Ultrasonographic Thyroid Nodule Classification Using a Deep Convolutional Neural Network with Surgical Pathology (original) (raw)
Abstract
Ultrasonography with fine-needle aspiration biopsy is commonly used to detect thyroid cancer. However, thyroid ultrasonography is prone to subjective interpretations and interobserver variabilities. The objective of this study was to develop a thyroid nodule classification system for ultrasonography using convolutional neural networks. Transverse and longitudinal ultrasonographic thyroid images of 762 patients were used to create a deep learning model. After surgical biopsy, 325 cases were confirmed to be benign and 437 cases were confirmed to be papillary thyroid carcinoma. Image annotation marks were removed, and missing regions were recovered using neighboring parenchyme. To reduce overfitting of the deep learning model, we applied data augmentation, global average pooling. And 4-fold cross-validation was performed to detect overfitting. We employed a transfer learning method with the pretrained deep learning model VGG16. The average area under the curve of the model was 0.916, and its specificity and sensitivity were 0.70 and 0.92, respectively. Positive and negative predictive values were 0.90 and 0.75, respectively. We introduced a new fine-tuned deep learning model for classifying thyroid nodules in ultrasonography. We expect that this model will help physicians diagnose thyroid nodules with ultrasonography.
Keywords: Deep convolutional neural network, Deep learning, Ultrasonography, Thyroid nodule classification
Background
The incidence of thyroid cancer has increased steeply worldwide over the past few decades [1]. The National Cancer Institute reported 56,870 new cases and 2010 thyroid cancer–specific deaths in 2017.
According to the American Thyroid Association guidelines, ultrasonography with fine-needle aspiration biopsy is the main method of thyroid cancer detection [2]. Thyroid ultrasonography is real-time and noninvasive; however, it is easily affected by echo perturbation and speckle noise. Further, there are various echo patterns of each thyroid nodule; thus, if ultrasonography is not performed by experienced physicians, it is prone to subjective interpretations and interobserver variabilities. To reduce these limitations and facilitate communication with other physicians, the Thyroid Imaging Reporting and Data System (TIRADS) was introduced for classification using several features for identification (composition, echogenicity, margins, calcifications, and shape) [3, 4]. However, the use of TIRADS for thyroid nodule evaluation is still time consuming and not consistently accurate because it is solely based on a physician’s knowledge and experience.
With the advance of machine learning technology, many computer-aided diagnosis systems have been devised to reduce operator dependency and help physicians diagnose thyroid nodules correctly [3, 5–7]. Although these studies have shown promising results, they mostly rely on handcrafted features extracted from images after preprocessing [8].
To overcome the limitations of early machine learning, researchers have started to apply deep learning for image identification. Through deep learning, artificial neural networks automatically extract the most discriminative features from the data and return an answer. Among the various deep learning systems, the deep convolutional neural network (DCNN), which mimics a biological neural network in the visual cortex, is especially suitable for medical image recognition [9]. Recently, DCNNs have been used to classify various medical images such as images of breast cancer histopathology, pulmonary nodules, and lymph nodes [10–12]. However, there have been few studies dealing with thyroid nodule identification using DCNN [8, 13].
The objective of this study is to develop a thyroid nodule classification system based on ultrasonography using a DCNN. To evaluate the proposed system, its results are compared with those of previous methods.
Methods
Patients and Datasets
In this study, transverse and longitudinal ultrasonographic thyroid images of 762 patients from the Korea Institute of Radiological & Medical Sciences were used for training and testing a deep learning model. Of all the cases, 325 were confirmed as benign (nodular hyperplasia, follicular adenoma, or cyst), and 437 were confirmed as papillary thyroid carcinoma after surgical biopsy. Each image has only one thyroid nodule, and an expert physician drew a rectangular region of interest around the pathologic nodule. The images were extracted from thyroid ultrasound video sequences captured with EPIQ 5G, HI VISION Ascendus, and EUB-7500 ultrasound devices. The images were in JPEG format and ranged from 640 × 480 to 1024 × 768 pixels in size. All devices employed 12-MHz convex and linear transductor settings. The extracted images were distributed into training, validation, and test sets in a ratio of 6:2:2, as summarized in Table 1.
Table 1.
The distribution of benign and cancer cases in training, validation, and test datasets with different ultrasound devices
| Benign | Cancer | |||||||
|---|---|---|---|---|---|---|---|---|
| Total | A | B | C | Total | A | B | C | |
| Training | 199 | 108 | 52 | 39 | 260 | 101 | 86 | 73 |
| Validation | 64 | 39 | 13 | 12 | 94 | 35 | 34 | 25 |
| Test | 62 | 33 | 15 | 14 | 83 | 34 | 26 | 23 |
Image Preprocessing
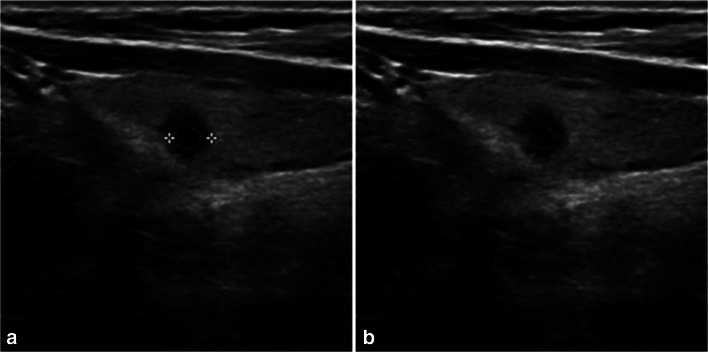
To remove annotations such as the caliper marks used to locate and measure nodule size as well as restore the gaps with the textures surrounding the annotations after removal, the input ultrasonographic images were processed as follows (Fig. 1a, b). All of the input images were rescaled to [0,1] for standardization. The Roberts cross operator implemented in the scikit-image library, which is a Python image-processing module, was used to detect edges [14]. Then, pixels greater than 0.25 in value were identified as artifacts. The pixel values of the detected artifacts were deleted in the original image, and the missing regions were recovered using the “inpaint_biharmonic” function from the scikit-image library with the default parameters [15]. And the mean of each image was set to zero using a Keras (version 2.1.5) built-in operation.
Fig. 1.
Original ultrasonographic images before and after the removal of caliper marks. a Original image with caliper marks. b Image with caliper marks removed and the missing region restored
Sample Augmentation
Our database consists of 1524 images in total, which is not large enough to avoid overfitting when fine-tuning an existing DCNN. Thus, the samples were augmented to enhance our deep learning model’s prediction capability. When augmenting the samples, we included normal nearby structures such as thyroid parenchyma and/or strap muscles because the echogenicity of a nodule should be evaluated relative to that of adjacent structures.
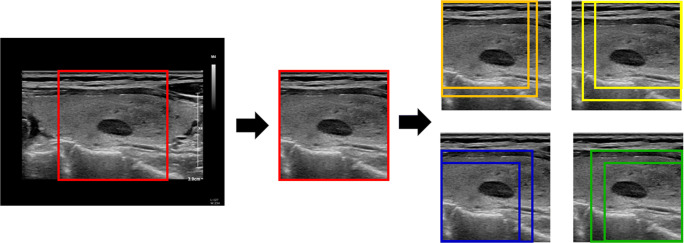
First, nodule-centered images were cropped to the largest square shape in each of the original transverse and longitudinal sonographic images. Two random-sized square images including a nodule were cropped from each vertex. Thus, eight square images including a nodule and one vertex were cropped from the original images (Fig. 2). In addition, considering the bilateral symmetry of the thyroid, horizontal flip was applied to these images. In summary, 18 transverse and 18 horizontal images per patient were created from the largest square image plus the eight smaller square images.
Fig. 2.
Process of cropping image and image augmentation. First, the nodule-centered images were cropped to the largest square shape in each original transverse and longitudinal sonographic image. Two random-sized square images including the nodule were cropped from each vertex. Thus, eight square images including a nodule and one vertex were cropped from the original images
All of patches were resized to 224 × 224 pixels using the “resize” function from the scikit-image library with Gaussian filter to suppress the artifacts. Furthermore, grayscale images were converted to RGB; however, each RGB channel image was equal to the others to conserve the grayscale image values. This is because the input image size of the deep learning model used in this study is 224 × 224 × 3.
Contrast Limited Adaptive Histogram Equalization (CLAHE)
Contrast limited adaptive histogram equalization (CLAHE) is a contrast enhancement technique often used to reduce speckle noise in medical sonographic images [16, 17]. CLAHE was applied to determine whether it improves the deep learning prediction results. It was performed using the “equalize_adapthist” function from the scikit-image library with the default parameters.
Transfer Learning by Fine-Tuning Modified VGG16 Model
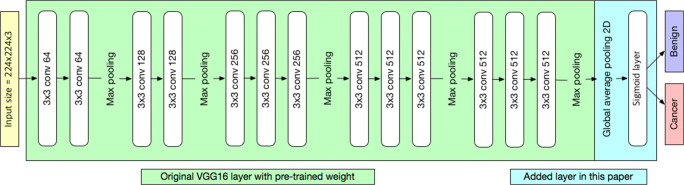
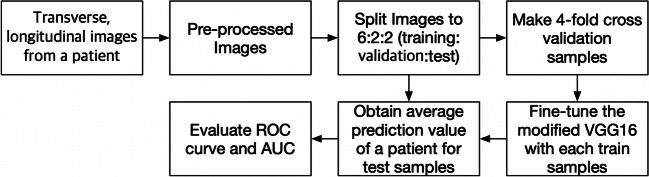
Because of the insufficient number of training samples, we employed a transfer learning method with a pretrained deep learning model. The model used in this study was VGG16, which is known for its good classification results in the ImageNet Large Scale Visual Recognition Challenge [18, 19]. We loaded a set of weights pretrained on ImageNet to VGG16 and modified VGG16 by replacing the fully connected layer with global average pooling and sigmoid layers (Fig. 3). The binary cross entropy function was used for the loss function, and the network was minimized by an Adam optimizer at an initial learning rate of 4 × 10−6 with a decay rate of 10−6. The batch size was set to three. All layers in the modified VGG16 were fine-tuned. The Keras (version 2.1.5) wrapper deep learning library with TensorFlow (version 1.7) was used as a backend with Python version 3.6.5 for implementing the modified VGG16 deep learning model. Our calculation was performed on a computer running 64-bit Windows 10 and equipped with one Geforce 1080 Ti. Four-fold cross-validation was performed to detect overfitting. The weights of the fine-tuned deep learning model were obtained at the lowest validation loss. Figure 4 shows the detailed process of fine-tuning the VGG16 model in this study.
Fig. 3.
Architecture of the convolutional neural network used in this work. The VGG16 was modified by replacing the fully connected layer with global average pooling and sigmoid layers
Fig. 4.
Process of fine-tuning VGG16. The images were preprocessed and split into training, validation, and test sets. Then, we performed 4-fold cross-validation, fine-tuned the VGG16 with each training sample, and evaluated the performance using receiver operating characteristic curves
Results
The sigmoid function results of all images from one patient were averaged and the performance of the model was evaluated for the test data set (145 patients) using the receiver operating characteristic curve, area under the curve, positive predictive value, negative predictive value, sensitivity, and specificity. Table 2 shows the performance of the fine-tuned deep learning model. Figures 5 and 6 show the four receiver operating characteristic curves of each fold and confusion matrix, respectively. The average area under the curve was 0.916 [0.907–0.922], and specificity and sensitivity were 0.70 and 0.92, respectively, for the test dataset. The area under the curve result of the CLAHE-preprocessed images was 0.873 [0.854–0.888].
Table 2.
Performance
| Area under the curve | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|
| 0.916 [0.907–0.922] | 0.92 [0.83–0.96] | 0.70 [0.61–0.89] | 0.79 [0.71–0.88] | 0.87 [0.84–0.94] |
Fig. 5.
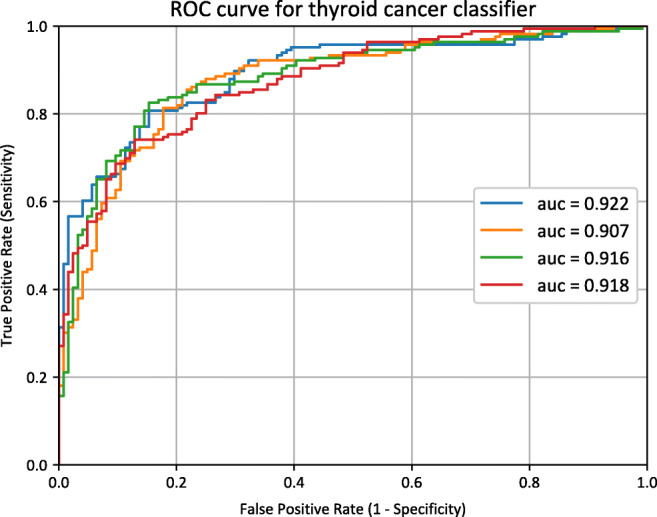
Receiver operating characteristic curves. Four receiver operating characteristic curves and the corresponding area under curve values of each fold
Fig. 6.
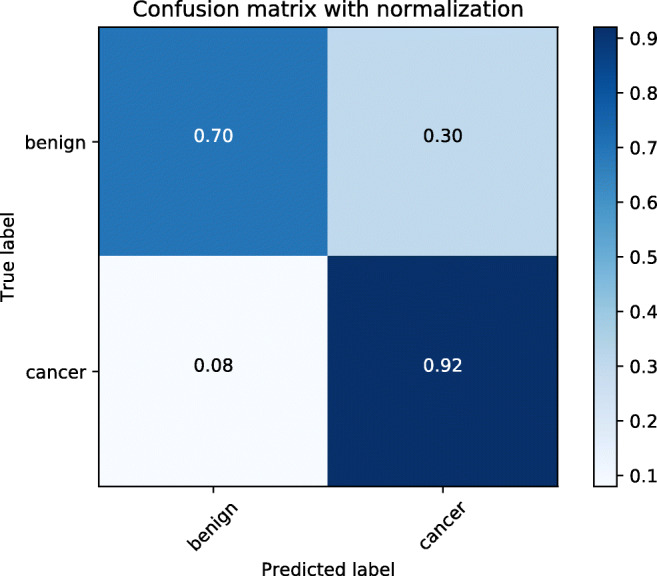
Confusion matrix. Confusion matrix of the result of the proposed deep learning model
PPV positive predictive value, NPV negative predictive value
Discussion
In this study, we described a new fine-tuned deep learning model for identifying thyroid malignancy in ultrasonographic images. We employed a DCNN in this research because it is a specialized machine learning model for identifying patterns of images. Compared to the traditional computer-aided diagnosis system using handcrafted feature extraction methods, a DCNN has several advantages: (1) it can automatically learn effective features for classifying images; (2) detection and identification with DCNNs are very powerful even when images have distortions from the camera lens, light source, angle, and other factors; and (3) the computational power, time, and cost are relatively low because the same coefficients are calculated repeatedly across the input images. Hence, DCNNs are being widely applied and modified for the analysis of medical images such as those obtained using computed tomography, magnetic resonance imaging, and ultrasonography [20–22].
We combined data from different devices into a single dataset because we found that performance results from combined dataset and those from datasets of each devices were not so different.
There are a few previously proposed thyroid nodular classification systems that use deep learning methods. A thyroid ultrasonographic image classification system that uses a random forest classifier by fine-tuning a pretrained ImageNet deep learning model has been proposed [13]. It obtained good results; however, its output data were in the TIRADS classification system format, which categorizes the thyroid nodule images subjectively according to the probability of malignancy, not the exact surgical pathology. They used binary classifiers, positive (TI-RAD 3, 4a, 4b, 5) and negative (TI-RAD 1 or 2). From the TI-RAD system, physician gets a help to identify a probability of final surgical pathology to be malignant or benign before surgery. TI-RAD category 3 stands for 1.7% of malignant surgical pathology probability, category 4, 3.3 ~ 72.4%, and category 5, 87.5% [2]. We believe that their classifier includes too broad spectrum of probability of malignancy respectively. Moreover, their model predicts only subjective physician’s findings. On the contrary, we trained our model to output the corresponding final surgical pathologies, malignancy or benignancy which all physicians want to predict with ultrasonography before surgery. We assume that surgical pathology is mandatory output for this kind of deep learning model to have a valuable clinical significance.
Another deep learning classification system to predict thyroid pathology uses an ensemble of two DCNN results with a softmax function, support vector machine classifier, and 10-fold cross-validation [8]. Its performance was good with an average area under the curve of 0.893 with the softmax classifier. However, we found that overfitting was possible because the validation and test datasets were not separated. Additionally, the input image data were only the thyroid nodules themselves and did not include the surrounding normal structures, which can be a cue to diagnosis. Hypoechogenicity, which is the most important feature for predicting thyroid malignancy, should be compared with nearby thyroid normal parenchyma. Moreover, annotations such as caliper marks were not removed during preprocessing.
To overcome the limitations of previous methods, we set the surgical pathology as the ground truth for each nodule; strictly divided the data into training, validation, and test datasets; included surrounding structures in input images; and removed artifacts such as caliper marks. To our best knowledge, although there have been a few studies regarding deep learning ultrasonographic image classification, our model has the strictest and clearest clinical settings and image data.
To reduce the overfitting of the deep learning model, we applied data augmentation, global average pooling. And 4-fold cross-validation was performed to detect overfitting. First, it has been reported that image augmentation improves the classification performance of a deep learning model [23]. The TIRADS and Korean TIRADS classify thyroid nodules using features such as composition, echogenicity, shape, orientation, margin, and calcification [24]. To take into account echogenicity, it was necessary to include nearby structures (the thyroid parenchyma and/or anterior neck muscle) in the preprocessed images because the evaluation of the echogenicity of a nodule should be relative to adjacent structures. Furthermore, the geometric transformations that are widely employed in augmentation, such as rotation and elastic transformation, were not used in this study because they can distort features such as the orientation and margins. In this research, data augmentation was performed by cropping and horizontal flipping of the square patches of the entire medical image to preserve these ultrasonographic features. Second, the global average pooling layer is known as a regularizer to reduce the overfitting of deep learning models, and it can replace the conventional fully connected layer [25]. Thus, the top layer of the original VGG16 was replaced by a global average pooling layer in this work (Fig. 3). Finally, we performed 4-fold cross-validation, which is a well-known validation technique in deep learning to evaluate the generalization ability of our deep learning model. We chose 4-folds due to the limited size of the dataset.
In this research, there are several limitations. First, we need more image samples with consequent surgical pathology. We were forced to fine-tune the pretrained transfer learning model VGG16 because of a lack of sufficient number of samples. However, if we had enough samples, we could make a completely new deep learning model suitable for the aims of this research after trialing some different models. Second, we included only papillary thyroid carcinoma and benign conditions such as nodular hyperplasia, follicular adenoma, and cyst. There are other pathologies such as follicular thyroid carcinoma, poorly differentiated carcinoma, and anaplastic thyroid carcinoma that were not considered. However, papillary thyroid carcinoma comprises most thyroid carcinoma, and nodules with other carcinoma exhibit features completely different from those of papillary cancer. Hence, relevant image samples with other pathologies are relatively difficult to collect and hard to analyze together with papillary thyroid carcinoma in this type of deep learning. In the future, if larger quantities of image samples with various pathologies are collected, we should try to train a model to classify each pathology. Third, validation with more images from different ultrasonographic devices obtained by other operators is necessary. Because ultrasonography is well known for its machine and operator dependency, we need to collaborate with other hospitals to validate the proposed system.
Currently, we are working to automate the process of finding nodules and drawing the regions of interest automatically. We anticipate the automation of preprocessing will improve the workload in this type of research.
Conclusion
In conclusion, we introduced a new fine-tuned deep transfer learning model for classifying thyroid nodules in ultrasonography. We expect this model will help physicians diagnose thyroid nodules with ultrasonography.
Acknowledgments
This study was supported by a grant of the Korea Institute of Radiological and Medical Sciences (KIRAMS), funded by Ministry of Science and ICT (MSIT), Republic of Korea (No. 50543-2019).
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Soon Woo Kwon and Ik Joon Choi contributed equally to this work.
References
- 1.Sanabria A, Kowalski LP, Shah JP, Nixon IJ, Angelos P, Williams MD, Rinaldo A, Ferlito A. Growing incidence of thyroid carcinoma in recent years: factors underlying overdiagnosis. Head Neck. 2018;40:855–866. doi: 10.1002/hed.25029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haugen BR, Alexander EK, Bible KC, Doherty GM, Mandel SJ, Nikiforov YE, Pacini Furio, Randolph GW, Sawka AM, Schlumberger M, Schuff KG, Sherman SI, Sosa JA, Steward DL, Tuttle RM, Wartofsky L: 2015 American Thyroid Association Management Guidelines for adult patients with thyroid nodules and differentiated thyroid cancer: The American Thyroid Association Guidelines Task Force on Thyroid Nodules and Differentiated Thyroid Cancer,2016 [DOI] [PMC free article] [PubMed]
- 3.Kwak JY, Han KH, Yoon JH, Moon HJ, Son EJ, Park SH, Jung HK, Choi JS, Kim BM, Kim EK. Thyroid imaging reporting and data system for US features of nodules: a step in establishing better stratification of cancer risk. Radiology. 2011;260:892–899. doi: 10.1148/radiol.11110206. [DOI] [PubMed] [Google Scholar]
- 4.Lingam RK, Qarib MH, Tolley NS. Evaluating thyroid nodules: predicting and selecting malignant nodules for fine-needle aspiration (FNA) cytology. Insights Imaging. 2013;4:617–624. doi: 10.1007/s13244-013-0256-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi YJ, Baek JH, Park HS, Shim WH, Kim TY, Shong YK, Lee JH. A computer-aided diagnosis system using artificial intelligence for the diagnosis and characterization of thyroid nodules on ultrasound: initial clinical assessment. Thyroid. 2017;27:546–552. doi: 10.1089/thy.2016.0372. [DOI] [PubMed] [Google Scholar]
- 6.Acharya UR, Vinitha Sree S, Krishnan MM, Molinari F, Garberoglio R, Suri JS. Non-invasive automated 3D thyroid lesion classification in ultrasound: a class of ThyroScan systems. Ultrasonics. 2012;52:508–520. doi: 10.1016/j.ultras.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 7.Acharya UR, Faust O, Sree SV, Molinari F, Suri JS. ThyroScreen system: high resolution ultrasound thyroid image characterization into benign and malignant classes using novel combination of texture and discrete wavelet transform. Comput Methods Prog Biomed. 2012;107:233–241. doi: 10.1016/j.cmpb.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Ma J, Wu F, Zhu J, Xu D, Kong D. A pre-trained convolutional neural network based method for thyroid nodule diagnosis. Ultrasonics. 2017;73:221–230. doi: 10.1016/j.ultras.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 9.Seeliger K, Fritsche M, Guclu U, Schoenmakers S, Schoffelen JM, Bosch SE, van Gerven MAJ. Convolutional neural network-based encoding and decoding of visual object recognition in space and time. Neuroimage. 2018;180:253–266. doi: 10.1016/j.neuroimage.2017.07.018. [DOI] [PubMed] [Google Scholar]
- 10.Nahid AA, Mehrabi MA, Kong Y. Histopathological breast cancer image classification by deep neural network techniques guided by local clustering. Biomed Res Int. 2018;2018:2362108. doi: 10.1155/2018/2362108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W, Cao P, Zhao D, Wang J. Pulmonary nodule classification with deep convolutional neural networks on computed tomography images. Comput Math Methods Med. 2016;2016:6215085. doi: 10.1155/2016/6215085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roth HR, Lu L, Seff A, Cherry KM, Hoffman J, Wang S, Liu J, Turkbey E, Summers RM. A new 2.5D representation for lymph node detection using random sets of deep convolutional neural network observations. Med Image Comput Comput Assist Interv. 2014;17:520–527. doi: 10.1007/978-3-319-10404-1_65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi J, Walia E, Babyn P, Wang J, Groot G, Eramian M. Thyroid nodule classification in ultrasound images by fine-tuning deep convolutional neural network. J Digit Imaging. 2017;30:477–486. doi: 10.1007/s10278-017-9997-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Walt S, Schonberger JL, Nunez-Iglesias J, Boulogne F, Warner JD, Yager N, Gouillart E, Yu T, Scikit-image contributors Scikit-image: image processing in Python. PeerJ. 2014;2:e453. doi: 10.7717/peerj.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damelin S, Hoang N: On surface completion and image inpainting by biharmonic functions: numerical aspects. Int J Math Math Sci 2018,2018
- 16.Tay PC, Garson CD, Acton ST, Hossack JA. Ultrasound despeckling for contrast enhancement. IEEE Trans Image Process. 2010;19:1847–1860. doi: 10.1109/TIP.2010.2044962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benzarti F, Amiri H. Speckle noise reduction in medical ultrasound images. Int J Comput Sci Issues. 2012;9:187–94. [Google Scholar]
- 18.Simonyan K, Zisserman A: Very deep convolutional networks for large-scale image recognition. In ICLR,2015
- 19.Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M. Imagenet large scale visual recognition challenge. Int J Comput Vis. 2015;115:211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- 20.Pereira S, Pinto A, Alves V, Silva CA. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging. 2016;35:1240–1251. doi: 10.1109/TMI.2016.2538465. [DOI] [PubMed] [Google Scholar]
- 21.Colevray M, Tatard-Leitman VM, Gouttard S, Douek P, Boussel L. Convolutional neural network evaluation of over-scanning in lung computed tomography. Diagn Interv Imaging. 2019;100:177–183. doi: 10.1016/j.diii.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Ko SY, Lee JH, Yoon JH, Na H, Hong E, Han K, Jung I, Kim EK, Moon HJ, Park VY, Lee E, Kwak JY. Deep convolutional neural network for the diagnosis of thyroid nodules on ultrasound. Head Neck. 2019;41:885–891. doi: 10.1002/hed.25415. [DOI] [PubMed] [Google Scholar]
- 23.Wong SC, Gatt A, Stamatescu V, McDonnell MD: Understanding data augmentation for classification: when to warp?. 2016 International Conference on Digital Image Computing: Techniques and Applications (DICTA), 2016, pp 1-6
- 24.Shin JH, Baek JH, Chung J, Ha EJ, Kim JH, Lee YH, Lim HK, Moon WJ, Na DG, Park JS, Choi YJ, Hahn SY, Jeon SJ, Jung SL, Kim DW, Kim EK, Kwak JY, Lee CY, Lee HJ, Lee JH, Lee JH, Lee KH, Park SW, Sung JY, Korean Society of Thyroid Radiology, Korean Society of Radiology Ultrasonography diagnosis and imaging-based management of thyroid nodules: revised Korean Society of Thyroid Radiology consensus statement and recommendations. Korean J Radiol. 2016;17:370–395. doi: 10.3348/kjr.2016.17.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin M, Chen Q, Yan S: Network in network. In ICLR,2014