellular site of viral replication (29) are visualized in Fig. 4, E and P. The nuclei in both organoids differed from nuclei in mock-infected organoids by a slightly rounder shape. The nuclear contour index (30) was 4.0+/−0.5 vs 4.3+/−0.5 for control set. There was more heterochromatin (4N), and one or two dense nucleoli in the center (4O).
Fig. 4. Transmission electron microscopy analysis of SARS-CoV-2 infected intestinal organoids.
(a to h) Overview of an intact organoid (a) showing the onset of virus infection (b to d) at different stages of the viral lifecycle, i.e., early double membrane vesicles (DMVs) [(e), asterisk], initial viral production in the Golgi apparatus [(f) and (g)] and complete occupation of virus particles inside the endomembrane system (h). (i to k) Extracellular viruses are observed in the lumen of the organoid (i), and are found at the basal (j) and apical side (k) alongside the microvilli (arrows). Scale bars represent 10 μm (a), 2.5 μm [(b) to (d)], 250 nm [(e), (f), and (h) to (k)] and 100 nm (g). (l to q) Overview of an organoid (l) showing severely infected cells [(m) and (o)], disintegrated cells (o) and stressed cells as evident from the atypical nucleoli (p). Intact cells reveal DMV areas of viral replication [(p), asterisks] and infected Golgi apparatus (q). (r) Extracellular clusters of viruses. Scale bars represent 10 μm (l), 2.5 μm [(m) to (p)] and 250 nm [(p) to (r)]. Data was deposited to the Image Data Resource (https://idr.openmicroscopy.org) under accession number idr0083.
RNA expression changes in infected enterocytes
We then performed mRNA sequence analysis to determine gene expression changes induced by SARS-CoV and SARS-CoV-2-infection of hSIOs cultured continuously in EXP medium and hSIOs cultured in DIF medium. Infection with SARS-CoV-2 elicited a broad signature of cytokines and interferon stimulated genes (ISGs) attributed to type I and III interferon responses (Fig. 5A and tables S1 and S2), as confirmed by Gene Ontology analysis (Fig. 5B). An overlapping list of genes appeared in SARS-CoV-2-infected DIF organoids (fig. S6 and table S3). RNA sequencing analysis confirmed differentiation of DIF organoids into multiple intestinal lineages, including ACE2 up-regulation (fig. S7). SARS-CoV also induced ISGs, yet to a much lower level (table S4). Figure 5C visualizes the regulation of SARS-CoV-2-induced genes in SARS-CoV infected organoids. This induction was similar to infections with other viruses like norovirus (31), rotavirus (32) and enteroviruses (33, 34). A recent study (35) describes an antiviral signature induced in human cell lines after SARS-CoV-2 infection. Whereas the ISG response is broader in intestinal organoids, the induced gene sets are in close agreement between the two datasets (fig. S8). One striking similarity was the low expression of Type I and III interferons: we only noticed a small induction of the Type III interferon IFNL1 in SARS-CoV-2 infected organoids. In SARS-CoV-infected organoids, we did not observe any type I or type III interferon induction. We confirmed these findings by ELISA on culture supernatant and qRT-PCR, which in addition to IFNL1 picked up low levels of type I interferon IFNB1 in SARS-CoV-2 but not in SARS-CoV infected organoids (fig. S9). The specific induction of IP-10/CXCL10 and ISG15 by SARS-CoV-2 was also confirmed by ELISA and qRT-PCR, respectively (fig. S10). As in (35), a short list of cytokine genes was induced by both viruses albeit it to modest levels. For a comparison with (35), see fig. S11. Altogether these data indicate that SARS-CoV-2 induces a stronger interferon response than SARS-CoV in HIOs.
Finally, the infection was repeated in a second experiment in the same ileal HIO line and analyzed after 72 hours. Analysis involved viral titration (fig. S12), confocal imaging (fig. S13), and RNA sequencing (fig. S14). This experiment essentially confirmed the observations presented above. A limited, qualitative experiment applying confocal analysis demonstrated infectability of two other lines available in the lab (one ileal, one duodenal) from independent donors (fig. S13). This study shows that SARS-CoV and SARS-CoV-2 infect enterocyte lineage cells in a human intestinal organoid model. We observed similar infection rates of enterocyte-precursors and enterocytes whereas ACE2 expression increases ~1000-fold upon differentiation at the mRNA level (fig. S2). This suggests that low levels of ACE2 may be sufficient for viral entry.
SARS-CoV-2 is the third highly pathogenic coronavirus (after SARS-CoV and MERS-CoV) to jump to humans within less than 20 years suggesting that novel zoonotic coronavirus spillovers are likely to occur in the future. Despite this, limited information is available on coronavirus pathogenesis and transmission. This is in part due to the lack of in vitro cell models that accurately model host tissues. Very recently, it was shown that human iPS cells differentiated toward a kidney fate support replication of SARS-CoV-2 (13). Our data imply that human organoids represent faithful experimental models to study the biology of coronaviruses.
Acknowledgments
We thank E. Eenjes and R. Rottier for providing human lung material, A. de Graaff and Hubrecht Imaging Centre (HIC) for microscopy assistance and Single Cell Discoveries for RNA library preparation, and the Utrecht Sequencing Facility (subsidized by the University Medical Center Utrecht, Hubrecht Institute, Utrecht University and NWO project 184.034.019). Funding: This work was supported by ERC Advanced Grant 67013 and by Lung Foundation Netherlands to HC, and by NWO Grant 022.005.032. K.K., J.Q.D., P.J.P., and R.B.G.R. received funding from the Dutch Technology Foundation STW (UPON 14207) and from European Union’s Horizon 2020 Programme, Grant Agreement No 766970 Q-SORT. Author contributions: M.L., J.B., and J.V. performed experiments and designed the study. K.K. and J.Q.D. prepared samples. K.K. and R.B.G.R. performed imaging. K.K., J.P.v.S., P.J.P., and R.G.B.R. interpreted results. T.B., A.M., S.R., D.S., and M.G. measured virus titers. J.P. analyzed RNAseq data. E.C. performed sequencing. M.K., B.H., and H.C. supervised. Competing interests: H.C. is inventor on patents held by the Royal Netherlands Academy of Arts and Sciences that cover organoid technology.: PCT/NL2008/050543, WO2009/022907; PCT/NL2010/000017, WO2010/090513; PCT/IB2011/002167, WO2012/014076; PCT/IB2012/052950, WO2012/168930;PCT/EP2015/060815, WO2015/173425; PCT/EP2015/077990, WO2016/083613; PCT/EP2015/077988, WO2016/083612; PCT/EP2017/054797,WO2017/149025; PCT/EP2017/065101, WO2017/220586; PCT/EP2018/086716, and GB1819224.5. H.C.’s full disclosure is given at https://www.uu.nl/staff/JCClevers/. Data and materials availability: Organoid lines may be requested directly from the non-profit HUB (https://huborganoids.nl/), which does not directly benefit from this research. RNA sequence data can be accessed through GEO GSE149312. This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using such material.
Supplementary Materials
References and Notes
- 1.Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R. A. M., Berger A., Burguière A.-M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.-C., Müller S., Rickerts V., Stürmer M., Vieth S., Klenk H.-D., Osterhaus A. D. M. E., Schmitz H., Doerr H. W., Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348, 1967–1976 (2003). 10.1056/NEJMoa030747 [DOI] [PubMed] [Google Scholar]
- 2.Guan W. J., Ni Z. Y., Hu Y., Liang W. H., Ou C. Q., He J. X., Liu L., Shan H., Lei C. L., Hui D. S. C., Du B., Li L. J., Zeng G., Yuen K.-Y., Chen R. C., Tang C. L., Wang T., Chen P. Y., Xiang J., Li S. Y., Wang J. L., Liang Z. J., Peng Y. X., Wei L., Liu Y., Hu Y. H., Peng P., Wang J. M., Liu J. Y., Chen Z., Li G., Zheng Z. J., Qiu S. Q., Luo J., Ye C. J., Zhu S. Y., Zhong N. S.; China Medical Treatment Expert Group for Covid-19 , Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 382, 1708–1720 (2020). 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang S., Du L., Shi Z., An emerging coronavirus causing pneumonia outbreak in Wuhan, China: Calling for developing therapeutic and prophylactic strategies. Emerg. Microbes Infect. 9, 275–277 (2020). 10.1080/22221751.2020.1723441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., Tan W.; China Novel Coronavirus Investigating and Research Team , A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 (2020). 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen K. G., Rambaut A., Lipkin W. I., Holmes E. C., Garry R. F., The proximal origin of SARS-CoV-2. Nat. Med. 26, 450–452 (2020). 10.1038/s41591-020-0820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W. J., Wang D., Xu W., Holmes E. C., Gao G. F., Wu G., Chen W., Shi W., Tan W., Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 395, 565–574 (2020). 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorbalenya A. E., Baker S. C., Baric R. S., de Groot R. J., Drosten C., Gulyaeva A. A., Haagmans B. L., Lauber C., Leontovich A. M., Neuman B. W., Penzar D., Perlman S., Poon L. L. M., Samborskiy D. V., Sidorov I. A., Sola I., Ziebuhr J.; Coronaviridae Study Group of the International Committee on Taxonomy of Viruses , The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 5, 536–544 (2020). 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H., Crackower M. A., Fukamizu A., Hui C. C., Hein L., Uhlig S., Slutsky A. S., Jiang C., Penninger J. M., Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature 436, 112–116 (2005). 10.1038/nature03712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., Bao L., Zhang B., Liu G., Wang Z., Chappell M., Liu Y., Zheng D., Leibbrandt A., Wada T., Slutsky A. S., Liu D., Qin C., Jiang C., Penninger J. M., A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 11, 875–879 (2005). 10.1038/nm1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walls A. C., Park Y. J., Tortorici M. A., Wall A., McGuire A. T., Veesler D., Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292.e6 (2020). 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan Y., Shang J., Graham R., Baric R. S., Li F., Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 94, e00127-20 (2020). 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wrapp D., Wang N., Corbett K. S., Goldsmith J. A., Hsieh C. L., Abiona O., Graham B. S., McLellan J. S., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020). 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R. A., Stahl M., Leopoldi A., Garreta E., Hurtado Del Pozo C., Prosper F., Romero J. P., Wirnsberger G., Zhang H., Slutsky A. S., Conder R., Montserrat N., Mirazimi A., Penninger J. M., Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell 10.1016/j.cell.2020.04.004 (2020). 10.1016/j.cell.2020.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi F., Qian S., Zhang S., Zhang Z., Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 526, 135–140 (2020). 10.1016/j.bbrc.2020.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Y. Zhao, Z. Zhao, Y. Wang, Y. Zhou, Y. Ma, W. Zuo, Single-cell RNA expression profiling of ACE2, the receptor of SARS-CoV-2. bioRxiv 2020.01.26.919985 [Preprint]. 9 April 2010. . 10.1101/2020.01.26.919985 [DOI] [PMC free article] [PubMed]
- 16.Jia H. P., Look D. C., Shi L., Hickey M., Pewe L., Netland J., Farzan M., Wohlford-Lenane C., Perlman S., McCray Jr. P. B., ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 79, 14614–14621 (2005). 10.1128/JVI.79.23.14614-14621.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The Human Protein Atlas, ACE2 protein expression summary (2020); https://www.proteinatlas.org/ENSG00000130234-ACE2.
- 18.Gu J., Han B., Wang J., COVID-19: Gastrointestinal manifestations and potential fecal-oral transmission. Gastroenterology 158, 1518–1519 (2020). 10.1053/j.gastro.2020.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cholankeril G., Podboy A., Aivaliotis V. I., Tarlow B., Pham E. A., Spencer S., Kim D., Hsing A., Ahmed A., High prevalence of concurrent gastrointestinal manifestations in patients with SARS-CoV-2: Early experience from California. Gastroenterology 10.1053/j.gastro.2020.04.008 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W., Detection of SARS-CoV-2 in different types of clinical specimens. JAMA (2020). 10.1001/jama.2020.3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holshue M. L., DeBolt C., Lindquist S., Lofy K. H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S. I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M. A., Weldon W. C., Biggs H. M., Uyeki T. M., Pillai S. K.; Washington State 2019-nCoV Case Investigation Team , First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 382, 929–936 (2020). 10.1056/NEJMoa2001191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H., Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology S0016-5085(20)30282-1 (2020). 10.1053/j.gastro.2020.02.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato T., Stange D. E., Ferrante M., Vries R. G. J., Van Es J. H., Van den Brink S., Van Houdt W. J., Pronk A., Van Gorp J., Siersema P. D., Clevers H., Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772 (2011). 10.1053/j.gastro.2011.07.050 [DOI] [PubMed] [Google Scholar]
- 24.Ettayebi K., Crawford S. E., Murakami K., Broughman J. R., Karandikar U., Tenge V. R., Neill F. H., Blutt S. E., Zeng X. L., Qu L., Kou B., Opekun A. R., Burrin D., Graham D. Y., Ramani S., Atmar R. L., Estes M. K., Replication of human noroviruses in stem cell-derived human enteroids. Science 353, 1387–1393 (2016). 10.1126/science.aaf5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beumer J., Artegiani B., Post Y., Reimann F., Gribble F., Nguyen T. N., Zeng H., Van den Born M., Van Es J. H., Clevers H., Enteroendocrine cells switch hormone expression along the crypt-to-villus BMP signalling gradient. Nat. Cell Biol. 20, 909–916 (2018). 10.1038/s41556-018-0143-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leung W. K., To K. F., Chan P. K. S., Chan H. L. Y., Wu A. K. L., Lee N., Yuen K. Y., Sung J. J. Y., Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology 125, 1011–1017 (2003). 10.1016/j.gastro.2003.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan W. S., Wu C., Chow S. C. S., Cheung T., To K. F., Leung W. K., Chan P. K. S., Lee K. C., Ng H. K., Au D. M. Y., Lo A. W. I., Coronaviral hypothetical and structural proteins were found in the intestinal surface enterocytes and pneumocytes of severe acute respiratory syndrome (SARS). Mod. Pathol. 18, 1432–1439 (2005). 10.1038/modpathol.3800439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faas F. G. A., Avramut M. C., van den Berg B. M., Mommaas A. M., Koster A. J., Ravelli R. B. G., Virtual nanoscopy: Generation of ultra-large high resolution electron microscopy maps. J. Cell Biol. 198, 457–469 (2012). 10.1083/jcb.201201140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knoops K., Kikkert M., Worm S. H., Zevenhoven-Dobbe J. C., van der Meer Y., Koster A. J., Mommaas A. M., Snijder E. J., SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLOS Biol. 6, e226 (2008). 10.1371/journal.pbio.0060226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNutt N. S., Crain W. R., Quantitative electron microscopic comparison of lymphocyte nuclear contours in mycosis fungoides and in benign infiltrates in skin. Cancer 47, 698–709 (1981). [DOI] [PubMed] [Google Scholar]
- 31.Hosmillo M., Chaudhry Y., Nayak K., Sorgeloos F., Koo B. K., Merenda A., Lillestol R., Drumright L., Zilbauer M., Goodfellow I., Norovirus replication in human intestinal epithelial cells is restricted by the interferon-induced JAK/STAT signaling pathway and RNA polymerase II-mediated transcriptional responses. mBio 11, e00215-20 (2020). 10.1128/mBio.00215-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saxena K., Simon L. M., Zeng X. L., Blutt S. E., Crawford S. E., Sastri N. P., Karandikar U. C., Ajami N. J., Zachos N. C., Kovbasnjuk O., Donowitz M., Conner M. E., Shaw C. A., Estes M. K., A paradox of transcriptional and functional innate interferon responses of human intestinal enteroids to enteric virus infection. Proc. Natl. Acad. Sci. U.S.A. 114, E570–E579 (2017). 10.1073/pnas.1615422114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drummond C. G., Bolock A. M., Ma C., Luke C. J., Good M., Coyne C. B., Enteroviruses infect human enteroids and induce antiviral signaling in a cell lineage-specific manner. Proc. Natl. Acad. Sci. U.S.A. 114, 1672–1677 (2017). 10.1073/pnas.1617363114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Good C., Wells A. I., Coyne C. B., Type III interferon signaling restricts enterovirus 71 infection of goblet cells. Sci. Adv. 5, eaau4255 (2019). 10.1126/sciadv.aau4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blanco-Melo D., Nilsson-Payant B. E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T. X., Oishi K., Panis M., Sachs D., Wang T. T., Schwartz R. E., Lim J. K., Albrecht R. A., Tenoever B. R., Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 10.1016/j.cell.2020.04.026 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sachs N., Papaspyropoulos A., Zomer-van Ommen D. D., Heo I., Böttinger L., Klay D., Weeber F., Huelsz-Prince G., Iakobachvili N., Amatngalim G. D., de Ligt J., van Hoeck A., Proost N., Viveen M. C., Lyubimova A., Teeven L., Derakhshan S., Korving J., Begthel H., Dekkers J. F., Kumawat K., Ramos E., van Oosterhout M. F., Offerhaus G. J., Wiener D. J., Olimpio E. P., Dijkstra K. K., Smit E. F., van der Linden M., Jaksani S., van de Ven M., Jonkers J., Rios A. C., Voest E. E., van Moorsel C. H., van der Ent C. K., Cuppen E., van Oudenaarden A., Coenjaerts F. E., Meyaard L., Bont L. J., Peters P. J., Tans S. J., van Zon J. S., Boj S. F., Vries R. G., Beekman J. M., Clevers H., Long-term expanding human airway organoids for disease modeling. EMBO J. 38, 100300 (2019). 10.15252/embj.2018100300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corman V. M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D. K., Bleicker T., Brünink S., Schneider J., Schmidt M. L., Mulders D. G., Haagmans B. L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J. L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M. P., Drosten C., Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 10.2807/1560-7917.ES.2020.25.3.2000045 (2020). 10.2807/1560-7917.ES.2020.25.3.2000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muñoz J., Stange D. E., Schepers A. G., van de Wetering M., Koo B.-K., Itzkovitz S., Volckmann R., Kung K. S., Koster J., Radulescu S., Myant K., Versteeg R., Sansom O. J., van Es J. H., Barker N., van Oudenaarden A., Mohammed S., Heck A. J. R., Clevers H., The Lgr5 intestinal stem cell signature: Robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 31, 3079–3091 (2012). 10.1038/emboj.2012.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hashimshony T., Senderovich N., Avital G., Klochendler A., de Leeuw Y., Anavy L., Gennert D., Li S., Livak K. J., Rozenblatt-Rosen O., Dor Y., Regev A., Yanai I., CEL-Seq2: Sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 17, 77 (2016). 10.1186/s13059-016-0938-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simmini S., Bialecka M., Huch M., Kester L., van de Wetering M., Sato T., Beck F., van Oudenaarden A., Clevers H., Deschamps J., Transformation of intestinal stem cells into gastric stem cells on loss of transcription factor Cdx2. Nat. Commun. 5, 5728 (2014). 10.1038/ncomms6728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H., Durbin R., Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaimal V., Bardes E. E., Tabar S. C., Jegga A. G., Aronow B. J., ToppCluster: A multiple gene list feature analyzer for comparative enrichment clustering and network-based dissection of biological systems. Nucleic Acids Res. 38, W96– W102 (2010). 10.1093/nar/gkq418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burel J.-M., Besson S., Blackburn C., Carroll M., Ferguson R. K., Flynn H., Gillen K., Leigh R., Li S., Lindner D., Linkert M., Moore W. J., Ramalingam B., Rozbicki E., Tarkowska A., Walczysko P., Allan C., Moore J., Swedlow J. R., Publishing and sharing multi-dimensional image data with OMERO. Mamm. Genome 26, 441–447 (2015). 10.1007/s00335-015-9587-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.