Multisystem inflammatory syndrome in children and COVID-19 are distinct presentations of SARS–CoV-2 (original) (raw)
Abstract
BACKGROUND
Initial reports from the severe acute respiratory coronavirus 2 (SARS–CoV-2) pandemic described children as being less susceptible to coronavirus disease 2019 (COVID-19) than adults. Subsequently, a severe and novel pediatric disorder termed multisystem inflammatory syndrome in children (MIS-C) emerged. We report on unique hematologic and immunologic parameters that distinguish between COVID-19 and MIS-C and provide insight into pathophysiology.
METHODS
We prospectively enrolled hospitalized patients with evidence of SARS–CoV-2 infection and classified them as having MIS-C or COVID-19. Patients with COVID-19 were classified as having either minimal or severe disease. Cytokine profiles, viral cycle thresholds (Cts), blood smears, and soluble C5b-9 values were analyzed with clinical data.
RESULTS
Twenty patients were enrolled (9 severe COVID-19, 5 minimal COVID-19, and 6 MIS-C). Five cytokines (IFN-γ, IL-10, IL-6, IL-8, and TNF-α) contributed to the analysis. TNF-α and IL-10 discriminated between patients with MIS-C and severe COVID-19. The presence of burr cells on blood smears, as well as Cts, differentiated between patients with severe COVID-19 and those with MIS-C.
CONCLUSION
Pediatric patients with SARS–CoV-2 are at risk for critical illness with severe COVID-19 and MIS-C. Cytokine profiling and examination of peripheral blood smears may distinguish between patients with MIS-C and those with severe COVID-19.
FUNDING
Financial support for this project was provided by CHOP Frontiers Program Immune Dysregulation Team; National Institute of Allergy and Infectious Diseases; National Cancer Institute; the Leukemia and Lymphoma Society; Cookies for Kids Cancer; Alex’s Lemonade Stand Foundation for Childhood Cancer; Children’s Oncology Group; Stand UP 2 Cancer; Team Connor; the Kate Amato Foundations; Burroughs Wellcome Fund CAMS; the Clinical Immunology Society; the American Academy of Allergy, Asthma, and Immunology; and the Institute for Translational Medicine and Therapeutics.
Keywords: COVID-19
Keywords: Cytokines
Introduction
Initial reports from the coronavirus disease 2019 (COVID-19) pandemic indicated that children were less severely affected by severe acute respiratory syndrome coronavirus 2 (SARS–CoV-2) infection than adults (1–3). The largest retrospective case series of 2135 children from China reported that less than 5% of children affected with SARS–CoV-2 had severe disease, with only 1 pediatric death (2).
On April 27, 2020, the National Health Service (NHS) in the United Kingdom issued an alert about pediatric patients presenting with a severe Kawasaki-like syndrome in the setting of recently proven or suspected SARS–CoV-2 infection, termed pediatric inflammatory multisystem syndrome temporally associated with SARS–CoV-2 (PIMS-TS) (4, 5). These patients presented with disease in the absence of preceding respiratory distress. A similar hyperinflammatory syndrome with features of Kawasaki disease (KD) was reported in Italy (6). On May 8, 2020, the Centers for Disease Control (CDC) in the United States issued an alert describing a new entity termed multisystem inflammatory syndrome in children (MIS-C) associated with COVID-19 (7). MIS-C is defined by clinically severe illness requiring hospitalization with fever, inflammatory marker elevation, and multisystem organ dysfunction in the setting of recent proven or probable SARS–CoV-2 infection, and in the absence of an alternative likely explanation (7).
KD is a medium vessel vasculitis that occurs almost exclusively in infants and children (8). It has a seasonal variation, implying an infectious trigger, and has been associated with viruses, including other coronaviruses (9–11). The clinical diagnosis is defined by fevers for 5 days and at least 4 of 5 cardinal exam findings (bilateral conjunctival injection, mucosal changes, peripheral extremity changes, polymorphous rash, and cervical lymphadenopathy) (12). Patients with at least 5 days of fever with fewer physical exam findings who meet laboratory criteria may be diagnosed with incomplete KD (8). If untreated, KD is associated with the development of coronary artery dilatation and aneurysms in approximately 20% of patients (8). Shock or vascular compromise can occur, but is rare, affecting less than 10% of patients (13). In contrast, the majority of patients with MIS-C reported to date have presented with shock requiring inotropic support (5, 6).
Initial reports of MIS-C are restricted to retrospective clinical case series. Moreover, they have not compared clinical or biologic features of MIS-C with other presentations of SARS–CoV-2. It is unknown if the processes that mediate MIS-C are similar to the processes that lead to life-threatening respiratory failure and shock with COVID-19. The reports of severe acute illness after SARS–CoV-2 in children are scant. Based on the clinical phenotype, we hypothesized that children with MIS-C are presenting with a syndrome that has not been previously described, distinct from both KD and severe COVID-19 infection. We used an extant multidisciplinary dysregulated immunity team to perform a systematic, comprehensive, prospective evaluation of clinical and laboratory biomarkers in children who presented with minimal COVID-19, severe COVID-19, and MIS-C. This evaluation included cytokine profiles, measures of viral burden, and markers of vascular and endothelial damage. This is a prospective report comparing life-threatening complications of SARS–CoV-2 in children, and it provides information on disease biology as well as clinical information that can help practitioners distinguish between the clinical syndromes.
Results
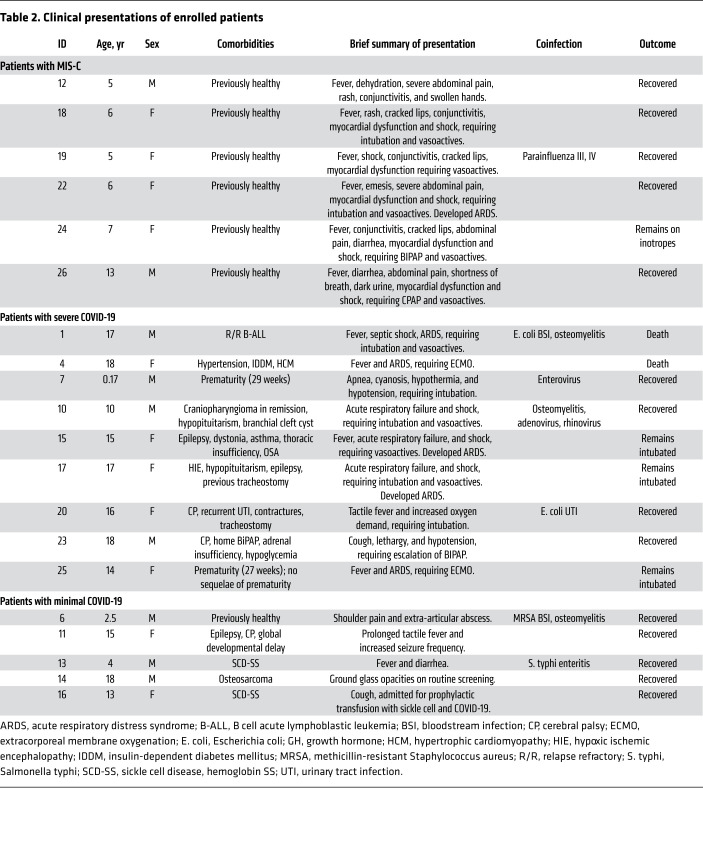
Between April 3, 2020, and May 15, 2020, 26 patients were enrolled. Twenty patients, on whom complete correlative data were available, were included in the final cohort (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI140970DS1). Demographics and narrative summaries of the MIS-C, severe COVID-19, and minimal COVID-19 cohorts are presented in Tables 1 and 2. Some clinical details of 3 patients in the MIS-C cohort have been reported in a previous case series (14).
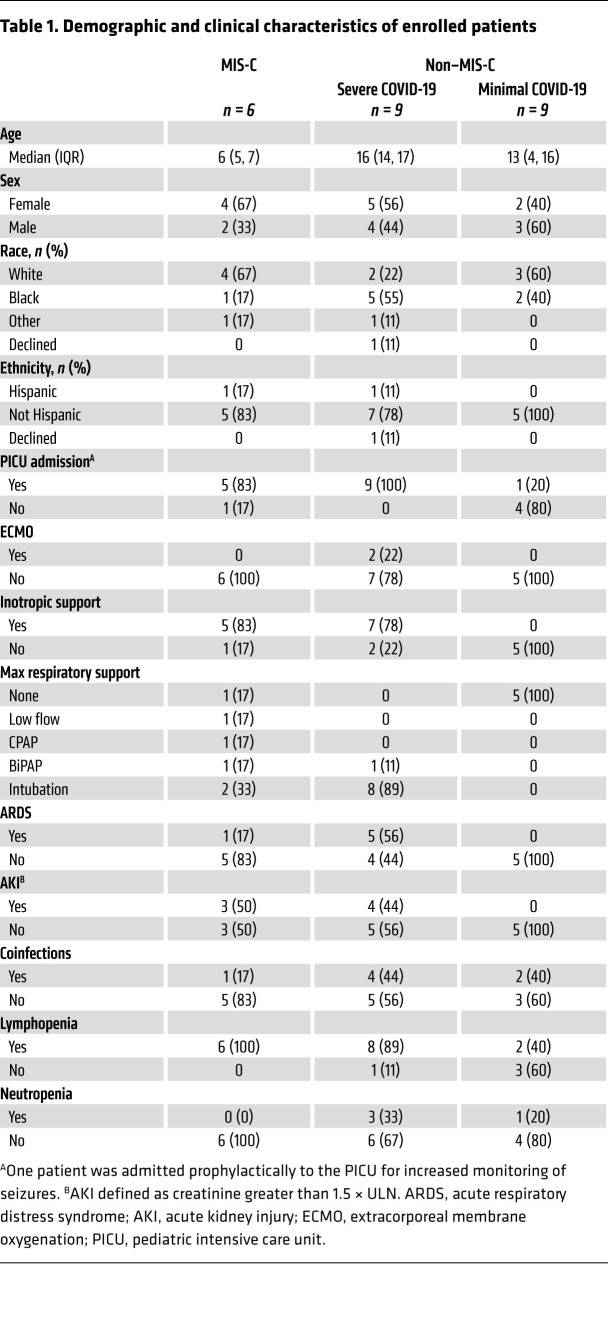
Table 1. Demographic and clinical characteristics of enrolled patients.
Table 2. Clinical presentations of enrolled patients.
Patients in both the MIS-C and severe COVID-19 cohorts had grossly abnormal laboratory values (Supplemental Table 1). Patients in the severe COVID-19 cohort had several clinical features associated with both macrophage activation syndrome (MAS) and shock, such as highly elevated ferritins, elevated transaminases, elevated lactate, thrombocytopenia, and prolonged partial thromboplastin times (PTT). Patients in the MIS-C cohort had findings associated with vascular injury and cardiac disease, as evidenced by highly elevated D-dimer and B-type natriuretic protein levels.
The MIS-C cohort patients were younger than the severe or minimal COVID-19 patients (median age 6 years versus 16 and 13 years, respectively). The majority of patients in the MIS-C and severe COVID-19 cohorts had lymphopenia during their hospital admission; only 2 patients in the minimal cohort had lymphopenia. Of note, 4 of 5 patients in the severe COVID-19 cohort had a coinfection with an additional organism compared with only 1 of 6 in the MIS-C cohort. There were no statistically significant differences in race or ethnic background between patients in each group; there was a higher rate of acute respiratory distress syndrome (ARDS) among Black children (n = 4) than children of other races (n = 2; 1 other, 1 declined, 0 White; P = 0.044, χ2 test). All children who developed ARDS had an existing comorbidity.
Two patients in the MIS-C cohort met classical criteria for KD and 2 met criteria for incomplete KD (Supplemental Tables 2 and 3). Five patients were treated with vasoactive infusions, 4 patients had evidence of myocardial dysfunction, and 1 had evidence of coronary artery dilatation. All patients were treated with intravenous immune globulin (IVIG) and intravenous methylprednisone.
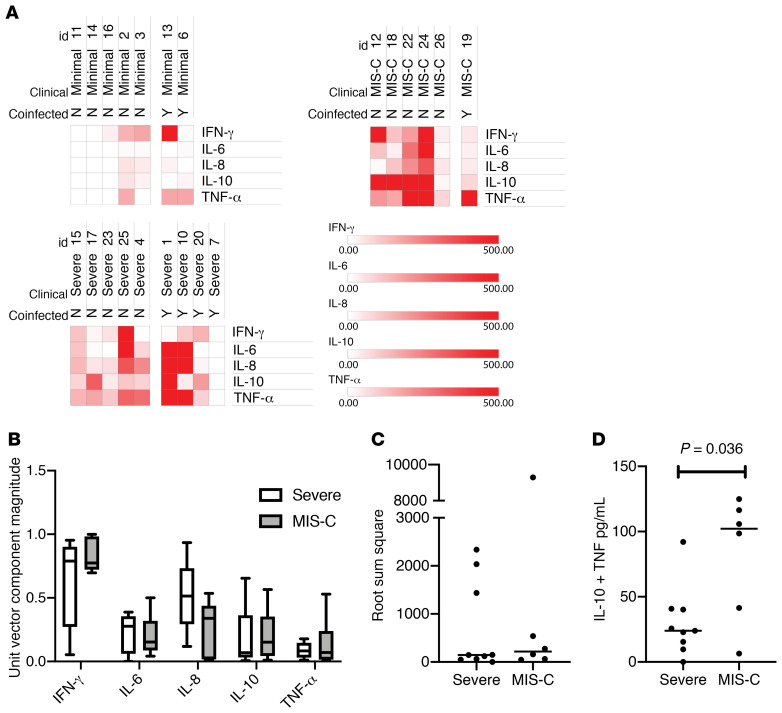
We sought to identify plasma cytokine patterns that reliably differentiated between MIS-C and severe COVID-19. Graphs of all cytokines are presented in Supplemental Figure 2. Five cytokines (IL-1R, IL-2, IL-4, IL-12p70, and IL-13) were excluded from further analyses because they were generally not elevated in patients. Importantly, this demonstrates that the hypercytokinemia observed in our patients reflects a specific hyperinflammatory process, and that significant elevations in IL-1R, IL-2, IL-4, IL-12p70, or IL-13 may indicate the presence of a different disease process. Several patients across all 3 cohorts had high IFN-γ.
Heatmaps of all patients separated based on the presence or absence of coinfection are presented in Figure 1A. In order to determine if cytokines could robustly classify the different phenotypes, subsequent analyses included all patients regardless of their coinfection status. We next considered the cytokine profile as a 5-dimensional vector, hypothesizing that either the direction or the magnitude of each vector may predict clinical cohort; neither were statistically significant (Figure 1, B and C). Based on exploratory multivariable logistic regression modelling, the sum of IL-10 and TNF-α levels was hypothesized to differentiate between MIS-C and severe non–MIS-C patients. The sum of IL-10 and TNF-α levels uniquely identified MIS-C from severe COVID-19 presentations (Figure 1D; mean [95% CI]; severe: 30.06 [9.54–50.6] versus MIS-C: 82.25 [32.5–132.0], P = 0.036).
Figure 1. Cytokine architecture associated with SARS–CoV-2 infections in children.
(A) Heatmap of the 5 most differentially present cytokines in the plasma of pediatric SARS–CoV-2 infections. Comparison of patients with and without coinfections for each of the 3 clinical phenotypes of pediatric SARS–CoV-2 infection (n = 20). Patient IDs are listed above each column for reference. (B) Cytokine profiles for each patient (n = 15) were treated as a 5-dimensional vector and converted into unit vectors by dividing each component by the root sum square of the vector. Box-and-whisker plot of each unit vector with median values of each for MIS-C versus severe COVID-19 presentations (line). Whiskers represent maximum and minimum and boxes represent the 25th to 75th percentiles. Differences between phenotypes or phenotype/cytokine interaction were not significant by 2-way ANOVA. (C) Vector magnitudes of each cytokine profile were computed as the root sum square with values plotted for severe COVID-19 and MIS-C phenotypes (n = 15). Bar represents the median of each group. Differences were not significant by Mann-Whitney U test. (D) The sum of the IL-10 and TNF-α concentrations were computed for each patient and plotted for severe COVID-19 and MIS-C phenotypes (n = 15). Bar represents the median of each group with P values computed by Mann-Whitney U test.
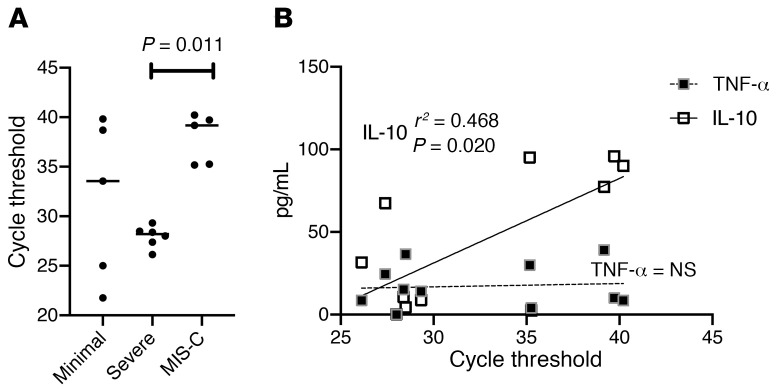
We hypothesized that MIS-C, as a putative postinfectious syndrome, should be associated with lower viral burden when compared with severe COVID-19. SARS–CoV-2 RT-PCR cycle threshold (Cts) from nasopharyngeal aspirates (which inversely correlate with viral burden) were predictive of the clinical cohorts. Patients with severe COVID-19 presented with low Cts whereas those with MIS-C presented with high Cts (mean [95% CI]; severe: 28.0 [26.8–29.1] versus MIS-C: 37.9 [34.8–41.0], P = 0.011; Figure 2). Univariate linear regression against plasma cytokine concentrations revealed a correlation with IL-10 levels (P = 0.02). In 4 patients, repeat cytokine panels were drawn to assess clinical response to immunomodulatory therapy (Supplemental Figure 3).
Figure 2. Viral Ct in pediatric SARS–CoV-2 infections.
(A) When available (n = 16), Cts for each patient are plotted. Bars represent mean values with P values computed using Dunn’s multiple comparisons test after Kruskal-Wallis testing. (B) Linear regression modelling of cycle time thresholds against IL-10 and TNF-α concentrations in plasma for all patients in the cohort (n = 16).
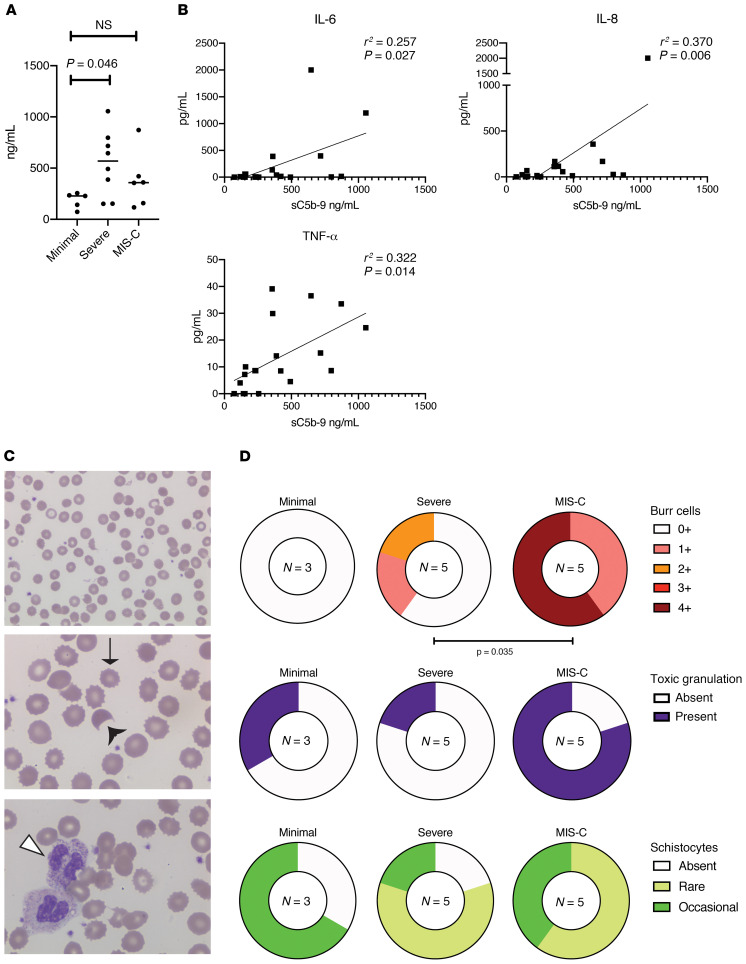
We postulated that immune activation related to SARS–CoV-2 was associated with endothelial dysfunction, and we measured soluble C5b-9 (sC5b-9) as a candidate biomarker. sC5b-9 represents the activation product of the terminal complement cascade, and elevations of this marker have been associated with microangiopathy in a number of settings (15, 16). Figure 3A demonstrates that sC5b-9 was significantly elevated in patients with severe COVID-19 relative to those with minimal COVID-19 (mean [95% CI]; minimal: 186.4 [91.16–281.6] versus severe: 555.0 [285.9–814.1], P = 0.046). While sC5b-9 values also trended higher for MIS-C (mean [95% CI]; 380.5 [97.7–663.3]), this did not reach statistical significance. Univariate linear regression modelling revealed that sC5b-9 concentrations correlated in a statistically significant manner with IL-6, IL-8, and TNF-α (Figure 3B). These data are consistent with previous reports of sC5b-9 being associated with endothelial damage and release of neutrophil-activating IL-8 (17).
Figure 3. Soluble C5b-9 measurements in pediatric SARS–CoV-2 infections.
(A) Soluble C5b-9 was successfully measured in 19 patients. Data are plotted for each of the 3 phenotypes; bars represent the mean values of each group. P value computed using Dunn’s multiple comparisons test after Kruskal-Wallis testing. (B) Linear regression modelling of sC5b-9 against IL-6, IL-8, and TNF-α concentrations in plasma for all patients in the cohort. (C) Representative photomicrographs of red blood cells and neutrophils from patients in the cohort. From top to bottom: ×50 field showing burr cells and schistocytes; ×100 field showing the same; ×100 field showing toxic granulation. Thin arrow denotes a burr cell, black arrowhead denotes a schistocyte, white arrowhead denotes toxic granulation. (D) Number of patients with degree of burr cells, toxic granulation, and schistocytes for each clinical phenotype; n = 13 patients with smears available. Number of assessable patients for each group is displayed in the center of each circle. Severe COVID-19 was compared with MIS-C using χ2 test for trend for burr cells; χ2 statistic was impossible to compute for the dichotomous outcome of toxic granulation due to low n. There were no statistically significant differences in schistocytes across the cohort.
Peripheral blood smears were available on 13 patients (3 minimal COVID-19, 5 severe COVID-19, and 5 MIS-C). Consistent with microangiopathy, examination revealed at least rare schistocytes in 67% of minimal COVID-19, 80% of severe COVID-19, and 100% of MIS-C patients (Figure 3, C and D). We note that 2 of the 3 minimal COVID-19 patients had sickle cell disease as a comorbidity which may confound the interpretation of schistocytes in this group (18). Burr cells were absent in patients with minimal disease but present in 40% of patients with severe disease. All patients with MIS-C had at least some burr cells present, with 60% of patients having 4 or more burr cells (Figure 3, C and D).
Patients with MIS-C were noted to have a marked increase in their white blood cell counts; longitudinal measures of absolute neutrophil count (ANC) and absolute lymphocyte count (ALC) for all severe COVID-19 and MIS-C patients are shown in Supplemental Figure 4A. Many of the patients with MIS-C were noted to have increased numbers of immature myeloid cells as well (data not shown). Consistent with this myeloid activation, we noted a remarkable number of neutrophils with toxic granulation in the MIS-C group, although these were also present in 1 minimal and 1 severe COVID-19 patient (Figure 3, C and D). The trajectory of change in ALC and ANC was approximated using the slopes of linear regression models, with no overall significant difference between patients with MIS-C and those with severe COVID-19 (Supplemental Figure 4B).
Discussion
We report on a prospective cohort of pediatric patients representing the spectrum of disease associated with SARS–CoV-2 from minimal to severe COVID-19, and MIS-C, and identify biomarkers that distinguish different manifestations of the same infection. We have made several observations that improve understanding of disease biology and impact clinical care. The positive but high Cts associated with MIS-C support a postinfectious etiology for this phenomenon that has been previously postulated but not demonstrated. We describe clinical and laboratory measures that discriminate between patients with MIS-C and those with severe COVID-19, specifically the combination of the cytokine levels for IL-10 and TNF-α, and the presence of burr cells and toxic granulation on peripheral blood smears. While clinical cytokine profiling is limited to tertiary care centers and central laboratories, peripheral blood smears are easily made in most clinics and hospitals.
Although initial reports have emphasized similarities between MIS-C and KD, consistent with our initial hypothesis, our findings suggest that MIS-C is a distinct entity that may have some overlapping features with KD. In our cohort, only 2 of 6 children with MIS-C met criteria for complete KD. As has been reported by other groups, most patients in our MIS-C cohort had cardiac dysfunction at presentation, a finding that is unusual in KD (5, 6). Conversely, despite their dramatic presentations, there was very limited coronary artery disease in patients with MIS-C. The cytokine profiles associated with the MIS-C group, particularly marked elevations in IL-10, are distinct from previously reported cytokine profiles in KD, which tend to be associated with mild elevations of IL-1, IL-2, and IL-6 (19). Differences between our cohort and previously published cohorts must be interpreted with caution due to methodologic differences such as differences in platform, substrates used, and timing of collection in disease course. However, TNF-α appears to play a key role in the pathogenesis of both MIS-C and KD (19, 20). Levels of IFN-γ, a cytokine not strongly associated with KD, were very high in many patients with SARS–CoV-2 infections, including children with minimal disease. There was wide variability of IFN-γ levels in patients included in the cohort, implying that IFN-γ signaling may be important in disease biology of COVID-19 and MIS-C, but is not necessary for development of either phenotype.
We also noted increases in markers of endothelial dysfunction in both MIS-C and severe COVID-19 with elevated sC5b-9 and altered red blood cell morphology. These findings correlated with elevations in the inflammatory cytokines IL-6, IL-8, and TNF-α, implying a connection to significant dysfunction of innate immunity. Further characterization of broader panels of cytokines, particularly sIL-2R, IL-18, and CXCL9, would be valuable and of interest. Research focused on characterizing these cytokines and on defining causal relationships is necessary. Other clinical markers also differed between the 2 groups, including markers of vascular injury, such as elevated d-dimers and B-natriuretic proteins, and markers of coagulopathy, such as prolonged PTT. Per institutional protocols, some patients in both the COVID-19 and MIS-C groups would have received prophylactic enoxaparin. Further studies will need to better characterize these differences between MIS-C and severe COVID-19.
Contrary to early reports that emphasized relatively mild disease outcomes in pediatric patients, we show that in some children SARS–CoV-2 appears to trigger a dysregulated hyperinflammatory pathophysiologic process (2, 3). Strikingly, all children with MIS-C in our cohort were previously healthy, in contrast to those in the severe COVID-19 cohort who had significant comorbidities, similar to disease in adult populations. A larger number of patients in our cohorts had ARDS based on what would be expected from previous reports of pediatric patients with respiratory COVID-19 (1–3, 21). In a report of pediatric patients with severe COVID-19 from Wuhan, only 2 patients required mechanical ventilation. The laboratory findings associated with severe COVID-19 presented in that report are less profoundly abnormal than those in our cohort (21). Congruent with adult reports, we note that patients who are Black may be at higher risk of more severe disease than patients of other races, with Black patients being overrepresented among those who developed ARDS (22). The extent to which these observed differences are mediated by social determinants of health and health care inequalities versus genetic or epigenetic factors warrants future investigation.
The small number of patients in our study precluded more detailed statistical analysis and modelling. We lacked the power to detect subtle differences between the groups in immunologic and cytokine changes. We additionally acknowledge the issues of multiplicity of analyses, particularly the cytokine correlations, for which our small data set precludes a complete multivariate analysis. Future work with a larger sample size and multiple cohorts for replications will allow for analysis similar to those we performed on cytokines with other clinical and laboratory variables. Due to the pandemic, we were only able to collect blood samples on hospitalized children, and therefore could not include nonadmitted patients with mild SARS–CoV-2 without comorbidities. We used a cohort of patients with minimal symptoms of disease as our control, however, these patients were all hospitalized with different comorbidities. Of note, 2 of these patients had sickle cell disease and may have fundamentally different hematologic parameters at baseline than other patients.
We believe that these results can inform hypotheses for future studies when larger cohorts are available, and we would caution against broad generalization of these results until such work is complete. Future work will need to examine why SARS–CoV-2 induces such a damaging inflammatory response in some children and not others, with a focus on potential genetic etiologies, as this has the potential to guide therapy. In our cohort of 6 patients, all children with MIS-C improved after treatment with IVIG, aspirin, and steroids. Optimal treatment strategies will need to be determined in clinical trials.
Pediatric patients with SARS–CoV-2 are at risk of developing severe inflammatory dysregulation with critical illness. The detailed evaluations we present begin to dissect the pathophysiologic processes in these mysterious complications of SARS–CoV-2 and open the door for targeted therapeutics. Examination of peripheral blood smears for evidence of schistocytes, burr cells, and neutrophil toxic granulations while awaiting cytokine profiling may aid in distinguishing between patients with MIS-C and those with severe COVID-19 and guide immunomodulatory therapy.
Methods
Study design and population.
We prospectively screened and enrolled patients admitted to the Children’s Hospital of Philadelphia (CHOP) between April 3, 2020, and May 15, 2020. Patients were enrolled if they had evidence of past or current SARS–CoV-2 infection with either SARS–CoV-2 + RT-PCR in blood, stool, or mucosa, or presence of serum immunoglobulin G to SARS–CoV-2. In order to prevent bias, patients were enrolled without regard to clinical presentation, and were only categorized after enrollment into the study.
Patients were categorized as having MIS-C if they had fever, clinically severe illness with multisystem organ involvement (>2 of cardiac, renal, respiratory, hematologic, gastrointestinal, dermatologic or neurological), no alternative plausible diagnosis, and positive SARS–CoV-2 infection by RT-PCR, serology, or COVID-19 exposure within the 4 weeks before the onset of symptoms, per the CDC case definition of MIS-C (7). Patients were categorized as having severe COVID-19 if they did not meet criteria for MIS-C and presented with a primarily respiratory process requiring an increase in positive pressure support above their baseline. Patients were classified as having minimal COVID-19 if they required hospitalization but did not otherwise meet criteria for MIS-C or have severe symptoms. Patients in the MIS-C cohort were evaluated for complete and incomplete KD. Coinfections were identified by chart review for microbiologically proven infections that were deemed clinically significant by a panel of infectious disease physicians.
Data collection.
Clinical data were abstracted from the medical record onto standardized case report forms using a REDCap database and were double-checked by a physician reviewer. Data collected included demographics, comorbid conditions, sources of coinfection, vasoactive infusions, mechanical ventilation, antiinfectives, immunomodulatory agents, and laboratory indicators of organ dysfunction or inflammation. Hematoxylin and eosin peripheral blood smears were independently examined by 2 reviewers (a hematologist and a hematopathologist). Presence of burr cells and schistocytes were scored using ordinal scales (burr cells: 1+, present but <25% cells; 2+, 25%–50% cells; 3+, 51%–75% cells; 4+, >75% of cells; schistocytes: absent, rare, or occasional).
Blood collection and assays.
Blood was collected at the first planned clinical blood draw after consent was obtained. Patients presented to our hospital at different time points in their clinical illness, and some patients were transferred from outside hospitals. For patients who remained admitted to the hospital, blood draws were obtained on a weekly basis after the initial collection.
Proinflammatory cytokine profiling.
Quantification of 10 proinflammatory cytokines was performed using V-Plex Proinflammatory Panel 1 Human Kits (catalog K15049D; Meso Scale Diagnostics). Cytokines assayed were IFN-γ, IL-1β, IL-2, IL-4, IL-6, IL-8, IL-10, IL-12p70, IL-13, and TNF-α. Samples were analyzed in duplicates and assays were performed per manufacturer protocol and read and analyzed on a QuickPlex SQ120 (Meso Scale Diagnostics).
Soluble C5b-9 assay.
Triplicates of plasma samples were assayed for sC5b-9 levels at 2 dilutions by using human C5b-9 ELISA set (catalog 558315; BD Biosciences) per manufacturer protocols. Standard curves were analyzed for best fit curves, and resulting equations were used to derive sC5b-9 sample concentrations.
Viral cycle thresholds.
Viral Cts for SARS–CoV-2 were measured in a Clinical Laboratory Improvement Amendments of 1988 — College of American Pathologist (CLIA-CAP) certified laboratory at CHOP using an RT-PCR assay for N2 viral RNA under Emergency Use Authorization (EUA) by the Food and Drug Administration (FDA).
Statistics.
Clinical characteristics were summarized using descriptive statistics. The analysis cohort was comprised of individuals for whom complete cytokine data were available. Analysis and visualizations were performed using Prism 8 (GraphPad) and Stata/IC (StataCorp). Heatmaps were generated using Morpheus (https://software.broadinstitute.org/morpheus).
For comparisons of groups, nonparametric tests were used with post hoc multiple comparison corrections when more than 2 groups were present. For the analyses presented in Supplemental Table 1, the Mann-Whitney test was used to estimate approximate P values. Multiple corrections were not performed, as the small sample size precluded this test. Two-way ANOVA was used to check for interaction effects of phenotype with size of unit vector components for cytokines. For count data, χ2 testing was used, using test for trends when ordinal variables were present. Univariate logistical regression was performed to determine correlations between continuous variables. A P value less than 0.05 was considered significant.
Study approval.
The protocol was approved by the CHOP Institutional Review Board. Due to the COVID-19 pandemic, verbal informed consent was obtained from a legally authorized representative as per the Declaration of Helsinki. Written informed consent was signed by the consenting physician and a copy was provided to participants.
Author contributions
EMB, DTT, and HB equally contributed to this work and are co–senior authors. CD, SEH, LAV, KOM, JC, EMB, HB, and DTT designed the study, analyzed data, and wrote the manuscript. KES, CJ, KC, ML, MEP, FB, DMB, BLB, KMB, AMB, BTF, JCF, JSG, KG, CG, SAG, RMH, TJK, AROJ, EJL, WP, CP, AFR, NDR, AS, DASP, and JV contributed to study design, analyzed data, and helped write the manuscript. CB and JHL performed experiments, provided data, and edited the manuscript. CD, JC, and KOM enrolled subjects. All authors contributed intellectually and reviewed and revised the manuscript.
Supplementary Material
supplemental data
ICMJE disclosure forms
Acknowledgments
We gratefully acknowledge the assistance of Jessica Perazzelli, Tiffaney Vincent, Richard Watson, Alex Felmeister, David Stokes, Susan Coffin, E. John Wherry, Scott Hensley, Richard Aplenc, Robert Berg, Barbara Engel, Lisa Sudell, Kriscynthia Smith, Meghan Martin, Margaret Tartaglione, Danielle Zambrano, Shannon Maude, Brandi Boland, Dan Fields, Jansen Weaver, Kienan O’Brian, Stephen P. Hunger, Joseph St. Geme III, and Bryan Wolf. We express our sincerest thanks to the physicians, nurses, respiratory therapists, and advanced practice providers who cared for these patients in the Special Isolation Unit and Pediatric Special Treatment Unit at the Children’s Hospital of Philadelphia, and our gratitude to the patients and families who so graciously agreed to participate. Financial support for this project was provided by CHOP Frontiers Program Immune Dysregulation Team (to DTT, EB, HB); National Institute of Allergy and Infectious Diseases (NIAID) R01AI121250 (to EMB), R01AI103280 (to AROJ), R01AI123433 (to AROJ), R21AI144472 (to AROJ), K08 AI136660 (to LVV), and K08AI135091 (to SEH); National Cancer Institute (NCI) R01CA193776, X01HD100702-01, 5UG1CA233249, and R01A1123538 (all to DTT); the Leukemia and Lymphoma Society (to DTT); Cookies for Kids Cancer (to DTT); Alex’s Lemonade Stand Foundation for Childhood Cancer (to DTT); Children’s Oncology Group (to DTT); Stand UP 2 Cancer (to DTT); Team Connor (to HB); and the Kate Amato Foundations (to HB). This work was also funded by Burroughs Wellcome Fund CAMS (to SEH and AROJ); the Clinical Immunology Society (to SEH); and the American Academy of Allergy, Asthma, and Immunology (to SEH). CD was supported by a scholarship from the Institute for Translational Medicine and Therapeutics at the University of Pennsylvania.
Version 1. 07/30/2020
In-Press Preview
Version 2. 10/05/2020
Electronic publication
Version 3. 11/02/2020
Print issue publication
Funding Statement
Financial support for this project was provided by CHOP Frontiers Program Immune Dysregulation Team (DTT, EB, HB), National Institute of Allergy and Infectious Diseases (NIAID): R01AI121250 (EMB), R01AI103280 (AOJ), R01AI123433 (AOJ), R21AI144472 (AOJ), K08 AI136660 (LVV), and K08AI135091(SH), National Cancer Institute (NCI) (US): R01CA193776 (DTT), X01HD100702-01(DTT), 5UG1CA233249) (DTT), and R01A1123538 (DTT), Leukemia and Lymphoma Society (DTT), Cookies for Kids Cancer (DTT), Alex’s Lemonade Stand Foundation for Childhood Cancer (DTT), Children’s Oncology Group (DTT), Stand UP 2 Cancer (DTT), Team Connor (HB), and Kate Amato Foundations (HB). Burroughs Wellcome Fund CAMS (SH, AOJ), Clinical Immunology Society (SH), and American Academy of Allergy, Asthma, and Immunology (SH).
Footnotes
Conflict of interest: DTT serves on advisory boards for Janssen, Amgen, La Roche, Sobi, and Humanigen. HB has stock ownership in CSL Behring. SH serves on the advisory board for Horizon Pharma. AOJ serves on the advisory board of Pluton Biosciences. MPL is an advisory board member for Octapharma and Shionogi, is a consultant for Amgen, Novartis, Shionogi, Dova, Bayer, and has received research funding from Sysmex, Novartis, and Astra Zeneca. KES has received personal fees from Elsevier and Immune Deficiency Foundation. SAG reports personal fees and grants from Novartis and CRISPR/Vertex, personal fees from Allogene, CBMG, Adaptimmune, TCR2, Juno, Jazz, Eureka, Cellectis, Roche, GSK, Cure Genetics, Humanigen, and Janssen/J&J, and grants from Kite and Servier.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(11):5967–5975.https://doi.org/10.1172/JCI140970.
Contributor Information
Caroline Diorio, Email: DIORIOC@EMAIL.CHOP.EDU.
Sarah E. Henrickson, Email: henricksons@email.chop.edu.
Laura A. Vella, Email: vellal@email.chop.edu.
Kevin O. McNerney, Email: mcnerneyk@email.chop.edu.
Chakkapong Burudpakdee, Email: BURUDPAKDC@EMAIL.CHOP.EDU.
Jessica H. Lee, Email: leej11@email.chop.edu.
Cristina Jasen, Email: jasenc@email.chop.edu.
Fran Balamuth, Email: Balamuthf@email.chop.edu.
David M. Barrett, Email: BARRETTD@email.chop.edu.
Kathrin M. Bernt, Email: berntk@email.chop.edu.
Allison M. Blatz, Email: blatza@email.chop.edu.
Kathleen Chiotos, Email: chiotosk@email.chop.edu.
Brian T. Fisher, Email: fisherbria@email.chop.edu.
Julie C. Fitzgerald, Email: fitzgeraldj@email.chop.edu.
Jeffrey S. Gerber, Email: gerberj@email.chop.edu.
Kandace Gollomp, Email: gollompk@email.chop.edu.
Christopher Gray, Email: GRAYC5@EMAIL.CHOP.EDU.
Stephan A. Grupp, Email: grupp@email.chop.edu.
Rebecca M. Harris, Email: harrisrm@email.chop.edu.
Todd J. Kilbaugh, Email: kilbaugh@email.chop.edu.
Audrey R. Odom John, Email: johna3@email.chop.edu.
Emily J. Liebling, Email: lieblinge@email.chop.edu.
Whitney Petrosa, Email: Petrosaw@email.chop.edu.
Anne F. Reilly, Email: reillya@email.chop.edu.
Julie Vardaro, Email: vardaroj@email.chop.edu.
David T. Teachey, Email: teacheyd@email.chop.edu.
Hamid Bassiri, Email: bassiri@email.chop.edu.
References
- 1.Liu W, et al. Detection of Covid-19 in children in early January 2020 in Wuhan, China. N Engl J Med. 2020;382(14):1370–1371. doi: 10.1056/NEJMc2003717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong Y, et al. Epidemiology of COVID-19 among children in China. Pediatrics. 2020;145(6):e20200702. doi: 10.1542/peds.2020-0702. [DOI] [PubMed] [Google Scholar]
- 3.Lu X, et al. SARS-CoV-2 infection in children. N Engl J Med. 2020;382(17):1663–1665. doi: 10.1056/NEJMc2005073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viner RM, Whittaker E. Kawasaki-like disease: emerging complication during the COVID-19 pandemic. Lancet. 2020;395(10239):1741–1743. doi: 10.1016/S0140-6736(20)31129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riphagen S, Gomez X, Gonzalez-Martinez C, Wilkinson N, Theocharis P. Hyperinflammatory shock in children during COVID-19 pandemic. Lancet. 2020;395(10237):1607–1608. doi: 10.1016/S0140-6736(20)31094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verdoni L, et al. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 2020;395(10239):1771–1778. doi: 10.1016/S0140-6736(20)31103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. Multisystem inflammatory syndrome in children (MIS-C) associated with coronavirus disease 2019 (COVID-19). https://emergency.cdc.gov/han/2020/han00432.asp Updated May 14, 2020. Accessed August 25, 2020.
- 8.McCrindle BW, et al. Diagnosis, treatment, and long-term management of Kawasaki disease: a scientific statement for health professionals from the American Heart Association. Circulation. 2017;135(17):e927–e99. doi: 10.1161/CIR.0000000000000484. [DOI] [PubMed] [Google Scholar]
- 9.Kitano N, Suzuki H, Takeuchi T. Patient age and the seasonal pattern of onset of Kawasaki’s disease. N Engl J Med. 2018;378(21):2048–2049. doi: 10.1056/NEJMc1804312. [DOI] [PubMed] [Google Scholar]
- 10.Shirato K, Imada Y, Kawase M, Nakagaki K, Matsuyama S, Taguchi F. Possible involvement of infection with human coronavirus 229E, but not NL63, in Kawasaki disease. J Med Virol. 2014;86(12):2146–2153. doi: 10.1002/jmv.23950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson RM, Little JR, Storch GA. Kawasaki-like syndromes associated with human immunodeficiency virus infection. Clin Infect Dis. 2001;32(11):1628–1634. doi: 10.1086/320523. [DOI] [PubMed] [Google Scholar]
- 12.Sundel RP. Kawasaki disease. Rheum Dis Clin North Am. 2015;41(1):63–73, viii. doi: 10.1016/j.rdc.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 13.Kanegaye JT, et al. Recognition of a Kawasaki disease shock syndrome. Pediatrics. 2009;123(5):e783–e9. doi: 10.1542/peds.2008-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiotos K, et al. Multisystem inflammatory syndrome in children during the coronavirus 2019 pandemic: a case series. J Pediatric Infect Dis Soc. 2020;9(3):393–398. doi: 10.1093/jpids/piaa069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wall SA, et al. Complement-mediated thrombotic microangiopathy as a link between endothelial damage and steroid-refractory GVHD. Blood Adv. 2018;2(20):2619–2628. doi: 10.1182/bloodadvances.2018020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bu F, et al. Soluble c5b-9 as a biomarker for complement activation in atypical hemolytic uremic syndrome. Am J Kidney Dis. 2015;65(6):968–969. doi: 10.1053/j.ajkd.2015.02.326. [DOI] [PubMed] [Google Scholar]
- 17.Kilgore KS, et al. Sublytic concentrations of the membrane attack complex of complement induce endothelial interleukin-8 and monocyte chemoattractant protein-1 through nuclear factor-kappa B activation. Am J Pathol. 1997;150(6):2019–2031. [PMC free article] [PubMed] [Google Scholar]
- 18.Schapkaitz E, Mezgebe MH. The clinical significance of schistocytes: a prospective evaluation of the International Council for Standardization in Hematology schistocyte guidelines. Turk J Haematol. 2017;34(1):59–63. doi: 10.4274/tjh.2016.0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stringer E, Yeung RSM. Pathogenesis of Kawasaki disease: the central role of TNF-α. Fut Rheumatol. 2008;3(1):69–77. doi: 10.2217/17460816.3.1.69. [DOI] [Google Scholar]
- 20.Jinkawa A, et al. Cytokine profile of macrophage activation syndrome associated with Kawasaki disease. Cytokine. 2019;119:52–56. doi: 10.1016/j.cyto.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 21.Sun D, et al. Clinical features of severe pediatric patients with coronavirus disease 2019 in Wuhan: a single center’s observational study. World J Pediatr. 2020;16(3):251–259. doi: 10.1007/s12519-020-00354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yancy CW. COVID-19 and African Americans. JAMA. 2020;323(19):1891–1892. doi: 10.1001/jama.2020.6548. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplemental data
ICMJE disclosure forms