Local externalization of phosphatidylserine mediates developmental synaptic pruning by microglia (original) (raw)
Abstract
Neuronal circuit assembly requires the fine balance between synapse formation and elimination. Microglia, through the elimination of supernumerary synapses, have an established role in this process. While the microglial receptor TREM2 and the soluble complement proteins C1q and C3 are recognized as key players, the neuronal molecular components that specify synapses to be eliminated are still undefined. Here, we show that exposed phosphatidylserine (PS) represents a neuronal “eat‐me” signal involved in microglial‐mediated pruning. In hippocampal neuron and microglia co‐cultures, synapse elimination can be partially prevented by blocking accessibility of exposed PS using Annexin V or through microglial loss of TREM2. In vivo, PS exposure at both hippocampal and retinogeniculate synapses and engulfment of PS‐labeled material by microglia occurs during established developmental periods of microglial‐mediated synapse elimination. Mice deficient in C1q, which fail to properly refine retinogeniculate connections, have elevated presynaptic PS exposure and reduced PS engulfment by microglia. These data provide mechanistic insight into microglial‐mediated synapse pruning and identify a novel role of developmentally regulated neuronal PS exposure that is common among developing brain structures.
Keywords: C1q, microglia, phosphatidylserine, synapse pruning, TREM2
Subject Categories: Immunology, Neuroscience
Exposed phosphatidylserine on pre‐ and postsynaptic membranes functions as an “eat‐me” signal contributing to microglia‐mediated synapse pruning.

Introduction
The development of a properly connected nervous system is a complex process involving the regulation of synapse formation, controlled elimination of supernumerary synapses, and maintenance of appropriate connections. Elucidating the mechanisms of synaptic pruning is important in understanding normal neurodevelopment and maturation, and how disruptions in this process may contribute to neurological dysfunction. The involvement of microglia, the CNS resident immune cells and phagocytes, in synaptic refinement has been widely established. Microglial processes make direct and transient connections with subsets of neuronal synapses, with evidence for presynaptic and postsynaptic elements inside microglial lysosomes (Nimmerjahn et al, 2005; Tremblay et al, 2010; Paolicelli et al, 2011; Schafer et al, 2012; Filipello et al, 2018; Weinhard et al, 2018; Basilico et al, 2019). Microglia‐mediated synaptic pruning is a highly regulated process that occurs across several developing brain regions. In addition to a role in developmental circuit refinement, this process can become activated in vulnerable brain regions in several disease models (Neher et al, 2012; Wang et al, 2015; Hong et al, 2016; Schafer et al, 2016; Vasek et al, 2016; Werneburg et al, 2020), suggesting that disease and development could share common regulators and mechanisms of pruning.
Microglial engulfment of immature synapses and axon terminals occurs through a variety of mechanisms, with data demonstrating that several receptors including microglial complement receptor CR3/Mac1 (Schafer et al, 2012) and the phagocytic receptor TREM2 (Filipello et al, 2018) contribute to this process. The cues specifying which neuronal structures are targeted by microglia, however, and whether these signals are region‐specific, remain largely unknown. For example, in the visual system, complement factors C1q and C3 label subsets of synaptic inputs for removal by phagocytic microglia in a CR3‐dependent mechanism (Stevens et al, 2007; Schafer et al, 2016). Why specific neuronal structures are targeted, given that these complement factors represent secreted soluble proteins, remains unclear. Complement‐independent microglial synaptic elimination has also been demonstrated in other regions, including the hippocampus (Paolicelli et al, 2011; Weinhard et al, 2018) and barrel cortex (Gunner et al, 2019). Finally, in the dLGN, astrocytes also contribute to developmental pruning through TAM receptor‐specific mechanisms (Chung et al, 2013). Taken together, multiple glial mechanisms of synaptic recognition and elimination may occur in a spatially and temporally specific manner. What, then, are the neuronal signals that mediate this process? Are there multiple, region‐specific cues or are there common underlying signals among developing regions capable of being recognized by a variety of microglial and/or astrocytic receptors?
Externalization of phosphatidylserine (PS) has been established as one of the first detectable events to occur in cells undergoing apoptosis; however, recent studies have uncovered transient, localized PS exposure events that occur in a non‐apoptotic manner (Smrz et al, 2007; Segawa et al, 2011). For example, in Drosophila, PS exposure can occur locally on injured dendrites, which are then targeted for elimination while sparing the remaining uninjured cell structures (Sapar et al, 2018). The recognition of exposed PS (ePS) by phagocytes, a process that is critical to the clearance of apoptotic cells and debris to prevent autoimmunity, is facilitated by numerous phagocytic receptors including microglial TREM2 (Park et al, 2007; Grommes et al, 2008; Graham et al, 2014; Shirotani et al, 2019). Further, complement binding to ePS, either directly or indirectly, can mediate engulfment in the periphery (Païdassi et al, 2008; Martin et al, 2012). Recent work has shown that ePS may be present ex vivo on isolated synaptosomes also tagged with C1q (Györffy et al, 2018); whether local externalization of PS occurs in vivo in a non‐injury‐associated manner and what role this type of signal would have are unclear.
We hypothesized that PS exposure occurs locally on synapses during normal development, acting as an “eat‐me” signal to enable recognition and removal by microglia. We found that liposome engulfment by isolated microglia was dependent on PS concentration and that, when co‐cultured with hippocampal neurons, microglial‐dependent synaptic reductions also required ePS. In vivo, synaptic PS occurred predominantly at presynaptic inputs in both the hippocampus and the dorsal lateral geniculate nucleus (dLGN), and was highest during established developmental periods of microglial‐mediated synapse elimination. In C1q knockout animals, we observed elevated PS‐labeled presynaptic inputs as well as a decrease in microglial engulfment of PS‐labeled material. Taken together, our data provide mechanistic insights into developmental circuit refinement by identifying a novel role of ePS on neurons in instructing microglial engulfment during developmental critical periods.
Results
Microglia phagocytose liposomes in a phosphatidylserine‐dependent manner
We investigated whether the amount of ePS correlates with microglia engulfment by taking advantage of uniformly sized synthetic liposomes containing different amounts of PS and cardiolipin, and labeled with the lipid fluorescent dye DiO (shown as percentage, PS99:1, PS50:49:1, and PS20:79:1, all numbers are mol%). Liposomes were administered to microglia, and their internalization was quantified (Fig 1A). As expected, liposomes were internalized by CD68‐positive phagolysosomal structures (Fig 1A–E). Of note, WT microglia displayed a reduced phagocytic activity when the amount of PS in liposomes was lower (phagocytic activity efficiency: PS99:1 > PS50:49:1 > PS20:79:1; Fig 1B, C, and F). Moreover, cloaking PS with Annexin V (ANXV), an innate molecule that binds with high affinity to PS‐bearing membranes, significantly reduced engulfment of PS99:1 liposomes (Fig 1B, D, and F). The specificity of PS‐dependent liposome engulfment is further supported by the evidence that the internalization of fluorescent beads by microglia was not affected by preincubation with ANXV (Fig EV1A).
Figure 1. Microglia engulf liposomes in PS‐dependent manner.
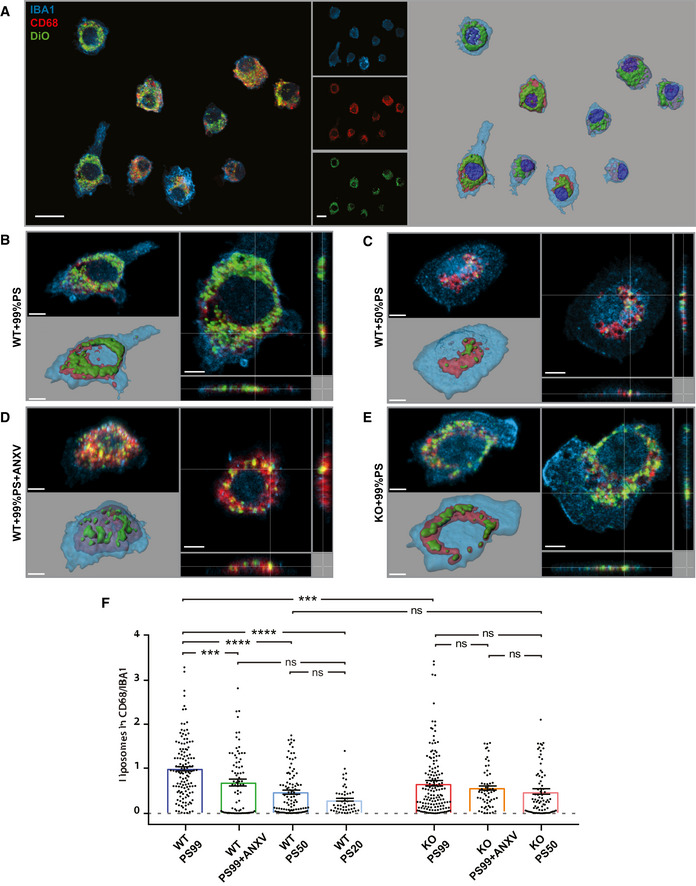
- A
Representative confocal images and 3D reconstruction of in vitro WT microglia fed with DiO‐labeled liposomes containing a controlled amount of phosphatidylserine (PS; 99, 50, 20%). Microglia were stained with Iba1 (blue) and lysosomes labeled with CD68 (red). Scale bar: 10 μm. - B–E
Representative images showing (B) WT microglia fed with 99% PS or (C) 50% PS liposomes, (D) WT microglia pre‐treated with ANXV and fed with 99% PS liposomes, and (E) Trem2 −/− microglia fed with 99% PS liposomes. - F
Histogram illustrating the quantification of the engulfed materials by microglia fed with liposomes (WT PS99: 1 ± 0.06, n = 129 cells analyzed; WT PS99 + ANXV: 0.70 ± 0.08, n = 76 cells analyzed; PS50: 0.49 ± 0.05, n = 99 cells analyzed; PS20: 0.31 ± 0.04, n = 50 cells analyzed; KO PS99: 0.68 ± 0.06, n = 146 cells analyzed; KO PS99 + ANXV: 0.57 ± 0.06, n = 76 cells analyzed; KO PS50: 0.48 ± 0.06, n = 77 cells analyzed). Bars represent mean ± SEM.
Data information: Scale bar in (B–E): 3 μm. At least three independent experiments (N) were performed; ***P < 0.001 and ****P < 0.0001, one‐way ANOVA with Dunn's multiple comparisons test.Source data are available online for this figure.
Figure EV1. In vitro treatment with Annexin V does not alter microglial viability or properties.
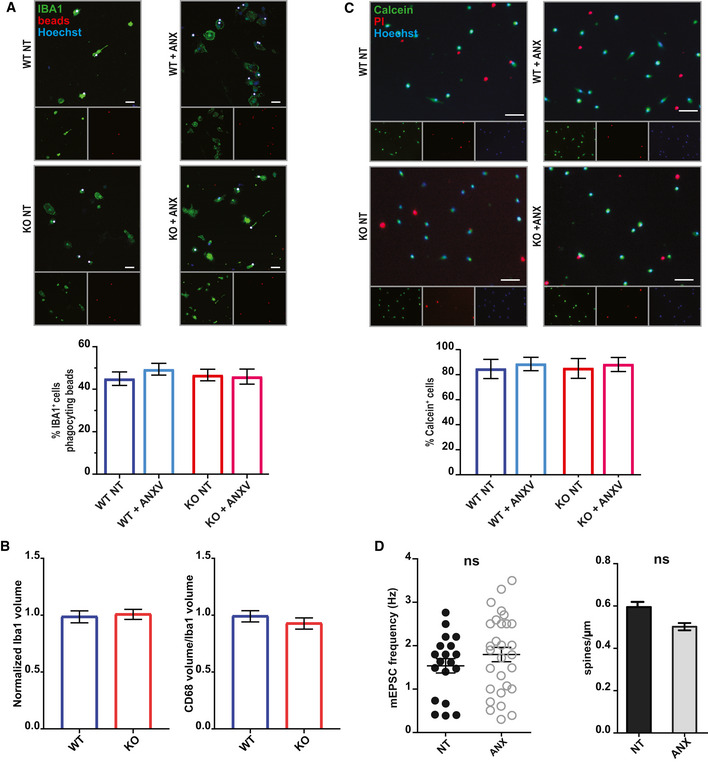
- Analysis of in vitro microglial phagocytic activity following ANXV treatment. Phagocytosis was assessed by measuring microglial engulfment of fluorescent beads (3 μm; Spherotech). No significant difference was observed between untreated and treated microglia (WT NT: 44.95 ± 3.19; WT + ANXV: 49.39 ± 2.76; KO NT: 46.68 ± 2.73; KO + ANXV: 45.93 ± 3.54). Three independent experiments were performed; scale bar 10 μm.
- Quantification of the cell volume (by Iba1 staining) and phagocytic compartment (by CD68 staining) reveals lack of differences between WT and Trem2−/− microglia (Iba1: 105 WT cells and 112 KO cells were analyzed. CD68: 98 WT cells and 128 KO cells were analyzed). Three independent experiments (N) were performed.
- Analysis of microglial viability in vitro following ANXV treatment. Viability was determined using the live marker Calcein combined with propidium iodide (PI) to label dying cells. No significant difference was observed between untreated and treated microglia (WT NT: 84.54% ± 7.67; WT + ANXV: 88.5% ± 5.36; KO NT: 84.94% ± 7.93; KO + ANXV: 88.12 ± 5.63). Three independent experiments (N) were performed; scale bar 30 μm.
- Analysis of mEPSC frequency and spine density in hippocampal neurons exposed to ANXV treatment shows the lack of significant differences between untreated and treated neurons for either parameter. Data reflect single‐cell measurements from three independent experiments (N). Unpaired _t_‐test was performed.
Data information: Bars represent mean ± SEM. Source data are available online for this figure.
The extent of PS99:1 liposome internalization was significantly lower when microglia were devoid of the phagocytic receptor TREM2 (Fig 1B, E, and F), which recognizes PS (Wang et al, 2015). This observation was not a result of underlying changes in CD68‐positive endolysosomal content between WT and Trem2 −/− microglia (Fig EV1B). Further, the lack of TREM2 did not affect the internalization of fluorescent beads by microglia (Filipello et al, 2018, and Fig EV1A), and the residual liposome engulfment by Trem2 −/− microglia was not significantly affected by either the reduction in PS amount or ANXV pre‐treatment (Fig 1F). These data point to a PS‐dependent, TREM2‐mediated liposome engulfment, with additional mechanisms, such as fluid‐phase uptake, likely contributing to the residual engulfment observed.
Microglial‐mediated synapse elimination is dependent on exposed PS in vitro
Microglia eliminate supernumerary synapses in the developing brain, regulating the dynamics of synaptic connections throughout life. It has been previously shown that co‐culturing microglial cells in contact with hippocampal neurons for 24‐h results in microglia‐mediated synapse reduction (Filipello et al, 2018), as indicated by the lower mushroom spine density (Fig 2A) and frequency of miniature excitatory postsynaptic currents (mEPSCs) (Fig 2B). To address whether PS exposure is required for microglia‐mediated synapse elimination, hippocampal neurons were exposed to ANXV, 15 min before being co‐cultured with WT microglia. Cloaking of ePS completely prevented both dendritic spine and mEPSC reduction (Fig 2A and B). Neuronal exposure to ANXV did not affect microglia viability, as assessed by calcein‐PI assay (Fig EV1C) or microglial phagocytic activity, as shown by bead phagocytosis assay (Fig EV1A). When microglia from TREM2‐deficient mice were cultured with WT neurons, no reduction in synapses was detected, in line with previous results (Filipello et al, 2018) (Fig 2C and D). In the latter case, cloaking of ePS with ANXV did not induce any modification in either spine density or miniature events (Fig 2C and D). As a further control, the exposure of pure neuronal cultures to ANXV did not alter either spine density or mEPSC frequency (Fig EV1D). These results indicate that masking ePS prevents microglia‐mediated synaptic engulfment.
Figure 2. In vitro microglia‐mediated synapse elimination is ePS‐dependent.
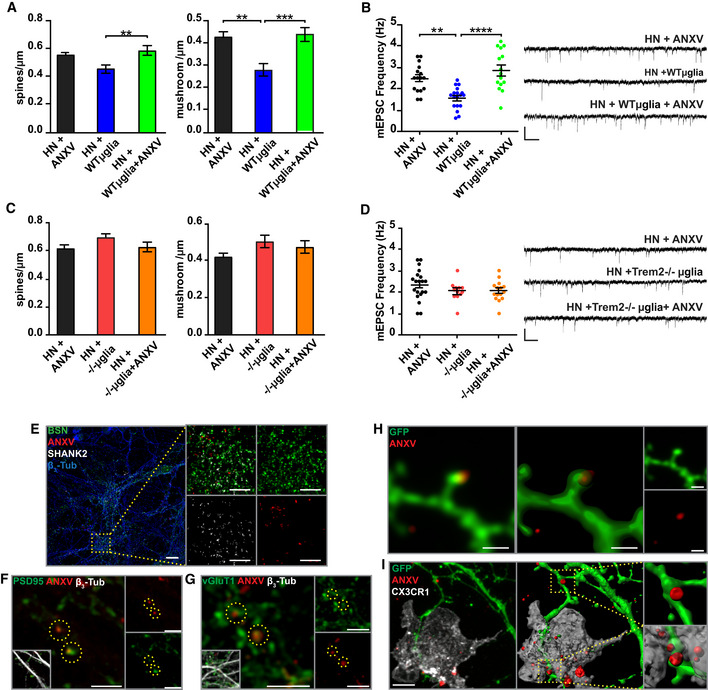
- A
Quantitative analysis of total and mushroom spines density in hippocampal neurons (HN) exposed to ANXV, HN co‐cultured with WT microglia, or HN exposed to ANXV and co‐cultured with WT microglia. HN + ANXV: spines/μm = 0.55 ± 0.03, mushroom/μm = 0.42 ± 0.03, number of examined dendrites: 47, n = 25 cells; HN + WT microglia: spines/μm = 0.45 ± 0.03, mushroom/μm = 0.28 ± 0.03, number of examined dendrites: 35, n = 19 cells; HN + WT microglia + ANXV: spines/μm = 0.58 ± 0.3, mushroom/μm = 0.44 ± 0.03, number of examined dendrites: 44, n = 18 cells. - B
Histogram and representative traces of mEPSC whole‐cell recordings showing miniature event frequency in the same conditions described for (A). mEPSC frequency: HN + ANXV: 2.48 ± 0.16 n = 16 cells, HN + WT microglia: 1.58 ± 0.12, n = 18 cells, HN + WT microglia + ANXV: 2.86 ± 0.24, n = 15 cells. Scale bars, 10 pA and 250 ms. - C
Quantitative analysis of total and mushroom spines density in HN exposed to ANXV, co‐cultured with Trem2 −/− microglia or HN co‐cultured with Trem2 −/− microglia and exposed to ANXV. No statistical differences were found among the different conditions. HN + ANXV: spines/μm = 0.62 ± 0.03, mushroom/μm = 0.42 ± 0.03, number of examined dendrites: 64, n = 8 cells; HN + −/−microglia: spines/μm = 0.70 ± 0.02, mushroom/μm = 0.50 ± 0.03, number of examined dendrites: 40, n = 8 cells; HN + −/−microglia + ANXV: spines/μm = 0.63 ± 0.3, mushroom/μm = 0.47 ± 0.03, number of examined dendrites: 17, n = 7 cells. - D
Histogram and representative traces of mEPSC whole‐cell recordings showing miniature event frequency in the same conditions described for (C). No statistical differences were found among the different conditions. mEPSC frequency: HN + ANXV: 2.34 ± 0.16, n = 20 cells, HN + −/−microglia: 2.04 ± 0.13, n = 12 cells, HN + −/−microglia + ANXV: 2.05 ± 0.12 n = 15 cells. Scale bars, 10 pA and 250 ms. - E
Representative confocal images of HN cultures labeled with ANXV and stained for the neuronal marker β3‐tubulin, the presynaptic marker Bassoon, and the postsynaptic marker Shank2. Scale bar: 5 μm - F, G
Representative confocal images of HN labeled with ANXV and stained for PSD95 (F) and vGlut1 (G). Co‐localization between ANXV‐positive puncta and PSD95/vGlut1 markers is highlighted by yellow dashed circles. No preferential association of ANXV with pre‐ or postsynaptic markers was detected. Scale bar: (F) 5 μm (G) 2 μm. - H
Representative confocal image of GFP‐expressing neurons labeled with ANXV. Note the presence of ANXV‐positive structures on the dendritic protrusion. Scale bar 1 μm. - I
Representative confocal image of microglia co‐cultured with GFP‐expressing neurons previously exposed to ANXV. Note the presence of ANXV‐positive structures on dendritic protrusions in the process to be engulfed by microglia. Scale bar 6 μm.
Data information: (A–D) At least three independent experiments (N) were performed for all conditions. **P < 0.01 and ****P < 0.0001, one‐way ANOVA followed by Tukey's multiple comparison test. Bars represent mean ± SEM. Source data are available online for this figure.
To investigate whether PS is externalized at synaptic sites, primary cultures of hippocampal neurons were exposed to ANXV‐568, fixed, and stained for the presynaptic markers Bassoon or vGluT1 and the postsynaptic markers Shank2 or PSD95. Confocal microscopy analysis followed by a deconvolution process indicated that ANXV labels a subpopulation of hippocampal synapses in vitro (Fig 2E–G). To obtain a better visualization of PS exposure, neuronal cultures were transfected with eGFP, which allows the detection of dendritic processes and spines, and subsequently exposed to ANXV. The presence of ePS in apposition to dendritic protrusions is shown in Fig 2H. ANXV‐positive structures on protrusions of GFP‐labeled dendrites were detectable while being engulfed by microglia growing in co‐culture (Fig 2I). These data indicate that PS is exposed at synaptic sites in vitro and that PS exposure is required for microglial synapse elimination.
PS is exposed at synapses in vivo in the developing hippocampus
To test whether PS exposure occurs locally at synapses in vivo, we utilized the commercially available PS‐binding probe PSVue (PSVue® 550). Once activated by zinc, PSVue has fast, calcium‐independent binding kinetics, is significantly smaller in size compared with Annexin‐based probes, and, similarly to ANXV, is cell impermeable, thus labeling only PS that has been externalized (Smith et al, 2011). While PSVue has been used previously to monitor cell death in vivo in several disease and injury models (Smith et al, 2012; Chan et al, 2015; Mazzoni et al, 2019), it remains unknown whether localized, non‐apoptotic PS exposure occurs in vivo.
To assess whether externalized PS is detectable in the developing CA1 hippocampal region, PSVue was intracerebroventricularly (ICV) injected into mice at postnatal day (P) 10 and P18, which represent the central time window for synapse refining in hippocampus (Paolicelli et al, 2011), and at P30 (young adult), when refinement is largely completed. Analysis was performed 3 h following injection (Dallérac et al, 2011; Walrave et al, 2016). Section preincubation with Sudan Black, which reacts against lipofuscin, an aggregate of oxidized proteins and lipids, reduced autofluorescence (Fig EV2). To directly test whether PS is exposed at synapses in CA1, brain sections were stained for the presynaptic marker Bassoon and the postsynaptic marker PSD‐95 (Fig 3A–B). A subset of synaptic sites in CA1 were labeled by PSVue, with a significantly higher co‐localization detected for Bassoon relative to PSD‐95 (Fig 3C and D). We examined synaptic PS exposure at relevant time points throughout this process, specifically P10, P18, and P30. Mice were injected 3 h prior to each time point with PSVue and analyzed. Results revealed the highest synaptic staining at P10, when hippocampal synapse elimination peaks, with progressively lower labeling at P18 and P30, which is at the end of the pruning period (Fig 3E and F).
Figure EV2. In vivo ePS signal is maintained after a specific background removal.
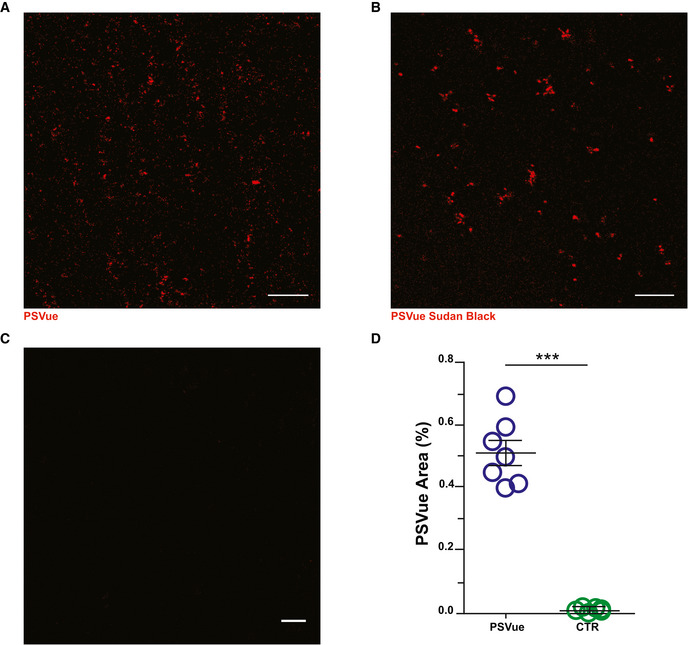
- A–C
Representative images of CA1 of WT P18 animals sacrificed 3 h after PSVue injection. (A) PsVue signal was tested by incubating slices with (B) Sudan Black. (C) The specificity of the signal was confirmed by comparing PsVue signal with slices stained with fluorophore only. Scale bar: 5 μm. - D
Quantification of PSVue puncta in WT P10 animals compared with unlabeled control. WT PSVue: 0.5149 ± 0.04 vs. CTRL: 0.011 ± 0.002; WT PSVue: N = 4 animals n = 7 fields; CTRL: N = 3 animals n = 6 fields; ***P < 0.001, unpaired _t_‐test. Bars represent mean ± SEM.
Source data are available online for this figure.
Figure 3. ePS occurs in vivo at synapses.
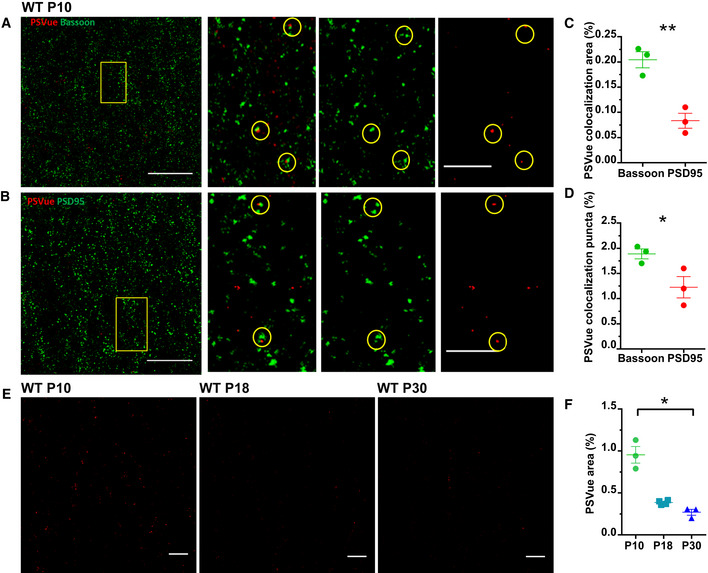
- A, B
Representative images of the CA1 region of WT P10 mouse brains stained for the presynaptic marker Bassoon (A) and the postsynaptic marker PSD95 (B). Mice were sacrificed 3 h after PSVue injection. Yellow boxes represent magnified image on the right. Co‐localization between ANXV‐positive puncta and Bassoon/PSD95 markers is highlighted by yellow dashed circles. Images taken at 63× magnification. Scale bar 5 μm. - C, D
Quantification of PSVue co‐localization with either presynaptic (Bassoon) or postsynaptic (PSD95) markers in CA1 region of WT P10 animals following PSVue injection (3 h of treatment). PSVue + Bassoon co‐localized area: 0.204 ± 0.1 vs. PSVue + PSD95 co‐localized area: 0.083 ± 0.01; PSVue + Bassoon co‐localized puncta 1.89% ± 0.1 vs. PSVue + PSD95 co‐localized puncta 1.22% ± 0.16; P10 N = 3 animals *P < 0.05 and **P < 0,01, unpaired _t_‐test. Bars represent mean ± SEM. - E
Representative images of the CA1 hippocampal region of WT P10, P18, and P30 animals sacrificed 3 h after PSVue injection. Images were acquired with a 63× magnification. Scale bar 5 μm. - F
Quantification of PSVue signal in the CA1 hippocampal region of WT P10, P18, and P30 animals sacrificed 3 h after PSVue injection. PSvue area: P10 0.954% ± 0.09 vs. P18 0.386% ± 0.01 vs. P30: 0.271% ± 0.030; P10 N = 3 animals; P18 N = 4 animals, P30 N = 3 animals. *P < 0.05, one‐way ANOVA with Kruskal–Wallis test. Bars represent mean ± SEM.
Source data are available online for this figure.
Exposed PS is developmentally regulated across periods of pruning in the visual system
To determine whether PS exposure occurred more broadly during periods of developmental pruning throughout the brain, we turned to the visual system, and specifically the dLGN, where microglial‐mediated pruning and synaptic refinement have been extensively studied (Stevens et al, 2007; Schafer et al, 2012; Lehrman et al, 2018). We performed ICV injections on C57/Bl6 animals at P4 with PSVue and conducted analyses 24 h later. Following ICV injection into the left ventricle, PSVue signal was assessed in the contralateral dLGN, where we observed robust, punctate PSVue labeling. . No signal was observed in the dLGN of animals injected with control PSVue non‐activated by zinc (Fig 4A and B).
Figure 4. In vivo exposed PS is developmentally regulated across periods of pruning in the visual system.
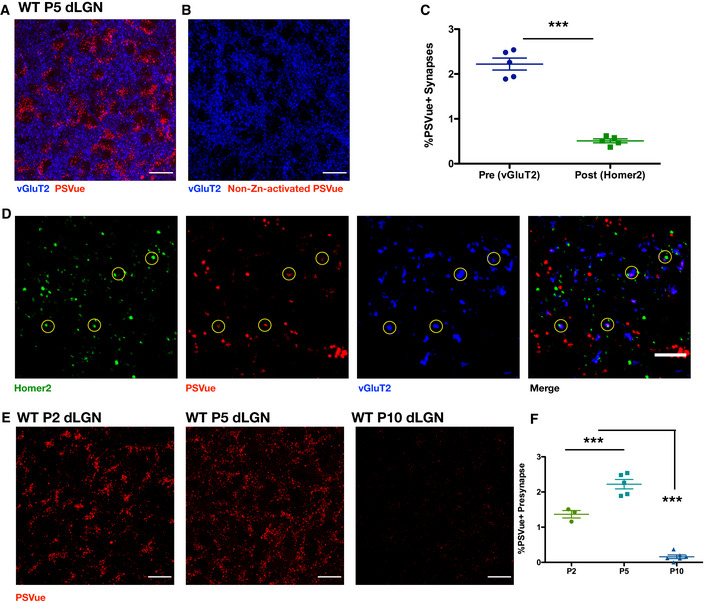
- A, B
Representative max intensity images of the dLGN following injection with either PSVue (A) or non‐zinc‐activated PSVue (B) in WT P4 C57/Bl6 mice 24 h prior. IHC for the presynaptic marker vGluT2 was performed. Images taken at 63× magnification; scale bar represents 15 μm. - C
Quantification of PSVue co‐localization at synapses in the dLGN of WT P5 mice injected with PSVue 24 h prior. Data represent the mean per animal ± SEM; N = 5 PSVue + presynaptic: 2.22% ± 0.134 vs. PSVue + postsynaptic: 0.51% ± 0.043. ***P < 0.0001, paired _t_‐test. - D
Representative images of presynaptic (vGluT2) and postsynaptic (Homer2) IHC performed in the dLGN of WT P5 animals following PSVue injection 24 h prior. Synapses were identified through co‐localization of pre‐ and postsynaptic markers in CellProfiler. Circles indicate synapses where PSVue co‐localization was observed. Images taken at 63× magnification; scale bar represents 5 μm. - E
Representative max intensity images of the dLGN following injection with PSVue 24 h prior in WT P2, P5, and P10 C57/Bl6 mice. Images taken at 63× magnification; scale bar represents 15 μm. - F
Quantification of PSVue co‐localization with vGluT2 at synapses in the dLGN of WT P2, P5, and P10 C57/Bl6 mice injected with PSVue 24 h prior. Data represent the mean per animal ± SEM; N = 3 (P2), N = 5 (P5), N = 6 (P10); P2: 1.36% ± 0.105 vs. P5: 2.22% ± 0.134 vs. P10: 0.16% ± 0.050. ***P < 0.0001, one‐way ANOVA with Tukey's multiple comparison test.
Source data are available online for this figure.
To directly test whether ePS occurred on synapses in the developing dLGN, we used immunohistochemistry to label presynaptic (vGluT2) and postsynaptic (Homer2) terminals of PSVue‐injected animals at P5. By defining synapses as the co‐localization of pre‐ and postsynaptic markers, we identified those that further co‐localized with PSVue and determined whether PSVue labeled the presynaptic or postsynaptic component. We found that, similar to the hippocampus (Fig 3A and B), PSVue labeled a subset of synapses in the dLGN and was largely localized to presynaptic rather than postsynaptic inputs (Fig 4C and D).
Finally, we analyzed ePS throughout the process of eye segregation in the dLGN, a well‐characterized period of developmental pruning where retinal ganglion cell (RGC) inputs from either eye segregate into discrete patches (Katz & Shatz, 1996; Jaubert‐Miazza et al, 2005; Huberman, 2007). In mice, this occurs between P2 and P8 and is largely completed by P10. We examined synaptic PS exposure at relevant time points throughout this process, specifically P2, P5, and P10. Wild‐type C57/Bl6 mice were injected 24 h prior to each time point with PSVue and analyzed. We found robust PSVue labeling at younger ages (P2, P5) that was considerably reduced by P10 (Fig 4E). Quantification of PSVue presynaptic (vGluT2) co‐localization peaked at P5 and was significantly diminished by P10 (Fig 4F). Taken together, our data demonstrate that PS is exposed in vivo at synapses, coinciding with periods of developmental synaptic refinement in several brain regions.
In vivo developmental PS exposure is not caspase 3‐dependent
Programmed RGC apoptosis occurs early postnatally, typically between P0 and P2, in a caspase 3‐dependent manner (Cellerino et al, 2000). To ensure that the ePS observed in vivo in the dLGN was not due to cells undergoing apoptosis, we performed staining for activated (cleaved) caspase 3 in PSVue‐injected animals. No signal for active caspase 3 was observed in either the dLGN or the retina (Fig EV3A and B; respectively). To further examine whether developmental PS exposure was downstream of caspase 3 activation, we performed PSVue ICV injections, as described above, in P4 caspase 3 WT and KO littermates (Jax #006233). When analyzed 24 h later at P5, no significant difference in PSVue labeling was observed in the dLGN between caspase 3 WT and KO littermates (Fig EV3C and D), revealing that PS exposure observed in vivo is not downstream of caspase 3‐mediated activation or apoptosis.
Figure EV3. In vivo PS exposure is not downstream of activated caspase 3.
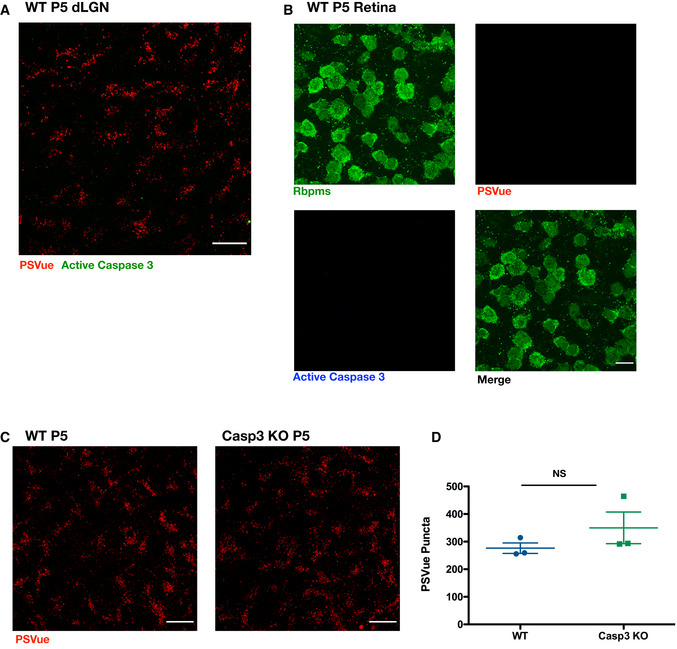
- Representative max intensity image of the dLGN following injection with PSVue 24 h prior in WT P5 C57/Bl6 mice. IHC for active caspase 3 was performed. Images taken at 63× magnification; scale bar represents 15 μm.
- Representative images of retinal whole mounts of WT P5 C57/Bl6 mice following injection with PSVue 24 h prior. IHC for the pan‐RGC marker Rpbms and active caspase 3 was performed. Images taken at 63× magnification; scale bar represents 15 μm.
- Representative max intensity images of the dLGN following injection with PSVue 24 h prior in WT or caspase 3 knockout (KO) P5 littermates. Images taken at 63× magnification; scale bar represents 15 μm.
- Quantification of PSVue in the dLGN of WT or caspase 3 KO P5 littermates injected with PSVue 24 h prior. Data represent the mean per animal ± SEM; N = 3; WT: 276.4 ± 19.01 vs. caspase 3 KO: 349.9 ± 57.29. P = 0.2904, unpaired _t_‐test.
Source data are available online for this figure.
Microglia engulf PSVue‐labeled material in vivo in the dLGN and hippocampus
The role of microglia in synaptic pruning has been established in several developing brain regions, including the visual system—both the dLGN (Stevens et al, 2007; Schafer et al, 2012; Lehrman et al, 2018) and primary visual cortex (Tremblay et al, 2010) —and the hippocampus (Paolicelli et al, 2011; Filipello et al, 2018; Weinhard et al, 2018). Given that we observed that (i) ePS was necessary for microglial‐mediated synaptic reductions in vitro (Fig 2), and (ii) PSVue labeling occurred in vivo at synapses in both the hippocampus (Fig 3) and dLGN (Fig 4) across periods of microglial‐mediated developmental pruning, we hypothesized that ePS may broadly act as a neuronal “eat‐me” signal, contributing to microglial recognition and elimination.
To test whether microglia engulf PSVue‐labeled material in the dLGN and hippocampus, we utilized our established method of in vivo engulfment analysis (Schafer et al, 2014) by measuring the volume of PSVue co‐localized with the microglial lysosomal marker CD68 within microglia. Wild‐type C57/Bl6 mice were injected 24 h prior to each time point with PSVue and analyzed by immunohistochemistry. We observed that microglia (labeled with Iba1 and P2yR12 antibodies) within the dLGN (Fig 5A and B) and hippocampus (Fig 5D and E) had internalized PSVue‐labeled material. Microglial engulfment was temporally regulated in both regions. In the dLGN, engulfment of PSVue‐labeled material was highest at P5 and was significantly reduced by P10 (Fig 5C), similar to what was observed previously when quantifying microglial engulfment of presynaptic inputs in the dLGN (Schafer et al, 2012).
Figure 5. Microglia engulf PSVue‐labeled inputs in vivo in the dLGN and hippocampus.
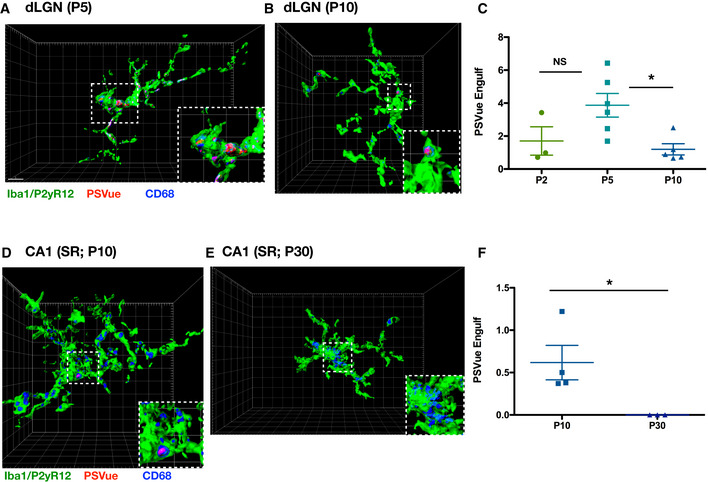
- A, B
Representative Imaris surface‐rendered images of microglia in the dLGN following injection with PSVue 24 h prior in WT P5 (A) and P10 (B) C57/Bl6 mice. Microglia are labeled by IHC with Iba1 and P2yR12, and lysosomes are labeled with CD68. Images taken at 63× magnification; scale bar represents 5 μm. - C
Quantification of the volume of engulfed PSVue material in dLGN microglia analyzed from WT P2, P5, and P10 C57/Bl6 mice injected with PSVue 24 h prior. Data represent the mean of 15–20 microglia per animal ± SEM; N = 3 (P2), N = 6 (P5), N = 5 (P10); P2: 1.703% ± 0.861 vs. P5: 3.868% ± 0.715 vs. P10: 1.196% ± 0.340. *P = 0.0237 (P5 vs. P10), one‐way ANOVA with Tukey's multiple comparison test. - D, E
Representative Imaris surface‐rendered images of microglia in the SR of CA1 following injection with PSVue 24 h prior in WT P10 (D) and P30 (E) C57/Bl6 mice. Microglia are labeled by IHC with Iba1 and P2yR12, and lysosomes are labeled with CD68. Images taken at 63× magnification. - F
Quantification of the volume of engulfed PSVue material in CA1 microglia analyzed from WT P10 and P30 C57/Bl6 mice injected with PSVue 24 h prior. Data represent the mean of 15–20 microglia per animal ± SEM; N = 4 (P10), N = 3 (P30); P10: 0.617% ± 0.203 vs. P30: 1.17e−5% ± 3.33e−7. *P = 0.05, one‐way ANOVA with Tukey's multiple comparison test.
Source data are available online for this figure.
In the hippocampus, we evaluated engulfment at P10 and P30, time points where microglial pruning has been observed (P10) and completed (P30) (Paolicelli et al, 2011; Filipello et al, 2018), in both CA1 (Fig 3C and D) and CA3 (Fig EV4A–C). Similar to the dLGN, engulfment was significantly higher in the hippocampus of P10 animals when compared to P30 (CA1 Fig 5F; CA3 Fig EV4D–F). Taken together, these data reveal that microglia engulf PSVue‐labeled material in vivo in a temporally regulated manner, coinciding with established periods of microglial pruning.
Figure EV4. Developmental PS exposure and engulfment in CA3.
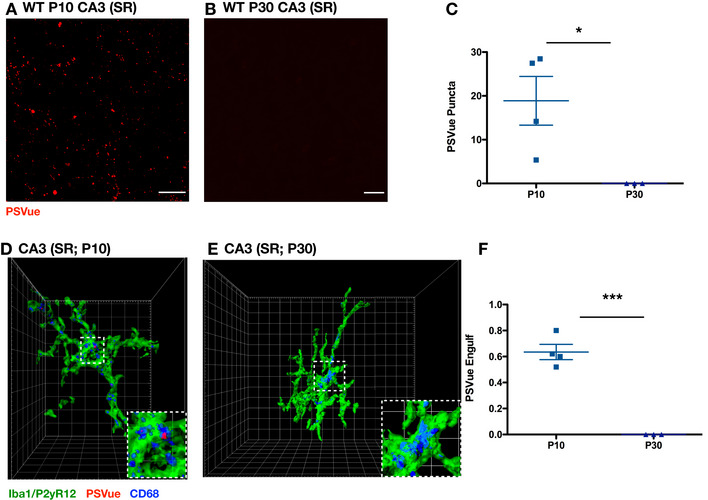
- A, B
Representative max intensity images of CA3 SR following injection with PSVue 24 h prior in WT P10 (A) and P30 (B) C57/Bl6 mice. Images taken at 63× magnification; scale bar represents 15 μm. - C
Quantification of PSVue in CA3 SR of WT P10 and P30 C57/Bl6 mice injected with PSVue 24 h prior. Data represent the mean per animal ± SEM; N = 4 (P10), N = 3 (P30); P10: 18.88 ± 5.55 vs. P30: 1.50e−5 ± 8.386e−6. *P = 0.0349, unpaired _t_‐test. - D, E
Representative Imaris surface‐rendered images of microglia in CA3 SR following injection with PSVue 24 h prior in WT P10 (D) and P30 (E) C57/Bl6 mice. Microglia are labeled by IHC with Iba1 and P2yR12, and lysosomes are labeled with CD68. Images taken at 63× magnification. - F
Quantification of the volume of engulfed PSVue material in CA3 microglia analyzed from WT P10 and P30 C57/Bl6 mice injected with PSVue 24 h prior. Data represent the mean of 15–20 microglia per animal ± SEM; N = 4 (P10), N = 3 (P30); P10: 0.635% ± 0.059 vs. P30: 1.033e−5% ± 6.386e−6. ***P = 0.0003, unpaired _t_‐test.
Source data are available online for this figure.
Increased PSVue labeling and microglial engulfment of ipsilateral RGC inputs during eye segregation
As eye segregation proceeds in the dLGN, contralateral and ipsilateral RGC inputs are segregated into two distinct regions. Although inputs from both eyes are removed, the area ultimately occupied by ipsilateral inputs is reduced to a greater extent (Jaubert‐Miazza et al, 2005; Ziburkus & Guido, 2006). To test whether PS exposure occurred preferentially on ipsilateral inputs, we performed intraocular injections with fluorescently labeled CTB, labeling contralateral (CTB‐488), and ipsilateral (CTB‐647) RGC inputs of P4 WT C57/Bl6 mice. Concurrent PSVue ICV injections were performed, and the dLGN was analyzed 24 h later at P5 (Fig 6A). Consistent with previous studies (Jaubert‐Miazza et al, 2005; Ziburkus & Guido, 2006), we found that the percentage of dLGN volume occupied by contralateral RGC inputs was significantly higher relative to ipsilateral RGC inputs (Fig 6B). However, when we calculated the percentage of PSVue‐labeled CTB inputs, we found that a higher percentage of ipsilateral inputs (CTB647) were co‐localized with PSVue (Fig 6C).
Figure 6. Increased PSVue labeling and microglial engulfment of ipsilateral RGC inputs during eye segregation.
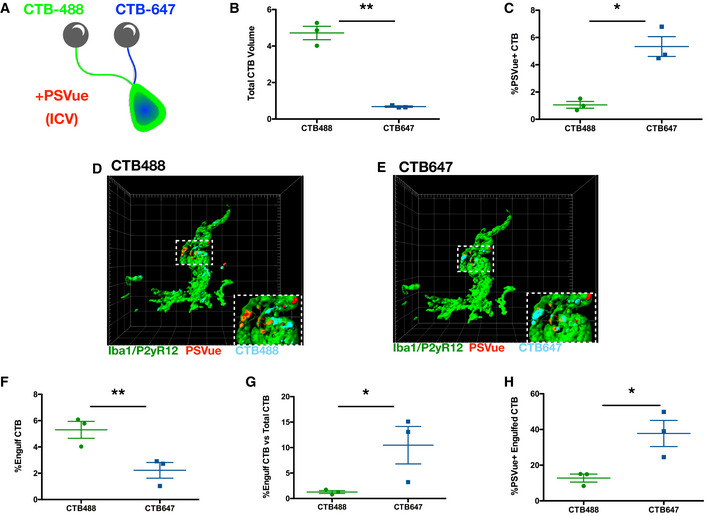
- A
Schematic of the experimental design for analysis of PSVue co‐localization onto retinal ganglion cell (RGC) inputs. Cholera toxin β‐subunit (CTB) conjugated to Alexa 488 or 647 was intraocularly injected into the contralateral or ipsilateral eyes of P4 WT mice, respectively. PSVue was then ICV‐injected, and animals were sacrificed 24 h later. - B
Quantification of the total volume of CTB‐488 or CTB‐647 RGC inputs in the dLGN of P5 mice injected with PSVue 24 h prior. Data represent mean volume per animal. N = 3 animals ± SEM; CTB‐488: 4.717% ± 0.367 vs. CTB‐647: 0.68% ± 0.035. **P = 0.0079, paired _t_‐test. - C
Quantification of the percentage of the volume of CTB‐488 or CTB‐647 RGC inputs that are co‐localized with PSVue. The volume of co‐localized signal was divided by the total volume of the inputs for either eye. N = 3 animals ± SEM; CTB‐488: 1.06% ± 0.247 vs. CTB‐647: 5.337% ± 0.73. *P = 0.0132, paired _t_‐test. - D, E
Representative Imaris surface‐rendered images of a microglia showing engulfed PSVue material along with engulfed CTB‐488 (D) or CTB‐647 (E) in the dLGN following injection with PSVue 24 h prior in WT P5 mice. Microglia are labeled by IHC with Iba1 and P2yR12. Images taken at 63× magnification. - F
Quantification of the volume of engulfed CTB‐488 or CTB‐647 material in dLGN microglia analyzed from WT P5 mice injected 24 h prior. Data represent the mean engulfment of either CTB‐488 or CTB‐647 from 15 to 20 microglia per animal ± SEM; N = 3, CTB‐488: 5.3% ± 0.641 vs. CTB‐647: 2.22% ± 0.597. **P = 0.0224, paired _t_‐test. - G
Quantification of the volume of microglial engulfed CTB‐488 or CTB‐647 normalized to the total volume of contralateral or ipsilateral CTB. Data represent normalized CTB engulfment of CTB‐488 or CTB‐647 from 15 to 20 microglia per animal ± SEM; N = 3, CTB‐488: 1.27% ± 0.280 vs. CTB‐647: 11.33% ± 2.83. *P = 0.0132, paired _t_‐test. - H
Quantification of the volume of co‐localized PSVue and CTB (488 vs. 647) engulfed by microglia normalized to the volume of engulfed CTB‐488 or CTB‐647. Data represent normalized PSVue/CTB engulfment of contralateral (488) vs. ipsilateral (647) from 15 to 20 microglia per animal ± SEM; N = 3, CTB‐488: 12.77% ± 2.231 vs. CTB‐647: 37.76% ± 07.331. *P = 0.0432, paired _t_‐test.
Source data are available online for this figure.
To determine whether elevated ipsilateral PS exposure translated into preferential microglial elimination, we directly compared PSVue‐labeled contralateral and ipsilateral engulfment within dLGN microglia (Fig 6D and E). We found that microglia engulfed significantly more contralateral inputs (CTB‐488) relative to ipsilateral (CTB‐647), when the volume of internalized CTB was normalized to microglia volume (Fig 6F). The total volume of ipsilateral inputs was significantly less than the volume of contralateral inputs, as described above. When we took this into account by normalizing the volume of engulfed CTB by the total CTB volume, we found that a higher proportion of ipsilateral inputs within the dLGN had been engulfed (Fig 6G). Furthermore, when we calculated the percentage of engulfed CTB that also co‐localized with PSVue, we found that significantly more engulfed ipsilateral inputs were positive for PSVue (Fig 6H). Although ipsilateral inputs account for less volume of the total RGC inputs in the developing dLGN, our data suggest that a higher proportion are labeled with PSVue and are ultimately targeted for elimination by microglia during eye segregation.
Loss of C1q leads to elevated PSVue‐positive presynaptic inputs and reduced microglial PSVue engulfment
Microglia engulf presynaptic RGC inputs within the developing dLGN in a complement‐ and CR3‐dependent manner (Schafer et al, 2012). In the periphery, C1q binds to PS—either directly (Païdassi et al, 2008) or indirectly (Martin et al, 2012)—to facilitate phagocytic removal, a mechanism that can involve CR3 (Ma et al, 2012). Given our findings that local synaptic PS exposure occurs during periods of microglial‐mediated pruning in the dLGN (P5‐P8; Schafer et al, 2012; Lehrman et al, 2018), we hypothesized that externalized PS may also contribute to C1q recruitment on RGC inputs.
Immunohistochemistry analysis of P5 WT C57/Bl6 mice injected with PSVue 24 h prior revealed that C1q and PSVue co‐localized at a subset of retinogeniculate presynaptic inputs (Fig 7A). There was no significant difference in the percentage of C1q co‐localization with vGluT2 when quantified across developmental pruning (Fig 7B), and only a subset of those C1q‐positive presynaptic inputs were also PSVue‐positive (Fig 7A and C). However, analysis across these developmental time points revealed that presynaptic co‐localization of PSVue with C1q peaked at P5 and was significantly reduced by P10 (Fig 7C and D).
Figure 7. Loss of C1q leads to increased PSVue‐labeled inputs and reduced microglial PSVue engulfment.
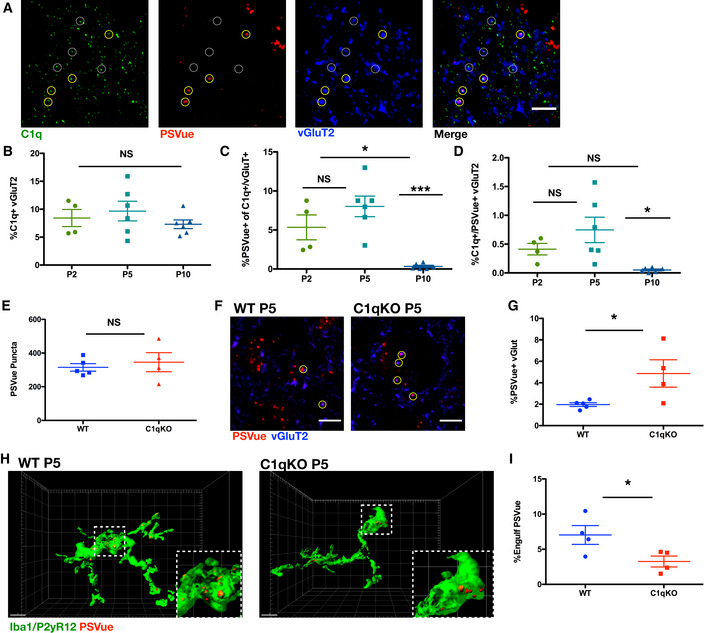
- A
Representative IHC images of C1q and vGluT2 in the dLGN of WT P5 animals following PSVue injection 24 h prior. Yellow circles indicate where PSVue co‐localization with C1q and vGluT2 was observed. Dashed white circles indicate vGluT2 and C1q co‐localization independent of PSVue labeling. Images taken at 63× magnification; scale bar represents 5 μm. - B–D
Quantification performed in the dLGN of WT P2, P5, and P10 C57/Bl6 mice injected with PSVue 24 h prior of (B) vGluT2 co‐localization with C1q. Data were calculated by quantifying the percentage of C1q/vGluT2 puncta normalized to total vGluT2 inputs. Data represent the mean per animal ± SEM; N = 4 (P2), N = 6 (P5), N = 6 (P10); P2: 8.425% ± 1.527 vs. P5: 9.662% ± 1.756 vs. P10: 7.3% ± 0.774 NS. P > 0.05, one‐way ANOVA with Tukey's multiple comparison test. (C) Quantification of PSVue co‐localization with C1q and vGluT2 inputs. Data were calculated by quantifying the percentage of PSVue/C1q/vGluT2 puncta normalized to C1q/vGluT2 inputs. Data represent the mean per animal ± SEM; N = 4 (P2), N = 6 (P5), N = 6 (P10); P2: 5.34% ± 1.591 vs. P5: 8.023% ± 1.323 vs. P10: 0.33% ± 0.148. *P < 0.05 (P2 vs. P10) and ***P < 0.0001 (P5 vs. P10), one‐way ANOVA with Tukey's multiple comparison test. (D) Quantification of PSVue and C1q co‐localization with vGluT2. Data were calculated by quantifying the percentage of PSVue/C1q/vGluT2 puncta normalized to total vGluT2. Data represent the mean per animal ± SEM; N = 4 (P2), N = 6 (P5), N = 6 (P10); P2: 0.412% ± 0.099 vs. P5: 0.746% ± 0.221 vs. P10: 0.051% ± 0.015. *P = 0.015 (P5 vs. P10), one‐way ANOVA with Tukey's multiple comparison test. - E
Quantification of PSVue in the dLGN of WT or C1qKO P5 littermates injected with PSVue 24 h prior. Data represent the mean per animal ± SEM; N = 5 (WT), N = 4 (C1qKO); WT: 315.0 ± 22.04 vs. C1qKO: 345.8 ± 56.47. P = 0.597, unpaired _t_‐test. - F
Representative images of the dLGN of P5 WT or C1qKO littermates following injection with PSVue 24 h prior. IHC was performed for the presynaptic marker vGluT2. Yellow circles indicate where co‐localization between PSVue and vGluT2 was observed. Images taken at 63× magnification; scale bar represents 5 μm. - G
Quantification of PSVue co‐localization with vGluT2 at synapses of P5 WT or C1qKO littermates following injection with PSVue 24 h prior. Data represent the mean per animal ± SEM; N = 5 (WT), N = 4 (C1qKO); WT: 1.962% ± 0.174 vs. C1qKO: 4.864% ± 1.277. *P = 0.0382, _t_‐test. - H
Representative Imaris surface‐rendered images of microglia in the dLGN following injection with PSVue 24 h prior in P5 WT or C1qKO littermates. Microglia are labeled by IHC with Iba1 and P2yR12. Images taken at 63× magnification; scale bar represents 5 μm. - I
Quantification of the volume of engulfed PSVue material in microglia of P5 WT or C1qKO littermates following injection with PSVue 24 h prior. Data represent the mean of 15–20 microglia per animal ± SEM; N = 4 (WT), N = 4 (C1qKO); WT: 6.978% ± 1.294 vs. C1qKO: 3.258% ± 0.775. *P = 0.0487, unpaired _t_‐test.
Source data are available online for this figure.
Mice lacking C1q have disrupted developmental refinement, resulting in impaired eye segregation and excessive retinal innervation of the dLGN (Stevens et al, 2007). To test whether PS exposure and microglia engulfment were also affected in these animals, we performed PSVue ICV injections into P4 WT and C1qKO littermates and analyzed 24 h later. We found that, although the total amount of PSVue was unchanged between WT and C1qKO (Fig 7E), a significantly higher percentage of vGluT2 presynaptic inputs were labeled with PSVue in C1qKO animals (Fig 7F and G). Moreover, analysis of P5 WT and C1qKO littermates revealed significantly reduced microglial PSVue engulfment in animals deficient in C1qKO (Fig 7H and I). Together, these data demonstrate that presynaptic co‐localization of C1q and ePS is developmentally regulated and that loss of C1q results in an increase in presynaptic PS‐positive inputs and reduced microglial PS engulfment. How the interaction of ePS and C1q at synapses confers temporal specificity, facilitating complement‐mediated synaptic pruning, and whether there are pruning‐independent roles of synaptic C1q (which likely binds to additional, currently unknown, synaptic molecules) remains an area for future investigation.
Discussion
Microglia eliminate synapses during developmental pruning, but the neuronal signals specifying which synapses are targeted remain unknown. Here, we demonstrate that local synaptic PS exposure occurs in a developmentally regulated manner across several brain regions, contributing to microglial recognition of neuronal structures and subsequent elimination in vitro and in vivo. We found that liposome engulfment by isolated microglia was dependent on PS concentration and TREM2 in vitro. In co‐culture with hippocampal neurons, microglial‐dependent synaptic reduction was also PS‐ and TREM2‐dependent. In vivo, we utilized the PS‐binding probe PSVue and found that PS exposure occurred locally at synapses in both the hippocampus and the dLGN early in development, and that microglia in both regions engulfed PSVue‐labeled material, a process that was also developmentally regulated. Neuronal PS exposure was not stochastic, as ipsilateral RGC inputs in the dLGN were preferentially labeled and targeted for engulfment by microglia. Finally, in the dLGN, presynaptic C1q and PSVue co‐localization peaked during developmental microglial pruning, and in C1qKO animals, we observed an increase in PSVue‐labeled presynaptic inputs as well as a decrease in microglial engulfment of PSVue‐labeled material. Our findings support a model in which local PS exposure is a common neuronal signal that interacts with complement and other proteins to promote microglial engulfment during developmental critical periods of synaptic refinement.
Emerging roles of PS localization and regulation in diverse biological functions
Phosphatidylserine is a ubiquitous lipid, estimated to represent 13–15% of all phospholipids in the human cerebral cortex (Svennerholm, 1968). Along with other anionic lipids, PS is typically asymmetrically distributed within the inner leaflet where several critical functions have been ascribed. PS participates in membrane translocation and activation of protein kinase C (Newton & Keranen, 1994), is involved in Akt signaling through interactions with PIP3 (Huang et al, 2011), and contributes to membrane curvature and vesicular transport (Xu et al, 2013). At the synaptic level, PS alters the interaction of SNARE proteins with membranes (Dennison et al, 2006), influences neurotransmitter vesicle fusion (Murray et al, 2004), binds to synaptotagmins (Fernandez et al, 2001; Zhang et al, 2009), and modulates AMPA glutamate receptors (Baudry et al, 1991).
While the function of inner leaflet plasma membrane PS continues to be an area of active research, emerging roles of PS exposed on the outer leaflet are also being discovered. PS exposure is classically associated with apoptosis, where loss of membrane asymmetry occurs early on in the cell death signaling cascade (Fadok et al, 1992; Martin et al, 1995; Hoffmann et al, 2001). Non‐apoptotic PS exposure has also been described in several biological contexts. In the blood, PS exposure occurs on activated platelets, where it contributes to the clotting response following injury (Lentz, 2003). In the immune system, transient PS exposure occurs during the activation of T lymphocytes, B lymphocytes, mast cells, and neutrophils (Martin et al, 2000; Callahan et al, 2003; Elliott et al, 2005). In neurons, PS exposure can occur locally on injured axons (Shacham‐Silverberg et al, 2018) or dendrites (Sapar et al, 2018), which are then targeted for elimination while sparing the remaining uninjured cell structures. Ectopic PS exposure on living neurons causes phagocytic cells to engulf distal neurites (Sapar et al, 2018), and direct masking of ePS signal reduces axonal debris engulfment (Shacham‐Silverberg et al, 2018). Our data reveal an additional novel role of locally externalized PS, where developmentally regulated local exposure contributes to microglial engulfment, a critical process important for the maturation of neuronal circuits.
An array of enzymes, namely flippases and scramblases, control PS membrane asymmetry and localization; the upstream mechanisms regulating these proteins, and whether apoptotic and local PS exposure are similarly controlled, remain largely unknown. Flippases, also known as type IV P‐type ATPases (P4‐ATPases), catalyze the movement of anionic phospholipid species from the extracellular leaflet to the cytosolic leaflet. Alternatively, scramblases function to disrupt membrane asymmetry in a calcium‐dependent, ATP‐independent manner. Many different flippases and scramblases have been identified to date, with much of the current understanding of upstream regulation focused on caspase‐mediated inactivation and activation, respectively. For example, PS is irreversibly exposed on apoptotic cells by the activation of a PS scramblase, Xk‐related protein 8 (Xkr8), through caspase cleavage (Suzuki et al, 2016), and inhibition of PS translocases ATP11A and ATP11C is also caspase‐dependent (Segawa et al, 2014). While caspase activation appears to be an important mechanism in apoptosis‐associated PS exposure, it is likely not the upstream mechanism driving the locally ePS that we observed in vivo, as revealed by our studies in caspase 3‐deficient mice (Fig EV3).
A critical question that remains is the identification of the upstream neuronal mechanisms that trigger local PS exposure at specific synaptic inputs during development. Importantly, we cannot rule out that PS exposure marks immature synapses more generally, as this may represent a transient and dynamic process. We observed only a small percentage of total synapses in both the hippocampus and dLGN that co‐localized with ePS at several early developmental time points, but it remains unclear how maturation may contribute to or control synaptic PS exposure or if PS marks specific synapses to be engulfed. Previous work has demonstrated that in some cells transient PS exposure relies on TMEM16 family PS scramblases and/or calcium inhibition of translocase ATP11A or ATP11C (Suzuki et al, 2010). Interestingly, Anoctamin 1 (ANO1)/TMEM16A, a Ca2+‐activated Cl− channel, has been shown to mediate process extension in RGCs, contributing to cortex development (Hong et al, 2019). Whether the TMEM16 family of scramblases could be involved in PS exposure at synapses is worth investigating.
The role of neuronal activity has been well‐established in synaptic refinement (Katz & Shatz, 1996; Hua & Smith, 2004; Hooks & Chen, 2006; Huberman, 2007; Burbridge et al, 2014) and microglial‐mediated synaptic pruning (Schafer et al, 2012), but how activity is translated into local cues mediating engulfment is not well‐understood. Of note, in both the hippocampus and dLGN, ePS was significantly higher during the first two postnatal weeks, but whether or how this developmental localization is regulated by neuronal activity is not yet clear. In the dLGN, although both contralateral and ipsilateral RGC inputs are pruned by microglia throughout the process of eye segregation, the area occupied by ipsilateral inputs is more greatly reduced. Interestingly, we found that ipsilateral RGC inputs were preferentially labeled with PSVue, and these inputs were disproportionately targeted for microglial elimination. If PS externalization occurs more frequently on ipsilateral inputs, this may increase the likelihood of PSVue binding following injection and further explain the observation that engulfed PSVue‐labeled ipsilateral material was significantly higher relative to contralateral engulfed material. However, as not all engulfed CTB material was co‐labeled with PSVue, it is likely that additional mechanisms contribute to engulfment of RGC inputs. For example, CD47, a well‐known “don't eat‐me” signal, mediates microglial pruning in an activity‐driven manner (Lehrman et al, 2018). How neuronal activity contributes to local synaptic PS exposure and concomitant loss of CD47, whether these signals are linked or represent two distinct mechanisms, and whether there are additional, currently unknown signals involved in these processes is an area of active investigation.
Neuronal PS exposure may represent a common signal involved in multiple mechanisms of glial synaptic pruning
Numerous receptors involved in the recognition of ePS (either directly or through adapter proteins) are expressed by microglia and astrocytes. For example, TREM2, GPR56, and BAI1 bind directly to ePS, while TAM surface receptor tyrosine kinase proteins TYRO3, AXL, and MER recognize ePS through interaction with their ligands Gas6 and protein S (reviewed in Lemke, 2019 and Nakano et al, 1997; Fadok et al, 2001; Park et al, 2007; Grommes et al, 2008; Païdassi et al, 2008; Neher et al, 2012; Chung et al, 2013; Brown & Neher, 2014; Graham et al, 2014;Wang et al, 2015; Shirotani et al, 2019; Li et al, 2020). Importantly, several of these receptors have also been implicated in synaptic elimination by glia (Chung et al, 2013; Filipello et al, 2018; Li et al, 2020). In microglia, TREM2 mediates synaptic pruning in the developing hippocampus, where loss results in compromised brain connectivity in adult mice (Filipello et al, 2018). TAM receptors are necessary for astrocyte‐mediated synaptic pruning in the visual system, but are dispensable for microglia (Chung et al, 2013). Most recently, microglial‐expressed GPR56 was demonstrated to contribute to visual system developmental pruning through a PS‐dependent interaction. A splicing isoform of GPR56 mediates microglia engulfment via PS binding. Similar to loss of C1q, loss of microglial GPR56 led to elevated synapses and decreased microglial engulfment of presynaptic inputs in the developing dLGN (Li et al, 2020). Whether GPR56, TREM2, and C1q all cooperate throughout developmental pruning remains unclear. In C1qKO animals, for example, microglia PS engulfment is reduced, but not eliminated (Fig 7H and I). The same occurs for PS‐liposome phagocytosis by TREM2KO microglia (Fig 1F), indicating that additional mechanisms of microglial PS recognition and elimination must exist. Understanding how these different pathways may be spatially or temporally regulated within glia will be important to determine how each contribute to synaptic pruning, in both development and disease.
Together, our findings suggest that locally ePS could be a common neuronal signal that occurs during developmental pruning, one that is recognized by several glial‐expressed receptors and secreted factors involved in this process. Whether and how these pathways cooperate, in a temporally or spatially specific manner, and whether astrocytes also contribute to PS removal, will be interesting to examine further. Determining the source of PS‐labeled material that has been engulfed will also be important in future studies, as we cannot exclude the possibility that some engulfed PS‐labeled material is derived from axonal or neuronal structures or debris. Current technical limitations surrounding antibody labeling make quantifying PS‐bound to synaptic elements within microglial endolysosomal compartments from fixed brain tissue difficult to interpret. In our current study, we observed synaptic PS exposure in vitro and in vivo, demonstrated that both PS and TREM2 were necessary for synaptic reductions in vitro, and found altered presynaptic PS exposure and reduced PS engulfment in C1qKO animals, a model known to have disrupted synaptic refinement. Development of new tools and models to label and image PS dynamics and microglia‐mediated engulfment in different contexts will be important next steps.
Potential role of exposed PS and microglial engulfment in neurological diseases
Emerging evidence suggests developmental mechanisms of synaptic pruning can become aberrantly “reactivated” and contribute to pathological synapse loss and dysfunction (Stephan et al, 2012; Salter & Stevens, 2017). In AD, for example, both the complement cascade and TREM2 have been implicated in pathology (Wang et al, 2015; Hong et al, 2016; Dejanovic et al, 2018; Parhizkar et al, 2019). Indeed, synaptic C1q is aberrantly elevated in several models of AD, contributing to early synapse loss through microglial‐mediated elimination (Hong et al, 2016; Dejanovic et al, 2018). In synaptosomes isolated ex vivo from AD patient tissue, elevated PS exposure has been reported (Bader Lange et al, 2008); whether this also occurs in vivo, facilitating C1q deposition and contributing to synapse loss, remains unknown. On the other hand, GWAS analyses have identified in AD patients several TREM2 variants, most associated with either reduced surface expression or reduced phagocytic activity (reviewed in (Ulrich et al, 2017)). Studies in TREM2‐deficient AD mouse models have revealed that the receptor is important for the microglial response to amyloid pathology and that impairment of TREM2 function accelerates early amyloidogenesis due to reduced phagocytic clearance of amyloid seeds (Parhizkar et al, 2019). As PS exposure is hypothesized to occur on plaques, TREM2 may have dual roles in AD, where activation through ePS binding may contribute to plaque containment and clearance, while synaptic PS exposure may contribute to aberrant synapse loss. Our findings suggest that synaptic PS exposure may be involved in both C1q‐ and TREM2‐mediated microglial elimination. Exploring the potential interactions between these two mechanisms, in both development and disease, will be an important area of future investigation.
Materials and Methods
Mouse strains
Humanitas
Mice were housed in the SPF animal facility of Humanitas Clinical and Research Center in individually ventilated cages. All experiments followed the guidelines established by the European Directive 2010/63/EU and the Italian D.Lg. 26/2014. The study was approved by the Italian Ministry of Health. All efforts were made to minimize the number of subjects used and their suffering. C57BL/6 Trem2−/− mice, generated as previously described (Turnbull et al, 2006), were provided by BioXell‐Cosmo Pharmaceutical (Milan, Italy) (Correale et al, 2013). P10, P18, and P30 male and female littermate animals were used for each experiment. Trem2−/− and littermate control WT mice (backcrossed 12 generations to the C57BL/6 background) were obtained from Marco Colonna (Turnbull et al, 2006), and the two strains were bred in parallel.
Boston Children's Hospital
All mice were used at the ages specified in the experimental procedures outlined below, and unless stated otherwise, male and female mice were included in all experiments. Wild‐type C57BL/6 mice were obtained from Charles River, unless otherwise specified below. caspase 3 KO mice were obtained from Jackson Labs (Jax #006233), and caspase 3+/− × caspase 3+/− crosses were used to generate wild‐type and caspase 3 KO littermates. C1qaKO mice are on a C57BL/6J background and were a kind gift from M. Botto (Imperial College, London). C1qa+/− × C1qa+/− crosses were used to generate wild‐type and C1qKO littermates. Animals were group‐housed in Optimice cages and maintained in the temperature range and environmental conditions recommended by AAALAC. All procedures were approved by the Boston Children's Hospital institutional animal care and use committee in accordance with NIH guidelines for the humane treatment of animals.
Neuronal cultures
Mouse hippocampal neurons were established from hippocampi of embryonic stage E17 fetal mice. Neurons were plated onto glass coverslips coated with poly‐l‐lysine at densities of 95 cells/mm2. Cells were maintained in Neurobasal medium (Invitrogen) with B27 supplement and antibiotics, 2 mM glutamine, and glutamate. All experiments were performed at 13–15 days in vitro (DIV). ANXV cloaking was performed for 15 min at a dilution of 1:20.
Microglial cultures
Primary microglia were obtained from mixed cultures established from telencephalon of P1–4 mice. Cells were maintained in DMEM containing 20% of heat‐inactivated fetal bovine serum (FBS), glucose 0.6%, sodium pyruvate 2 mM, and antibiotics. Microglia were isolated at DIV10–15 by shaking flasks at 245 rpm for 45 min, and then, cells were seeded on poly‐l‐ornithine pre‐coated slides of 18 mm diameter. For the microglia–neuron co‐culture experiments, WT or Trem2−/− microglia were added to hippocampal neurons (13–14 DIV) at a microglia to neuron ratio of 1.5:1 for 24 h. In a distinct set of experiments, ANXV blocking treatment was performed by pre‐incubating hippocampal neurons with ANXV (1:20) for 15′. After ANXV wash, microglia were added to neurons for 24 h. To visualize neuronal processes and dendritic spines, hippocampal neurons were transfected with GFP using Lipofectamine 2000 (Invitrogen) at DIV11.
For the experiments of bead phagocytosis, microglia were incubated with ANXV (1:20) for 15′ and fluorescent particles (3 μm; Spherotech) were added to microglial cells at a 1:1 microglia/bead ratio for 2 h. Cells were then washed with ice‐cold PBS, fixed and stained with IBA1 antibody, and analyzed. To assess microglial viability following treatment with Annexin V, microglia were incubated with ANXV (1:20) for 15′, then washed with PBS, and tested for viability after 24 h using the live marker Calcein combined with propidium iodide (PI) to label dying cells.
Phagocytosis assay of liposomes
Liposomes were prepared by mixing chloroform stocks of cardiolipin and PS (both 18:1, Avanti Polar Lipids), supplemented with 1 mol% DiO (Thermo Fisher Scientific) in given molar ratios (79:20:1; 49:50:1; 0:99:1) followed by drying of the lipid film with a N2 stream and desiccation under 15‐ to 25‐m bar for more than 1 h. Lipid film was then rehydrated with a buffer containing HEPES 20 mM (pH 7.4) and KCl 150 mM with agitation on a ThermoMixer heated to 72°C for at least 1 h. This vesicle mixture was then extruded 25× through polycarbonate membranes with pore size of 400 and 100 nm, consecutively. Uniform liposome size among different lipid compositions was confirmed with dynamic light scattering (DynaPro Titan, Wyatt Technology). If needed, blocking of liposomes with ANXV was performed at room temperature for 15 min before cell incubation. Microglia were seeded in DMEM containing 5% of heat‐inactivated FBS, and after 1 h, it was lowered to 1%. Liposomes were then added to medium in a final lipid concentration of 0.1 mM, and after 1 h of incubation, cells were fixed and stained for IBA1 and CD68 in order to perform confocal microscopy.
Immunocytochemistry and cell imaging
Cells were fixed for 10 min at room temperature (RT) in 4% (w/v) PFA, 4% (w/v) sucrose, 20 mM NaOH, and 5 mM MgCl2 in PBS, pH 7.4. For microglia intracellular staining, cells were permeabilized and blocked for 45 min at RT in 15% (w/v) goat serum, 0.3% (v/v) saponin, 450 mM NaCl, 20 mM phosphate buffer, pH 7.4, and incubated at 4°C overnight with primary antibodies Iba‐1 (1:750, Wako); CD68 (1:1,000 BioLegend) diluted in blocking buffer. Following, cells were incubated with secondary antibodies (1:400, Alexa Fluor®‐conjugated, Invitrogen) at room temperature for 2 h, counterstained with DAPI, and mounted in FluorSave (Calbiochem). For neuronal staining, if needed, cells were stained by treating with ANXV (1/80) for 7 min before being fixed, and thereafter, they were permeabilized and blocked in 15% (w/v) goat serum, 0.3% (v/v) Triton X‐100, 450 mM NaCl, 20 mM phosphate buffer, pH 7.4, and stained for SHANK2 (1:1,000), VGluT1 (1:2,000), beta‐tubulin (1:80), and PSD‐95 (1:500).
Confocal images were acquired with a laser scanning LEICA SP8 STED3X confocal microscope, using a HC PL APO 100X/1.40 oil white objective (Leica). Emission filter bandwidths and sequential scanning acquisition were set up, in order to avoid any possible spectral overlap between fluorophores. Images shown in Fig 2 were deconvolved using Huygens Professional software (SVI, scientific volume imaging) and subsequently processed using Imaris software (Bitplane).
In vitro liposome engulfment quantification
Liposomes, CD68, and Iba‐1 volumes were quantified by applying 3D surface rendering of confocal stacks in their respective channels, using identical settings (fix thresholds of intensity and voxel) within each experiment. For quantification of liposome engulfment by microglia, single cells were considered separately. Only liposome signals present within microglial CD68+ structures were considered, in order to guarantee that only liposomes completely phagocytosed by microglia were included in the analysis. To this aim, a new channel for “CD68‐liposomes” was created, by using the mask function in Imaris, masking the DiO signal within CD68+ structures. Quantification of volumes for “engulfed liposome” was performed following the “3D Surface rendering of engulfed material” protocol, previously published by Schafer et al (2014). To account for variations in body size, for each single cell the amount of “engulfed liposome” was normalized to the volume of the same microglia cell (given by Iba‐1+ volume). All data were normalized to WT + PS99% values within each experiment.
Dendritic spine analysis
Images of primary hippocampal cultures were acquired using a UPLSAPO 60×/1.35 oil objective (Olympus). Each image consisted of a stack of images taken through the _z_‐plane of the GFP‐transfected hippocampal neurons. Spine analysis was performed with ImageJ. Spines were classified as mushroom when they present a well‐defined neck and a very voluminous head (Harris & Stevens, 1989). At least four dendritic branches were analyzed for each neuron. The number of analyzed neurons is reported in each figure legend. At least three independent replications were performed for each experimental setting.
Electrophysiology: cell culture recordings
Whole‐cell voltage‐clamp recordings were performed on WT embryonic hippocampal neurons maintained in culture for 13–15 DIV. During recordings, cells were bathed in a standard external solution containing (in mM) 125 NaCl, 5 KCl, 1.2 MgSO4, 1.2 KH2PO4, 2 CaCl2, 6 glucose, and 25 HEPES‐NaOH, pH 7.4. Recording pipettes (resistances of 3–5 MU) were filled with a standard intracellular solution containing (in mM) 135 K+‐gluconate, 1 EGTA, 10 HEPES, 2 MgCl2, 4 MgATP, and 0.3 Tris–GTP (pH 7.4). For mEPSC recordings, 1 mM tetrodotoxin, 20 mM bicuculline, and 50 mM AP5 were added to the standard extracellular solution to block spontaneous action potential propagation by GABA‐A and NMDA receptors. Recordings were performed in voltage‐clamp mode at a holding potential of −70 mV using a MultiClamp 700B amplifier and pCLAMP 10 software (Axon Instruments, Foster City, CA). Series resistance ranged from 10 to 20 MU and was monitored for consistency during recordings. Cells in culture with leak currents > 100 pA were excluded from the analysis. Signals were amplified, sampled at 10 kHz, filtered to 2 or 3 KHz, and analyzed using the pCLAMP 10 data acquisition and analysis program.
Intracerebroventricular PSVue injection
For all injections, a volume of 2 μl of 1 mM PSVue® 550 (PSVue; P‐1005 Molecular Targeting Technologies; prepared following manufacturer's instructions) was utilized. No gross toxic effects, overt signs of distress or pain, or any other behavioral changes were observed as expected with ICV injection procedures.
Neonates (P0–P9)
Neonatal pups (P0–P5) were anesthetized by hypothermia or isoflurane (P9; 2–4% exposure followed by 1.5% throughout the experiment). Once anesthetized, as determined by lack of toe pinch reflex, PSVue was injected at a location approximately 0.7–1.0 mm lateral to the sagittal suture and 0.7–1.0 mm caudal from the neonatal bregma with a 27G needle inserted 2 mm into the tissue perpendicular to the skull surface. PSVue was injected slowly, and the needle was removed 10–20 s after discontinuing the plunger movement to prevent backflow. Following injection, neonates were warmed on a warm water circulating heating pad, and once pink and breathing were returned to the dam. Animals were sacrificed 24 h later for analysis as described below.
P10/P18/P30 (hippocampal synaptic analysis)
Mice were anesthetized using a ketamine (100 mg/kg)/xylazine (20 mg/kg) cocktail. PSVue or vehicle (zinc nitrate solution) with fluorophore only was stereotaxically (KOPF Instruments) injected into left lateral ventricle of two distinct group of animals via a Hamilton syringe at 0.5 μl/min (Gauge 33 Hamilton Cat# 549‐0700). The stereotaxic coordinates were the following: P10: 0.4 mm anteroposterior (AP); 1 mm mediolateral (ML); and 1.8 mm dorsoventral (DV) from bregma; P18/P30: 0.5 mm AP, 1.1 mm ML, and 2.5 mm DV from bregma. Upon completion of injection (< 15 min), mice were allowed to recover from anesthesia and returned to their home cage. Animals were sacrificed 3 h later for analysis as described below.
P30 (microglial engulfment analysis)
Once anesthetized using isoflurane (2–4% exposure followed by 1.5% throughout the experiment), PSVue was stereotaxically injected into left lateral ventricles (−0.4 mm anteroposterior, 1 mm mediolateral, and −2.5 mm dorsoventral) via a Hamilton syringe at 0.5 μl/min similar to ICV procedures described previously (Hong et al, 2016). Upon completion of injection (< 15 min), mice were allowed to wake up from anesthesia and returned to their home cage. Animals were sacrificed 24 h later for analysis as described below.
Immunohistochemistry
Hippocampal immunohistochemistry
Mice were anesthetized with a ketamine (100 mg/kg)/xylazine (20 mg/kg) cocktail and perfused transcardially with 0.9% saline, and brains were collected and post‐fixed in 4% paraformaldehyde for 24 h, then washed, and transferred to PBS solution for 24–48 h at 4°C. Fifty millimeter‐mm‐thick coronal sections were cut at the level of the dorsal hippocampus with a VT1000S Vibratome. Immunofluorescent staining was carried out on free‐floating sections. Sections were processed by 45 min RT blocking in 10% normal goat serum and 0.2% Triton X‐100 and incubated overnight with specific primary antibodies VGLUT1 (1:1,000, SySy, Cat# 135 304) and Bassoon (1:400, SySy) followed by incubation with secondary antibodies (1:400, Alexa Fluor‐conjugated, Invitrogen), counterstained with DAPI, and mounted in FluorSave (Calbiochem). For PSD‐95 (1:300, Thermo Fisher, Cat# 516900) staining, slices were permeabilized at RT in 0.5% Triton X‐100 (Sigma), followed by 1‐h RT blocking in 2% BSA and 0.5% Triton X‐100 and overnight incubation with primary antibody.
Sections were treated for 7 min with Sudan Black at a concentration of 0.1% in 70% ethanol and then thoroughly washed to reduce tissue autofluorescence and background, while preserving the specific fluorescence signal.
dLGN Immunohistochemistry
Mice were anesthetized with Avertin (240 mg/kg, i.p.) and transcardially perfused with PBS. Brains and eyes were harvested and post‐fixed in 4% PFA (Electron Microscopy Sciences, Catalog #15710) for 2 h, then washed, and transferred to 30% sucrose solution for 24–48 h at 4°C. Cryosections were prepared from tissue embedded in a 2:1 mixture of 30% sucrose: OCT (Sakura Finetek, Catalog #4583) onto X‐tra slides (Leica Microsystems, Catalog #3800200). Tissue sections were dried, washed in PBS, and blocked for 2 h at room temperature with slow agitation in 20% normal goat serum (Sigma‐Aldrich, Catalog #G9023–10ML) with 0.3% TX‐100 (Sigma‐Aldrich, Catalog #T8787–100ML) in PBS. Primary antibody (see below) was applied overnight, in 10% normal goat serum with 0.3% TX‐100 in PBS, at 4°C, with slow agitation. Tissue sections were then washed 3 × 10 min with PBS and incubated with the appropriate Alexa Fluor‐conjugated secondary antibody (1:200; Invitrogen/Thermo Fisher Scientific) for 2 h, in 10% normal goat serum with 0.3% TX‐100 in PBS, at room temperature with slow agitation. Finally, tissue sections were mounted with VECTASHIELD with DAPI (Vector Laboratories, Catalog #H‐1000). Images were acquired on a LSM880 confocal microscope (Zeiss) at 63× magnification. At least two _z_‐stack images were taken (0.3‐μm slices) per section, and two sections per animal were analyzed.
Synaptic labeling
Brains were sectioned at 15 μm on a cryostat, and primary antibodies (guinea pig anti‐vGluT2, 1:1,000 Millipore #AB2251; rabbit anti‐Homer2, 1:500 Cedarlane Labs #160203(Sy); mouse anti‐Bassoon, 1:300 Enzo Life Sciences # ADI‐VAM‐PS003‐F) for synaptic staining were performed as described above.
Active caspase 3
Brains were sectioned at 15 μm on a cryostat, and primary antibody labeling (rabbit anti‐cleaved caspase 3 (Asp175) 1:500 Cell Signaling Technology #9661S) was performed as described above.
Microglia
Brains were sectioned at 40 μm on a cryostat, and primary antibodies (rabbit anti‐Iba1, 1:500 Wako #016‐26461; rabbit anti‐P2yR12, 1:500 AnaSpec #AS‐55043A; rat anti‐CD68, 1:200 Serotec #MCA1957) for microglial labeling were performed as described above.
C1q synaptic labeling
Brains were sectioned at 15 μm on a cryostat, and primary antibodies (guinea pig anti‐vGluT2, 1:1,000 Millipore #AB2251; rabbit anti‐C1q, 1:500 Abcam #ab182451) for synaptic staining were performed as described above.
Retinal whole mounts
Whole‐mount retina preparations were performed by dissecting out the retinas of PSVue‐injected mice following transcardial perfusion. Retinas were blocked in staining buffer (10% normal goat serum and 2% Triton X‐100 in PBS) for 1 h before incubation with the pan‐RGC marker Rbpms (guinea pig anti‐Rbpms 1:500 HuaBio #HM0401) and active caspase 3 (rabbit anti‐cleaved caspase 3 (Asp175) 1:500 Cell Signaling Technology #9661S) at 4°C for three days. After 3 × 10 min washes in PBS, retinas were incubated in appropriate Alexa Fluor‐conjugated secondary antibodies (1:200; Invitrogen/Thermo Fisher Scientific) at room temperature for 1 h. Retinas were washed 3 × 10 min and mounted using glycerol.
Image analysis
dLGN analysis
Confocal images were processed using the open‐source programs ilastik (Berg et al, 2019; v1.3.2) and CellProfiler (McQuin et al, 2018; v3.0). ilastik is machine‐learning software for image processing that can be trained to recognize various features in a semi‐automated manner. Representative images were used for training to generate channel‐specific ilastik projects for segmentation. Binary probability images were then generated from single‐channel images processed using the appropriate project. These images were then further processed in CellProfiler, cell image analysis software that we utilized to identify objects and generate co‐localization data. For caspase 3 WT and KO and C1q WT and experiments, all analyses were performed blind to genotype. Specific analyses are described below.
PSVue quantification
PSVue puncta quantification of segmented probability images was performed using CellProfiler, where objects between 3 and 100 pixels were enumerated for each image. The mean per image (two _z_‐stacks per section, two sections per animal) of PSVue puncta was calculated.
PSVue synaptic co‐localization
To determine PSVue co‐localization on synapses, binary probability images were generated for each independent immunostaining in Ilastik and further processed in CellProfiler. PSVue objects were then masked onto either presynaptic (vGluT2) or postsynaptic (Homer2) images. Co‐localized objects were then further masked onto postsynaptic or presynaptic images, respectively, to both identify synapses (co‐localization of pre‐ and postsynaptic markers) and to determine specific PSVue localization on synapses. The percentage of pre‐ or postsynaptic PSVue co‐localization was calculated by dividing the number of triple‐positive masked objects by the number of co‐localized pre‐ and postsynaptic objects. The mean percentage per image (two _z_‐stacks per section, two sections per animal) was then calculated.
C1q presynaptic co‐localization
To determine C1q co‐localization with the presynaptic marker vGluT2, binary probability images were generated for each independent immunostaining in Ilastik and further processed in CellProfiler. C1q objects were masked onto vGluT2 objects. The percentage of presynaptic C1q co‐localization was calculated for each image (two _z_‐stacks per section, two sections per animal) by dividing the number of C1q/vGluT2 masked objects by the number of vGluT2 objects.
C1q and PSVue presynaptic co‐localization
To determine C1q and PSVue co‐localization with the presynaptic marker vGluT2, binary probability images were generated for each independent immunostaining in Ilastik and further processed in CellProfiler. C1q objects were masked onto vGluT2 objects to determine the percentage of presynaptic C1q co‐localization. PSVue objects were masked onto C1q objects, and co‐localized objects were then further masked onto presynaptic images. The percentage of presynaptic C1q and PSVue co‐localization was calculated for each image (two _z_‐stacks per section, two sections per animal) by dividing the number of triple‐positive objects by the number of vGluT2 objects or by dividing the number of triple‐positive by vGluT2/C1q masked objects.
Hippocampal analysis
Confocal images were acquired with the laser scanning confocal microscope LEICA SP8, using a HC PL APO 63×/NA 1.40 oil objective. Acquisition parameters (i.e., laser power, gain, and offset) were kept constant among different conditions in each single experiment. Images were acquired in stratum radiatum of the CA1 subfield of the hippocampus, as indicated. Each image consisted of a stack of images taken through the _z_‐plane (0.5 μm). Confocal images were processed and analyzed using ImageJ program and Fiji (NIH software).
PSVue 550 quantification
Confocal images were modified as eight‐bit images and processed applying fixed color balance and threshold mean value by using the image adjust function of ImageJ. The resulting images were analyzed by choosing the measure or analyze particle function of ImageJ or Fiji. Area fraction is the percentage of pixels in the image or selection that have been highlighted in red using the image adjust threshold function divided by the total number of pixels in the image.
Co‐localization analysis
Co‐localization of PSVue with presynaptic marker Bassoon or postsynaptic marker PSD95 as indicated in Fig 3 was measured using the Boolean function “and” for the selected channels by choosing the image calculator process function of ImageJ. The resulting image was binarized and used as a co‐localization mask to be subtracted from a single channel. Area fraction and/or the number of puncta resulting from co‐localization mask subtraction were measured for each marker (Menna et al, 2013). PSVue/Bassoon co‐localization or PSVue/PSD‐95 co‐localization puncta were calculated divided by the number of co‐localization puncta by the total number of Bassoon and PSD‐95 puncta, respectively.
Engulfment analysis
PSVue engulfment
Mice were injected with PSVue (as described above), and their brains were harvested 24 h later using the same methods as for immunohistochemistry. Brains were cryosectioned and 40‐μm sections were immunostained for microglial markers Iba‐1 and P2yR12, as well as for the microglial lysosomal marker CD68 (as described above). For each animal, two sections were imaged and used for analysis. Images were acquired on a LSM880 confocal microscope at 63× with 0.3‐μm _z_‐steps. For engulfment analysis in the dLGN, at least 4 cells were imaged in the ipsilateral territory and at least four cells were imaged in the contralateral territory (minimum eight cells per dLGN, 16–20 cells per animal). Images were processed and analyzed as described previously using the 3D surface rendering software Imaris (Schafer et al, 2014). Data were used to calculate percent engulfment (volume engulfed PSVue/volume of the cell). All experiments were performed blind to genotype.
CTB and PSVue engulfment
P4 mice were injected with anterograde tracers (CTB‐488 and CTB‐594, Thermo Fisher Scientific (C‐22841, C‐22842)) in the left and right eyes, respectively. PSVue ICV injections were also performed as described above, and their brains were harvested 24 h later at P5. Brains were sectioned, stained, and imaged as described above. For each microglia quantified, the percent engulfment of CTB‐488 and CTB‐647 alone and co‐localized with PSVue was calculated, along with the input volumes (total volume CTB inputs/volume of field of view).
Statistical analysis
Data are displayed as individual dots and mean ± SEM, except for Figs 1F and, 2A and 2C where the data are presented as bars and scattered dots with mean ± SEM. For each graph, the number of biological replicates (mice) is indicated with “N,” except for Figs 1 and 2 where the number of observations (the number of cells) is indicated with “n_” and the number of cultures these observations derives from with “_N.”
PRISM (GraphPad Software) was used to perform Student's paired or unpaired _t_‐test and analysis of variance (ANOVA), as appropriate. In order to control for multiple comparisons, we used Tukey's multiple comparison test and Dunn's multiple comparisons test. Specific statistical analyses can further be found in figure legends.
Author contributions
NS‐H, FP, BS, and MMat designed the study and wrote the manuscript, with help from all authors. NS‐H, FP, and RM performed most experiments and data analysis; FF and LP contributed to experimental design and analysis; ME performed confocal analysis; AW and RJ designed and executed the liposome preparation; LTS performed liposomes treatments and imaging analysis; MMah, SM, and MB performed immunohistochemistry and microglial engulfment analysis, EF and MT performed stereotactic ICV injections; LTS and MB performed hippocampal immunocytochemistry; and AC performed retinal whole mounts and immunohistochemistry.
Conflict of interest
B.S. serves on the Scientific Advisory Board of Annexon and is a minor shareholder of Annexon. The remaining authors declare no competing financial interests.
Supporting information
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Acknowledgements
We thank Yvanka de Soysa and Kevin Mastro for critical comments on the manuscript and Elisabetta Menna (IN‐CNR), Davide Pozzi (Humanitas University), and Chandana Kondapalli (BCH) for discussion. We thank Sivapratha Nagappan Chettiar for technical assistance in stereotactic ICV injections of adult animals; Cassandra‐Victoria Innocent of the Cellular Imaging Core at Boston Children's Hospital (C. Chen; NIH U54 HD090255) for technical support with imaging; Milanka Stevanovic for contributions to microglial engulfment quantifications; and Arnaud Frouin and Joel Cuadrado for help with mouse colony maintenance (BCH). This work was supported by NIH T32‐AG000222 (N.S.H.), NIH R01‐NS071008 (B.S), NIH Conte Center (B.S.), NIH R01‐NS092578 (B.S), Cure Alzheimer's FP01017732 (B.S), PRIN (Ministero dell'Istruzione dell'Università e della Ricerca, #2017A9MK4R), Regione Lombardia “NeOn”, POR‐FESR 2014‐2020, ID 239047, CUP E47F17000000009, Fondazione Pisa 107/16, and MinSal FR 2016 (Ministero della Salute #RF‐201602361571) to M.Mat and CARIPLO grant 2018 (#2018‐0364) to F.F. F.P. was supported by Fondazione Umberto Veronesi “2017 grant,” and M.T is supported by Fondazione Veronesi Fellowship 2019. Summary schematic was created with BioRender.com.
The EMBO Journal (2020) 39: e105380
Contributor Information
Beth Stevens, Email: beth.stevens@childrens.harvard.edu.
Michela Matteoli, Email: michela.matteoli@hunimed.eu.
References
- Bader Lange ML, Cenini G, Piroddi M, Abdul HM, Sultana R, Galli F, Memo M, Butterfield DA (2008) Loss of phospholipid asymmetry and elevated brain apoptotic protein levels in subjects with amnestic mild cognitive impairment and Alzheimer disease. Neurobiol Dis 29: 456–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basilico B, Pagani F, Grimaldi A, Cortese B, Di Angelantonio S, Weinhard L, Gross C, Limatola C, Maggi L, Ragozzino D (2019) Microglia shape presynaptic properties at developing glutamatergic synapses. Glia 67: 53–67 [DOI] [PubMed] [Google Scholar]
- Baudry M, Massicotte G, Hauge S (1991) Phosphatidylserine increases the affinity of the AMPA/quisqualate receptor in rat brain membranes. Behav Neural Biol 55: 137–140 [DOI] [PubMed] [Google Scholar]
- Berg S, Kutra D, Kroeger T, Straehle CN, Kausler BX, Haubold C, Schiegg M, Ales J, Beier T, Rudy M_et al_ (2019) ilastik: interactive machine learning for (bio)image analysis. Nat Methods 16: 1226–1232 [DOI] [PubMed] [Google Scholar]
- Brown GC, Neher JJ (2014) Microglial phagocytosis of live neurons. Nat Rev Neurosci 15: 209–216 [DOI] [PubMed] [Google Scholar]
- Burbridge TJ, Xu HP, Ackman JB, Ge X, Zhang Y, Ye MJ, Zhou ZJ, Xu J, Contractor A, Crair MC (2014) Visual circuit development requires patterned activity mediated by retinal acetylcholine receptors. Neuron 84: 1049–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan MK, Halleck MS, Krahling S, Henderson AJ, Williamson P, Schlegel RA (2003) Phosphatidylserine expression and phagocytosis of apoptotic thymocytes during differentiation of monocytic cells. J Leukoc Biol 74: 846–856 [DOI] [PubMed] [Google Scholar]
- Cellerino A, Bähr M, Isenmann S (2000) Apoptosis in the developing visual system. Cell Tissue Res 301: 53–69 [DOI] [PubMed] [Google Scholar]
- Chan MM, Gray BD, Pak KY, Fong D (2015) Non‐invasive in vivo imaging of arthritis in a collagen‐induced murine model with phosphatidylserine‐binding near‐infrared (NIR) dye. Arthritis Res Ther 17: 50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WS, Clarke LE, Wang GX, Stafford BK, Sher A, Chakraborty C, Joung J, Foo LC, Thompson A, Chen C_et al_ (2013) Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature 504: 394–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correale C, Genua M, Vetrano S, Mazzini E, Martinoli C, Spinelli A, Arena V, Peyrin‐Biroulet L, Caprioli F, Passini N_et al_ (2013) Bacterial sensor triggering receptor expressed on myeloid cells‐2 regulates the mucosal inflammatory response. Gastroenterology 144: 346–356.e343 [DOI] [PubMed] [Google Scholar]
- Dallérac G, Zerwas M, Novikova T, Callu D, Leblanc‐Veyrac P, Bock E, Berezin V, Rampon C, Doyère V (2011) The neural cell adhesion molecule‐derived peptide FGL facilitates long‐term plasticity in the dentate gyrus in vivo. Learn Mem 18: 306–313 [DOI] [PubMed] [Google Scholar]
- Dejanovic B, Huntley MA, De Mazière A, Meilandt WJ, Wu T, Srinivasan K, Jiang Z, Gandham V, Friedman BA, Ngu H_et al_ (2018) Changes in the synaptic proteome in tauopathy and rescue of tau‐induced synapse loss by C1q antibodies. Neuron 100: 1322–1336.e1327 [DOI] [PubMed] [Google Scholar]
- Dennison SM, Bowen ME, Brunger AT, Lentz BR (2006) Neuronal SNAREs do not trigger fusion between synthetic membranes but do promote PEG‐mediated membrane fusion. Biophys J 90: 1661–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott JI, Surprenant A, Marelli‐Berg FM, Cooper JC, Cassady‐Cain RL, Wooding C, Linton K, Alexander DR, Higgins CF (2005) Membrane phosphatidylserine distribution as a non‐apoptotic signalling mechanism in lymphocytes. Nat Cell Biol 7: 808–816 [DOI] [PubMed] [Google Scholar]
- Fadok VA, Voelker DR, Campbell PA, Cohen JJ, Bratton DL, Henson PM (1992) Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J Immunol 148: 2207–2216 [PubMed] [Google Scholar]
- Fadok VA, Xue D, Henson P (2001) If phosphatidylserine is the death knell, a new phosphatidylserine‐specific receptor is the bellringer. Cell Death Differ 8: 582–587 [DOI] [PubMed] [Google Scholar]
- Fernandez I, Araç D, Ubach J, Gerber SH, Shin O, Gao Y, Anderson RG, Südhof TC, Rizo J (2001) Three‐dimensional structure of the synaptotagmin 1 C2B‐domain: synaptotagmin 1 as a phospholipid binding machine. Neuron 32: 1057–1069 [DOI] [PubMed] [Google Scholar]
- Filipello F, Morini R, Corradini I, Zerbi V, Canzi A, Michalski B, Erreni M, Markicevic M, Starvaggi‐Cucuzza C, Otero K_et al_ (2018) The microglial innate immune receptor TREM2 is required for synapse elimination and normal brain connectivity. Immunity 48: 979–991.e978 [DOI] [PubMed] [Google Scholar]
- Graham DK, DeRyckere D, Davies KD, Earp HS (2014) The TAM family: phosphatidylserine sensing receptor tyrosine kinases gone awry in cancer. Nat Rev Cancer 14: 769–785 [DOI] [PubMed] [Google Scholar]
- Grommes C, Lee CY, Wilkinson BL, Jiang Q, Koenigsknecht‐Talboo JL, Varnum B, Landreth GE (2008) Regulation of microglial phagocytosis and inflammatory gene expression by Gas6 acting on the Axl/Mer family of tyrosine kinases. J Neuroimmune Pharmacol 3: 130–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunner G, Cheadle L, Johnson KM, Ayata P, Badimon A, Mondo E, Nagy MA, Liu L, Bemiller SM, Kim KW et al (2019) Sensory lesioning induces microglial synapse elimination via ADAM10 and fractalkine signaling. Nat Neurosci 22: 1075–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Györffy BA, Kun J, Török G, Bulyáki É, Borhegyi Z, Gulyássy P, Kis V, Szocsics P, Micsonai A, Matkó J_et al_ (2018) Local apoptotic‐like mechanisms underlie complement‐mediated synaptic pruning. Proc Natl Acad Sci USA 115: 6303–6308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM, Stevens JK (1989) Dendritic spines of CA 1 pyramidal cells in the rat hippocampus: serial electron microscopy with reference to their biophysical characteristics. J Neurosci 9: 2982–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann PR, deCathelineau AM, Ogden CA, Leverrier Y, Bratton DL, Daleke DL, Ridley AJ, Fadok VA, Henson PM (2001) Phosphatidylserine (PS) induces PS receptor‐mediated macropinocytosis and promotes clearance of apoptotic cells. J Cell Biol 155: 649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Beja‐Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, Merry KM, Shi Q, Rosenthal A, Barres BA_et al_ (2016) Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352: 712–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong GS, Lee SH, Lee B, Choi JH, Oh SJ, Jang Y, Hwang EM, Kim H, Jung J, Kim IB_et al_ (2019) ANO1/TMEM16A regulates process maturation in radial glial cells in the developing brain. Proc Natl Acad Sci USA 116: 12494–12499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks BM, Chen C (2006) Distinct roles for spontaneous and visual activity in remodeling of the retinogeniculate synapse. Neuron 52: 281–291 [DOI] [PubMed] [Google Scholar]
- Hua JY, Smith SJ (2004) Neural activity and the dynamics of central nervous system development. Nat Neurosci 7: 327–332 [DOI] [PubMed] [Google Scholar]
- Huang BX, Akbar M, Kevala K, Kim HY (2011) Phosphatidylserine is a critical modulator for Akt activation. J Cell Biol 192: 979–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman AD (2007) Mechanisms of eye‐specific visual circuit development. Curr Opin Neurobiol 17: 73–80 [DOI] [PubMed] [Google Scholar]
- Jaubert‐Miazza L, Green E, Lo FS, Bui K, Mills J, Guido W (2005) Structural and functional composition of the developing retinogeniculate pathway in the mouse. Vis Neurosci 22: 661–676 [DOI] [PubMed] [Google Scholar]
- Katz LC, Shatz CJ (1996) Synaptic activity and the construction of cortical circuits. Science 274: 1133–1138 [DOI] [PubMed] [Google Scholar]
- Lehrman EK, Wilton DK, Litvina EY, Welsh CA, Chang ST, Frouin A, Walker AJ, Heller MD, Umemori H, Chen C_et al_ (2018) CD47 protects synapses from excess microglia‐mediated pruning during development. Neuron 100: 120–134.e126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke G (2019) How macrophages deal with death. Nat Rev Immunol 19: 539–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz BR (2003) Exposure of platelet membrane phosphatidylserine regulates blood coagulation. Prog Lipid Res 42: 423–438 [DOI] [PubMed] [Google Scholar]
- Li T, Chiou B, Gilman CK, Luo R, Koshi T, Yu D, Oak HC, Giera S, Johnson‐Venkatesh E, Muthukumar AK_et al_ (2020) A splicing isoform of GPR56 mediates microglial synaptic refinement via phosphatidylserine binding. EMBO J e104136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Rai V, Hudson BI, Song F, Schmidt AM, Barile GR (2012) RAGE binds C1q and enhances C1q‐mediated phagocytosis. Cell Immunol 274: 72–82 [DOI] [PubMed] [Google Scholar]
- Martin SJ, Reutelingsperger CP, McGahon AJ, Rader JA, van Schie RC, LaFace DM, Green DR (1995) Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl‐2 and Abl. J Exp Med 182: 1545–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Pombo I, Poncet P, David B, Arock M, Blank U (2000) Immunologic stimulation of mast cells leads to the reversible exposure of phosphatidylserine in the absence of apoptosis. Int Arch Allergy Immunol 123: 249–258 [DOI] [PubMed] [Google Scholar]
- Martin M, Leffler J, Blom AM (2012) Annexin A2 and A5 serve as new ligands for C1q on apoptotic cells. J Biol Chem 287: 33733–33744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni F, Müller C, DeAssis J, Lew D, Leevy WM, Finnemann SC (2019) Non‐invasive in vivo fluorescence imaging of apoptotic retinal photoreceptors. Sci Rep 9: 1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuin C, Goodman A, Chernyshev V, Kamentsky L, Cimini BA, Karhohs KW, Doan M, Ding L, Rafelski SM, Thirstrup D_et al_ (2018) Cell Profiler 3.0: next‐generation image processing for biology. PLoS Biol 16: e2005970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menna E, Zambetti S, Morini R, Donzelli A, Disanza A, Calvigioni D, Braida D, Nicolini C, Orlando M, Fossati G_et al_ (2013) Eps8 controls dendritic spine density and synaptic plasticity through its actin‐capping activity. EMBO J 32: 1730–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J, Cuccia L, Ianoul A, Cheetham JJ, Johnston LJ (2004) Imaging the selective binding of synapsin to anionic membrane domains. ChemBioChem 5: 1489–1494 [DOI] [PubMed] [Google Scholar]
- Nakano T, Ishimoto Y, Kishino J, Umeda M, Inoue K, Nagata K, Ohashi K, Mizuno K, Arita H (1997) Cell adhesion to phosphatidylserine mediated by a product of growth arrest‐specific gene 6. J Biol Chem 272: 29411–29414 [DOI] [PubMed] [Google Scholar]
- Neher JJ, Neniskyte U, Brown GC (2012) Primary phagocytosis of neurons by inflamed microglia: potential roles in neurodegeneration. Front Pharmacol 3: 27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton AC, Keranen LM (1994) Phosphatidyl‐L‐serine is necessary for protein kinase C's high‐affinity interaction with diacylglycerol‐containing membranes. Biochemistry 33: 6651–6658 [DOI] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, Helmchen F (2005) Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308: 1314–1318 [DOI] [PubMed] [Google Scholar]
- Païdassi H, Tacnet‐Delorme P, Garlatti V, Darnault C, Ghebrehiwet B, Gaboriaud C, Arlaud GJ, Frachet P (2008) C1q binds phosphatidylserine and likely acts as a multiligand‐bridging molecule in apoptotic cell recognition. J Immunol 180: 2329–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L_et al_ (2011) Synaptic pruning by microglia is necessary for normal brain development. Science 333: 1456–1458 [DOI] [PubMed] [Google Scholar]
- Parhizkar S, Arzberger T, Brendel M, Kleinberger G, Deussing M, Focke C, Nuscher B, Xiong M, Ghasemigharagoz A, Katzmarski N_et al_ (2019) Loss of TREM2 function increases amyloid seeding but reduces plaque‐associated ApoE. Nat Neurosci 22: 191–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Tosello‐Trampont AC, Elliott MR, Lu M, Haney LB, Ma Z, Klibanov AL, Mandell JW, Ravichandran KS (2007) BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 450: 430–434 [DOI] [PubMed] [Google Scholar]
- Salter MW, Stevens B (2017) Microglia emerge as central players in brain disease. Nat Med 23: 1018–1027 [DOI] [PubMed] [Google Scholar]
- Sapar ML, Ji H, Wang B, Poe AR, Dubey K, Ren X, Ni JQ, Han C (2018) Phosphatidylserine externalization results from and causes neurite degeneration in Drosophila. Cell Rep 24: 2273–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, Stevens B (2012) Microglia sculpt postnatal neural circuits in an activity and complement‐dependent manner. Neuron 74: 691–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Heller CT, Stevens B (2014) An engulfment assay: a protocol to assess interactions between CNS phagocytes and neurons. J Vis Exp 88: 51482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Heller CT, Gunner G, Heller M, Gordon C, Hammond T, Wolf Y, Jung S, Stevens B (2016) Microglia contribute to circuit defects in Mecp2 null mice independent of microglia‐specific loss of Mecp2 expression. Elife 5: e15224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa K, Suzuki J, Nagata S (2011) Constitutive exposure of phosphatidylserine on viable cells. Proc Natl Acad Sci USA 108: 19246–19251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa K, Kurata S, Yanagihashi Y, Brummelkamp TR, Matsuda F, Nagata S (2014) Caspase‐mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science 344: 1164–1168 [DOI] [PubMed] [Google Scholar]
- Shacham‐Silverberg V, Sar Shalom H, Goldner R, Golan‐Vaishenker Y, Gurwicz N, Gokhman I, Yaron A (2018) Phosphatidylserine is a marker for axonal debris engulfment but its exposure can be decoupled from degeneration. Cell Death Dis 9: 1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirotani K, Hori Y, Yoshizaki R, Higuchi E, Colonna M, Saito T, Hashimoto S, Saido TC, Iwata N (2019) Aminophospholipids are signal‐transducing TREM2 ligands on apoptotic cells. Sci Rep 9: 7508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BA, Xiao S, Wolter W, Wheeler J, Suckow MA, Smith BD (2011) In vivo targeting of cell death using a synthetic fluorescent molecular probe. Apoptosis 16: 722–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BA, Xie BW, van Beek ER, Que I, Blankevoort V, Xiao S, Cole EL, Hoehn M, Kaijzel EL, Löwik CW_et al_ (2012) Multicolor fluorescence imaging of traumatic brain injury in a cryolesion mouse model. ACS Chem Neurosci 3: 530–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrz D, Dráberová L, Dráber P (2007) Non‐apoptotic phosphatidylserine externalization induced by engagement of glycosylphosphatidylinositol‐anchored proteins. J Biol Chem 282: 10487–10497 [DOI] [PubMed] [Google Scholar]
- Stephan AH, Barres BA, Stevens B (2012) The complement system: an unexpected role in synaptic pruning during development and disease. Annu Rev Neurosci 35: 369–389 [DOI] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B_et al_ (2007) The classical complement cascade mediates CNS synapse elimination. Cell 131: 1164–1178 [DOI] [PubMed] [Google Scholar]
- Suzuki J, Umeda M, Sims PJ, Nagata S (2010) Calcium‐dependent phospholipid scrambling by TMEM16F. Nature 468: 834–838 [DOI] [PubMed] [Google Scholar]
- Suzuki J, Imanishi E, Nagata S (2016) Xkr8 phospholipid scrambling complex in apoptotic phosphatidylserine exposure. Proc Natl Acad Sci USA 113: 9509–9514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svennerholm L (1968) Distribution and fatty acid composition of phosphoglycerides in normal human brain. J Lipid Res 9: 570–579 [PubMed] [Google Scholar]
- Tremblay M, Lowery RL, Majewska AK (2010) Microglial interactions with synapses are modulated by visual experience. PLoS Biol 8: e1000527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull IR, Gilfillan S, Cella M, Aoshi T, Miller M, Piccio L, Hernandez M, Colonna M (2006) Cutting edge: TREM‐2 attenuates macrophage activation. J Immunol 177: 3520–3524 [DOI] [PubMed] [Google Scholar]
- Ulrich JD, Ulland TK, Colonna M, Holtzman DM (2017) Elucidating the role of TREM2 in Alzheimer's disease. Neuron 94: 237–248 [DOI] [PubMed] [Google Scholar]
- Vasek MJ, Garber C, Dorsey D, Durrant DM, Bollman B, Soung A, Yu J, Perez‐Torres C, Frouin A, Wilton DK_et al_ (2016) A complement‐microglial axis drives synapse loss during virus‐induced memory impairment. Nature 534: 538–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walrave L, Vinken M, Albertini G, De Bundel D, Leybaert L, Smolders IJ (2016) Inhibition of Connexin43 hemichannels impairs spatial short‐term memory without affecting spatial working memory. Front Cell Neurosci 10: 288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cella M, Mallinson K, Ulrich JD, Young KL, Robinette ML, Gilfillan S, Krishnan GM, Sudhakar S, Zinselmeyer BH_et al_ (2015) TREM2 lipid sensing sustains the microglial response in an Alzheimer's disease model. Cell 160: 1061–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhard L, di Bartolomei G, Bolasco G, Machado P, Schieber NL, Neniskyte U, Exiga M, Vadisiute A, Raggioli A, Schertel A_et al_ (2018) Microglia remodel synapses by presynaptic trogocytosis and spine head filopodia induction. Nat Commun 9: 1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werneburg S, Jung J, Kunjamma RB, Ha SK, Luciano NJ, Willis CM, Gao G, Biscola NP, Havton LA, Crocker SJ_et al_ (2020) Targeted complement inhibition at synapses prevents microglial synaptic engulfment and synapse loss in demyelinating disease. Immunity 52: 167–182.e167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Baldridge RD, Chi RJ, Burd CG, Graham TR (2013) Phosphatidylserine flipping enhances membrane curvature and negative charge required for vesicular transport. J Cell Biol 202: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hui E, Chapman ER, Jackson MB (2009) Phosphatidylserine regulation of Ca2+‐triggered exocytosis and fusion pores in PC12 cells. Mol Biol Cell 20: 5086–5095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziburkus J, Guido W (2006) Loss of binocular responses and reduced retinal convergence during the period of retinogeniculate axon segregation. J Neurophysiol 96: 2775–2784 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7