CHOP-Dependent Stress-Inducible Expression of a Novel Form of Carbonic Anhydrase VI (original) (raw)
Abstract
CHOP (also called GADD153) is a stress-inducible nuclear protein that dimerizes with members of the C/EBP family of transcription factors and was initially identified as an inhibitor of C/EBP binding to classic C/EBP target genes. Subsequent experiments suggested a role for CHOP-C/EBP heterodimers in positively regulating gene expression; however, direct evidence that this is the case has so far not been uncovered. Here we describe the identification of a positively regulated direct CHOP-C/EBP target gene, that encoding murine carbonic anhydrase VI (CA-VI). The stress-inducible form of the gene is expressed from an internal promoter and encodes a novel intracellular form of what is normally a secreted protein. Stress-induced expression of CA-VI is both CHOP and _C/EBP_β dependent in that it does not occur in cells deficient in either gene. A _CHOP_-responsive element was mapped to the inducible CA-VI promoter, and in vitro footprinting revealed binding of CHOP-C/EBP heterodimers to that site. Rescue of CA-VI expression in _c/ebp_β−/− cells by exogenous C/EBPβ and a shorter, normally inhibitory isoform of the protein known as LIP suggests that the role of the C/EBP partner is limited to targeting the CHOP-containing heterodimer to the response element and points to a preeminent role for CHOP in CA-VI induction during stress.
Stress is associated with adaptive alterations in cellular gene expression programs. These in turn are coordinated by transcription factors that serve as transducers of stress signals to the nucleus. The GADD153 or CHOP gene encodes the transcription factor CHOP, which is highly responsive to certain forms of stress. Multiple toxins and states of metabolic deprivation serve to activate CHOP gene expression (3, 5, 7, 14, 16, 19, 26) and modulate the activity of the CHOP protein (29, 32). These correlative observations suggest that CHOP may play a role in regulating target genes in response to stress.
Early biochemical studies of the CHOP protein indicated that it forms stable dimers with transcription factors of the C/EBP family and that such dimers are incapable of binding to classical C/EBP sites. Therefore, under certain circumstances CHOP functions as an inhibitor of C/EBP proteins (9, 11, 22). This simple picture of CHOP as a stress-inducible inhibitor of C/EBP proteins was complicated by the finding that CHOP-C/EBP heterodimers are capable of binding unique DNA sequences that are distinct from classical C/EBP sites (29). Furthermore, it was recently demonstrated that CHOP forms stable dimers with a non-C/EBP transcription factor, ATF3. The latter is encoded by a gene that is itself stress inducible, raising the possibility of stress signaling by CHOP-ATF3 heterodimers (6, 15). In an effort to determine if CHOP plays a role in activating gene expression in response to stress, we recently sought to identify genes differentially expressed as a result of stress in cells that do and do not contain CHOP. This analysis led to the identification of three distinct genes that are absolutely dependent on CHOP for their induction by stress (31).
The cloning of such CHOP-dependent, stress-inducible genes has allowed us to examine the role of CHOP and its dimerization partners in the activation of gene expression. Here we present data indicating that the most stress-inducible of the CHOP-dependent genes identified in the aforementioned screen encodes a novel intracellular form of the normally secreted carbonic anhydrase VI protein (CA-VI). We perform a functional analysis of the role of CHOP and its dimerization partners in the stress-inducible activation of this novel CA-VI gene, and we speculate on the possible physiological role that an intracellular form of carbonic anhydrase may play in cellular adaptation to stress.
MATERIALS AND METHODS
Cell culture, transfection, and treatment.
_chop_−/− mice and _c/ebp_β−/− mice have been previously described (24, 34). Mouse embryonic fibroblasts with wild-type or mutant genotypes were generated from day 14.5 mouse embryos (20). _chop_−/− or _c/ebp_β−/− 3T3 fibroblasts were produced from the mouse embryonic fibroblasts by serial passage as described previously (28) and were studied here between passages 22 and 27. NIH 3T3, COS1, and 293T cell lines were originally obtained from the American Type Culture Collection. All cells were cultured in Dulbecco modified Eagle medium in the presence of 10% fetal bovine serum (Intergen) except during methionine starvation conditions, where cells were cultured in methionine-deficient Dulbecco modified Eagle medium with 10% dialyzed fetal bovine serum for 16 h. Unless stated otherwise in the legends, tunicamycin (Sigma) was used at 2 μg/ml for 10 h. Retroviral vectors bearing genes encoding ATF3, ATF3ΔLZ, CHOP, and the different C/EBP isoforms were constructed by introducing the corresponding cDNAs into the pBABE-puromycin retroviral vector (17). C/EBPα12K and LIP have been previously described (8, 30). Retroviral vectors, together with the expression vectors pCMV-HIV Tat, pSV-VSV-G, and pJK3 (bears the genes that encode the retroviral Gag and Pol proteins), were cotransfected into 293T cells by the calcium phosphate method; recombinant retroviral particles were harvested 48 h after transfection, and the viral supernatants were used to infect the mutant 3T3 fibroblasts. Forty-eight hours after infection, the cells were placed in selection medium containing puromycin (2 μg/ml) for 10 additional days.
RNA isolation, Northern blot analysis, and RT-PCR.
Total RNA was prepared from cultured cells or tissues by the phenol-guandinium isothiocyanate method (ULTRASPEC; Biotex Lab, Inc.). For poly(A)+ RNA isolation, total RNA was purified by additional CsCl ultracentrifugation, followed by double-passing the RNA over an oligo(dT) cellulose column. For Northern blots, total RNA was fractionated on a formaldehyde agarose gel and transferred onto HyBond-N nylon membranes (Amersham). All cDNA probes were labeled by random priming in the presence of [α-32P]dCTP and hybridized at 65°C in Church solution (7% sodium dodecyl sulfate [SDS], 1 mM EDTA, 0.5 M sodium phosphate buffer [pH 7.4]). Murine CA-VI (DOC1), CHOP, C/EBPβ, BiP, and α-tubulin were used as probes as described previously (31). To analyze the expression of CA-VI by reverse transcription (RT)-PCR, 1 μg of total RNA was primed with oligo(dT) to synthesize first-strand cDNA with reverse transcriptase. The reaction mixture was diluted in Tris-EDTA, and 5% of the reaction mixture was used for PCR. PCR conditions were 94°C (1 min), 62°C (45 s), and 72°C (1 min) for 30 cycles. CA-VI type A mRNA was detected by using primers 3S and 6AS, whereas the type B mRNA was detected by using primers 1S and 6AS. For primer sequence, see Fig. 1B. The translated in liposarcoma (TLS) control PCR was performed by using primers with the following sequences: GAT CAA GGA TCT CGT CAT GAT TC for the sense primer and CCT CAC CCT TCA ACT TGC CAG for the antisense primer.
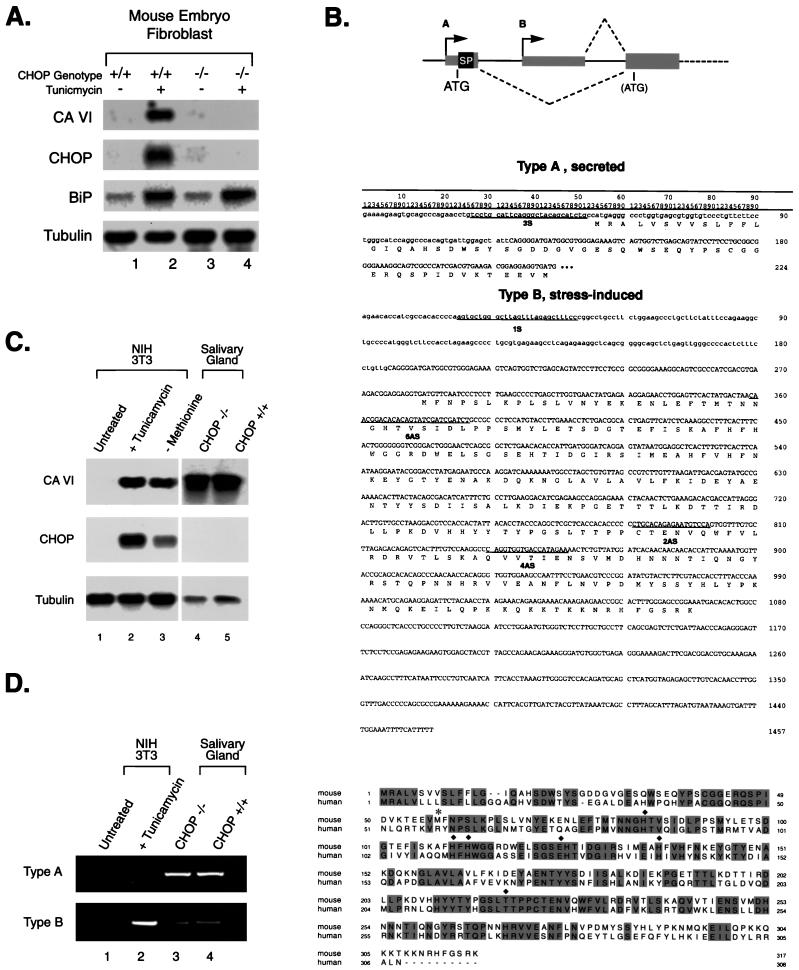
FIG. 1.
A gene downstream of CHOP encodes a novel form of carbonic anhydrase VI. (A) Northern blot of total cellular RNA prepared from untreated and tunicamycin (2 μg/ml)-treated (10 h) chop+/+ and _chop_−/− fibroblasts. The blot was hybridized sequentially with a _Dpn_II fragment of CA-VI obtained by representational-difference analysis of genes downstream of CHOP (31), followed by hybridization with CHOP and BiP (which served to document that a stress response had been induced) and Tubulin as a control for integrity of the RNA. (B) (Top) Diagram depicting the structure of the 5′ end of the murine CA-VI gene. A indicates the promoter that is active in salivary glands and that directs the expression of a secreted form of the protein with a signal peptide (SP). B indicates the internal, stress-induced, and CHOP-dependent promoter that encodes an intracellular form of the protein. Coding regions are indicated by wide boxes, and noncoding regions are indicated by thin boxes. Introns are presented as thin lines. The positions of the two initiating methionines are indicated. (Middle) Nucleotide sequences of the mRNAs encoding the secreted (type A) and stress-induced (type B) proteins. Lowercase letters are used to indicate nucleotides contributed by exons unique to either type of mRNA, and uppercase letters are used for those residues common to both forms. The methionine at codons 222 to 224 of the type A sequence corresponds to codons 285 to 287 of the type B mRNA. The positions of the PCR primers used in performing the RACE and RT-PCR analyses are indicated. The _Dpn_II fragment constituting _DOC_1/CA-VI corresponds to nucleotides 870 to 1394 of the type B mRNA. (Bottom) Alignment between the amino acid sequences of the secreted forms of mouse and human CA-VI. Identical residues are shaded, the asterisk denotes the initiation codon for the type B protein, and the diamonds denote residues that are essential for enzymatic activity and conserved between all carbonic anhydrases. (C) Northern blot of total RNA from NIH 3T3 cells left untreated, treated with tunicamycin, or exposed to methionine-deficient medium for 16 h (left blots) and RNAs from the salivary glands of mice with the indicated CHOP genotypes (right blots). The blot was sequentially hybridized with CA-VI, CHOP, and Tubulin probes. The exposure of the left blot was approximately five times longer than that of the right blot. (D) RT-PCR analysis of the RNAs from panel C with primers specific for the type A and type B mRNAs.
Western blot analysis, immunocytochemistry, and immunoprecipitation.
Whole-cell extracts were prepared in SDS buffer, electrophoresed on an SDS–12% polyacrylamide gel (or 10% polyacrylamide for C/EBPβ immunoblots), transferred to nitrocellulose filters (Micron Separations, Inc.), and then reacted with the antisera indicated. CHOP and the Myc epitope were detected with the murine monoclonal antibodies 9C8 and 9E10, respectively, as previously described (4), while C/EBPβ and TLS were detected with rabbit polyclonal antibodies (21, 33). The immunoreactive protein species were visualized by the enhanced-chemiluminescence detection system (DuPont, NEN). For the glutathione _S_-transferase (GST) pull-down experiments followed by Western blotting (see Fig. 4D), plasmids bearing genes expressing CHOP and a fusion protein containing GST and ATF3 or ATF3ΔLZ were cotransfected into COS1 cells. All proteins were tagged with the Myc epitope. Whole-cell extracts were isolated, and the GST fusion proteins were purified on agarose beads conjugated with glutathione as described previously (23). The presence of CHOP and ATF3 in the GST-pulled-down protein complexes was determined by Western blotting with a mixture of 1:10-diluted 9C8 and 9E10 antibodies (the anti-CHOP 9C8 antibody is required because 9E10-CHOP is poorly reactive with the 9E10 antibody on Western blots). 3′ Myc-tagged versions of type A and B CA-VI were constructed by a patch PCR with 9E10.CA6 (AGA GAT CAG CTT CTG CTC GCC CCC AAA GTG CCG GTT CTT C) and 9E10.U (GGG GCT CGA GTC ACA GAT CCT CCT CAG AGA TCA GCT TCT GCT C) primers at a 1:10 ratio. PCR conditions were 94°C (1 min), 50°C (1 min), and 72°C (2 min) for 33 cycles. Immunocytochemistry and immunoprecipitation with the 9E10 antibody were performed as previously described (32), with the modification that the 9E10 hybridoma supernatant was used at a dilution of 1:5 in the immunocytochemistry experiments.
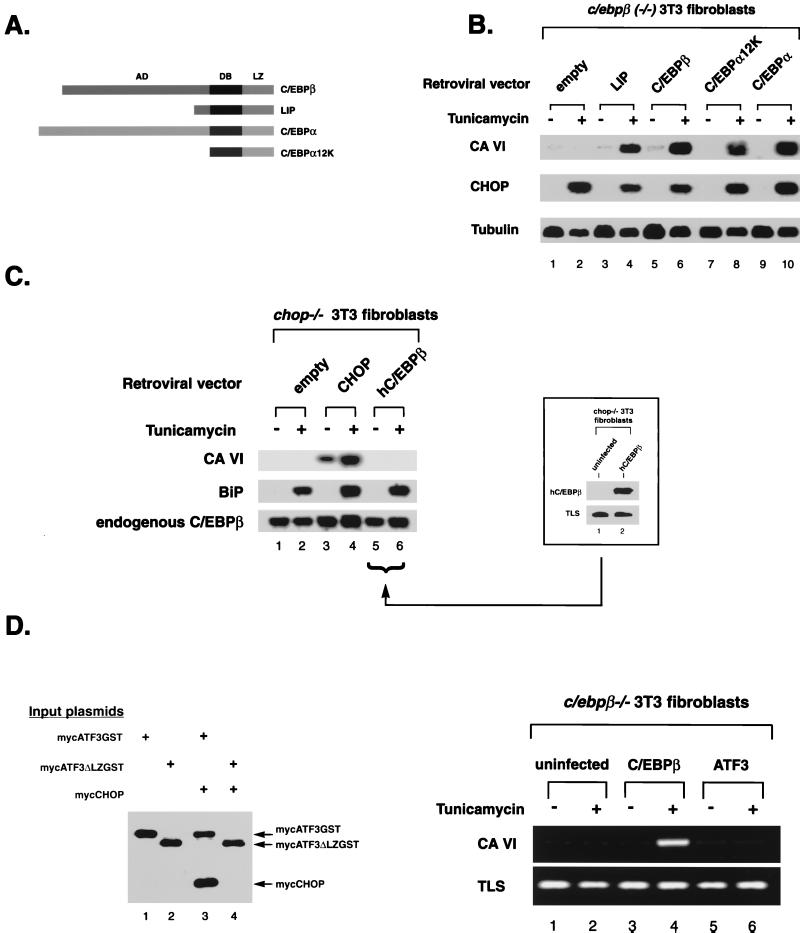
FIG. 4.
CA-VI induction by CHOP is C/EBP dependent but is indifferent to the C/EBP isoforms available. (A) Schematic representation of the C/EBP proteins used in the experiment. AD, activation domain; DB, DNA-binding basic region; LZ, leucine zipper domain. (B) Northern blot analysis of total RNA from _c/ebp_β−/− fibroblasts transduced with retroviral vectors bearing genes encoding the indicated C/EBP proteins. Cells were treated with tunicamycin or left untreated. The blot was sequentially hybridized with labeled CA-VI, CHOP, and Tubulin cDNAs. (C) _C/EBP_β overexpression does not activate CA-VI in the absence of CHOP. Shown is a Northern blot of RNA from _chop_−/− fibroblasts infected with the indicated retroviral vectors and either left untreated or treated with tunicamycin. The blot was hybridized sequentially to labeled CA-VI, BiP, and _C/EBP_β cDNAs. _C/EBP_β served as the internal control while BiP served to document the stress response in the _chop_−/− cells. A Western blot (inset) documents the expression of human C/EBPβ in the cells transduced by that virus. TLS, an abundantly expressed nuclear protein, served as the internal control. (D) The bZIP protein ATF3, which is unrelated to C/EBPs but capable of dimerizing with CHOP, is incapable of replacing C/EBPβ in the stress-induced activation of CA-VI. (Left blot) Soluble GST proteins from a lysate of COS1 cells transfected with the indicated expression plasmids were subjected to purification on glutathione-agarose beads. The isolated protein complexes were boiled in SDS and resolved by SDS-polyacrylamide gel electrophoresis, blotted to a nitrocellulose membrane, and reacted with a mixture of anti-Myc (9E10) and anti-CHOP (9C8) antibodies. The positions of the migrations of the proteins are indicated by arrows on the right. (Right gels) RT-PCR estimation of CA-VI expression in _c/ebp_β−/− fibroblasts, uninfected or infected with C/EBPβ or ATF3 retroviral vectors and treated or not treated with tunicamycin. TLS expression served as an internal control for the integrity of the RNA.
Isolation of type A and B CA-VI clones and functional characterization of the type B promoter.
Rapid amplification of 5′ cDNA ends (5′RACE) to clone type A (secreted form) CA-VI was performed on poly(A)+ RNA from mouse salivary glands. RT reaction mixtures were primed with the 4AS oligonucleotide (see Fig. 1B) followed by synthesis of a poly(A) tail and PCR amplification between 2AS and the T.Ad primer as described previously (13). To isolate the type B (stress-induced) version of CA-VI, a λ-Zap (Stratagene) library made from tunicamycin-induced NIH 3T3 cells was screened with a 160-bp _Aat_II-_Xba_I fragment of the representational-difference analysis product DOC1.3 (31). Twenty-nine positive plaques were identified after screening of 3 × 105 recombinants; of these, eight were further characterized by restriction analysis and the two largest clones were sequenced. The type B promoter was isolated by screening a murine S129 genomic library in λ-Fix II (Stratagene), with the cloned cDNA as a probe. A 2.8-kb _Apa_I-_Nco_I subclone containing both the type A and type B first exons was obtained (see Fig. 3A). DNase I footprint analysis was performed with bacterially expressed C/EBPβ and CHOP as previously described (29). The probe consisted of a 479-bp _Sau_3A-_Pml_I fragment of the type B CA-VI promoter labeled at the 5′ end of the sense strand by [γ-32P]ATP and T4 kinase. To assay promoter activation by CHOP, the genomic fragment from −2.8 kb to +94 of the type B transcription start site was fused upstream of the luciferase gene in the pGL vector (Promega) and cotransfected into _chop_−/− 3T3 cells or NIH 3T3 cells with or without expression plasmids bearing CHOP, CHOP lacking the leucine zipper domain, or CHOP lacking the DNA-binding basic region (29). The luciferase assay was performed on duplicate samples 24 to 36 h after transfection, and tunicamycin was introduced 20 h after transfection. 5′ sections of this promoter were serially deleted by using the restriction sites shown in Fig. 3A to generate genomic fragments of 1.2, 0.6, 0.4, 0.35, and 0.2 kb in length. Replacement of the TGCAAT sequence with a _Bam_HI site in the context of the 600-bp _Sau_3A reporter was carried out by “sewing” PCR using CA6.M1.S (TGT ACT GGA TCC CCT CCT GCC TCT ACC TCA) and a CA6.M1.AS (AGG AGG GGA TCC AGT ACA AGG TCA TCC TCT). Transfection efficiency was monitored by cotransfecting a β-galactosidase reporter driven by the cytomegalovirus promoter, whose activity varied by less than 10% between the samples in a given assay.
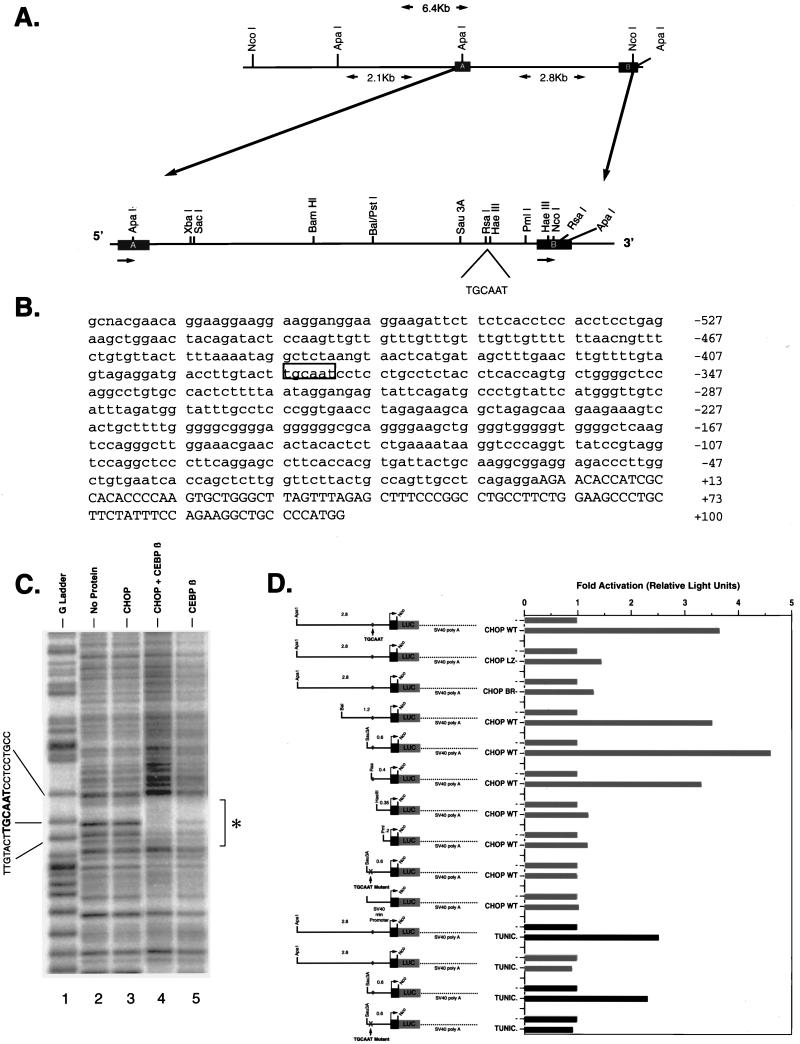
FIG. 3.
Identification of a _cis_-acting element that mediates CHOP induction of the type B CA-VI promoter. (A) Schematic representation of the type B CA-VI promoter region in the S129 mouse genome. The thin line represents the 5′ flanking region, and the thick line represents the transcribed region corresponding to exon 1 of the type A and type B mRNAs. The position of the CHOP response element, TGCAAT, is indicated. (B) Type B CA-VI promoter sequence from −527 to +100 with respect to the position of the transcription start site. The CHOP-C/EBP binding site is boxed, and the transcribed region is denoted by capital letters. (C) DNase I footprint analysis of the CA-VI type B promoter with the indicated bacterially expressed proteins. The probe was the 479-bp _Sau_3A-_Pml_I fragment labeled on the sense strand at the 5′ end. The footprinted region is indicated by the asterisk. (D) Functional analysis of the type B CA-VI promoter with luciferase reporter genes transiently transfected into wild-type NIH 3T3 cells (dark bars) or _chop_−/− 3T3 cells (light bars). Where indicated, cells were cotransfected with wild-type CHOP (CHOP WT) or mutant CHOP (CHOP LZ−, with leucine zipper deleted; CHOP BR−, with basic region deleted) expression plasmids or treated with tunicamycin (TUNIC; 5 μg/ml, 4 h). The position of the CHOP response element is indicated by diamonds. An X is used to indicate that the CHOP binding site was replaced by a _Bam_HI linker (TGCAAT mutant). The simian virus 40 (SV40) minimal promoter served to control for any nonspecific effects CHOP might have.
Nucleotide sequence accession numbers.
The sequences of the mRNAs encoding the secreted (type A) and stress-induced (type B) CA-VI proteins have been deposited in GenBank under accession no. AF079835 and AF079834, respectively. The sequence of the type B CA-VI promoter from −527 to +100 with respect to the position of the transcription start site has been given accession no. AF079836.
RESULTS
Genes dependent on CHOP are predicted to be expressed in stressed wild-type cells but not in stressed cells derived from chop knockout mice.
Representational-difference analysis of mRNAs expressed in tunicamycin-treated (stressed) chop+/+ and _chop_−/− mouse embryo fibroblasts identified several cDNA fragments from differentially expressed genes (31), two of which (DOC1 and DOC3) proved, upon sequencing, to be fragments of the mouse homologue of the carbonic anhydrase VI gene (Fig. 1A and B). In several mammalian species, including human and sheep, CA-VI is highly expressed in the salivary and lachrymal glands and encodes a secreted form of carbonic anhydrase that accumulates in saliva and tears (10). The differentially expressed cDNA fragment was used as a hybridization probe to isolate several full-length murine CA-VI cDNAs from a tunicamycin-treated mouse fibroblast library. Sequencing revealed that the encoded protein, while being highly similar to the human and bovine CA-VIs from amino acid 60 on, lacked a signal peptide and is not predicted to encode a secreted protein (Fig. 1B). To further explore the relationship between the tunicamycin-induced CA-VI cDNA isolated from fibroblasts and the predicted murine homologue of the secreted form of the enzyme, the tunicamycin-induced form was hybridized at high stringency to a Northern blot of mouse submandibular salivary gland mRNAs derived from chop+/+ and _chop_−/− mice. A very strong hybridization signal was obtained in both samples (Fig. 1C, lanes 4 and 5). This result indicates that the tunicamycin-induced mRNA has significant nucleotide identity with murine salivary gland CA-VI and furthermore that CHOP plays no role in the expression of the secreted form of the enzyme. In fact, the hybridization signal in the salivary glands was much stronger than that obtained in the stressed fibroblasts (Fig. 1C, compare lanes 2 and 3 with lanes 4 and 5), consistent with the reported enormous abundance of the CA-VI protein in saliva (10).
5′RACE with primers located in the portion of the tunicamycin-induced CA-VI cDNA that is homologous to the human CA-VI cDNA was used to obtain the 5′ end of the salivary gland version of the murine mRNA (Fig. 1B). Sequencing of the RACE products revealed that the salivary gland form of CA-VI encodes a protein that has a long hydrophobic N-terminal stretch consistent with a signal peptide. The predicted amino acid sequence immediately downstream of this hydrophobic domain corresponded exactly to that obtained by N-terminal sequencing of purified mouse salivary carbonic anhydrase protein (10). Comparison of the salivary gland (type A) and tunicamycin-induced (type B) forms of CA-VI revealed the presence of divergent N-terminal sequences in proteins that were otherwise identical, indicating that both the secreted form and the stress-induced form of CA-VI are likely to be products of the same gene (Fig. 1B). PCR analysis of cDNA from untreated and tunicamycin-treated fibroblasts revealed that only the type B CA-VI was expressed in response to stress in these cells and that the type A CA-VI (secreted form) was restricted to the salivary glands. A small amount of type B transcript was also present in the salivary gland (Fig. 1D). Since the active site of the enzyme (beginning at histidine 84 of the secreted form [25]) is encoded by sequences shared by the two forms, it is likely that both proteins are active carbonic anhydrases.
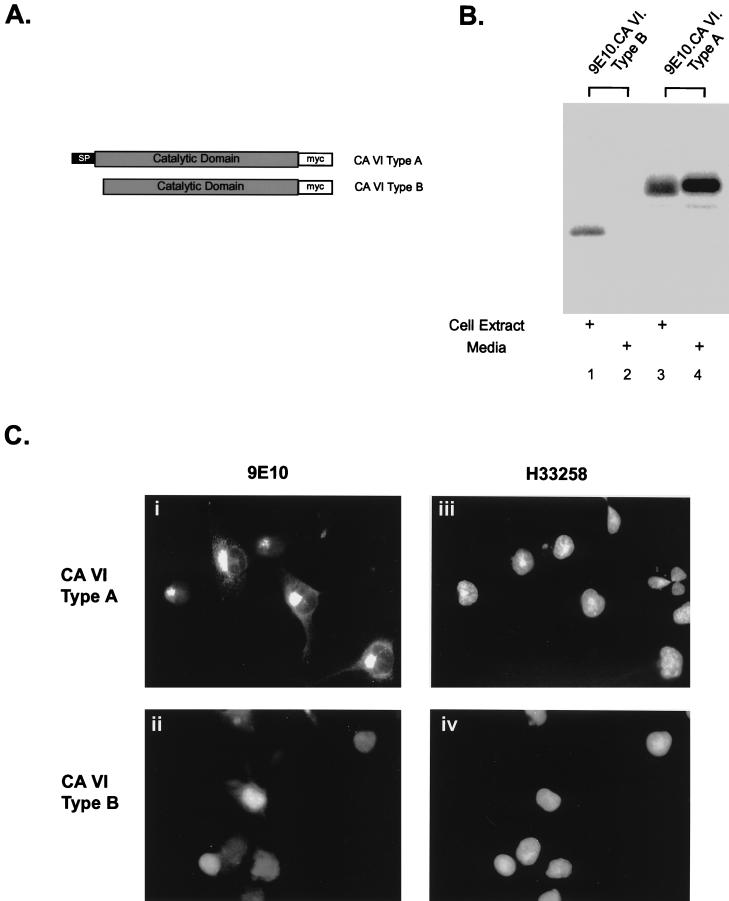
Based on the predicted peptide sequence, the type A mRNA encodes a secreted protein whereas the type B mRNA is expected to be retained in the cell. To examine this issue experimentally, both forms of CA-VI were tagged at their C termini with an epitope tag and expression vectors for the corresponding proteins were transfected into COS1 cells (Fig. 2A). Metabolic labeling followed by immunoprecipitation of the tagged proteins from the cell lysate or the culture supernatant revealed that the type B protein was entirely intracellular but that the type A protein accumulated in the medium (Fig. 2B). Immunofluorescence microscopy with an antibody directed against the epitope tag revealed that staining of cells transfected with an expression plasmid encoding the type A protein was restricted to a focal eccentric structure, presumably reflecting protein in transit through the endomembrane system, but that staining of cells expressing the type B protein was present in the nucleus and to a lesser degree in the cytoplasm (diffuse staining) (Fig. 2C). These experiments strongly suggest that the stress-induced form of CA-VI encodes an intracellular carbonic anhydrase.
FIG. 2.
The stress-inducible form of CA-VI encodes an intracellular protein. (A) Diagram depicting the proteins encoded by the type A and type B CA-VI mRNAs, which were tagged with the myc (9E10) epitope at their C termini. (B) COS1 cells transiently expressing the tagged proteins were metabolically labeled for 3 h, followed by immunoprecipitation of the tagged proteins from the cell extract (lanes 1 and 3) and the medium (lanes 2 and 4) and then resolving by SDS-polyacrylamide gel electrophoresis and autoradiography. The smaller type B protein product is present only in the cell extract (lane 1), whereas the type A protein is observed as a stable protein present in both the cellular extracts and in the medium (lanes 3 and 4). (C) COS1 cells transiently expressing the epitope-tagged forms of CA-VI were fixed, permeabilized, and immunostained with a monoclonal antibody to the Myc tag, 9E10 (left images), and the nucleophilic dye H33258 (right images). Type A CA-VI can be visualized, in transit, as punctuate staining that outlines the endosomal compartment (i), whereas type B CA-VI can be visualized as an intracellular, predominantly nuclear pattern of staining (ii).
To determine the genomic basis for the diversity in CA-VI mRNAs, mouse genomic clones containing the region corresponding to both the type A and type B cDNA 5′ ends of CA-VI were isolated. Restriction fragment analysis and Southern blotting combined with genomic PCR and sequencing revealed that the type A 5′ sequence that encodes the signal peptide is derived from a separate 5′ first exon but that the type B mRNA 5′ end is derived from a different exon and arises from transcription initiated from an alternative promoter, contained within a large first intron of the gene (Fig. 1B and 3A).
5′RACE of the tunicamycin-induced mRNA confirmed that the cDNA sequence presented in Fig. 1B is full length and thus that its 5′ end defines the transcription start site of the putative tunicamycin-induced promoter. Sequencing the genomic clone 5′ of the tunicamycin-induced first exon revealed, at a position 386 nucleotides upstream of the transcription start site, the presence of a perfect match for a CHOP-C/EBP binding site, as previously defined by in vitro site selection (29) (Fig. 3B). DNase I footprint analysis of this region with purified, bacterially expressed CHOP and C/EBP proteins showed that this sequence is a binding site for both C/EBP homodimers and C/EBP-CHOP heterodimers; the latter gave rise to a stronger and more extended footprint than that of the C/EBP homodimers (Fig. 3C, compare lanes 4 and 5). CHOP by itself does not alter the footprint pattern of this region (lane 3).
To address the functional role of this CHOP-C/EBP binding site, reporter genes were constructed by fusing the genomic fragment from −2.8 kb to +94 to a luciferase encoding cDNA. Serial deletions of the 5′ sequence were also performed, and a mutant reporter was constructed by replacing the CHOP-C/EBP binding site at −386 with a _Bam_HI restriction endonuclease site. Wild-type reporter genes containing a minimum of 400 bp of promoter sequence were responsive to cotransfected CHOP. Deletion of an additional 45 bp containing the footprinted region led to significant loss of CHOP responsiveness. The mutant reporter gene in which the CHOP-C/EBP binding site had been deleted by introduction of a _Bam_HI linker at −386 was likewise unresponsive to CHOP. Mutant forms of CHOP that lack either the leucine zipper dimerization domain or the DNA-binding basic region did not activate the reporter (Fig. 3D). These experiments established a correlation between the presence of a CHOP-C/EBP binding site in the type B (tunicamycin-inducible) CA-VI promoter and its activation by the CHOP protein. We also measured the response of the type B CA-VI reporter constructs to tunicamycin stimulation. In chop+/+ cells, tunicamycin treatment led to a modest but highly reproducible twofold activation of the wild-type reporters. The mutant reporter lacking the CHOP-C/EBP binding site was not induced by tunicamycin, and a wild-type reporter was not inducible in _chop_−/− cells. While these results do not fully recapitulate the strong stress dependence of the activation of the endogenous CA-VI gene by the CHOP protein (31) (Fig. 4C, compare lanes 3 and 4), they do indicate that this experimental system is capable of determining the CHOP inducibility of the CA-VI promoter.
CA-VI was identified as a CHOP target gene in mouse embryonic fibroblasts. In fibroblasts C/EBPβ is the major dimerization partner of CHOP (2, 31, 33). CHOP induction of CA-VI maps to a promoter element that interacts with CHOP only in the context of a CHOP-C/EBP heterodimer. Consistent with these facts we find that CA-VI is not inducible in _c/ebp_β−/− cells (Fig. 4B). To analyze the role of the C/EBP dimerization partner in CA-VI induction by CHOP, we rescued the _c/ebp_β−/− fibroblasts with a C/EBPβ-expressing retrovirus and with viruses expressing various derivatives of C/EBP. Stress-induced expression of CA-VI did not require the activation domain of C/EBPβ, as LIP, an isoform of C/EBP that lacks an activation domain (8), was still capable of rescuing CA-VI expression. An even smaller derivative of a C/EBP protein, consisting entirely of the DNA-contacting basic region and leucine zipper dimerization domain of C/EBPα, was also capable of rescuing CA-VI expression as was full-length C/EBPα (Fig. 4B, lanes 8 and 10). Rescue of CA-VI expression by all these forms of C/EBP was strictly CHOP dependent, as reflected in the requirement for stress (Fig. 4B, compare odd- and even-numbered lanes) and in the fact that the expression of C/EBPβ alone in _chop_−/− cells could not induce CA-VI (Fig. 4C, lanes 5 and 6). These experiments indicate that the activation domain of the C/EBPs does not play an essential role in CA-VI activation by stress, implying that activation is contributed by the CHOP portion of the heterodimer.
The transcription factor ATF3, a member of the CREB-ATF family of bZIP proteins, has recently been shown to associate with CHOP (6). ATF3 is not normally expressed in fibroblasts and is not induced in these cells by stress (data not shown). The DNA-binding domain of ATF3 and its target sequences are very different from those of the C/EBP family. It was of interest, therefore, to determine if ATF3 could also rescue CA-VI expression in _c/ebp_β−/− cells. We first confirmed that the ATF3 and CHOP proteins expressed in our system form stable heterodimers that coprecipitate in vivo (Fig. 4D, left gel) and then measured CA-VI induction by stress in ATF3-rescued cells; none was evident (Fig. 4D, right gel). We thus conclude that CA-VI is a specific target gene of CHOP-C/EBP heterodimers and that CHOP dimerization partners that do not belong to the C/EBP family cannot substitute for the essential role of the C/EBPs.
DISCUSSION
The identification of a novel stress-inducible form of CA-VI downstream of CHOP and the demonstration of the presence of a functional CHOP-C/EBP binding site in its stress-inducible promoter provide the first direct evidence for CHOP’s activity as a DNA-binding transcription factor capable of positively regulating a direct cellular target gene. These experiments provide evidence for a dual role for CHOP in the regulation of cellular gene expression: both as an inhibitor of C/EBP binding to classical C/EBP target genes (9, 11, 22) and as an activator of genes that have CHOP-C/EBP binding sites; CA-VI is prototypical of the latter group. This new evidence for a positive component to CHOP action fits nicely with previous experimental results in which it has been found that the DNA-contacting basic region of CHOP is essential for eliciting CHOP-dependent phenotypes (2, 33). Furthermore, the existence of a positively regulated direct CHOP target gene provides a teleological explanation for the presence of a strong and regulated transcriptional activation domain in the CHOP protein (32).
The functional CHOP-C/EBP binding site in the CA-VI promoter is very similar to the CHOP-C/EBP consensus site selected in vitro in that both contain the TGCAAT hexanucleotide motif. Both sites are also capable of binding C/EBP homodimers as well as CHOP-C/EBP heterodimers, and the footprints obtained from the selected consensus site and the functional site from the CA-VI gene are similar in that the CHOP-C/EBP heterodimer consistently gives rise to a stronger protection pattern than that observed with C/EBP homodimers (Fig. 3C) (29). CHOP’s participation in the DNA-binding complex at the tunicamycin-inducible CA-VI promoter is also supported by the observation that a basic-region mutant of CHOP is incapable of activating a reporter gene driven by the promoter (Fig. 3D) and by the inability of the basic-region mutant of CHOP to rescue CA-VI expression in response to stress in _chop_−/− cells (data not shown). In spite of the compelling genetic evidence, the mechanistic aspects of CHOP interaction with DNA are far from clear. The TGCAAT motif, common to the in vitro-selected sites and the CA-VI promoter, represents little more than a C/EBP half-site, GCAAT preceded by a T (18, 30). This suggests that it is the C/EBP partner that provides most of the sequence-specific information for the interaction of the heterodimer with DNA. Indeed, the rescue experiments with _c/ebp_β−/− cells suggest that the C/EBP dimerization partner contributes nothing more than a DNA-binding component in that endogenous C/EBPβ can be replaced not only by C/EBPα but also by nonactivating isoforms such as LIP or truncated versions of C/EBPα that entirely lack a transactivation domain (8, 12). The specific role played by the C/EBP component in DNA binding in CA-VI activation by stress is demonstrated by the fact that ATF3 (a protein that dimerizes avidly with CHOP but has a different ATF-type DNA-binding domain) is incapable of rescuing CA-VI expression in _c/ebp_β−/− cells. In spite of the essential role of the C/EBP dimerization partner in directing the heterodimer to the response element, the inability of CHOP to activate through classic C/EBP target sites (22), together with the complete dependence of activation of CA-VI on an intact CHOP basic region, suggests that CHOP too plays some role in mediating protein-DNA interactions. The structural basis for this activity of CHOP remains to be defined.
Activation of the tunicamycin-inducible promoter of CA-VI as well as activation of two other genes downstream of CHOP (DOC4 and DOC6) requires that, in addition to CHOP being expressed, a stress signal be provided to the cell. This is revealed by CHOP rescue experiments in _chop_−/− cells in which the induction of CHOP and stress are effectively unlinked (Fig. 4C) (31). This stress dependence of CA-VI expression is not recapitulated by the reporter gene we have constructed (the latter is, however, responsive to CHOP). Furthermore, the full magnitude of the tunicamycin inducibility of even the largest reporter construct we have tested (2.8 kb) is increased only ∼2-fold compared to the >20-fold increase in the expression of the endogenous gene. Stable integration of the 2.8-kb reporter does not remedy this low-level inducibility (data not shown). Collectively, these results suggest that portions of the gene lying outside the fragment used in constructing the reporter contribute to the inducibility of CA-VI by stress. We have no way to know if these putative additional regulatory elements are CHOP responsive or if they are used to allow the convergence of a parallel, _chop_-independent stress-inducible pathway on the CA-VI promoter. The fact that stress induction of CA-VI (DOC1) in _chop_−/− cells can be rescued by a mutant CHOP protein that lacks the stress-inducible phosphorylation sites (31) suggests that at least part of this second signal in CA-VI induction is CHOP independent. Either way, CHOP appears to exert an essential permissive role on the induction of the stress-inducible CA-VI promoter, and this permissive effect correlates with the presence of a direct binding site in the type B promoter-proximal region. A model for the role of CHOP and its dimerization partner in the induction of CA-VI is diagramed in Fig. 5.
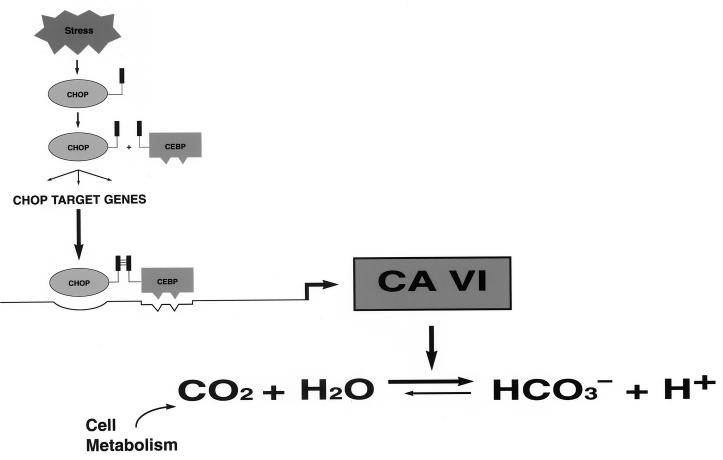
FIG. 5.
Diagram depicting components of the signaling pathway hypothesized to play a role in cellular stress-mediated induction of type B CA-VI. Cellular stress leads to the induction of the chop gene and accumulation of CHOP protein. The latter forms a dimer with a member of the C/EBP family, and this activates the type B CA-VI promoter. Intracellular accumulation of CA-VI is predicted to increase the proton concentration in the cell by promoting the hydration of CO2 produced by the metabolic activity of the cell.
The CHOP-dependent, novel form of CA-VI induced in response to stress encodes a protein that is retained intracellularly. This fact is in marked contrast to the major exocrine-gland-specific product of the CA-VI gene which is a constitutively secreted form of carbonic anhydrase (expression of the latter is not CHOP dependent; compare lanes 4 and 5 in Fig. 1C). There is every reason to believe that, as with the secreted form of CA-VI, the stress-induced form is a protein with carbonic anhydrase and esterase activities; this assumption is based on the fact that both forms contain most of the residues conserved among other known carbonic anhydrases and all of the residues that are known to contribute to the active site of the enzyme (25). Carbonic anhydrase catalyzes the reversible hydration of CO2 to H2CO3 (27). Inside the cell, where a net production of CO2 takes place, the cell’s CO2 hydration may have the effect of acidifying the intracellular milieu (this is because of the disassociation of H2CO3 into H+ and HCO3−). We do not at present have measurements for the magnitude of this hypothesized CHOP- and CA-VI-dependent stress-inducible acidification of the cell, and therefore, the physiological significance of the expression of the stress-induced form of CA-VI is not known. However, changes in intracellular pH, even those occurring within the physiological range, may significantly affect many cellular processes. It is intriguing to speculate, therefore, that CHOP-dependent expression of CA-VI, by altering intracellular pH, may contribute in some way to the CHOP-dependent component of the response of cells to stress. Cells from chop knockout mice exhibit less programmed cell death in response to stress than cells from wild-type mice (34). Recently, it has been demonstrated that the membrane pore-forming activity of the proapoptotic regulator Bax is pH dependent, increasing with decreasing pH (1). We propose, as a hypothesis for future testing, that during stress, CHOP- and CA-VI-dependent intracellular acidification may contribute to apoptosis by increasing the pore-forming activity of proapoptotic mediators such as Bax.
ACKNOWLEDGMENTS
We are indebted to Valeria Poli for the gift of C/EBPβ knockout mice; to Ross Fernley and Thomas Maren for advice on carbonic anhydrases; to Tsonwin Hai for discussions on ATF3; to Edward Skolnik, Lennart Philipson, and members of their lab as well as the other members of the Ron lab for criticism and advice; and to Jue Yee Kim for assistance with the figure layouts.
This work was supported by NIH grants DK47119 and ES08681. J.S. is a trainee in the NYU MSTP program and supported by NIGMS grant GM07308. D.R. is a Stephen Birnbaum Scholar of the Leukemia Society of America.
REFERENCES
- 1.Antonsson B, Conti F, Ciavatta A, Montessuit S, Lewis S, Martinou I, Bernasconi L, Bernard A, Mermod J J, Mazzei G, Maundrell K, Gambale F, Sadoul R, Martinou J C. Inhibition of Bax channel-forming activity by Bcl-2. Science. 1997;277:370–372. doi: 10.1126/science.277.5324.370. [DOI] [PubMed] [Google Scholar]
- 2.Barone M V, Crozat A Y, Tabaee A, Philipson L, Ron D. CHOP (GADD153) and its oncogenic variant, TLS-CHOP, differ in their ability to induce G1/S arrest. Genes Dev. 1994;8:453–464. doi: 10.1101/gad.8.4.453. [DOI] [PubMed] [Google Scholar]
- 3.Bartlett J, Luethy J, Carlson S, Sollott S, Holbrook N. Calcium ionophore A23187 induces expression of the growth arrest and DNA damage inducible CCAAT/enhancer-binding protein (C/EBP)-related gene, gadd153. J Biol Chem. 1992;267:20465–20470. [PubMed] [Google Scholar]
- 4.Batchvarova N, Wang X-Z, Ron D. Inhibition of adipogenesis by the stress-induced protein CHOP (GADD153) EMBO J. 1995;14:4654–4661. doi: 10.1002/j.1460-2075.1995.tb00147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson S G, Fawcett T W, Bartlett J D, Bernier M, Holbrook N J. Regulation of the C/EBP-related gene gadd153 by glucose deprivation. Mol Cell Biol. 1993;13:4736–4744. doi: 10.1128/mcb.13.8.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen B P C, Wolfgang C D, Hai T. Analysis of ATF3, a transcription factor induced by physiological stresses and modulated by gadd153/Chop10. Mol Cell Biol. 1996;16:1157–1168. doi: 10.1128/mcb.16.3.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Q, Yu K, Holbrook N J, Stevens J L. Activation of the growth arrest and DNA damage-inducible gene gadd153 by nephrotoxic cysteine conjugates and dithiothreitol. J Biol Chem. 1992;267:8207–8212. [PubMed] [Google Scholar]
- 8.Descombes P, Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- 9.Fawcett T W, Eastman H B, Martindale J L, Holbrook N J. Physical and functional association between GADD153 and CCAAT/enhancer-binding protein beta during cellular stress. J Biol Chem. 1996;271:14285–14289. doi: 10.1074/jbc.271.24.14285. [DOI] [PubMed] [Google Scholar]
- 10.Fernley R, Darling P, Aldred P, Wright R, Coghlan J. Tissue and species distribution of the secreted carbonic anhydrase isoenzyme. Biochem J. 1989;259:91–96. doi: 10.1042/bj2590091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman A D. GADD153/CHOP, a DNA damage-inducible protein, reduced CAAT/enhancer binding protein activities and increased apoptosis in 32D c13 myeloid cells. Cancer Res. 1996;56:3250–3256. [PubMed] [Google Scholar]
- 12.Friedman A D, McKnight S L. Identification of two polypeptide segments of CCAAT/enhancer-binding protein required for transcriptional activation of the serum albumin gene. Genes Dev. 1990;4:1416–1426. doi: 10.1101/gad.4.8.1416. [DOI] [PubMed] [Google Scholar]
- 13.Frohman M A, Dush M K, Martin G R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gately D, Jones J, Christen R, Barton R, Los G, Howell S. Induction of the growth arrest and DNA damage-inducible gene GADD153 by cisplatin in vitro and in vivo. Br J Cancer. 1994;70:1102–1106. doi: 10.1038/bjc.1994.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu J-C, Laz T, Mohn K, Taub R. Identification of LRF-1, a leucine-zipper protein that is rapidly and highly induced in regenerating liver. Proc Natl Acad Sci USA. 1991;88:3511–3513. doi: 10.1073/pnas.88.9.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelsey G, Schutz G. Lessons from albino lethal mice. Curr Opin Genet Dev. 1993;3:259–264. doi: 10.1016/0959-437x(93)90032-k. [DOI] [PubMed] [Google Scholar]
- 17.Morgenstern L, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olive M, Krylov D, Echlin D R, Gardner K, Taparowsky E, Vinson C. A dominant negative to activation protein-1 (AP1) that abolishes DNA binding and inhibits oncogenesis. J Biol Chem. 1997;272:18586–18594. doi: 10.1074/jbc.272.30.18586. [DOI] [PubMed] [Google Scholar]
- 19.Price B, Calderwood S. Gadd45 and Gadd153 messenger RNA levels are increased during hypoxia and after exposure of cells to agents which elevate the levels of glucose-regulated proteins. Cancer Res. 1992;52:3814–3817. [PubMed] [Google Scholar]
- 20.Robertson E. Teratocarcinomas and embryonic stem cells. A practical approach. Oxford, United Kingdom: IRL Press; 1987. [Google Scholar]
- 21.Ron D, Brasier A R, McGehee R E, Habener J F. Tumor necrosis factor-induced reversal of adipocytic phenotype of 3T3-L1 cells is preceded by a loss of nuclear CCAAT/enhancer binding protein (C/EBP) J Clin Investig. 1992;89:223–233. doi: 10.1172/JCI115566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ron D, Habener J F. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant negative inhibitor of gene transcription. Genes Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- 23.Sánchez I, Hughes R, Mayer B, Yee K, Woodget J, Avruch J, Kyriakis J, Zon L. Role of SAPK/ERK kinase-1 in the stress-activated pathway regulating transcription factor c-Jun. Nature. 1994;372:794–797. doi: 10.1038/372794a0. [DOI] [PubMed] [Google Scholar]
- 24.Screpanti I, Romani L, Musiani P, Modesti A, Fattori E, Lazzaro D, Sellitto C, Scarpa S, Bellavia D, Lattanzio G, Bistoni F, Frati L, Cortese R, Gulino A, Ciliberto G, Constantini F, Poli V. Lymphoproliferative disorder and imbalanced T-helper response in C/EBPβ-deficient mice. EMBO J. 1995;14:1932–1941. doi: 10.1002/j.1460-2075.1995.tb07185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sly W S, Hu P Y. Human carbonic anhydrases and carbonic anhydrase deficiencies. Annu Rev Biochem. 1995;64:375–401. doi: 10.1146/annurev.bi.64.070195.002111. [DOI] [PubMed] [Google Scholar]
- 26.Sylvester S L, ap Rhys C M J, Luethy-Martindale J D, Holbrook N J. Induction of GADD153, a CCAAT/enhancer-binding protein-related gene during the acute-phase response in rats. J Biol Chem. 1994;269:20119–20125. [PubMed] [Google Scholar]
- 27.Tashian R E. The carbonic anhydrases: widening perspectives on their evolution, expression and function. Bioessays. 1989;10:186–192. doi: 10.1002/bies.950100603. [DOI] [PubMed] [Google Scholar]
- 28.Todaro G, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ubeda M, Wang X-Z, Zinszner H, Wu I, Habener J, Ron D. Stress-induced binding of transcription factor CHOP to a novel DNA-control element. Mol Cell Biol. 1996;16:1479–1489. doi: 10.1128/mcb.16.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vinson C R, Sigler P B, McKnight S L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989;246:911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- 31.Wang X-Z, Kuroda M, Sok J, Batchvarova N, Kimmel R, Chung P, Zinszner H, Ron D. Identification of novel stress-induced genes downstream of chop. EMBO J. 1998;17:3619–3630. doi: 10.1093/emboj/17.13.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X-Z, Ron D. Stress-induced phosphorylation and activation of the transcription factor CHOP (GADD153) by p38 MAP-kinase. Science. 1996;272:1347–1349. doi: 10.1126/science.272.5266.1347. [DOI] [PubMed] [Google Scholar]
- 33.Zinszner H, Albalat R, Ron D. A novel effector domain from the RNA-binding proteins TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 1994;8:2513–2526. doi: 10.1101/gad.8.21.2513. [DOI] [PubMed] [Google Scholar]
- 34.Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot R T, Remotti H, Stevens J L, Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]