The Two Forms of Karyogamy Transcription Factor Kar4p Are Regulated by Differential Initiation of Transcription, Translation, and Protein Turnover (original) (raw)
Abstract
Kar4p is a transcription factor in Saccharomyces cerevisiae that is required for the expression of karyogamy-specific genes during mating, for the efficient transit from G1 during mitosis, and for essential functions during meiosis. Kar4p exists in two forms: a constitutive slower-migrating form, which predominates during vegetative growth, and a faster-migrating form, which is highly induced by mating pheromone. Transcript mapping of KAR4 revealed that the constitutive mRNA was initiated upstream of two in-frame ATG initiation codons, while the major inducible mRNA originated between them. Thus, the two forms of Kar4p are derived from the translation of alternative transcripts, which possess different AUG initiation codons. Site-directed mutations were constructed to inactivate one or the other of the initiation codons, allowing the expression of the two Kar4p forms separately. At normal levels of expression, the constitutive form of Kar4p did not support wild-type levels of mating. However, the two forms of Kar4p could also be expressed separately from the regulatable GAL1 promoter, and no functional difference was detected when they were expressed at equivalent levels. Pulse-chase experiments showed that the induced form of Kar4p was highly expressed and stable during mating but rapidly turned over in vegetative cells. In contrast, the constitutively expressed longer form showed the same rate of turnover regardless of the growth condition. Furthermore, overexpression of either form of Kar4p in vegetative cells was toxic. Thus, the elaborate regulation of the two forms of Kar4p at the levels of transcription, translation, and protein turnover reflects the requirement for high levels of the protein during mating and for low levels during the subsequent phases of the cell cycle.
In the yeast Saccharomyces cerevisiae, mating entails both cellular and nuclear fusion of two haploid cells of opposite mating type, MATa and MATα (for a review, see reference 29). Mating initiates after the reciprocal exchange of the cell-type- specific pheromones, α-factor and a-factor. Binding of the pheromone to its cognate receptor triggers a mitogen activated protein (MAP) kinase cascade that mediates a series of cellular changes in preparation for mating (for reviews, see references 11, 16, 26, and 27). One of the primary downstream targets of the MAP kinase cascade is the transcriptional induction of a variety of mating-specific genes. The process is mediated by the transcriptional activator Ste12p, alone or in concert with other proteins, which binds upstream of pheromone-induced genes via sequences known as pheromone response elements, or PREs (for a review, see reference 42). Important cellular changes in response to pheromone include agglutination via surface glycoproteins, arrest in the G1 phase of the cell cycle, and polarized growth toward a selected partner. The two partner cells adhere tightly to form prezygotes and ultimately fuse at the site of close apposition. Concurrently, microtubules emanating from the spindle pole body (SPB) move the haploid nuclei up to the site of polarized growth (31). Cell fusion occurs with the breakdown of the intervening cell wall and plasma membranes. Nuclear fusion, or karyogamy, begins when microtubules extending from the SPBs become interconnected by the kinesin-related motor protein, Kar3p (30), and its associated light chain, Cik1p (34). Kar3p produces antiparallel microtubule movement, thereby bringing the nuclei together (30), in a process termed nuclear congression (24). The nuclei initiate fusion along the edges of the SPBs (7), and nuclear membrane fusion occurs (4, 24), resulting in the formation of a diploid cell.
KAR4 is one of eight genes initially identified as part of a genetic screen for bilateral mating mutants (24, 25). KAR4 expression is significantly induced during exposure to pheromone. The induced expression is mediated by Ste12p, which presumably binds to the PREs found upstream of the KAR4 coding region. Kar4p is a karyogamy-specific transcription factor that acts in combination with Ste12p to promote the pheromone induction of KAR3 and CIK1. Thus, Kar4p is essential for nuclear congression during karyogamy (25).
Kar4p also acts during mitosis and meiosis. In mitotically growing cells, KAR4 mRNA is specifically expressed during G1/S. Furthermore, kar4 mutants exhibit a G1 pause during vegetative growth. Similarly, KAR4 mRNA is induced during meiosis, and homozygous kar4 diploids fail to sporulate (25).
Previous work revealed two forms of Kar4p with different electrophoretic mobilities that are present in both vegetative and mating cells (25). The slower-migrating form corresponds to a 38.5-kDa protein (Kar4p-long), which is constitutively expressed and predominates during vegetative growth. The faster-migrating form corresponds to a 35.5-kDa protein (Kar4p-short), which is highly induced upon exposure to pheromone. Although Kar4p is a phosphoprotein (25), treatment with phosphatases did not alter the relative mobilities of the two forms of Kar4p (our unpublished observations). Therefore, simple phosphorylation models do not account for the two forms.
In this paper, we explore the origins of the two forms of Kar4p and the functional basis for their differential regulation. Examination of the DNA sequence showed two potential in-frame ATG translational start codons, 90 bp apart, at the 5′ end of the KAR4 open reading frame (25). We show that the two species of Kar4p have their ultimate origins in differential transcription from the KAR4 locus. The different transcripts then produce two distinct proteins initiating at the different AUGs. Although high levels of Kar4p are required for efficient karyogamy, overexpression is toxic in vegetatively growing cells. Accordingly, we find that the two forms of Kar4p have different stabilities in vegetative cells but not in mating cells, allowing for tighter regulation of the levels of Kar4p during mating and the subsequent reentry into mitosis.
MATERIALS AND METHODS
Strains and microbiological techniques.
A list of all the yeast strains and plasmids used in this study is found in Table 1. Yeast media and genetic techniques used were essentially as described previously (36). Sporulation experiments were performed as described in reference 23. MY4166 and MY4239, Gal+ _kar4_Δ strains, were produced by crossing MY3375 (Gal+) with MS3212 (kar4Δ::HIS3) (25). Tetrads were dissected, and MATa His+ spores were scored for growth on galactose and for the kar4 mating defect. Furthermore, a PGAL-KAR4 construct (pMR3291), described below, complemented the mating defect upon galactose induction.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmida | Genotype or yeast markers | Reference |
|---|---|---|
| Strains | ||
| MS1919 | MATa his3Δ200 ura3-52 ade2-101 trp1Δ1 | This work |
| MS2710 | matΔ::LEU2 ura3-52 trp1Δ1 his3::TRP1 leu2-3,112 kar4-2150 [_MATα URA3 CEN ARS_] | 24 |
| MS3216 | MATa leu2-3,112 his3Δ200 ura3-52 ade2-101 kar4Δ::HIS3 | 25 |
| MS3212 | MATα leu2-3,112 his3Δ200 ura3-52 ade2-101 kar4Δ::HIS3 | 25 |
| MY3371 | MATa ura3-52 leu2Δ1 | This work |
| MY3375 | MATa ura3-52 leu2Δ1 his3Δ200 | This work |
| MY4166 | MATa ura3-52 leu2Δ1 his3Δ200 kar4Δ::HIS3 | This work |
| MY4239 | MATα ura3-52 leu2Δ1 his3Δ200 kar4Δ::HIS3 | This work |
| MY5792 | MATa ura3-52 leu2Δ1 his3Δ200 KAR4::HA | This work |
| Plasmids | ||
| pMR2516 | KAR4 CEN ARS URA3 | 25 |
| pMR2654 | KAR4::HA CEN ARS URA3 | 25 |
| pMR2973 | PGAL-KAR4-long CEN ARS URA3 | This work |
| pMR3291 | PGAL-KAR4-short CEN ARS URA3 | This work |
| pMR3356 | PGAL-KAR4::HA-short CEN ARS URA3 | This work |
| pMR3357 | KAR4::HA (ATG1→AAG) CEN ARS URA3 | This work |
| pMR3359 | KAR4::HA (ATG2→AAG) CEN ARS URA3 | This work |
| pMR3459 | PGAL-KAR4::HA-long CEN ARS URA3 | This work |
| pRS416 | CEN ARS URA3 | 40 |
Galactose inductions of Kar4p.
For all galactose inductions, strains were first pregrown in synthetic complete medium lacking uracil with 2% raffinose to mid-exponential phase. For galactose induction during limited mating tests, MATa _kar4_Δ cells harboring the appropriate plasmid were spotted onto a lawn of MATα kar4-2150 cells (MS2710). The cells were allowed to mate on synthetic complete plates with 2% raffinose and 2% galactose for 3 h and then replica-plated onto the appropriate medium to select for the diploid.
For glucose-modulated galactose induction, strains were induced with 2% galactose in combination with various concentrations of glucose (range, 0.1 to 0.6%) for 3 h. Limited plate mating tests using glucose-modulated galactose induction were conducted as described above, except that synthetic complete plates containing glucose and galactose concentrations equivalent to those of the liquid cultures were used. The exponentially growing cultures were spotted onto the MATα kar4 lawn and mating was allowed to proceed for 4 h. The mating plates were then replica printed to the appropriate medium to select for the diploids.
For PGAL shutoff experiments, cells were grown to early exponential phase in synthetic complete medium lacking uracil with 2% raffinose and induced with 2% galactose plus 0.35% glucose (for Kar4p-long) and 2% galactose plus 0.2% glucose (for Kar4p-short). Inductions were for 3 h. The cells were then washed and resuspended in synthetic medium lacking uracil with 2% glucose, and if applicable, synthetic α-factor (Princeton Synthesizing/Sequencing Facility) was added to 5.6 μM.
Molecular techniques.
DNA manipulations, including isolation of plasmid DNA, cloning procedures, electroporation into bacteria, PCR, and colony lifts, were performed as described in reference 37. DNA was also prepared by the STET protocol (36). Oligonucleotides were obtained from the Princeton Synthesizing/Sequencing Facility.
Total RNA was isolated from wild-type (MS1919) and kar4Δ (MS3216) yeast cultures treated with α-factor (5.6 μM) dissolved in methanol or mock treated with an equivalent volume of methanol. The RNA was extracted as described previously (36) except that 1% sodium dodecyl sulfate was included in the initial lysis buffer. Primer extension was as described previously (12) with minor modifications. A total of 7.5 ng of the 16-mer oligonucleotide PREXT1, 5′ GGA TAG CCA TCA ACC C 3′, was end labeled with 50 μCi of [γ-32P]ATP and hybridized to 42 μg of total yeast RNA for each sample. One unit of RNasin (Promega Biotech Corp., Madison, Wis.) was used for each sample, as was 60 U of avian myoblastosis virus reverse transcriptase (United States Biochemicals, Cleveland, Ohio). Reaction products were ethanol precipitated, resuspended in sample buffer, and then run on a denaturing polyacrylamide-urea gel in parallel with sequencing reactions (see below).
S1 nuclease mapping of mRNA 5′ start sites was performed as described previously (2) by using the single-stranded probe protocol. The primer used was the same as that used for primer extension, PREXT1. The primer was end labeled and hybridized to the KAR4 DNA fragment. After extension with the Klenow fragment from DNA polymerase (Boehringer Mannheim, Indianapolis, Ind.), the product was cleaved with _Sac_I to delineate the 3′ end of the probe. After hybridization of the probe with the yeast RNA, 400 U of S1 nuclease (Boehringer Mannheim) was used. The S1 reactions were run in parallel with sequencing reactions on a sequencing gel.
Sequencing reactions were carried out according to the Sequenase, version 2.0, protocol (Sequenase DNA sequencing kit; United States Biochemicals). PREXT1 was the primer, and pMR2516 was used as the KAR4 template. The denaturing polyacrylamide-urea gel was poured, loaded, and run according to the Sequenase protocol.
Site-directed mutagenesis.
Site-directed mutagenesis was carried out with the Muta-Gene phagemid in vitro mutagenesis kit (Bio-Rad Laboratories, Richmond, Calif.). Two 16-mer oligonucleotides to independently change either the first or the second ATG to an AAG were synthesized. The primer used to change the first ATG (ATG1→AAG) was 5′ GAA TGC CTT CTT AAT A 3′. The primer used to alter the second ATG (ATG2→AAG) was 5′ AGA TTT CTT TTC TAT T 3′. The changes were made to a triple hemagglutinin (HA)-tagged version of KAR4 (pMR2654) to allow for visualization of the protein products. The mutagenesis protocol used was essentially as described by the manufacturers, except that R408 helper phage was used instead of M13KO7 and no gene 32 product was used. The ATG1→AAG sequence change introduced an _Xmn_I site, which was used as a detector of positive colonies. The site-directed mutants were sequenced to confirm the change.
Engineering PGAL-KAR4::HA constructs to separately express the forms of Kar4p.
PGAL-KAR4 constructs were constructed by using a pRS416-based vector (40) that contained a 750-bp _Eco_RI/_Bam_HI fragment containing the GAL1 promoter (PGAL). The KAR4 coding region was amplified by PCR from pMR2516 by using a 5′ primer near the first ATG (sequence encoding the long protein) or the second ATG (sequence encoding the short protein) in combination with a 3′ primer near the TAA stop codon. The primers used were KAR4-N1 (long), 5′ GCG GAT CCG AGA AGT GAG AAT ACT AT 3′; KAR4-N2 (short), 5′ GCG GAT CCG CCA AAC CAG GAA ACA AT 3′; and KAR4-C, 5′ GCG GAT CCG AGC TAA GCA AGG ATT TA 3′. The final cloned KAR4 region in the constructs consisted of a _Bam_HI to _Blp_I fragment. The engineered _Bam_HI site is 5′ of the ATG, and the _Blp_I site is found 3′ of KAR4. To allow for the immunological detection of Kar4p, a 140-bp _Xba_I fragment from pMR2654 (25), containing the triple HA epitope, was cloned into the _Xba_I site of KAR4 on the PGAL-KAR4-short (pMR3291) and PGAL-KAR4-long (pMR2973) constructs to form PGAL-KAR4::HA-short (pMR3356) and PGAL-KAR4::HA-long (pMR3459), respectively. The _Xba_I site of KAR4 is found at the NH2-terminal coding region of the gene, just downstream of the second ATG codon. These epitope-tagged constructs fully complemented the kar4Δ strains MS3216 (gal2) and MY4166 (GAL2) for mating upon galactose induction.
Detection of epitope-tagged Kar4p.
Previous work (25) and most of the experiments reported here made use of an epitope-tagged form of the protein in which the triple HA epitope was inserted after amino acid 12 of the short form (or amino acid 42 of the long form). For the pulse-chase experiment, we constructed strain MY5792, which contains a chromosomal version of KAR4 with the triple HA epitope inserted at the extreme carboxyl terminus of the coding region. The epitope tagging technique used is described elsewhere (38). The DNA fragment used to direct the integration was amplified by PCR by using the following primer combination: 5′ CTT ATT TAC TAG TAT ATT TAA TTG AGC TAA GCA AGG ATT TAT GTG TTG ATG CTT TAC TAT AGG GCG AAT TGG and 5′ CCA TTA AAA AAT GAG ATT GAG CTG TTA AGA CCA AGA AGT CCA GTA CAA AAA GCA CAA AGG GAA CAA AAG CTG G. The integration was confirmed by PCR and Western blot analysis. The KAR4::HA strain, MY5792, was fully functional for mating.
Total yeast protein was extracted as described by Ohashi et al. (33). Proteins were separated on 10 to 12.5% polyacrylamide minigels run at 100 V and transferred onto nitrocellulose at 100 V for 90 min. Western blotting was performed by using a 1:2,500 dilution of the monoclonal antibody 12CA5 directed against the HA epitope (Princeton Monoclonal Facility). After incubation with a 1:2,500 dilution of the secondary anti-mouse immunoglobulin G antibody (Promega Biotech), the Kar4::HA proteins were detected with the ECL kit (Amersham, Arlington Heights, Ill.) system.
Assays for Kar4p function.
All microscopic analyses of zygotes and sporulation assays were conducted as described previously (24, 25). To analyze the karyogamy defect, limited filter matings of MS2710 (kar4-2150) and MS3216 (kar4Δ::HIS3), which harbored either the ATG1→AAG construct (pMR3357), the ATG2→AAG construct (pMR3359), the vector (pRS416) (40), or KAR4 (pMR2654), were conducted. The matings were for 2 h on yeast extract-peptone-dextrose (YEPD). For each mating, 100 zygotes were scored for their class I karyogamy defect or wild-type morphology (24).
The G1 pause assays were conducted by examining cells after release from pheromone arrest as follows. MATa cells were grown to early exponential phase and treated with 5.6 μM α-factor for 2 h. The kar4Δ strain (MS3216) contained either the ATG1→AAG mutation construct (pMR3357), the ATG2→AAG mutation construct (pMR3359), the pRS416 vector (40) as a negative control, or the wild-type KAR4 construct (pMR2654) as a positive control. After 80 to 100% of the cells were G1 arrested (unbudded cells), the cells were washed with prewarmed medium and returned to a 30°C water bath. Over a time course of 80 min, cells were collected every 10 min and fixed in 3:1 methanol-acetic acid mixture. Nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole) as described previously (24), and for each strain, 250 cells were scored for a budded versus unbudded morphology.
Determination of the half-lives of the forms of Kar4p.
35S pulse-chase experiments were performed as described previously (13). MY5792 and MY3375 cultures were first grown in synthetic complete medium to early exponential phase. The cultures were split, and half of each was exposed to 5.6 μM α-factor for 40 min. The cells were then starved for cysteine and methionine in the presence or absence of pheromone for 30 min prior to labeling. The cultures were labeled with a 35S-Translabel (Amersham) pulse at 200 μCi per OD600 for 10 min. Immunoprecipitations were conducted with anti-HA monoclonal antibody 12CA5 at a dilution of 1:1,000 in the presence of cold competitor extract from the control strain, MY3375. Kar4p levels were quantitated by using a PhosphorImager and ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.). Decay curves were plotted by using Microsoft Excel software (Microsoft Co., Seattle, Wash.).
RESULTS
KAR4 5′ transcript mapping.
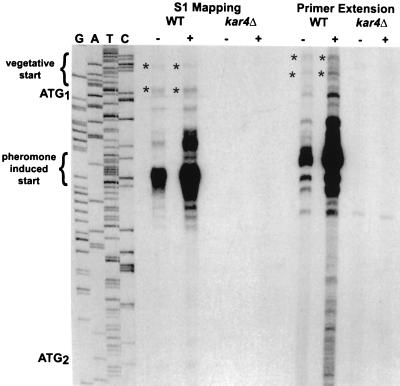
We tested if a differential transcription-translation mechanism was responsible for the production of the two forms of Kar4p. That is, we examined the possibilities that two sets of mRNA transcripts with different 5′ ends are produced from the KAR4 coding region and that translation of these two mRNA species gives rise to the two forms of Kar4p. To accomplish this, we mapped the start sites of KAR4 mRNA by two different techniques, primer extension and S1 nuclease protection. A kar4Δ strain served as a negative control to identify nonspecific bands (Fig. 1).
FIG. 1.
Mapping of the 5′ mRNA start sites of KAR4. Two separate techniques, S1 nuclease protection and primer extension, were used to map the 5′ ends of KAR4 mRNA, as described in Materials and Methods. Total yeast RNA was isolated from a wild-type (WT) strain (MS1919) and an isogenic kar4Δ strain (MS3216) grown in the absence (−) or presence (+) of α-factor. Sequencing was performed with a WT KAR4 plasmid (pMR2516) as the template. The products from S1 nuclease protection, primer extension, and the sequencing were run in parallel on the same sequencing gel. The sequencing lanes are labeled in the order G A T C. The first (ATG1) and second (ATG2) translational start codons are indicated in the margin but appear on the sequencing gel as the complementary sequence TAC. The pheromone-induced transcriptional initiation sites are bracketed. Asterisks indicate the constitutively expressed transcriptional initiation sites.
In the wild type, an abundant set of KAR4 transcripts originated approximately 20 to 30 bp downstream from the first ATG codon (Fig. 1). Upon treatment with pheromone, there was a 2.7-fold increase in the level of these transcripts (Fig. 1). Because these transcripts map to the region between the two potential initiation ATGs, they presumably encode the shorter form of Kar4p, which is strongly induced upon pheromone treatment (25). The results of the two methods of mapping agreed within 5 or 6 bp on the start sites of the major induced transcripts. Uninduced lanes (Fig. 1) as well as lighter exposures of the autoradiograph (not shown) indicated that there is one preferred start site, 27 bp after the first ATG.
Our analysis also showed that two constitutively expressed transcripts mapped upstream of the first ATG. These transcripts were expressed at 6- to 10-fold lower levels than the pheromone-induced transcripts. The constitutive transcripts could encode the longer form, which predominates in vegetative cultures (25). Given the relative abundance of the longer transcripts, the first AUG must have a greater translational efficiency than the second AUG to account for the steady-state protein levels observed previously (see Discussion).
Site-directed mutagenesis of the two potential initiation codons.
To confirm that alternative usage of the AUG codons is responsible for the two forms of Kar4p, we used site-directed mutagenesis to engineer two epitope-tagged KAR4 constructs, one with the first ATG (ATG1) mutated and one with the second ATG (ATG2) mutated. Each ATG was changed to AAG, thereby eliminating the start codon and substituting a lysine codon.
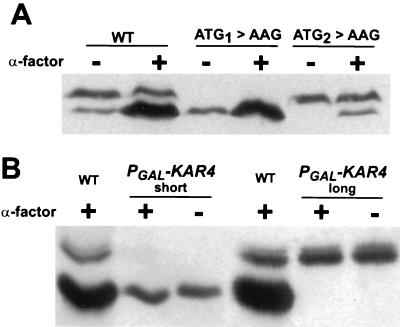
The ATG mutants were transformed into a kar4Δ strain, and their protein profiles were analyzed to determine which form(s) of Kar4p were expressed. Strains harboring the ATG1→AAG mutation expressed exclusively the faster-migrating inducible form (Kar4p-short) in both the absence and presence of pheromone (Fig. 2A), and the levels of expression of Kar4p-short were identical to those in the wild type for both the vegetative and pheromone-induced cultures (Fig. 2A). This analysis established that mutating the first ATG codon is sufficient to terminate the expression of Kar4p-long.
FIG. 2.
The separate expression of the two forms of Kar4p. (A) Expression patterns of the ATG mutants. The kar4Δ::HIS3 (MS3216) strain harbored either a wild-type (WT) KAR4 construct (pMR2654), the ATG1→AAG construct (pMR3357), or the ATG2→AAG construct (pMR3359). The cells were grown in the absence (−) or presence (+) of α-factor, as indicated. The 12CA5 monoclonal antibody was used to detect the triple HA epitope in the Kar4 proteins. The long form of Kar4::HAp migrates at 41.5 kDa, and the short form migrates at 38.5 kDa. (B) Differential expression of Kar4p by using a galactose-inducible promoter. Galactose induction of the PGAL-KAR4::HA constructs in the presence (+) and absence (−) of pheromone (α-factor) is shown. A kar4Δ strain, MS3216, harbored the galactose-inducible promoter (PGAL) constructs to produce the Kar4p-long (pMR3459) or the Kar4p-short (pMR3356) form. WT Kar4p (WT) expression from pMR2654 is shown as a reference for the mobility of the two forms. For this experiment, the induction conditions were 2% galactose plus 0.35% glucose for Kar4p-long and 2% galactose plus 0.2% glucose for Kar4p-short. For the controls, we confirmed that for strains carrying both constructs, Kar4p was not detected when the cells where not under galactose induction conditions (not shown).
The second ATG mutant (ATG2→AAG) expressed only Kar4p-long when grown vegetatively (Fig. 2A), and Kar4p-long predominated when this strain was grown in the presence of α-factor (Fig. 2A). As in the wild type, the levels of the long form did not change upon pheromone induction. However, under these conditions, a small amount of a shorter form of Kar4p with an electrophoretic mobility similar to that of Kar4p-short was expressed (Fig. 2A). Subsequent analysis presented below ruled out the possibility that the faster-migrating form was a proteolytic cleavage product of Kar4p-long. One likely source of this shorter form of Kar4p is initiation at an alternate non-AUG start codon slightly upstream from AUG2 (see Discussion). Thus, despite the presence of a small amount of a faster-migrating band in the pheromone-induced culture for the ATG2→AAG mutant, we conclude that the differential use of the AUG initiation codons dictates which form of Kar4p is produced.
Separate expression of the two forms of Kar4p from PGAL-KAR4.
To separately produce the forms of Kar4p, we put KAR4 under independent regulation using two PGAL-KAR4 promoter fusions. In these two plasmids, _PGAL-KAR4_-long and _PGAL-KAR4_-short, the galactose-induced mRNAs start upstream of the first or the second ATG initiation codon, respectively. When the strains were induced on galactose, we observed that each plasmid expressed only one form of Kar4p, depending upon which was the first AUG on the predicted transcript (Fig. 2B). Thus, even though both AUGs are present on the _PGAL-KAR4_-long mRNA, only the long form of the protein is produced. By inference, the minor amount of the Kar4p short form observed in the ATG2→AAG mutant (see above) must arise from transcripts initiating downstream of the first ATG. These results confirm the hypothesis that the two forms of Kar4p arise from differential transcription initiation followed by translation beginning at the first available AUG. Furthermore, we note that the presence or absence of α-factor had no effect on the form of Kar4p produced from these PGAL-KAR4 constructs, effectively ruling out pheromone-dependent modification (including proteolysis) of Kar4p as an explanation for the different migrations (Fig. 2B).
The two forms of Kar4p function equally well in meiosis and in the mitotic cell cycle.
Using the ATG mutants and the PGAL-KAR4 constructs, we next determined whether the two forms of Kar4p had different cellular functions. Previous work showed that kar4Δ/kar4Δ diploids fail to undergo meiosis when placed in sporulation medium (25). Analysis of the forms of Kar4p produced during sporulation determined that while both forms of the protein were present, the longer form of Kar4p predominated (our unpublished observations), as in vegetative cells. Hence, the ATG mutants could serve to address the issue of functionality of the two forms of Kar4p during meiosis and mitosis.
To evaluate whether both ATG mutants could rescue the sporulation defect, we used a kar4Δ/kar4Δ diploid that harbored either the vector alone or constructs with the wild-type or mutated KAR4 gene. As expected, no sporulation products were seen in the vector control, while the KAR4 wild-type construct exhibited 18% sporulation (Table 2). We found that both ATG mutants supported sporulation equally well (11% sporulation for ATG1→AAG and 13% for ATG2→AAG). Although the efficiency of sporulation in the ATG mutants was somewhat reduced compared to that in the wild-type control, the data clearly demonstrated that the two forms of Kar4p were equally capable of providing the essential meiotic function.
TABLE 2.
Sporulation of the ATG mutantsa
| Construct | No. of: | % Sporulation | n | |||
|---|---|---|---|---|---|---|
| Single cells | Dyads | Triads | Tetrads | |||
| Vector | 800 | 0 | 0 | 0 | 0 | 800 |
| KAR4 | 571 | 39 | 32 | 52 | 18 | 694 |
| ATG1→AAG | 790 | 35 | 35 | 30 | 11 | 890 |
| ATG2→AAG | 675 | 30 | 32 | 40 | 13 | 777 |
We next determined whether either of the ATG mutants displayed the G1 defect observed in kar4 mutants during the mitotic cell cycle (25). To assay the G1 pause, we conducted a time course experiment using cultures that had been synchronized with mating pheromone. The results are reported in Table 3. Wild-type cultures began budding between 20 and 30 min after release from pheromone arrest and reached 50% budded by 60 min. In contrast, the kar4 cultures began budding much later, between 40 and 50 min, and only 20% of the cells were budded by 60 min. Over time, kar4 cultures consistently lagged about 20 min behind the wild type (our unpublished experiments and reference 25). We observed no significant differences in the times that the ATG mutants either initiated or reached 50% budding compared to the wild type. These results showed that either form of Kar4p could perform the vegetative function necessary during the cell cycle. This is consistent with the observation that, under vegetative conditions, the two mutants express equivalent levels of Kar4p (Fig. 2A). We conclude that there is no significant difference in the functions of the two forms of Kar4p with respect to the mitotic cell cycle.
TABLE 3.
Recovery of ATG mutants from G1 arrest
| Plasmida | % Budded cells atb: | ||||||
|---|---|---|---|---|---|---|---|
| 0 min | 10 min | 20 min | 30 min | 40 min | 50 min | 60 min | |
| Vector | 3 | 3 | 0 | 5 | 6 | 17 | 20 |
| KAR4 | 1 | 3 | 0 | 28 | 29 | 37 | 50 |
| ATG1→AAG | 2 | 6 | 4 | 38 | 43 | 42 | 56 |
| ATG2→AAG | 2 | 1 | 1 | 15 | 20 | 50 | 49 |
Analysis of the mating function of the ATG mutants.
We next wanted to assess whether the two forms of Kar4p function equally well during mating, by measuring the ability of the ATG mutants to rescue the kar4 nuclear fusion defect. We performed quantitative filter matings and analyzed the zygotes microscopically to measure the efficiency of karyogamy. MATa kar4 strains that harbored the ATG→AAG mutations or the appropriate controls were mated to a MATα kar4 partner. In the control matings, where the MATa kar4 strain contained the plasmid vector, zygotes exhibited only 5% nuclear fusion. In the matings where the strain contained the wild-type KAR4 plasmid, zygotes exhibited 96% nuclear fusion. We found that the ATG1→AAG mutant conferred mating proficiency to a level comparable to that of the wild type (90% nuclear fusion). However, in matings with the ATG2→AAG mutant, only 45% of the zygotes successfully completed nuclear fusion. These results suggest that expression of the short form of Kar4p is required for efficient nuclear fusion. The lower karyogamy efficiency of the ATG2→AAG mutant could be due either to the lower total expression of Kar4p after pheromone induction (Fig. 2A) or to inherent differences in the functions of the proteins. To further address the issue we sought to express the different forms of Kar4p to equivalent levels to determine whether both forms function equally well during mating.
The long and short forms of Kar4p are equally functional for mating.
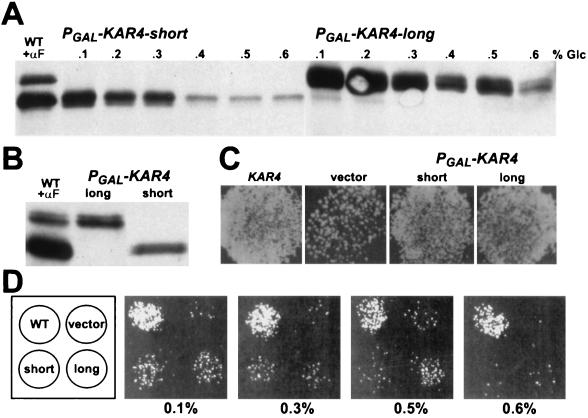
The GAL1 promoter constructs provided the means to separately express each Kar4p species to levels comparable to those observed during pheromone induction. However, under conditions of full induction, Kar4p-long was much more abundant than Kar4p-short (Fig. 3A). To express the two forms of Kar4p to similar levels, we used glucose to modulate the level of galactose induction (Fig. 3A). The two proteins could be expressed to equivalent levels by the addition of different concentrations of glucose to the galactose medium. Typically, we found that 0.2 to 0.3% glucose for _PGAL-KAR4_-short and 0.3 to 0.5% glucose for _PGAL-KAR4_-long generally allowed expression of the proteins to equivalent levels (Fig. 3B).
FIG. 3.
Modulation of Kar4p to sufficient levels for mating. (A) Western blot showing the modulation of Kar4p expression by glucose repression of galactose induction. A kar4Δ strain, MS3216, harbored the galactose-inducible promoter (PGAL) constructs to produce the Kar4p-long (pMR3459) or the Kar4p-short (pMR3356) forms. Wild-type (WT) Kar4p expression from pMR2654 in the presence of α-factor (+αF) is shown as a reference. PGAL induction was carried out in synthetic complete medium lacking uracil with 2% raffinose and 2% galactose and with various concentrations of glucose (% Glc). (B) Western blot showing comparable levels of Kar4p-long (long) and Kar4p-short (short) expression upon modulation of the galactose induction with glucose. For the strain harboring the Kar4p-long PGAL construct, 2% galactose and 0.35% glucose were used. For the Kar4p-short PGAL construct, 2% galactose and 0.2% glucose were used. (C) Both forms of Kar4p complement the mating defect of kar4Δ when fully expressed. Shown is a plate mating in which a MATa kar4Δ strain (MS3216) carried a plasmid expressing the wild-type KAR4 gene (pMR2654), a vector control (pRS416) (vector), or the galactose-inducible promoter constructs to produce Kar4p-long (pMR3459) (long) or Kar4p-short (pMR3356) (short). These strains were pregrown on synthetic complete plates lacking uracil with 2% raffinose and then mated to a MATα kar4-2150 lawn (MS2710) for 4 h on synthetic complete plates with 2% raffinose and 2% galactose. The mating plate was then replica printed to diploid-selective plates. (D) Mating efficiency decreases as Kar4p is repressed. The same strains as described for panel C were used. The left square is a key to the pattern of cells on the mating plates. The modulated mating assay is described in Materials and Methods. For the plates shown, the matings were conducted at glucose concentrations ranging from 0.1% to 0.6% to modulate the expression of Kar4p-long or Kar4p-short.
Using the modulated expression of Kar4p, we tested the ability of the two forms to complement the kar4 mating defect. Plate matings were performed with a kar4Δ strain carrying the vector plasmid, the wild-type KAR4 construct, _PGAL-KAR4_-long plasmid, or _PGAL-KAR4_-short plasmid. Both the long and short forms of Kar4p complemented the defect to near wild-type levels when fully induced (Fig. 3C). As increased levels of glucose reduced protein expression, the two PGAL-KAR4 plasmids generally supported similar levels of complementation commensurate with the Kar4 protein level (Fig. 3A and D). The efficiency of mating for the strains harboring the PGAL constructs fell as Kar4p levels were reduced (Fig. 3A and D). Under conditions of strong repression (0.5% glucose for the short form and 0.6% glucose for the long form), neither form of Kar4p complemented the defect yet protein was still detectable (Fig. 3A). Taken together, these data demonstrate that when expressed to sufficient levels, either form can satisfy the requirement for Kar4p during mating. Furthermore, there must be a threshold Kar4p level that needs to be attained for efficient mating to occur.
Long Kar4p and short Kar4p are turned over at different rates.
As described above, when each form was transcribed from the GAL1 promoter, Kar4p-long was expressed to higher levels than Kar4p-short (Fig. 3A). Preferential expression of Kar4p-long might arise from a more favorable context for the initiation codon, from differences in protein stability, or both. To explore the reasons for the differential expression, we performed a galactose shutoff experiment to investigate the rates of turnover of the two proteins. In this experiment, the two forms were expressed from the PGAL constructs to similar levels, in the presence of α-factor. Subsequently, transcription initiation was repressed by removing galactose and adding glucose. Upon analysis of the proteins, we found that Kar4p-long persisted far longer in the cell (up to 3 h) than did Kar4p-short (0.5 to 1 h) (data not shown). However, when expressed from the PGAL constructs, the two Kar4p species are translated from mRNAs differing by about 90 nucleotides at their 5′ ends. Thus, it remained a possibility that the observed difference in protein persistence resulted from differences in mRNA stability rather than from differences in protein turnover. To address this issue, we next examined the half-lives of the proteins directly with a pulse-chase analysis.
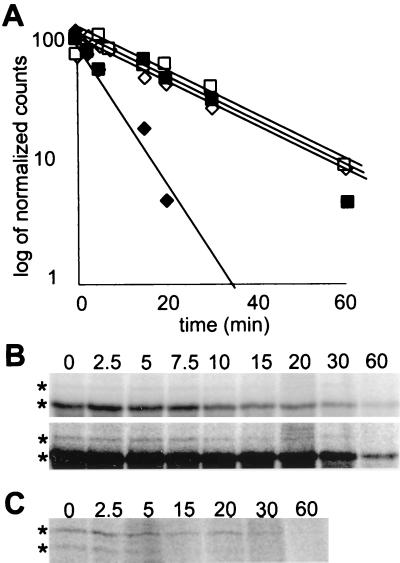
We were also concerned that the triple HA epitope near the NH2 terminus might differentially affect the stability of the two proteins. Therefore, to determine the turnover of the two proteins directly, we assayed wild-type Kar4 proteins, which were epitope tagged at their carboxy termini and expressed from their own promoter. After a 10-min pulse with 35S-labeled amino acids followed by various chase intervals, we immunoprecipitated the two forms of Kar4p and assessed their half-lives. Furthermore, we determined whether pheromone altered the relative stability of the proteins. In the presence of pheromone, the short form was expressed at much higher levels than the long form, as expected from the levels of the two sets of mRNA. In addition, we determined that under these conditions, the short and long forms had very similar rates of turnover (half-lives of approximately 15 min [Fig. 4A and B]). During vegetative growth, the long and short forms were synthesized to the same level, in spite of the 6- to 10-fold greater abundance of the short mRNA (compare Fig. 1 with Fig. 4C, 0 time point), confirming that the long form is translated more efficiently.
FIG. 4.
Differential stabilities of the two Kar4p species. (A) Decay curves for Kar4p-short (diamonds) and Kar4p-long (squares). A KAR4::HA strain (MY5792) was pulsed for 10 min in the presence (open symbols) and absence (black symbols) of pheromone. The samples were chased for the times indicated, and the extracts were immunoprecipitated with anti-HA antibody as described in Materials and Methods. Kar4p bands were quantified, corrected for background, and normalized to the level at t = 0 min (100%) by processing with a PhosphorImager and ImageQuant software. The curves were fitted to the data points by exponential regression with Microsoft Excel software. (B) Two exposures of the pulse-chase gel in the presence of pheromone. The top panel is included to show the decay of Kar4p-short, and the bottom panel is included to show the decay of Kar4p-long. (C) The gel of the forms of Kar4p in the absence of pheromone.
Remarkably, in vegetative cells, the two forms of the protein had significantly different rates of turnover (Fig. 4A and C). Kar4p-short had a half-life of approximately 5 min, whereas Kar4p-long had the same turnover rate as pheromone-treated cells (Fig. 4A and C). Thus, in the absence of pheromone, Kar4p-short is degraded approximately three times faster than Kar4p-long. Therefore, we conclude that in vegetative cells the levels of Kar4p-short are low because of a combination of inefficient translation and increased protein degradation.
Vegetative overexpression of Kar4p causes a growth defect.
Given that high levels of Kar4p are required for mating and that several mechanisms ensure low levels in vegetative growth, we next wanted to determine the selective advantage of the elaborate regulation of KAR4. One hypothesis is that high constitutive levels of Kar4p would cause inappropriate expression of genes that are ordinarily expressed only during mating or G1. To determine whether overexpression of Kar4p might be toxic in vegetative cells, we induced high levels of Kar4p using the GAL1 promoter constructs. If either form of Kar4p was overexpressed, growth was significantly retarded compared to that of the wild type (Fig. 5). After two generations in the presence of overexpressed Kar4p, the cultures had accumulated cells in both the G1 and G2/M stages of the cell cycle (Table 4). The overexpression toxicity does not appear to be caused by inappropriate regulation of Kar4p’s mating-specific targets, because the growth inhibition was not dependent on an intact STE12 gene (data not shown), which is required for the expression of KAR3 and CIK1 (25). Thus, down regulation of Kar4p is required for normal passage through the cell cycle, most likely to prevent the inappropriate expression of genes regulated by Kar4p during vegetative growth.
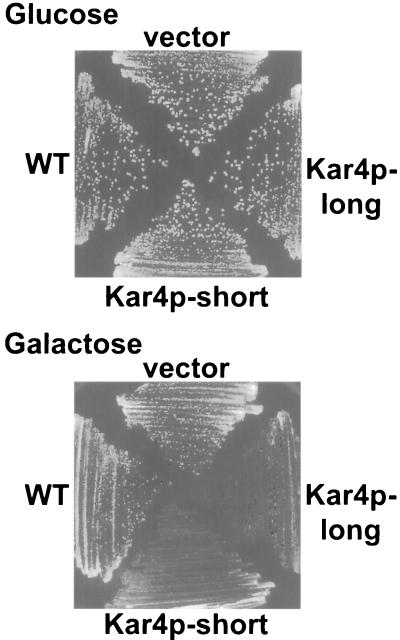
FIG. 5.
Vegetative growth defect caused by overexpression of Kar4p. Strains were streaked for single colony growth on 2% glucose (PGAL repression) and 2% galactose (PGAL induction). A wild-type strain (MY3375) carried either a plasmid expressing the wild-type (WT) KAR4 gene (pMR2654), a vector control (pRS416) (vector), or the galactose-inducible promoter construct to produce the Kar4p-long (pMR3459) or the Kar4p-short (pMR3356) form of Kar4p.
TABLE 4.
Cell cycle distribution of cultures overexpressing Kar4p
| Plasmida | Time (h) after induction | % of cells that wereb: | ||||
|---|---|---|---|---|---|---|
| Unbudded (G1) | Small budded | Medium budded | Large budded, single nucleus (G2/M) | Large budded, two nuclei | ||
| KAR4 (wild type) | 0 | 37 | 9 | 41 | 6 | 7 |
| _PGAL-KAR4_-short | 0 | 39 | 8 | 38 | 4 | 11 |
| _PGAL-KAR4_-long | 0 | 37 | 11 | 33 | 4 | 15 |
| KAR4 (wild type) | 6 | 46 | 7 | 31 | 5 | 11 |
| _PGAL-KAR4_-short | 6 | 55 | 2 | 18 | 16 | 9 |
| _PGAL-KAR4_-long | 6 | 55 | 6 | 13 | 13 | 13 |
DISCUSSION
The work presented in this paper elucidates multiple levels of regulation of a yeast transcription factor gene, KAR4. The expression of KAR4 is regulated at the levels of transcription, translation, and protein turnover. The two forms of Kar4p originate from differential transcription and translation. Transcription mapping and site-directed mutagenesis demonstrated that the two proteins originate from different initiation codons, separated by 90 bp. A highly expressed pheromone-induced transcript initiates between the ATG start codons, forcing the use of the second start codon and resulting in expression of the shorter form of the protein. A high level of expression was necessary for efficient mating, but either form of Kar4p could function, if expressed to sufficient levels. The reason that two forms of Kar4p are produced appears to be related to the rapid degradation of the short form in vegetative cells. During exposure to pheromone, the short form is both induced and stabilized. However, high levels of either form of Kar4p are toxic during mitotic growth. Therefore, the cell must have an effective way of eliminating the high levels of Kar4p after mating has occurred. We propose that the specific induction of a more labile form of the protein accomplishes this task.
Transcription dictates which form of Kar4p is produced.
In this work we showed that in wild-type cells, the form of Kar4p produced is controlled by which in-frame AUG codon is first on the transcript. We observed that when the second in-frame ATG was mutagenized (ATG2→AAG), a small amount of a shorter form of Kar4p was produced upon pheromone induction. We hypothesized that in the ATG2→AAG mutant, infrequent spurious initiation would be driven by the abundance of pheromone-induced KAR4 transcripts that lack a standard in-frame AUG codon. For these transcripts, the first AUG would be out of frame with the KAR4 coding region, thus resulting in a failure to express Kar4p. In order for detectable levels of Kar4p to be produced from these transcripts, a non-AUG codon would have to serve to initiate translation.
Previous work has shown that in S. cerevisiae, certain non-AUG codons, including AUA, can serve as translational initiators at low efficiencies (9, 10, 47; for a review, see reference 19). Initiation from nonstandard codons is enhanced by a favorable context, in particular if A is found in the −3 position (47). Analysis of the KAR4 coding region shows that an AUA with A in the −3 position is found two codons upstream from AUG2, and initiation at the AUA would lead to a protein indistinguishable from the Kar4p-short form. In confirmation of this view, a short form is not produced from the ATG2→AAG mutant when the 5′ ends of the mRNA are controlled by use of promoter fusions.
The translational efficiencies of the two in-frame AUG codons.
The expression of the two forms of Kar4p reflects an additional difference beyond transcriptional control. We observed that in vegetative cells the constitutive transcripts for Kar4p-long were 6- to 10-fold less abundant than the transcripts for Kar4p-short, yet pulse-chase data showed that the proteins were expressed to equivalent levels. The same relative difference in translational efficiency was observed in pheromone-treated cells. Thus, the efficiency of translation from the longer transcript must be at least 6- to 10-fold higher than that from the shorter transcript. The different translational efficiencies are likely to reside in the contexts of the initiating AUG codons. For KAR4, the first AUG (A at −3 and G at +4) has a more favorable context for initiation than the second AUG (G at −3 and A at +4) (reviewed in reference 19). Furthermore, because the two sets of transcripts have different 5′ regions, they may also have different secondary structures that have an impact on the efficiency of initiation.
Comparisons with other differential initiation regulatory systems.
The differential AUG usage mechanism for Kar4p regulation is similar to that seen for a handful of other genes in S. cerevisiae, including SUC2, HTS1, VAS1, LEU4, MOD5, TRM1, and CCA1 (5, 6, 8, 32, 35, 43, 46; for a review, see reference 19). For all of these genes, differential transcription and translation initiation produces different proteins from the same gene. For example, SUC2 produces both secreted and intracellular forms of yeast invertase, while HTS1 encodes both cytoplasmic and mitochondrial histidine tRNA synthetases on transcripts of different lengths. For all of these genes, the additional protein sequence found on the longer form includes a cellular localization signal. In the case of invertase, the NH2-terminal extension contains a secretory signal sequence. For the other proteins, the extensions contain mitochondrial import signals. In contrast, we found no specific subcellular localization of Kar4p in either vegetative or pheromone-induced wild-type cells (our unpublished observations). Thus, the regulatory role of the 30 additional NH2-terminal amino acids in Kar4p appears to be novel. The presence of the NH2-terminal extension serves to stabilize the protein during vegetative growth.
The role of differential protein turnover rates.
We propose that the differential stability of the long and short forms reflects the different regulatory needs of the cell during vegetative growth and during mating. Upon pheromone induction, a burst of transcription and translation of the short form of Kar4p would rapidly raise the total concentration of Kar4p above the threshold required for the pheromone-induced transcription of KAR3 and CIK1. Since the induced form of Kar4p is short-lived, a return to vegetative growth and reduced levels of transcription would be quickly followed by a return to the decreased vegetative levels of the protein.
Many regulatory proteins have short in vivo half-lives, which allows for rapid adjustment of their intracellular concentrations. Relevant examples of rapidly degraded regulatory proteins include the MATα_2 repressor (22) and p40_SIC1, a specific inhibitor of S-phase and M-phase cyclin-kinase complexes (39). Degradation of both of these proteins is modulated by ubiquitination. The ubiquitin pathway for protein degradation is highly conserved and selective (for reviews, see references 15, 21, and 41). Proteins targeted by this pathway are ubiquitinated following recognition of the NH2-terminal residues. A ubiquitin moiety is covalently attached to an internal lysine residue, and the proteins are then rapidly degraded by the proteasome. The N-end rule for the ubiquitin pathway relates the in vivo half-life of a protein to the identity of its NH2-terminal amino acid (for reviews, see references 44 and 45).
In S. cerevisiae, most amino acids at the NH2 terminus have been classified as either protective against or promoters of proteolytic degradation. Using reporter constructs in yeast, Bachmair et al. found that NH2-terminal alanine is protective, while lysine promotes rapid proteolytic degradation; the representative half-lives were >20 h and about 3 min, respectively (3). A good correlation exists between the relative half-lives of the two Kar4p species and their NH2-terminal amino acids, according to the N-end rule. After removal of the initial methionine, the more-stable form of Kar4p would have an NH2-terminal alanine residue, whereas the less-stable form would have a lysine. A second indication that the N-end rule might be involved in Kar4p degradation came from the effects of placing the epitope tag at different positions in the protein. When the 39-residue triple HA epitope tag was inserted near the NH2 terminus, Kar4p-long became significantly stabilized, such that the half-life went from 15 to 40 min (data not shown). Because ubiquitin moieties are placed at internal lysines located within a favorable proximity of the NH2 terminus, the epitope tag might have hindered the recognition of the long form. Thus, the identity of the NH2-terminal amino acids in the two forms and the effects of the epitope near the NH2 terminus of the long form are consistent with modulation of Kar4p by the ubiquitin pathway.
Pheromone control of proteolysis?
The presence of pheromone lengthened the half-life of Kar4p-short threefold. The effect of pheromone on protein stability represents a novel level of regulation in the mating pathway. Our data raise the possibility of a regulatory link between the pheromone response pathway and the proteolytic pathway. In one model, the elements of the ubiquitin-dependent protein degradation pathway are specifically down regulated by pheromone response. The discovery of a number of specificity-conferring ubiquitin-conjugating enzymes (E2s) and substrate recognition factors (E3s) (20) raises the possibility that pheromone-specific factors exist.
In this regard, we note that two other components of the pheromone-response pathway are degraded via ubiquitin-dependent pathways. First, degradation of Gpa1p, the alpha subunit of the trimeric G protein, follows the N-end rule pathway (28). Although degradation is independent of the pheromone response per se, it does require functional Sst2p (28), its cognate GTPase-activating protein (1). Second, the endocytosis of pheromone receptor and its eventual degradation in the vacuole are mediated by ubiquitination (17, 18). However, both the pheromone-stimulated endocytosis and the constitutive endocytosis appear to proceed via the same pathway. Thus, neither case represents a clear example of pheromone-regulated ubiquitin-dependent protein degradation.
Kar4p’s vegetative-growth role.
We observed that KAR4 overexpression causes a growth defect, strengthening the hypothesis that KAR4 has a role in vegetative growth. Previous work showed that KAR4 expression is regulated in the cell cycle, such that it is maximally expressed at G1/S. Furthermore, kar4 mutants pause in the G1 phase of the cell cycle. Here we found that overexpression of Kar4p is toxic and results in accumulation of cells in the G1 and G2/M phases of the cell cycle. The overexpression toxicity was not dependent upon the presence of Ste12p, suggesting that the effect was not a consequence of inducing KAR3 and CIK1 at the wrong time in the cell cycle. One possible explanation for the toxicity is a phenomenon known as squelching (14), where an overabundance of a transcriptional activator titrates other transcription factors. Alternatively, it could be the case that Kar4p temporally regulates the induction of cell cycle-specific genes. In keeping with this, as cells traverse G1/S or G2/M, the inappropriate expression of a subset of genes regulated by Kar4p might create a pause in the cell cycle. Given the role of Kar4p in both vegetative growth and the pheromone response pathways, the need for its regulation is even more apparent. Progression through distinct cell cycle and developmental phases undoubtedly requires the careful temporal regulation of a diverse set of functionally related proteins to ensure that inefficient and possibly deleterious interactions between pathways do not occur.
ACKNOWLEDGMENTS
We thank Laurie Jo Kurihara and members of the laboratory for helpful discussions.
A National Institutes of Health grant (GM37739) to M.D.R. supported this work. For part of this work, A.E.G. was supported by the Jane Coffin Childs Memorial Fund for Cancer Research.
REFERENCES
- 1.Apanovitch D M, Slep K C, Sigler P B, Dohlman H G. Sst2 is a GTPase-activating protein for Gpa1: purification and characterization of a cognate RGS-Gα protein pair in yeast. Biochemistry. 1998;37:4815–4822. doi: 10.1021/bi9729965. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M. Current protocols in molecular biology. Brooklyn, N.Y: Greene Publishing Associates; 1987. [Google Scholar]
- 3.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 4.Beh C T, Brizzio V, Rose M D. KAR5 encodes a novel pheromone-inducible protein required for homotypic nuclear fusion. J Cell Biol. 1997;139:1063–1076. doi: 10.1083/jcb.139.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beltzer J P, Morris S R, Kohlhaw G B. Yeast LEU4 encodes mitochondrial and nonmitochondrial forms of alpha-isopropylmalate synthase. J Biol Chem. 1988;263:368–374. [PubMed] [Google Scholar]
- 6.Boguta M, Hunter L A, Shen W-C, Gillman E C, Martin N C, Hopper A K. Subcellular locations of MOD5 proteins: mapping of sequences sufficient for targeting to mitochondria and demonstration that mitochondrial and nuclear isoforms commingle in the cytosol. Mol Cell Biol. 1994;14:2298–2306. doi: 10.1128/mcb.14.4.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byers B, Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatton B, Walter P, Ebel J P, Lacroute F, Fasiolo F. The yeast VAS1 gene encodes both mitochondrial and cytoplasmic valyl-tRNA synthetases. J Biol Chem. 1988;263:52–57. [PubMed] [Google Scholar]
- 9.Cigan A M, Pabich E K, Donahue T F. Mutational analysis of the HIS4 translational initiator region in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2964–2975. doi: 10.1128/mcb.8.7.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donahue T F, Cigan A M. Genetic selection for mutations that reduce or abolish ribosomal recognition of the HIS4 translational initiator region. Mol Cell Biol. 1988;8:2955–2963. doi: 10.1128/mcb.8.7.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Errede B, Cade R M, Yashar B M, Kamada Y, Levin D E, Irie K, Matsumoto K. Dynamics and organization of MAP kinase signal pathways. Mol Reprod Dev. 1995;42:477–485. doi: 10.1002/mrd.1080420416. [DOI] [PubMed] [Google Scholar]
- 12.Gammie A E, Crosa J H. Co-operative autoregulation of a replication protein gene. Mol Microbiol. 1991;5:3015–3023. doi: 10.1111/j.1365-2958.1991.tb01861.x. [DOI] [PubMed] [Google Scholar]
- 13.Gammie A E, Kurihara L J, Vallee R B, Rose M D. DNM1, a dynamin-related gene, participates in endosomal trafficking in yeast. J Cell Biol. 1995;130:553–566. doi: 10.1083/jcb.130.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- 15.Hershko A. Roles of ubiquitin-mediated proteolysis in cell cycle control. Curr Opin Cell Biol. 1997;9:788–799. doi: 10.1016/s0955-0674(97)80079-8. [DOI] [PubMed] [Google Scholar]
- 16.Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 17.Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 18.Hicke L, Zanolari B, Riezman H. Cytoplasmic tail phosphorylation of the α-factor receptor is required for its ubiquitination and internalization. J Cell Biol. 1998;141:349–358. doi: 10.1083/jcb.141.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinnebusch A G, Liebman S W. Protein synthesis and translational control in Saccharomyces cerevisiae. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis, and energetics. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 627–735. [Google Scholar]
- 20.Hochstrasser M. Ubiquitin and intracellular protein degradation. Curr Opin Cell Biol. 1992;4:1024–1031. doi: 10.1016/0955-0674(92)90135-y. [DOI] [PubMed] [Google Scholar]
- 21.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 22.Hochstrasser M, Ellison M J, Chau V, Varshavsky A. The short-lived MATα2 transcriptional regulator is ubiquitinated in vivo. Proc Natl Acad Sci USA. 1991;88:4606–4610. doi: 10.1073/pnas.88.11.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kassir Y, Simchen G. Monitoring meiosis and sporulation in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:94–110. doi: 10.1016/0076-6879(91)94009-2. [DOI] [PubMed] [Google Scholar]
- 24.Kurihara L J, Beh C T, Latterich M, Schekman R, Rose M D. Nuclear congression and membrane fusion: two distinct events in the yeast karyogamy pathway. J Cell Biol. 1994;126:911–923. doi: 10.1083/jcb.126.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurihara L J, Stewart B G, Gammie A E, Rose M D. Kar4p, a karyogamy-specific component of the yeast pheromone response pathway. Mol Cell Biol. 1996;16:3990–4002. doi: 10.1128/mcb.16.8.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leberer E, Thomas D Y, Whiteway M. Pheromone signalling and polarized morphogenesis in yeast. Curr Opin Genet Dev. 1997;7:59–66. doi: 10.1016/s0959-437x(97)80110-4. [DOI] [PubMed] [Google Scholar]
- 27.Levin D E, Errede B. The proliferation of MAP kinase signaling pathways in yeast. Curr Opin Cell Biol. 1995;7:197–202. doi: 10.1016/0955-0674(95)80028-x. [DOI] [PubMed] [Google Scholar]
- 28.Madura K, Varshavsky A. Degradation of G α by the N-end rule pathway. Science. 1994;265:1454–1458. doi: 10.1126/science.8073290. [DOI] [PubMed] [Google Scholar]
- 29.Marsh L, Rose M D. The pathway of cell and nuclear fusion during mating in S. cerevisiae. In: Pringle J R, Broach J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces: cell cycle and cell biology. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 827–888. [Google Scholar]
- 30.Meluh P B, Rose M D. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- 31.Miller R K, Rose M D. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J Cell Biol. 1998;140:377–390. doi: 10.1083/jcb.140.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Natsoulis G, Hilger F, Fink G R. The HTS1 gene encodes both the cytoplasmic and mitochondrial histidine tRNA synthetases of S. cerevisiae. Cell. 1986;46:235–243. doi: 10.1016/0092-8674(86)90740-3. [DOI] [PubMed] [Google Scholar]
- 33.Ohashi A, Gibson J, Gregor I, Schatz G. Import of proteins into mitochondria. The precursor of cytochrome c1 is processed in two steps, one of them heme-dependent. J Biol Chem. 1982;257:13042–13047. [PubMed] [Google Scholar]
- 34.Page B D, Satterwhite L L, Rose M D, Snyder M. Localization of the Kar3 kinesin heavy chain-related protein requires the Cik1 interacting protein. J Cell Biol. 1994;124:507–519. doi: 10.1083/jcb.124.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rose A M, Joyce P B, Hopper A K, Martin N C. Separate information required for nuclear and subnuclear localization: additional complexity in localizing an enzyme shared by mitochondria and nuclei. Mol Cell Biol. 1992;12:5652–5658. doi: 10.1128/mcb.12.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rose M D, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 38.Schneider B L, Seufert W, Steiner B, Yang Q H, Futcher A B. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- 39.Schwob E, Bohm T, Mendenhall M D, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 40.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith S E, Koegl M, Jentsch S. Role of the ubiquitin/proteasome system in regulated protein degradation in Saccharomyces cerevisiae. Biol Chem. 1996;377:437–446. [PubMed] [Google Scholar]
- 42.Sprague G F, Thorner J W. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces: gene expression. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 657–744. [Google Scholar]
- 43.Taussig R, Carlson M. Nucleotide sequence of the yeast SUC2 gene for invertase. Nucleic Acids Res. 1983;11:1943–1954. doi: 10.1093/nar/11.6.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varshavsky A. The N-end rule. Cold Spring Harbor Symp Quant Biol. 1995;60:461–478. doi: 10.1101/sqb.1995.060.01.051. [DOI] [PubMed] [Google Scholar]
- 45.Varshavsky A. The N-end rule. Cell. 1992;69:725–735. doi: 10.1016/0092-8674(92)90285-k. [DOI] [PubMed] [Google Scholar]
- 46.Wolfe C L, Lou Y C, Hopper A K, Martin N C. Interplay of heterogeneous transcriptional start sites and translational selection of AUGs dictate the production of mitochondrial and cytosolic/nuclear tRNA nucleotidyltransferase from the same gene in yeast. J Biol Chem. 1994;269:13361–13366. [PubMed] [Google Scholar]
- 47.Zitomer R S, Walthall D A, Rymond B C, Hollenberg C P. Saccharomyces cerevisiae ribosomes recognize non-AUG initiation codons. Mol Cell Biol. 1984;4:1191–1197. doi: 10.1128/mcb.4.7.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]